Significance
A key link to understand human history in Island Southeast Asia is the Philippine archipelago and its poorly investigated genetic diversity. We analyzed the most comprehensive set of population-genomic data for the Philippines: 1,028 individuals covering 115 indigenous communities. We demonstrate that the Philippines were populated by at least five waves of human migration. The Cordillerans migrated into the Philippines prior to the arrival of rice agriculture, where some remain as the least admixed East Asians carrying an ancestry shared by all Austronesian-speaking populations, thereby challenging an exclusive out-of-Taiwan model of joint farming–language–people dispersal. Altogether, our findings portray the Philippines as a crucial gateway, with a multilayered history, that ultimately changed the genetic landscape of the Asia-Pacific region.
Keywords: Philippines, human population genetics, Austronesian, Negrito, ISEA
Abstract
Island Southeast Asia has recently produced several surprises regarding human history, but the region’s complex demography remains poorly understood. Here, we report ∼2.3 million genotypes from 1,028 individuals representing 115 indigenous Philippine populations and genome-sequence data from two ∼8,000-y-old individuals from Liangdao in the Taiwan Strait. We show that the Philippine islands were populated by at least five waves of human migration: initially by Northern and Southern Negritos (distantly related to Australian and Papuan groups), followed by Manobo, Sama, Papuan, and Cordilleran-related populations. The ancestors of Cordillerans diverged from indigenous peoples of Taiwan at least ∼8,000 y ago, prior to the arrival of paddy field rice agriculture in the Philippines ∼2,500 y ago, where some of their descendants remain to be the least admixed East Asian groups carrying an ancestry shared by all Austronesian-speaking populations. These observations contradict an exclusive “out-of-Taiwan” model of farming–language–people dispersal within the last four millennia for the Philippines and Island Southeast Asia. Sama-related ethnic groups of southwestern Philippines additionally experienced some minimal South Asian gene flow starting ∼1,000 y ago. Lastly, only a few lowlanders, accounting for <1% of all individuals, presented a low level of West Eurasian admixture, indicating a limited genetic legacy of Spanish colonization in the Philippines. Altogether, our findings reveal a multilayered history of the Philippines, which served as a crucial gateway for the movement of people that ultimately changed the genetic landscape of the Asia-Pacific region.
The Philippines is an archipelago of 7,641 islands situated in Island Southeast Asia (ISEA) at the crossroads of past human migrations in the Asia-Pacific region. Until the end of the Last Glacial Period (∼11.7 thousand years ago [kya]), the Philippine islands were mostly contiguous as one large landmass, separated from Sundaland by the Mindoro Strait and the Sibutu Passage (SI Appendix, section 2). Hominins have inhabited the Philippine islands since at least 67 kya (1, 2). While the ancestors of the ethnic groups that self-identify as Negritos are widely regarded as the first human inhabitants (3, 4), their precise relationships to archaic humans, to other early Asian groups, and to subsequent colonizations remain poorly explored and disputable.
Earlier investigations using uniparental markers and/or autosomal data have attempted to resolve the various potential migration events into ISEA (5–11), without arriving at a clear consensus. The lack of resolution may be due to insufficient representation of the diverse ethnic groups of the Philippines and the limited density of the genomic data used. To address these issues, we collected and analyzed the most comprehensive set of population-genomic data thus far for the Philippines: 1,028 individuals representing 115 distinct cultural communities from all geographical regions (Fig. 1A and SI Appendix, Fig. S1A), genotyped for ∼2.5 million single-nucleotide polymorphisms (SNPs). To better understand the history of migrations from the East Asian mainland during the early Holocene, we produced genomic data from two ancient individuals from Liangdao in the Taiwan Strait, with calibrated radiocarbon dates of 8,060 to 8,320 before present (B.P.) and 7,510 to 7,590 B.P., respectively (Fig. 1A).
Fig. 1.
Overview of Philippine population structure. (A) Location of indigenous cultural communities included in the study. The populations are color-coded to represent ethnic group clusters. (B) PCA with worldwide populations, with sample size matched for each regional group. (C) PCA restricted to Philippine ethnic groups. (D) Clustering of Asia-Pacific populations (315,692 SNPs) with an Inset Graph showing clustering of 115 Philippine populations (2.3 million SNPs), assuming K number of genetic ancestry components.
Our analyses indicate that the Philippines was populated by at least five major human migrations: Northern and Southern Negrito branches of a Basal Australasian group, who likely admixed independently with local Denisovans within the Philippines, plus Papuan-related groups, as well as Manobo, Sama, and Cordilleran branches of Basal East Asians. Cordillerans, who remain the least admixed branch of Basal East Asians, likely entered the Philippines prior to established dates for the agricultural transition and carried with them a genetic ancestry that is widespread among all Austronesian (AN)-speaking populations. This complex demographic history underscores the importance of the Philippines as a migration gateway that profoundly influenced the genetic makeup of populations in the Asia-Pacific region.
