ISSN: 1837-9664Journal of Cancer
J Cancer 2011; 2:515-526. doi:10.7150/jca.2.515 This volume Cite
Research Paper
Regulatory Effects of microRNA-92 (miR-92) on VHL Gene Expression and the Hypoxic Activation of miR-210 in Clear Cell Renal Cell Carcinoma
1. Translational Surgical Pathology Section, Laboratory of Pathology;
2. Urologic Oncology Branch, Center for Cancer Research, National Cancer Institute, National Institutes of Health, Bethesda MD 20892, USA.
Abstract
Background & Aims: In order to understand the role of miRNAs in renal tumorigenesis, we undertook a stepwise approach that included a comprehensive differential miRNA expression analysis for the most common histological subtypes of human renal neoplasms appearing in either sporadic or hereditary forms. We also aimed to test the hypothesis that microRNAs can act as an alternative mechanism of VHL gene inactivation and therefore might be correlated with tumorigenesis in ccRCC. Finally, we wanted to explore whether the well-known hypoxic activation of ccRCC is followed by a specific pattern of miRNA expression.
Methods: Tumor and normal adjacent kidney parenchyma from patients with RCC were tested for microRNA expression. Twenty cases of different histologies were used for profiling by PCR miRNA arrays. For validation, a separate cohort of samples used to test specifically miR92a expression and its involvement in VHL gene mRNA silencing. Finally, miR210 as a marker of hypoxia was evaluated. Expression values were correlated with important clinicopathologic features from the patients.
Results: We identified unique miRNA expression signatures for each histologic subtype of kidney tumors. Expression values for downregulated miRNAs ranged from 0.3-fold (in VHL-clear cell RCC) up to 0.393 fold (in papillary type II (HLRCC) tumors). For the upregulated miRNAs, fold-changes ranged from 2.1 up to 290-fold. Specific patterns together with type-specific profiles were observed. Twenty-three miRNAs were found to be differentially expressed in both sporadic and VHL-dependent ccRCC. Sporadic clear cell tumors showed a unique pattern of 14-miRNA that were absent from the VHL-dependent tumors. These also showed 15 miRNAs specific to the hereditary type. Common miRNAs to both sporadic and hereditary forms included miR-92a and miR-210. For miR-92a, and a striking inverse correlation with VHL mRNA levels was found. For the hypoxia-regulated miR-210, clear cell tumors showed significantly higher expression levels when compared to tumor of non-clear cell histology (9.90-fold vs. 1.36, p<0.001).
Conclusions: microRNA expression seems to be involved in every step of RCC pathogenesis: both as an element for tumor development as well as a consequence of or in response to the initial malignant transformation and part of tumor progression. Our data show consistent disregulation of miRNAs in human kidney cancer, some of which are potentially involved in critical gene silencing in RCC and others that are activated as part of the pathophysiological response in these tumors.
Keywords: miRNA, VHL, RCC, ccRCC, BHD, HLRCC, TSC, BHD.
Introduction
Renal cell carcinoma (RCC) is the most common epithelial malignancy of the kidney in adults, representing over 90 percent of the primary renal neoplasms and accounts for approximately 3% of all adult malignancies in western countries. In the United States, 58,000 new cases were diagnosed in 2010 with approximately 13,000 deaths (1). The worldwide incidence and mortality rate is in the order of 270,000 cases and 120,000 deaths respectively (2).
RCC is also a complex and partially understood disease. However, significant advances in the classification of the disease during the last decade have resulted from the combined application of molecular biology techniques and thorough histological examination of tumors. These advances have shown that renal epithelial neoplasms are not a single tumor but rather a group of distinguishable entities (3).
In several types of human cancer, gene and protein expression studies have demonstrated to be useful for classifying tumors and identifying unique molecular biomarkers (4-7). More recently, microRNA (miRNA) expression studies have appeared trying to establish molecular classification of tumors (8-11). microRNAs are a class of small RNAs that are phylogenetically conserved and play important roles in the regulation of cell survival, proliferation and differentiation through post-transcriptional regulation of gene expression (12). Although the functions of most of the identified miRNAs have yet to be determined, their use as potential biomarkers has been considered in several human diseases and cancers (13, 14).
microRNA-mediated regulation of tumorigenesis has emerged as a new paradigm in the field of cancer biology. Accumulated studies show that miRNAs are frequently deregulated in hematologic and solid human malignancies and may function as either oncogenes or tumor suppressor genes (15). This class of microRNAs has been termed oncomiRs, and alteration in their expression has been causatively linked to cancer development and disease outcome. Thus, the clinical value of these miRNAs as novel biomarkers for different aspect of RCC management is being actively investigated.
