If we want to retrieve "matching" profiles from a large collection of image-based profiling experiments (for example to find similar drugs, similar genes, or drug-gene or drug-disease combinations), how do we ensure that the profiles are aligned well enough? The CytoData 2018 Challenge addresses this, featuring batch effect correction and cross dataset profile matching 💿 🔀 📀. The challenge involves the transformation of signatures using machine learning 👾 or statistical methods 📊. You will be given two datasets of image-based signatures 💿 ➕ 📀 acquired at different times 📅 🕜 and with different experimental conditions 💊 💉 with the goal of retrieving correct matches accurately 🎯. See http://cytodata.org/ for details of the event.
👽 : What is image-based profiling?
😎 : In the study of biological systems, microscopy images are used to measure the response of cells to treatments or perturbations. Cell state can be observed and quantitatively measured using images by following a computational workflow known as profiling. Single cells are first identified in all images, and then their main characteristics are represented in feature vectors. The information of a population of cells is aggregated into a single vector, also called profile, containing summary statistics of the features of all cells. These profiles encode the morphological changes of cell populations exposed to treatments. Image-based profiles can be used to compare the response of cells to different treatments, and to map their similarities.
👽 : Is image-based profiling the same as image-based screening?
😎 : Screening and profiling are different. Screening uses images to identify phenotype(s) of interest known beforehand. Profiling measures as many cell properties as possible, using all the phenotypes to identify relationships among multiple different samples.
👽 : What are the applications of image-based profiling?
😎 : Image-based profiles can be used for drug discovery and functional genomics applications. There are many types of biological studies that can be conducted using image-based profiling. In the CytoData challenge, we use data from chemical and genetic perturbation experiments (see below).
👽 : What imaging assays can be used for profiling?
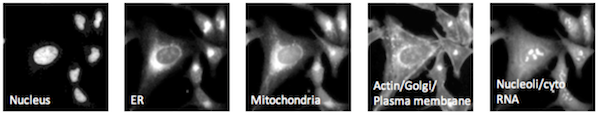
😎 : Virtually any imaging assay can be used for profiling, especially high-content assays. In the 2018 CytoData challenge, we use an imaging assay called Cell Painting, that paints the cells with 6 stains, imaged in 5 channels, highlighting 8 cellular compartments. This is an unbiased, general purpose assay that maximizes information content for profiling, but the assay can be adapted to meet the needs of a research project.
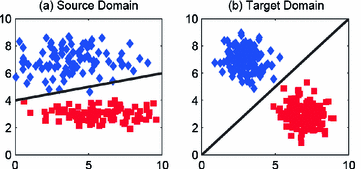
As in many biological experiments, imaging data may be subject to batch effects and undesired artifacts 😱.
More specifically, given two batches of microscopy images with the same treatments 💊, but acquired under different technical conditions
The goal of the challenge 🏁 is to analyze the profiles of two different batches of data
↗️ ↗️ Replicate correlation- 🔝🔄 Enrichment of biologically relevant matches in the top connections
- 🆔✅ Correct association of treatment type
From the data analysis perspective, the problem can be formulated in various ways, including manifold learning, domain adaptation, subspace alignment, and transfer learning.
We are glad to announce that four datasets will be provided during the CytoData 2018 Challenge 🎉🎉🎉🎉. All of them were acquired using the Cell Painting assay, at high-throughput, in 384 well plates 🔬, as part of the research conducted in the Broad Institute of MIT and Harvard. The following table describes the experimental details of each dataset.
| Dataset 📀 | Type 💉 💊 | Number of treatments #️⃣ | Cell line ♋ |
|---|---|---|---|
| BBBC037 | Genetic perturbations. ORF over-expression | 200 wild type genes | U2OS |
| BBBC043 | Genetic perturbations. ORF over-expression | 596 alleles of 53 genes | A549 |
| BBBC022 | Chemical perturbations. Bioactive compounds | 1,600 compounds | U2OS |
| BBBC036 | Chemical perturbations. Bioactive compounds | 5,000 compounds | U2OS |
Notice that two datasets represent genetic perturbations and the other two represent chemical perturbations. The challenge will consider the cross-dataset matching problem across each of the two pairs 💿🔀📀, i.e, profiles in BBBC037 have to be matched with profiles in BBBC043 because both contain genetic perturbations. Similarly, profiles in BBBC022 have to be matched with profiles in BBBC036 because both contain chemical perturbations.