Results
Distinct Population Stratification across the Philippines Reveals Multiple Ancestral Sources.
Investigation by principal component analysis (PCA) demonstrates that all Philippine ethnic groups cluster together with Asia-Pacific populations in a global comparison (Fig. 1B and SI Appendix, Fig. S1 B and C). While Negritos form a distinctive cline that aligns between Papuans and non-Negritos, Cordillerans interestingly lie to the edge that defines the East Asian cluster on principal component (PC) 1, even more extreme than Native American groups and Oceanian groups (Fig. 1B). There is a clear dichotomy between Negritos and non-Negritos, indicating the deep divergence between Basal East Asian ancestry best represented by Cordillerans and Basal Australasian ancestry represented by Negrito-AustraloPapuans (Fig. 1B and SI Appendix, Fig. S1 B and C). Looking into the fine-scale analysis of Asia-Pacific populations, non-Negritos clearly separate into groups either affiliated with Cordillerans or with Mainland Southeast Asia (MSEA) ethnic groups (such as Htin and Mlabri, or Malay non-Negritos) (SI Appendix, Fig. S1 D–G).
Further analyses reveal distinct genetic structure among Philippine Negrito groups, as will be described later, and stratification among non-Negrito groups, exemplified by Cordillerans, Mangyans, Manobos, and Sama Dilaut groups (Fig. 1 C and D). These observations are consistent with inferred ancestry components (SI Appendix, Fig. S2 B–D). Briefly, stratification commences with a dichotomy between AustraloPapuan-related Negritos vs. non-Negritos, subsequent clustering of non-Negritos into Cordilleran vs. MSEA-affiliated populations, and stratification of Negritos into Ayta and Agta groups and non-Negritos into Cordilleran, Mangyan, Manobo, and Sama-related populations.
Philippine Negritos Exhibit Deep Divergences.
Given the geographical barriers and the likely long history of isolation between populations, some degree of differentiation is expected among Negrito groups. For instance, PCA restricted to Negrito groups reveals a gradient between Central Luzon Negrito groups and Southern Negrito groups (PC1), and an east-to-west clustering of Negritos in the northern Philippines along PC2 (SI Appendix, Fig. S4A). The Northern Negritos additionally exhibit deep population structure, separating into three clusters: Central Luzon Negritos (all Ayta Negritos), Southeastern Luzon Negritos (Agta groups of Bicol region and Quezon province), and Northeast Luzon Negritos (Agta, Atta, and Arta Negritos of Cagayan region) (SI Appendix, Fig. S4B).
Negritos of the northern Philippines are an outgroup to both Australians and Papuans (Fig. 2A and SI Appendix, Figs. S4 D, E, G–I and S5D). Using a coalescent-based split-time estimation approach and assuming a population divergence model (12, 13), we estimate the ancestors of Negritos of the northern Philippines to have diverged ∼46 kya (95% CI: 45.5 to 46.8 kya) from a common ancestral Australasian population (Basal Sunda). The divergence likely happened in the old continental landmass of Sundaland, prior to the Australian-Papuan divergence ∼25 kya (95% CI: 24.7 to 26.7 kya), possibly as a consequence of migration into Luzon to become present-day Negritos of the northern Philippines (SI Appendix, Figs. S4J and S5D).
Fig. 2.
Inferred admixture graph models for Australasians and East Asians. Inferred admixture graph models based on a combination of qpGraph and f statistical analyses presented in SI Appendix, Figs. S4–S8. (A) Topology of Australasian cluster indicating pulses of Denisovan introgression events, and estimation of divergence time between Philippine Negritos and AustraloPapuans. (B) Topology of the East Asian cluster showing relationships between Cordillerans, Manobo, Sama, and mainland Asian ethnic groups with inferred admixture events and divergence dates. See main text for divergence time confidence intervals.
In contrast to the north, the AustraloPapuan-like genetic signal is distinctively higher in Southern Negritos (e.g., Mamanwa) than in other Negrito groups (Fig. 2A and SI Appendix, Fig. S4 K–M). The Mamanwa Negritos also appear as an outgroup to Papuans and Australians (SI Appendix, section 4.3), indicating that the ancestral Mamanwa is an offshoot group of Basal Oceanian that diverged ∼37 kya (95% CI: 36.2 to 38.7 kya), by which entered Mindanao Island, likely via the Sulu Archipelago, prior to the Australian-Papuan divergence (Fig. 2A and SI Appendix, Figs. S4J and S5D). However, an alternative model where Southern Negritos form a clade with Northern Negritos is not rejected (SI Appendix, section 4.3 and Fig. S5G). Given this, we are unable to exclude a scenario where a common ancestral Negrito population entered the Philippines exclusively through a single port of entry, either via Palawan or via the Sulu archipelago, and subsequently diverged within the Philippines to become the Northern and Southern Negritos.