Clear cell RCC (ccRCC) is the most common histologic subtype of kidney cancer, and accounts for approximately 70% of the cases (16), and loss of function of the von Hippel Lindau (VHL) tumor suppressor gene is a well-known mechanism in the pathogenesis of hereditary and sporadic ccRCCs (17). Allelic inactivation of the VHL gene has been shown to occur through mutation, DNA methylation, and/or chromosomal loss in the majority of ccRCCs (18), which leads to enhanced transcriptional activity of Hypoxia Inducible Factor 1 alpha (HIF-1α) and the resulting hypoxic pattern of gene expression (19).
Tumor hypoxia has been shown to be a prognostic factor in solid tumors (20-22). Many non-renal cancers are characterized by hypoxia, enhanced HIF-1α levels and increased expression of hypoxia-regulated genes, which correlate both with tumor progression and patient outcome (23, 24). microRNAs have also been shown to be altered in response to hypoxia (25). Particularly, the hypoxia-responsive miR-210 has been described to be upregulated in hypoxic tumors (26-28).
In order to understand the role of miRNA in renal tumorigenesis, we undertook a stepwise approach that included a comprehensive differential miRNA expression analysis for the most common histological subtypes of human renal neoplasms including clear cell RCC (ccRCC), papillary RCC, chromophobe RCC and the closely related benign tumor oncocytomas, some of them appearing in either sporadic or hereditary forms. We also aimed to test the hypothesis that microRNAs can act as an alternative mechanism of VHL gene inactivation and therefore might be correlated with tumorigenesis in ccRCC. Finally, we wanted to explore whether the well-known hypoxic activation of ccRCC is followed by a specific pattern of miRNA expression.
Materials and Methods
Tissues and sample preparation: Fresh frozen tumor and normal kidney samples from 20 RCC patients were used for miRNA expression profiling. Morphology included clear cell (both sporadic and VHL), papillary type I, chromophobe, hybrid oncocytic (Birt-Hogg-Dube (BHD)), oncocytomas, papillary type II (HLRCC), Tuberous Sclerosis Complex (TSC), and Succinate Dehydrogenase B (SDHB) kidney tumors. For validation, a separate cohort of forty-three FFPE tumor samples and corresponding normal kidney from 39 patients was used to assess miRNAs targeting the VHL gene and hypoxia-driven miR-210 expression. These samples included 21 sporadic clear cell RCCs, fifteen CCRCC arising in VHL syndrome, and 2 clear cell tumors in BHD patients. Three hybrid oncocytic tumors, one papillary type I and one tumor with sarcomatoid differentiation were also included for analysis. Five micron-thick samples mounted on glass slides were needle microdissected and two protocols for either total RNA or miRNA extraction were used in the samples.
Cell lines: Two normal epithelial cell lines (HRCE (Lonza, Basel, Switzerland), HK-2 (ATCC, Manassas, VA), one embryonic kidney cell line (HEK293T (ATCC)) and 2 clear cell kidney cancer cell lines (786.O (ATCC), UOK117) were used. Cells were grown in DMEM supplemented with 10% heat-inactivated FBS in a 5% CO2 atmosphere. At 80% confluence, cells were harvested after trypsinization for simultaneous total and microRNA extraction (miRNAeasy®, Invitrogen, Carlsbad CA). Quality and concentration of total RNA and microRNA were assessed by 260/280 spectrophotometry ratio (Nanodrop®, Thermo Scientific, Wilmington, DE) and by the Agilent bioanalyzer® (Agilent, Santa Clara, CA).
miRNA arrays and miRNA analysis: Extracted total or miRNA (10 ng/µl) was reverse transcribed using universal poly-A tailing of all microRNAs following manufacturer's recommendations (SA Biosciences, Frederick MD). PCR-based arrays containing a panel of 88 miRNAs and 4 small nucleolar RNAs (snoRNA) used for normalization. miRNA-specific stem-loop primers (Applied Biosystems, Foster City CA) were used for microRNAs targeting VHL and for miR-210 analysis. One µl was used in a 12 µl final volume reaction for Real-time PCR analysis on an Applied Biosystems Step-One Plus® real time PCR system. Two small nucleolar RNAs, RNU6B and Z30 were used as internal controls. Relative amounts were calculated by the ΔCT method and further normalized to the values of either the normal cell lines or the corresponding normal kidney for the tissue samples.
