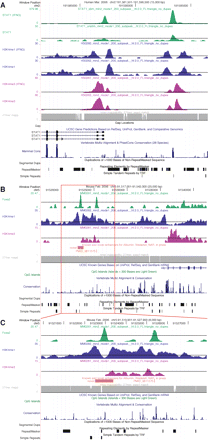
Examples of TF, histone modification, and read mappability profiles. (A) A 15-kb UCSC hg18 genome browser view around the 5′-end of the STAT1 gene (chr2:191,581,501–191,596,500). The gene is transcribed from right to left. Custom tracks are XSET coverage profiles for (from the top) STAT1, H3K4me1, and H3K4me3. Pairs of profiles show IFNG-stimulated cells above and unstimulated cells below. (B,C) Overall 25-kb (chr5:91,517,001–91,542,000) and detailed 8-kb (chr5:91,519,901–91,527,900) UCSC mm8 genome browser views that include a known and a novel enhancer for the albumin (Alb1) gene (Wederell et al. 2008). The gene is transcribed from left to right. ChIP-seq coverage profiles are (from the top) FOXA2, H3K4me1, and H3K4me3 in mouse adult liver. (A–C) The final custom track is a profile for (gray) 27-mer exact match mappability; (black horizontal lines) coverages (heights) that correspond to profile-specific FDR thresholds of ∼0.01 (Table 1).