Both Northern and Southern Negritos subsequently admixed with Cordilleran-related populations, and, interestingly, Southern Negritos received an additional gene flow from Papuan-related populations after Australian-Papuan divergence (SI Appendix, Fig. S5 D and G). This previously unappreciated northwest gene flow of Papuan-related ancestry had its greatest impact on eastern Indonesia, as well as ethnic groups of the southeastern Philippines, such as Sangil and Blaan (SI Appendix, section 4.5).
Ancestral Manobos and Ancestral Sama Entered the Southern Philippines Earlier Than the Expansion of Cordilleran-Related Populations.
The ethnic groups of the southern Philippines exhibit a ubiquitous ancestry that is non–AustraloPapuan-related and which is generally absent among non-Negrito groups of the northern Philippines. This unique genetic signature, heretofore designated as “Manobo ancestry,” is highest among inland Manobo groups of Mindanao Island (SI Appendix, Fig. S6A). When we masked Cordilleran and Southern Negrito ancestry and retained only the Manobo ancestry, the Manobo component became more apparent among other ethnic groups of Mindanao (SI Appendix, Fig. S6B). In addition to Manobo ancestry, another distinct ancestry was identified in the southwestern Philippines. This genetic signal is highest among Sama sea nomads of the Sulu Archipelago and is designated as “Sama ancestry” (SI Appendix, Fig. S6G). When we mask all other ancestries and retain only Sama ancestry, the Sama component becomes more evident among ethnic groups of Zamboanga Peninsula, Palawan, Basilan, Sulu, and Tawi-Tawi islands, even among populations who do not self-identify as Sama or speak a Sama-related language (SI Appendix, Fig. S6H).
Ethnic groups with high Sama ancestry exhibit significantly higher genetic affiliation with Austroasiatic-speaking ethnic groups of MSEA, such as Mlabri and Htin, relative to the least admixed Manobo group, Manobo Ata (SI Appendix, Figs. S6K and S7 A–D, J, and K). This Htin/Mlabri-related genetic signal is not only found in Sama Dilaut and inland Sama groups, but also in Palawanic and Zamboanga peninsula ethnic groups of the southwestern Philippines. These findings are consistent with previous observations where a Htin/Mlabri-related genetic signal was detected among ethnic groups of western Indonesia (10). In our analysis, we find that this genetic signal also extends beyond western Indonesia and into the southwestern Philippines.
Both Manobo and Sama genetic ancestries diverge from a common East Asian ancestral gene pool (∼15 kya [95% CI: 14.8 to 15.4 kya]) earlier than the estimated divergence between the indigenous peoples of Taiwan and Cordillerans (Fig. 2B and SI Appendix, Fig. S7E). Surprisingly, both of these ancestries (Manobo and Sama) diverged from the common East Asian branch before Han, Dai, and Kinh split from Amis, Atayal, or Cordillerans (Fig. 2B and SI Appendix, Figs. S6 E, F, and L). Hence, our findings indicate that Ancestral Manobo and Ancestral Sama, together with other Htin/Mlabri-related ethnic groups, form a branch that diverged from Basal East Asians ∼15 kya even before the expansion of Han, Dai, Japanese, Kinh, Amis, and Atayal (Fig. 2B and SI Appendix, Figs. S6 E, F, and L and S7E).
Sama forms a clade with Htin relative to Manobo Ata (SI Appendix, Fig. S7 A–D and sections 5.5 and 5.6). The common ancestor of Sama and Htin/Mlabri populations was estimated to have diverged from Ancestral Manobo ∼12 kya (95% CI: 11.4 to 12.6 kya). Given the geographic distribution of the Htin/Mlabri-related genetic signal today, their ancestors likely expanded into western Indonesia and the southwestern Philippines, via Sundaland, before the expansion of Cordilleran-related populations (14). Interestingly, the above estimated divergences (15 kya and 12 kya) coincide with the major geological changes in ISEA, inferred from reconstructions of Sundaland at the end of the Last Glacial Period (SI Appendix, Fig. S3 E and F). Accordingly, climate-driven changes in ISEA may have prompted postglacial movements and isolation of populations, which led to the differentiation of ethnic groups in the region.
Cordillerans Represent the Least Admixed Basal East Asian Group in a Worldwide Set of Populations.
Cordilleran ethnic groups reside in the only landlocked region of the Philippines, across the Cordilleran mountain range in north-central Luzon. Historically, Cordillerans were known to resist direct colonization and Christianization by the Spanish and hence were able to retain many of their distinct cultural practices (15). This geographic and cultural isolation may have played a role in the high linguistic diversity of the region and the low levels of genetic admixture displayed by some groups. In addition to the previously reported Kankanaey (16), and following admixture and combination of f3 admixture and f4 statistical analyses, we found Bontoc, Balangao, Tuwali, Ayangan, Kalanguya, and Ibaloi as being among the least admixed populations bearing Basal East Asian ancestry (SI Appendix, Figs. S2 B–D and S8 A–C and section 6.1).