Differential miRNA expression profiling: Differentially expressed miRNAs for each of the histologic subtypes as compared to normal kidney were defined as those with a two-fold change and a false discovery rate <5%. Supervised and unsupervised clustering was used to evaluate the role of these histology-specific miRNAs for the diagnosis of RCC.
Database search and VHL-targeting miRNA identification: Three miRNA prediction databases (MIRANDA [http://www.microrna.org/microrna/], SANGER [http://microrna.sanger.ac.uk/] and TARGETSCAN [http://www.targetscan.org/]) were interrogated for microRNAs having the VHL gene as a target. Integrated outputs for common overlapping miRNAs were identified by a intersecting the list through Venn diagrams.
VHL gene mRNA expression: Two gene-specific primers and FAM-MGB fluorescent probes sets were used for VHL mRNA quantification (Taqman® gene expression assays) by Real-time PCR. Relative expression to normal cell lines or normal adjacent kidney was used, normalized to cyclophilin A (PPIA) expression as internal control.
Statistical analysis: A two-fold change (up or down) in either microRNA or gene expression relative to normal kidney was considered as significant. MicroRNA expression was also correlated to the clinicopathologic features of the tumors. The relationship between expression values and these variables was evaluated by the Mann-Whitney or Kruskall-Wallis non-parametric tests. For categorical variables, Chi square tests were used with Fisher's correction where appropriate. Statistical significance was defined as p<0.05.
Results
Renal cell carcinomas show unique miRNA expression profiles
We identified unique miRNA expression signatures for each histologic subtype of kidney tumors. The clinicopathologic characteristics of the patients and samples used for miRNA profiling are shown in Table 1. Expression values for downregulated miRNAs ranged from 0.3-fold (in VHL-clear cell RCC) up to 0.393 fold (in papillary type II (HLRCC) tumors). Commonly lost miRNAs in clear cell tumors (both sporadic and clear cell) included miR-184 and miR-206, while miR-122 and -143 were the most commonly downregulated in Papillary type I tumors. For the upregulated miRNAs, fold-changes ranged from 2.058 up to 290-fold. Most differentially expressed genes were from HLRCC Pap-II cases. Overexpressed miRNAs included miR-92a in clear cell RCC (2.28-fold, p<0.001), miR-183 in papillary type I (29-fold), miR-10a and -10b in papillary type II tumors (240-fold and 290-fold), miR-203 in chromophobe tumors, and miRNas 210/let7i in TSC2 renal tumors.
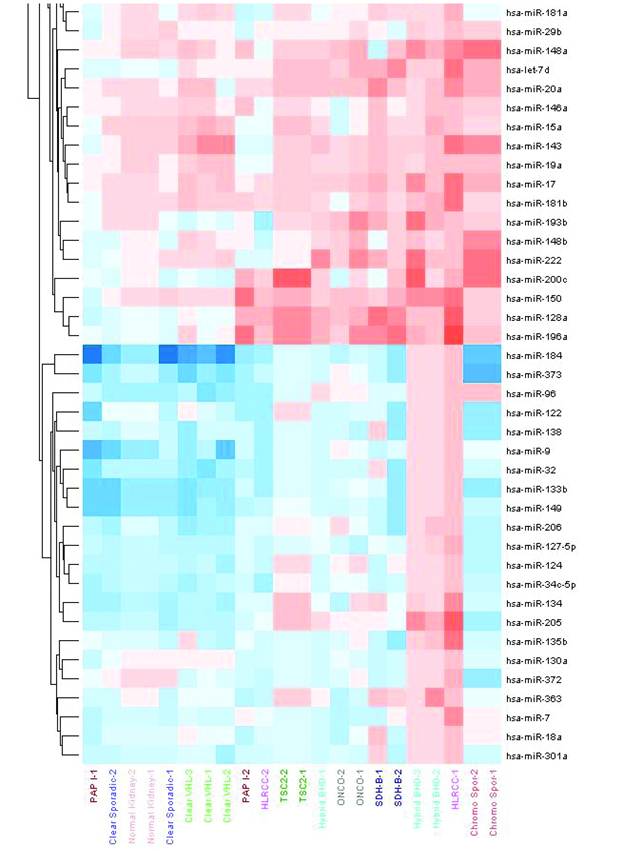
Unsupervised hierarchical clustering of tumors based on miRNA expression values showed two major clusters of samples (Figure 1). The first cluster grouped both the normal kidney and tumors derived from the proximal nephron (i.e. clear cell and papillary type I). Sporadic ccRCC as well as those tumors arising within VHL syndrome showed very similar expression pattern. The second major cluster contained tumors that are thought arise from the distal nephron, including benign oncocytomas and chromophobe RCCs.