The imaging data for all three datasets is more than 3TB of data 💥, which will be available to everyone during and after the challenge. However, to facilitate the analysis of treatment profiles and to focus on the cross-dataset matching problem, all the datasets have been processed before-hand using the profiling workflow described above 😎. In particular, two versions of well-level population profiles will be available during the challenge:
- Classical features computed with the CellProfiler software using pipelines optimized for Cell Painting images.
- Deep learning features computed with a convolutional neural network pretrained on ImageNet.
All data is available as Amazon Public Data Set on https://registry.opendata.aws/cell-painting-image-collection/ and be accessed at
s3://cytodata/
All image data and extracted single cell features and aggregated profiles can be found in s3://cytodata/datasets/. This folder has the structure
.
├── Bioactives-BBBC022-Gustafsdottir
│ ├── profiles
│ │ └── Bioactives-BBBC022-Gustafsdottir
│ ├── images
│ │ └── Bioactives-BBBC022-Gustafsdottir
│ └── metadata
│ └── Bioactives-BBBC022-Gustafsdottir
├── CDRPBIO-BBBC036-Bray
│ ├── profiles
│ │ └── CDRPBIO-BBBC036-Bray
│ ├── images
│ │ └── CDRPBIO-BBBC036-Bray
│ └── metadata
│ └── CDRPBIO-BBBC036-Bray
├── LUAD-BBBC041-Caicedo
│ ├── profiles
│ │ └── LUAD-BBBC041-Caicedo
│ ├── images
│ │ └── LUAD-BBBC041-Caicedo
│ └── metadata
│ └── LUAD-BBBC041-Caicedo
└── TA-ORF-BBBC037-Rohban
├── profiles
│ └── TA-ORF-BBBC037-Rohban
├── images
│ └── TA-ORF-BBBC037-Rohban
└── metadata
└── TA-ORF-BBBC037-Rohban
The subfolder contain the following information:
- the directory
imagescontain Cell Painting images as tiff files - the directory
profilescontains single cell data in sqlite format and profiles aggregated to replicate level as csv files (aggregated as mean profiles per well) - the metadata directory contains information about the platemaps and the used perturbations.
The CytoData 2018 challenge will be a collaborative hackathon ✨💻, with participants forming teams to discuss and implement solutions to the problem.
The challenge will run for two days only, so participants are encouraged to investigate and plan some solutions before the event starts 📝.
In order to meet other participants, we will provide a slack channel to make general announcements and allow participants to organize teams and exchange ideas 💡.
It's also a great idea to start discussing methods here in this GitHub repository ![]() :
:
add issues with relevant links if you want to suggest a methodology and discuss it with other participants!
Teams will have no fewer than three 3️⃣ and no more than five 5️⃣ participants, ideally from different institutions.
Teams will compete with each other ![]() to improve the three performance metrics mentioned above 🎳.
Participants of the team will be able to upload solutions to a scoreboard to check that everything is running properly and to get feedback on performance 👌.
The best performing solutions will win prizes provided by our sponsors! 🏆👏
to improve the three performance metrics mentioned above 🎳.
Participants of the team will be able to upload solutions to a scoreboard to check that everything is running properly and to get feedback on performance 👌.
The best performing solutions will win prizes provided by our sponsors! 🏆👏
The following resources will be provided during the challenge:
- 📡 Internet connection.
- 📀 Access to all files of the four datasets, including pre-computed profiles.
 A toolkit, written in R and Python, to load the pre-computed profiles, run a baseline model and create a submission.
A toolkit, written in R and Python, to load the pre-computed profiles, run a baseline model and create a submission.- 📈 An account in the scoreboard to evaluate the generated submissions.
- 💻 Teams will be given access to pre-configured virtual machines in the Amazon Cloud to run experiments.
Participants of the challenge can make use of their own computational resources (laptops, servers, etc) to run experiments during the challenge.