The homogenous ancestry among Cordillerans observed in all our Admixture analysis could be explained by the presence of strong genetic drift. However, a previous analysis (11) and our investigation on the number and tract length of runs of homozygosity do not support a recent bottleneck or extensive inbreeding among Cordillerans (SI Appendix, Fig. S1 H–J and section 3.8). Moreover, formal tests using f3 admixture and f4 statistics provide direct evidence that Central Cordillerans retained to be the only ethnic groups within the Philippines who did not receive gene flow from Negritos (SI Appendix, Fig. S8 A–C). This is unexpected, given the series of migrations and periods of colonization in the surrounding area of the Cordillera region and the documented records of trade and historical interactions with Negrito and non-Negrito groups of Luzon (17). All other ethnic groups in the Asia-Pacific region are admixed with Andaman, Papuan, Negrito, Htin/Mlabri, or northern East Asian (nEA)-related genetic ancestries (SI Appendix, Figs. S2 B–D and S9 D, E, and J and section 6.1). Hence, we find Cordilleran groups to consistently define the axis of PC1 in world PCA, in polar opposite to the African Khoe-San ethnic groups at the other extreme, even after we control for sample-size differences between regional groups (Fig. 1B and SI Appendix, section 6.1 and Fig. S1 B and C).
Cordillerans, and not Amis or Atayal of Taiwan, serve as the best modern-day surrogate for the least admixed genetic signal for the expansion of AN-speaking populations (11). Both Amis and Atayal exhibit admixture with genetic components resembling Htin/Mlabri-related (or “Austroasiatic-related”) and nEA-related (SI Appendix, Figs. S8 D and F–H and S9 D, E, and J). Additionally, all Philippine ethnic groups share more alleles with Cordillerans than with Amis or Atayal (SI Appendix, Table S7H). Moreover, Cordillerans, aside from Amis and Atayal, share the most alleles with Malaysians, Indonesians, and Oceanians, and even among ancient individuals from peninsular Malaysia and Oceanian Lapita (SI Appendix, Tables S7 J–M and Fig. S8 I and J (16)).
The 8,000-y-Old Liangdao-2 Woman Anchors the Migration of Cordilleran-Related Populations from Mainland Asia to the Philippines.
The two individuals from Liangdao of the Matsu archipelago in the Taiwan Strait (SI Appendix, Table S8A) share the highest levels of genetic drift with Cordillerans, Amis, Atayal, and ancient individuals from the northern Philippines, Malaysia, Taiwan, and Lapita (SI Appendix, Fig. S9 F–I). Given the location of Liangdao, close to mainland China (26 km and 167 km from Taiwan), the ∼7,000- to 8,000-y-old Liangdao-2 individual represents the oldest link of “Cordilleran” ancestry to mainland Asia. As expected, both Liangdao-1 and Liangdao-2 did not display admixture with Basal Sunda ancestry (SI Appendix, Table S8S). Both Liangdao-1 and Liangdao-2 exhibit clear evidence of shared ancestry/admixture with nEA, similar to present-day and ancient populations/individuals of mainland East Asia and Taiwan ((18) (SI Appendix, Figs. S9 D, E, and J)). This is consistent with the recent analysis of ancient individuals from East Asia, which revealed gene flow between populations across various points of time (18–20) and showed that there is some genetic affiliation between nEAs and southern East Asians, including Liangdao individuals (19).
Present-day Cordillerans, however, do not display this nEA ancestry component (SI Appendix, Figs. S2 B–D and S9 D, E, and J and Tables S8 V and X), thereby providing a minimum bound for the divergence between Cordilleran-related groups and ethnic groups of mainland East Asia and Taiwan at ∼8,000 B.P. This finding, taken together with the earliest archaeological evidence of Neolithic assemblages dated to ∼3 to 4 kya in the northern Philippines (21), suggests that the earliest Cordillerans were, like other groups in coastal East Asia at this time (22), complex hunter-gatherers rather than settled agriculturists. The nEA ancestry must have arrived later, originating from the coastal China/Taiwan area and dispersing into Batanes Islands and coastal regions of Luzon (SI Appendix, Table S8X). If the presence of nEA ancestry is indeed a genetic signal for the spread of agriculture, its general absence among Cordillerans suggests that the Neolithic transition among these groups was a consequence of cultural, rather than demic, diffusion.
The Holocene Migrations and the Disentangling of Language and the Neolithic Package.
Two contrasting models have been forwarded for the Holocene migrations into the Philippines. One is the out-of-Taiwan hypothesis, which espouses a unidirectional, north-to-south spread of the Neolithic package by ocean navigators from Taiwan, bringing with them the AN languages, red-slipped pottery, and cereal agriculture (23). On the other hand is the out-of-Sundaland hypothesis, a complex south-to-north movement of population groups into the Philippines since the early Holocene, preceded by a maritime trading network, and by population dispersals from Sundaland following a climate change-driven inundation of previously habitable lands (24, 25).