Specific patterns together with type-specific profiles were observed. Twenty-three miRNAs were found to be differentially expressed in both sporadic and VHL-dependent ccRCC. Sporadic clear cell tumors showed a unique pattern of 14-miRNA that were absent from the VHL-dependent tumors. These also showed 15 miRNAs specific to the hereditary type. Common miRNAs to both sporadic and hereditary forms included miR-92a and miR-210 (Table 2).
Unsupervised hierarchical clustering of RCC samples based on microRNA expression values. Two major clusters of samples were generated: a “proximal tubule origin” group that included clear cell RCCs either sporadic or hereditary, and papillary type I RCC. The second group included “distal tubule origin” samples that consisted of benign oncocytomas, hybrid oncocytic tumors from BHD patients and sporadic chromophobe tumors. This clustering indeed follows our current understanding of the biologic and anatomical origin of these tumors.

Clinicopathologic characteristics of the patients and samples used for miRNA profiling.
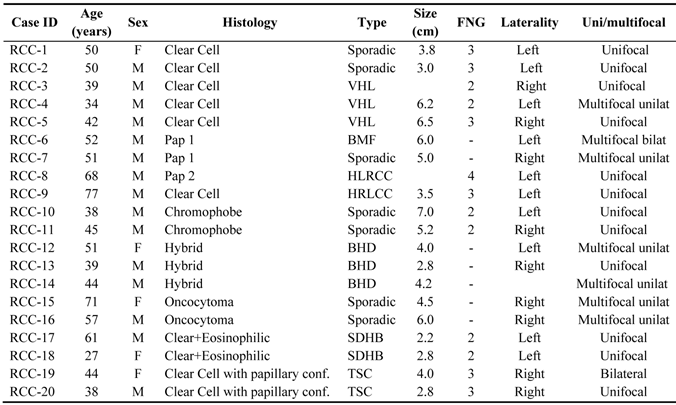
Abbreviations: VHL: Von-Hippel Lindau; BDH: Birt-Hogg-Dube; BMF: Bilateral Multifocal; HLRCC: Hereditary Leiomyomatosis Renal Cell Carcinoma; SDHB: Succinate Dehydrogenase B; TSC: Tuberous Sclerosis Complex.
Common and unique miRNAs to both sporadic and hereditary forms of clear cell RCCs.

Another major finding was the differential expression of two closely related histologic subtypes: renal oncocytomas and chromophobe tumors. The comparison demonstrated that these entities share most of the miRNAs expressed. Chromophobe tumors (77 miRNAs), hybrid oncocytic lesions (77 miRNAs) and renal oncocytomas (68 miRNas) had 57 miRNAs in common, and only a subset of 6, 2, and 1 miRNAs respectively were unique to each group (Figure 2).
Venn diagram showing the common and unique microRNAs expressed in the closely related chromophobe tumors, hybrid oncocytic and benign oncocytomas. Upregulated microRNAs are highlighted in red, whereas downregulated ones are in blue.

miR-92a has negative regulatory effects over VHL gene
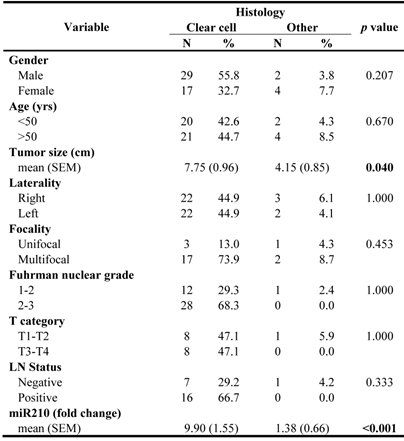
To test the hypothesis that miRNAs may have regulatory effects on the VHL gene and therefore might be implicated in ccRCC tumorigenesis, three major miRNA databases with alternative algorithms to determine potential miRNA-mRNA interactions and containing expected targets were interrogated to find miRNAs targeting the VHL gene. Four sequences were identified: miR-92a, miR-143, miR-320 and miR-519d with several sites of miRNA interactions throughout the VHL gene sequence. It is noteworthy that miR-92a came out as a common denominator of clear cell tumors in the profiling of all RCCs (Table 2). Forty-three ccRCCs were evaluated for the expression of these 4 miRNAs together with VHL gene mRNA and protein. The clinicopathologic characteristics of these samples including the genetic evaluation for VHL mutational status are shown in Table 3. Both groups of ccRCCs were similar except for tumor size that was larger in sporadic tumors. Mutations were equally distributed throughout the sequence, and no particular gene phenotype association was found.