Our analyses indicate that, after Negrito, Manobo, and Sama-like northward migrations, gene flow of Cordilleran-related ancestry from the Southern China/Taiwan area into the Philippines may have occurred in multiple pulses after ∼10 kya. This may explain the inferred (genetic) divergence between Amis and central Cordillerans being younger than the divergence between Cordillerans and various groups of the central and northern Philippines (SI Appendix, Figs S7E and S8 K–R). Furthermore, we do not observe a north-to-south gradient of chronology for Negrito/Papuan–Cordilleran admixture (SI Appendix, Fig. S8U and Table S7Q), which would be expected in a simplistic and stepwise unidirectional movement of Cordilleran-related ancestry from Luzon to Mindanao. On the contrary, the oldest dates for Negrito/Papuan–Cordilleran admixture are scattered throughout the archipelago, indicating a complex nonuniform movement of populations from a putative South China/Taiwan source area into the Philippines.
Despite the series of migrations from diverse genetic ancestries, the linguistic landscape of the Philippines is remarkably less diverse, in view of the fact that all Philippine ethnic groups speak a language that falls within the Malayo-Polynesian (MP) branch of the AN language family ((26), SI Appendix, section 8). One possible explanation for the linguistic-genetic dissonance is the dominant influence of migratory AN-speaking populations who precipitated widespread linguistic replacement throughout the archipelago and beyond. It may have not been complete replacement as some words may have been retained from the original non-AN language. For instance, some Negrito groups speak languages that potentially contain certain lexical elements that are not completely accounted for by any other AN etymologies (27). Likewise, Land Dayak AN-speakers of Borneo have some evidence of retained Austroasiatic lexical items in their language (28, 29).
The various lines of genetic and archeological evidence suggest that farming was not linked to the initial arrival of Cordilleran-related populations in the Philippines. In addition to the presence of an nEA ancestral component in ∼8,000-y-old Liangdao individuals, but not in Cordillerans, the divergence between Cordillerans and Amis/Atayal (8.4 kya [95% CI: 8.1 to 8.8 kya] and 8.9 kya [95% CI: 8.3 to 9.5 kya]) predates the arrival of agriculture into Taiwan and the Philippines. The divergence time is even older (∼17 kya [95% CI: 9.5 to 25 kya]) when we use publicly available genome sequence data and apply the Two-Two-outgroup (TTo) method (30), based on computing sample configurations in a population divergence model. Furthermore, archeological evidence indicates that communities in coastal South China and north Vietnam from 7 to 5 kya were fishermen and hunter-gatherers, and not farmers (22), and that paddy field rice agriculture was only established in southern China and ISEA late in the Holocene, around 2 to 3 kya (31). This is consistent with the recent comprehensive phylogenetic analysis of Oryza japonica where it was shown that rice entered ISEA only after 2.5 kya, indicating that rice agriculture in the Philippines likely commenced more recently (32). Moreover, the latter study does not provide support for derivation of Philippine and Indonesian rice varieties from Taiwan, nor does it provide strong support for a predominant out-of-Taiwan dispersal of rice.
Hence, the impetus for migration of Cordilleran-related groups may have been catalyzed by, instead of farming, a climate change-driven geological change in the ancient landmass between Taiwan and southern China, which resulted in the gradual submergence of the coastal plain from 12 to 7 kya (SI Appendix, Fig. S3G). This is around the time of an estimated drop in the effective population size of Cordillerans (SI Appendix, Fig. S8 S and T) and fits with the dates of divergence among Cordilleran-related populations and Amis/Atayal (SI Appendix, Fig. S8 Q and R). Thus, our findings, accompanied by previously argued lines of evidence (22, 33), do not lend support to a unitary model of farming–language–people dispersal in the context of Philippine and ISEA prehistory.
Evidence of Recent South Asian and Spanish Admixture into Some Philippine Ethnic Groups.
From ∼2 kya until precolonial times, cultural communities in ISEA actively participated in the region-wide Indian Ocean Trading Network (34). Along this long-distance transoceanic exchange route lie two sequential Hindu-Buddhist kingdoms, Srivijaya and Majapahit, which have ruled over a wide geographic area covering coastal MSEA, western Indonesia, Malaysia, and as far as Sulu Archipelago of the Philippines. The demographic impact of this extensive multilateral trade is evident today in lowlander Malays and some ethnic groups of Indonesia, provided by their detectable South Asian genetic signal (11, 35, 36). In addition to Javanese, Balinese, and Sumatran Indonesians, the seafaring Sama-related populations Kotabaru Bajo and Derawan Bajo also exhibit detectable levels of South Asian ancestry (37). Thus, though it is not unexpected, this report shows that Sama Dilaut sea nomads of Sulu Archipelago and Sama coastal dwellers of Zamboanga Peninsula exhibit evidence of gene flow from South Asians (SI Appendix, Table S6M).
The Philippines was a Spanish Colony for 333 y from 1565 until 1898. However, we only observe significant population-level signals of European admixture in some urbanized lowlanders, Bicolanos, and Spanish Creole-speaking Chavacanos (SI Appendix, Table S7Y). Some individuals from Bolinao, Cebuano, Ibaloi, Itabayaten, Ilocano, Ivatan, Kapampangan, Pangasinan, and Yogad groups also presented low levels of European admixture (SI Appendix, Table S7Y). This admixture is estimated to have taken place 100 to 450 y ago, which falls within the Spanish Colonial Period (SI Appendix, Table S7Z). In contrast to several other Spanish-colonized regions, Philippine demography appears to have remained largely unaffected by admixture with Europeans.
Summary
The nuanced demographic history of the Philippines described here does not fit exclusively with the basic models of either the out-of-Taiwan or out-of-Sundaland hypothesis. In summary, we demonstrate that the Philippines were populated by at least five major waves of ancient human migrations for the past ∼50,000 y (Fig. 3 A–D). The first two are characterized by the entry after ∼46 kya of Paleolithic hunter-gatherer groups, linked genetically to the Basal Australasian branch of modern humans. Of these, an earlier Negrito group entered the northern Philippines, likely via Palawan and Mindoro islands, and a subsequent branch (Basal Oceanians) (Figs. 2A and 3A), represented by the Mamanwa, entered the southern Philippines via Sulu Archipelago. In addition, it is likely that Denisovans or other related archaic humans were already present in the Philippines upon entry of Negritos, resulting in an independent and localized archaic admixture event (1, 3, 38). The archaic introgression signal remains evident until today, given the detectable levels of Denisovan ancestry among the Negrito ethnic groups (3, 38). Moreover, in contrast to European demographic history, where the original hunter-gatherer genetic ancestry, together with their foraging practices, was largely replaced and/or diluted due to subsequent migrations (39), the Negrito ancestry in the Philippines is still largely present until today. This is accompanied by, as observed in some groups until today, a practice of a hunter-gathering mode of subsistence.
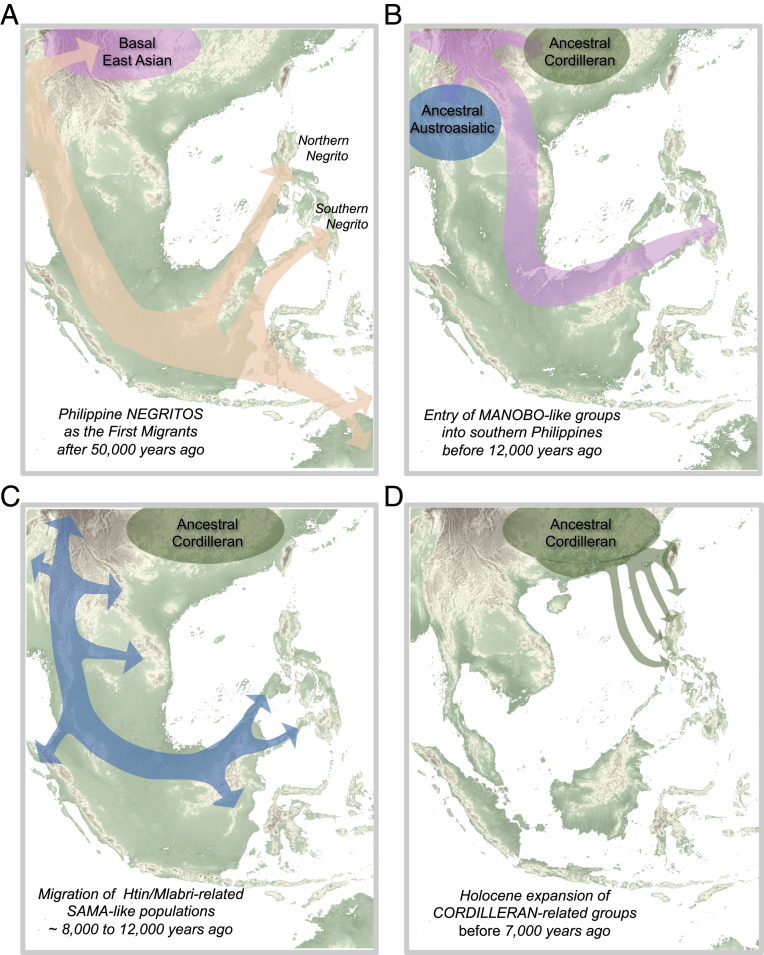
Fig. 3.
Model for Philippine demographic history. Suggested models for the five major migration events into the Philippines, (A) starting with entry of Northern Negritos and Southern Negritos into the Philippines from Sundaland, (B and C) followed by south-to-north movement of Manobo-related and Sama-related populations during the end of Last Glacial Period, and (D) culminating with the Holocene expansion of Cordilleran-related populations.