As a proof of principle, several cell lines were tested for VHL mRNA expression. The results demonstrated that 786.O and UOK117 tumor cell lines having VHL mutations had indeed low transcript levels compared to the two normal kidney cell lines HRCE and HK-2 (Figure 3A). Likewise, VHL-/- ccRCC tumors had much lower transcript levels when compared to sporadic tumors (Figure 3B), and a striking inverse correlation of miR-92a with VHL mRNA levels was found (Figure 4A). A trend of higher miR-92a and miR-519d expression was seen in high Fuhrman nuclear grade tumors (Figure 4B-C).
VHL transcript evaluation in clear cell RCC (VHL-derived and sporadic) and cell lines. A. Two clear cell tumor-derived cell lines with different mutations showing decreased expression for VHL mRNA. B. Clear cell tumors arising in hereditary form had significantly lower transcript levels compared to sporadic cases.

A. Influence of VHL-targeting microRNAs in VHL mRNA transcript levels. An inverse correlation between miR-92 expression and VHL mRNA levels was seen. B-C. Higher levels of miR-92a and miR-519d were seen in higher grade clear cell tumors.

Hypoxia activation in clear cell RCC is followed by miR-210 overexpression
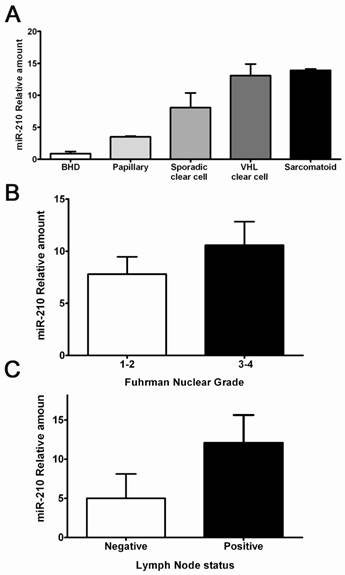
A hallmark of ccRCCs is the activation of hypoxia signaling mediated by HIF-1α accumulation. To test the hypothesis that the hypoxia-regulated microRNA-210 (miR-210) is part of the ccRCC phenotype, we evaluated its expression in a series of clear cell tumors and other histologic types supposedly hypoxia-independent. The clinicopathologic characteristics of these samples are shown in Table 4. Clear cell tumors showed significantly higher miR-210 expression levels when compared to tumor of non-clear cell histology (9.90-fold vs. 1.36, p<0.001). Tumors with sarcomatoid changes had the largest expression levels, followed by both sporadic and hereditary ccRCC. Papillary and hybrid oncocytic tumors showed significantly lower expression levels when compared to ccRCCs (Figure 5A). Interestingly, VHL mutations in exon 1, deletions, stop mutations and cases having more than 1 mutation in the gene had increased miR-210 expression (data not shown). Similarly, higher Fuhrman nuclear grade tumors and tumors with lymph node metstasis showed increased expression levels of miR-210 (Figure 5B-C).
Hypoxia-regulated miR-210 expression in RCCs and its correlation with some clinicopathologic parameters. A. Clear cell tumors including some with sarcomatoid differentiation showed the highest expression levels compared to RCC tumors of non-clear cell histology. B-C. Higher miR-120 expression was seen in high grade tumors and in lesions with positive lymph nodes.
Clinicopathologic characteristics of the patients with clear cell RCC (both VHL and sporadic).

Discussion
We have determined initially genome-wide expression changes of microRNAs in a group of kidney tumors representing a spectrum of RCC lesions. miRNA profiling of these tumors demonstrated unique expression patterns for each histologic subtype. These results are in agreement and expand previous attempts on the subject. Using different platforms, two of the earliest publications (10, 29) identified a subset of miRNAs that were mostly under-expressed in tumor tissues, including miR-141 and miR-200c, in a group of samples from patients with clear cell and chromophobe RCCs. Several publications have followed (11, 30-34), many of them replicating such initial findings despite variations in the technology used for expression analysis or statistical approaches.
We decided to use a medium throughput, PCR array-based analysis which allowed increased sensitivity for the detection of differentially expressed microRNAs for the initial profiling of our samples. We were able to detect microRNAs that are common denominators for the most frequent type of kidney cancer, i.e. clear cell RCC. From these, miR-92a, miR-210 and miR-200c appeared as significantly and differentially expressed between tumor cells and normal kidney parenchyma, indicating a putative role in disease pathogenesis. Both miR-210 and miR-200c have appeared in previous attempts to profile microRNA expression in clear cell RCC (10, 31, 32, 34, 35).