Following Negritos is a sequential ancestral “Manobo”-like and ancestral “Sama”-like gene flow into the Philippines after ∼15 kya, which occurred likely via a southern route, at a time of major geological changes in ISEA at the end of the Last Glacial Period (Figs. 2B and 3 B and C). Thereafter or concurrently, there was a westward expansion of Papuan-related populations who had left a genetic legacy among the ethnic groups of the southeastern Philippines. Lastly, prior to the minor effects of South Asian gene flow into Sama ethnic groups and recent European admixture with some urbanized lowlanders (SI Appendix, sections 4.5, 5.7, and 6.7), the most recent major migration event was the movement of Cordilleran-related groups from the South China-Taiwan greater area into the Philippines as early as ∼8 to 10 kya (Figs. 2B and 3D). The expansion likely happened in pulses whereby ancient seafaring groups brought with them an enduring legacy of linguistic dominance resonant in present-day populations of the Philippines and beyond, characterized by the spread of AN languages.
We find no genetic evidence for an association between currently established dates for the arrival and expansion of paddy field rice agriculture (∼2 to 3 kya) (31, 32) and the demographic movement of people from the South China-Taiwan greater area into the Philippines. Given the genetic makeup of the Liangdao-2 woman and other ancient Taiwanese and southern East Asian individuals (18) who all present some nEA ancestry, the most parsimonious explanation is that Cordillerans entered the Philippines prior to the nEA gene flow. This observation sets a boundary for the date of arrival of Cordillerans in the Philippines, at least ∼7 to 8 kya or earlier, indicating that early Cordillerans in the Philippines were mobile hunter-gatherers. All other Cordilleran-like groups eventually admixed with local populations at various points in time, as seen from the complex admixture among Philippine groups. Accordingly, this places the Cordillerans in a unique position in human demographic history, being revealed as the least admixed descendants of Basal East Asians.
Materials and Methods
Sampling and Ethical Considerations (SI Appendix, Sections 1.1 and 1.2).
This research project, with the aim of establishing baseline scientific data for the genetic diversity, interrelatedness, and migration history of Philippine indigenous cultural communities, is recognized by, and implemented in partnership with, the National Commission for Culture and the Arts (NCCA) of the Philippines, in accordance with the provisions of Philippine Republic Act 7356, or the Law Creating the NCCA. Saliva samples were collected with the use of the Oragene Saliva Collection Kit (DNA Genotek Inc., Ottawa, ON, Canada). Consent was secured from each individual and, whenever necessary, from each respective Indigenous Cultural Community Council. The consent process, sampling, and/or subsequent validation were performed in coordination with the NCCA and, in some regional areas, with local partners or agencies, including nongovernmental and cultural organizations, local educational institutions, Indigenous Cultural Community Councils, local government units, and/or regional offices of the National Commission on Indigenous Peoples (NCIP) (SI Appendix, section 10). The processing of samples and analysis of data were approved by the Regional Ethical Review Board in Uppsala, Sweden (Dnr 2016/103).
Sample Processing and Data Generation (SI Appendix, Sections 1.3–1.5).
The saliva samples were processed for DNA isolation using the prepIT DNA isolation kit (DNA Genotek Inc., Ottawa, ON, Canada). The purified 1,094 DNA samples were sent to the SNP&SEQ Technology Platform at Uppsala University for genotyping of SNP markers with the Illumina Infinium assay (San Diego, CA) using the InfiniumOmni2-5Exome-8v1-3 Bead Chip (2,612,357 SNPs). The generated data were filtered for SNPs with 10% missingness, SNPs that were not in Hardy–Weinberg equilibrium, indels, duplicates, and nonautosomal and unmapped SNPs. The dataset was also filtered for related individuals (resulting in the exclusion of 62 individuals) (SI Appendix, section 1.4), producing a final Phil_2.35M dataset with 1,028 individuals and 2,359,167 SNPs. The base dataset was merged with publicly available datasets to produce the Phil_1KGP_SGDP_1.69M dataset with 3,331 individuals and 1,690,499 SNPs, the Phil_AsiaPacific_315K dataset with 5,132 individuals and 315,692 SNPs, and the Phil_HO_201K dataset with 5,402 individuals and 201,387 SNPs.
Population Genetic Analysis (SI Appendix, Section 1.6).
Measures of basic statistics and measures of genetic diversity, including runs of homozygosity and inbreeding coefficient, were computed using the–het,–homozyg, and–ibc functions of PLINK v1.9 (40) (SI Appendix, Fig. S1 I and J and section 3.8). PCA and calculation of between-population FST were performed using EIGENSOFT v7.1 (41, 42), and an FST-based neighbor-joining tree was plotted using MEGA7 (43). Mantel tests were performed to determine statistical significance on correlations between genetic and linguistic and between genetic and geographic distances.