Clinicopathologic features of patients and samples analyzed for miR-210 expression.
Another major contribution of this work is that we have evaluated a genetically defined subset of RCCs for microRNA expression which, as a difference with in previous work, has frequently resulted from pooling of samples (30, 34). While the microRNA signature can reflect differences in the underlying and unique molecular changes for each histologic type, its major clinical application could be on the classification of normal and cancerous tissues as well as tumor categorization. This feature can prove useful for supporting the differential diagnosis or when definitive classification is needed (36, 37).
Consistent with bioinformatic approaches, experimental data supported the hypothesized involvement of miR-92a in clear cell tumors. ccRCC expressed increased amount of miR92a and these tumors showed decreased levels of VHL mRNA, supporting the concept that microRNAs may influence transcript abundance of protein-coding target genes in RCC. miR-92a is a member of the 13q31 miRNA polycistron (17-92 miRNA cluster) which also includes miR-17, miR-18a, miR-19a, miR-19b and miR-20a (38, 39). Only few reports are available on the role of miR-92 specifically in cancer (40-42) or about its potential targeted genes. Ghosh et al. (41) described for the first time the potential role of miR-92 in chronic lymphocytic leukemia (CLL) linking miR-92 and hypoxia signaling through HIF-1α activation. miR-92 was found overexpressed in CLL B cells targeting the VHL transcript, repressing its expression in this model. Almost simultaneously we started our initial work on the regulation of VHL by miR-92a (42). More recently, miR-92 has also been described to be involved in estrogen receptor beta (ERβ1) regulation in breast cancer (40). In that report, a negative correlation between ERβ1 expression and miR-92 in a series of breast cancer tissues and cell lines was found, further implicating a role for miR-92 in breast cancer pathogenesis.
Our data is also consistent with the results from independent groups analyzing clear cell RCCs that demonstrate increased expression of other members of the miR17-92 cluster at significantly higher levels than normal kidney (35). In a series of fifteen paired tumor and normal tissues from patients with clear cell RCC, Chow et al. (35) demonstrated not only that tumors overexpressed miR-17-5p, 18a and 20a but also that they have specific roles in the regulation of cell proliferation in vitro. Also in line with our results of an overexpression of miR-92a in clear cell kidney cancer, miR-17-5p and its cluster miR-17-92 have been observed overexpressed in human lung, breast, colon, pancreas, and prostate cancers (14, 38). Furthermore, overexpression of miR-17-5p has been associated to c-Myc amplification, accelerating tumor development in a mouse B-cell lymphoma model, and thus indicating an oncogenic role for this microRNA (39).
Lastly, we found evidence that the activation of some hypoxia-responsive microRNAs are part of the cascade of events following VHL gene inactivation, and that the expression of such microRNAs may be related to a more aggressive phenotype. Clear cell tumors, either in sporadic or hereditary forms, showed significantly higher levels of miR-210 when compared to other histologic types, and higher levels were seen in higher grade tumors of lesions with lymph node metastasis. Kulshreshtha et al. (25) documented for the first time the expression pattern of microRNAs in cell lines as induced by hypoxia. The study uncovered many hypoxia-regulated miRNAs, several of them -miR210, miR21- also described as upregulated in clear cell RCC. In kidney cancer, four out of 8 studies (31, 32, 35, 43), including our own, show miR210 in the top list of upregulated microRNAs in clear cell RCC.
This finding is not unique to RCC. Camps et al. (28) identified and validated that in low oxygen levels, miR-210 had the most significant change in breast cancer. They also showed evidence that this response is mediated through the Hypoxia Inducible Factor-1 alpha (HIF-1α)/Von Hippel Lindau (VHL) tumor suppressor system. Several other studies now point to miR-210 as the most ubiquitously upregulated microRNA by HIF-1α in different tumor types and cell lines (44).
In summary, our data show consistent disregulation of miRNAs in human kidney cancer, some of which are potentially involved in critical gene silencing in RCC and others that are activated as part of the pathophysiological response in these tumors, implicating a major role for microRNA expression in every step of RCC pathogenesis. The results of this study uncover an etiological contribution of miRNAs in RCC and validate their use for future studies on the role of individual miRNAs in disease progression that could be used for development of miRNA-based therapeutic strategies. A more detailed understanding of the molecular mechanisms and regulatory pathways at work in RCC will enormously assist in improving the design and target selection of therapeutic strategies.
Conflict of Interest
The authors have declared that no conflict of interest exists.
References
1. Jemal A, Siegel R, Xu J, Ward E. Cancer Statistics, 2010. CA: a cancer journal for clinicians. 2010
2. Ferlay J, Shin HR, Bray F. et al. GLOBOCAN 2008, Cancer Incidence and Mortality Worldwide: IARC CancerBase No.10 [Internet]. International Agency for Research on Cancer. 2010.
3. Linehan WM, Bratslavsky G, Pinto PA. et al. Molecular diagnosis and therapy of kidney cancer. Annual review of medicine. 2010;61:329-43
4. Jones TD, Eble JN, Cheng L. Application of molecular diagnostic techniques to renal epithelial neoplasms. Clinics in laboratory medicine. 2005;25:279-303
5. Schuetz AN, Yin-Goen Q, Amin MB. et al. Molecular classification of renal tumors by gene expression profiling. J Mol Diagn. 2005;7:206-18
6. Valera VA, Li-Ning TE, Walter BA, Roberts DD, Linehan WM, Merino MJ. Protein expression profiling in the spectrum of renal cell carcinomas. Journal of Cancer. 2010;1:184-96
7. Young AN, Amin MB, Moreno CS. et al. Expression profiling of renal epithelial neoplasms: a method for tumor classification and discovery of diagnostic molecular markers. The American journal of pathology. 2001;158:1639-51
8. Fridman E, Dotan Z, Barshack I. et al. Accurate Molecular Classification of Renal Tumors Using MicroRNA Expression. The Journal of Molecular Diagnostics. 2010;12:687-96
9. Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6:857-66
10. Nakada C, Matsuura K, Tsukamoto Y. et al. Genome-wide microRNA expression profiling in renal cell carcinoma: significant down-regulation of miR-141 and miR-200c. The Journal of pathology. 2008;216:418-27
11. Chow TF, Youssef YM, Lianidou E. et al. Differential expression profiling of microRNAs and their potential involvement in renal cell carcinoma pathogenesis. Clinical biochemistry. 2009;43:150-8
12. Croce CM. Causes and consequences of microRNA dysregulation in cancer. Nat Rev Genet. 2009;10:704-14
13. Calin GA, Liu CG, Sevignani C. et al. MicroRNA profiling reveals distinct signatures in B cell chronic lymphocytic leukemias. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:11755-60
14. Volinia S, Calin GA, Liu CG. et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:2257-61
15. Esquela-Kerscher A, Slack FJ. Oncomirs - microRNAs with a role in cancer. Nat Rev Cancer. 2006;6:259-69
16. Grignon DJ, Che M. Clear cell renal cell carcinoma. Clinics in laboratory medicine. 2005;25:305-16
17. Cohen HT, McGovern FJ. Renal-cell carcinoma. The New England journal of medicine. 2005;353:2477-90
18. Linehan WM, Rubin JS, Bottaro DP. VHL loss of function and its impact on oncogenic signaling networks in clear cell renal cell carcinoma. The international journal of biochemistry & cell biology. 2009;41:753-6
19. Wiesener MS, Munchenhagen PM, Berger I. et al. Constitutive activation of hypoxia-inducible genes related to overexpression of hypoxia-inducible factor-1alpha in clear cell renal carcinomas. Cancer research. 2001;61:5215-22
20. Fischer U, Radermacher J, Mayer J, Mehraein Y, Meese E. Tumor hypoxia: Impact on gene amplification in glioblastoma. International journal of oncology. 2008;33:509-15
21. Kirkpatrick JP. Tumor hypoxia and prognosis in human gliomas. Cancer journal (Sudbury, Mass. 2006;12:451-4
22. Vaupel P, Mayer A, Hockel M. Tumor hypoxia and malignant progression. Methods in enzymology. 2004;381:335-54
23. Hutchison GJ, Valentine HR, Loncaster JA. et al. Hypoxia-inducible factor 1alpha expression as an intrinsic marker of hypoxia: correlation with tumor oxygen, pimonidazole measurements, and outcome in locally advanced carcinoma of the cervix. Clin Cancer Res. 2004;10:8405-12
24. Beasley NJ, Leek R, Alam M. et al. Hypoxia-inducible factors HIF-1alpha and HIF-2alpha in head and neck cancer: relationship to tumor biology and treatment outcome in surgically resected patients. Cancer research. 2002;62:2493-7
25. Kulshreshtha R, Ferracin M, Wojcik SE. et al. A microRNA signature of hypoxia. Molecular and cellular biology. 2007;27:1859-67
26. Ho AS, Huang X, Cao H. et al. Circulating miR-210 as a Novel Hypoxia Marker in Pancreatic Cancer. Translational oncology. 2010;3:109-13
27. Gee HE, Camps C, Buffa FM. et al. hsa-mir-210 is a marker of tumor hypoxia and a prognostic factor in head and neck cancer. Cancer. 2010;116:2148-58
28. Camps C, Buffa FM, Colella S. et al. hsa-miR-210 Is induced by hypoxia and is an independent prognostic factor in breast cancer. Clin Cancer Res. 2008;14:1340-8
29. Gottardo F, Liu CG, Ferracin M. et al. Micro-RNA profiling in kidney and bladder cancers. Urologic oncology. 2007;25:387-92
30. Huang Y, Dai Y, Yang J. et al. Microarray analysis of microRNA expression in renal clear cell carcinoma. Eur J Surg Oncol. 2009;35:1119-23
31. Juan D, Alexe G, Antes T. et al. Identification of a microRNA panel for clear-cell kidney cancer. Urology. 2010;75:835-41
32. Jung M, Mollenkopf HJ, Grimm C. et al. MicroRNA profiling of clear cell renal cell cancer identifies a robust signature to define renal malignancy. Journal of cellular and molecular medicine. 2009;13:3918-28
33. Petillo D, Kort EJ, Anema J, Furge KA, Yang XJ, Teh BT. MicroRNA profiling of human kidney cancer subtypes. International journal of oncology. 2009;35:109-14
34. Yi Z, Fu Y, Zhao S, Zhang X, Ma C. Differential expression of miRNA patterns in renal cell carcinoma and nontumorous tissues. Journal of cancer research and clinical oncology. 2009;136:855-62
35. Chow T-fF, Mankaruos M, Scorilas A. et al. The miR-17-92 Cluster is Over Expressed in and Has an Oncogenic Effect on Renal Cell Carcinoma. The Journal of Urology. 2010;183:743-51
36. Schaefer A, Stephan C, Busch J, Yousef GM, Jung K. Diagnostic, prognostic and therapeutic implications of microRNAs in urologic tumors. Nature reviews. 2010;7:286-97
37. Valera VA, Merino MJ. Misdiagnosis of clear cell renal cell carcinoma. Nature reviews. 2011;8:321-33
38. Hayashita Y, Osada H, Tatematsu Y. et al. A polycistronic microRNA cluster, miR-17-92, is overexpressed in human lung cancers and enhances cell proliferation. Cancer research. 2005;65:9628-32
39. He L, Thomson JM, Hemann MT. et al. A microRNA polycistron as a potential human oncogene. Nature. 2005;435:828-33
40. Al-Nakhle H, Burns PA, Cummings M. et al. Estrogen receptor {beta}1 expression is regulated by miR-92 in breast cancer. Cancer research. 2010;70:4778-84
41. Ghosh AK, Shanafelt TD, Cimmino A. et al. Aberrant regulation of pVHL levels by microRNA promotes the HIF/VEGF axis in CLL B cells. Blood. 2009;113:5568-74
42. Valera VA, Walter BA, Linehan WM, Sobel ME, Merino MJ. microRNA Regulation of VHL Gene in Clear Cell Renal Cell Carcinomas. Mod Pathol. 2009;22:1709
43. Valera VA, Walter BA, Sobel M, Linehan M, Merino MJ. miR-210 (Hypoxia-Responsive) a Marker of Poor Prognosis in Clear Cell Renal Cell Carcinoma. San Antonio, TX: United States & Canadian Academy of Pathology 100th Annual Meeting. 2011
44. McCormick R, Buffa FM, Ragoussis J, Harris AL. The role of hypoxia regulated microRNAs in cancer. Current topics in microbiology and immunology. 2010;345:47-70
Author contact
![]() Corresponding author: Maria J. Merino M.D, Address: 10 Center Drive, Building 10 MSC Room 2B44, Bethesda MD 20892. Phone: (301) 496-2441; Fax: (301) 480-1458; E-mail: mjmerinonih.gov
Corresponding author: Maria J. Merino M.D, Address: 10 Center Drive, Building 10 MSC Room 2B44, Bethesda MD 20892. Phone: (301) 496-2441; Fax: (301) 480-1458; E-mail: mjmerinonih.gov
Received 2011-10-2
Accepted 2011-10-20
Published 2011-10-21

 Global reach, higher impact
Global reach, higher impact