ADMIXTURE v1.3 (44) and CLUMPP (45) were used to analyze population structure, which was subsequently visualized using Pong v1.4. Outgroup f3 statistics and formal tests of admixture were performed using qp3Pop and qpDstat of the AdmixTools v5.0 package (46). We used qpF4Ratio and qpAdmin of the AdmixTools v5.0 package for estimating admixture proportions in populations, and qpGraph for fitting populations in an admixture graph with baseline framework based on earlier publications (47). For estimating dates of admixture, we utilized a weighted linkage disequilibrium (LD) statistic-based method, MALDER (48), which also allows detection of multiple admixture events. To infer local ancestries and subsequent masking in admixed populations, we utilized RFMix (49), which employs a conditional random field parameterized by random forests trained on reference panels of least admixed populations.
Detection of identity-by-descent (IBD) segments was implemented using Beagle v4.1 (50), and IBDne was used for the estimation of recent effective population size based on IBD (51). Using a coalescent model and a maximum likelihood framework (12, 13, 52), divergence time was estimated with the following formula: Divergence Time = T × 2Ne × g, where T is the drift parameter, Ne is the effective population size, and g is the generation time (30 y).
Laboratory Processing of Ancient Samples (SI Appendix, Section 1.7).
Two phalanges were used for DNA extraction in a dedicated ancient DNA (aDNA) laboratory. Blunt-end Illumina libraries were generated for both Liangdao-1 and Liangdao-2 DNA samples, and, additionally, uracil–DNA–glycosylase (UDG)-treated libraries were generated for a higher quality Liangdao-2 sample. The libraries were shotgun sequenced using Illumina HiSeqX with a 150-base pair (bp) paired-end length and v2.5 chemistry at the SNP&SEQ Technology Platforms at Uppsala University.
Processing of aDNA Data (SI Appendix, Section 1.8).
For processing of aDNA data, paired-end reads were merged, and their adapters were trimmed and subsequently mapped to the human reference genome using BWA (53). PCR duplicates with identical start and end coordinates were collapsed into consensus sequences (54). Both Liangdao-1 and Liangdao-2 possessed the characteristic deamination pattern toward the read fragment-ends (55). The level of contamination was estimated using the Green (56), Contamix (56), and Schumtzi (57) methods for mitochondrial DNA and VerifyBamID for autosomal DNA.
Supplementary Material
Acknowledgments
We thank all of the volunteers and indigenous cultural communities who participated in this research. The project is recognized by, and implemented in partnership with, the NCCA of the Philippines, in accordance with the provisions of Philippine Republic Act 7356, or the Law Creating the NCCA. The project was reviewed and approved by the Regional Ethical Review Board in Uppsala, Sweden (Dnr 2016/103). We thank Felipe Mendoza de Leon Jr., NCCA chairperson 2010 to 2016, for his support. We also thank the NCIP, local government units, and regional cultural organizations for facilitating fieldwork. We thank Suei-Sheng Yang, former Lienchiang County Governor, and the Lienchiang County Cultural Affairs Department, and Marie Lin (Mackay Memorial Hospital) for the provision of Liangdao archaeological samples. See SI Appendix, section 10 for a complete list of acknowledgments. This work was supported by Swedish Research Council Grant 642-2013-8019 (to M.J.) and Grant 2020-04789 (to M.L.), and the Knut and Alice Wallenberg Foundation (M.J.). The computations were performed at the Swedish National Infrastructure for Computing (SNIC-UPPMAX).
Footnotes
The authors declare no competing interest.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2026132118/-/DCSupplemental.
Data Availability
Anonymized SNP genotype data have been deposited in the European Genome-Phenome Archive (EGAS00001005083), and will be made available upon request from the Data Access Committee, provided that the request is intended for academic purposes only and is in accordance with the consent provided by the study participants. Ancient DNA sequence data have been deposited as publically available in the European Nucleotide Archive (PRJEB43078).
References
- 1.Détroit F., et al., A new species of Homo from the Late Pleistocene of the Philippines. Nature 568, 181–186 (2019). [DOI] [PubMed] [Google Scholar]
- 2.Mijares A. S., et al., New evidence for a 67,000-year-old human presence at Callao Cave, Luzon, Philippines. J. Hum. Evol. 59, 123–132 (2010). [DOI] [PubMed] [Google Scholar]
- 3.Jinam T. A., et al., Discerning the origins of the negritos, first Sundaland people: Deep divergence and archaic admixture. Genome Biol. Evol. 9, 2013–2022 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Reid L. A., Who are the Philippine negritos? Evidence from language. Hum. Biol. 85, 329–358 (2013). [DOI] [PubMed] [Google Scholar]
- 5.Soares P. A., et al., Resolving the ancestry of Austronesian-speaking populations. Hum. Genet. 135, 309–326 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Trejaut J. A., et al., Taiwan Y-chromosomal DNA variation and its relationship with Island Southeast Asia. BMC Genet. 15, 77 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Abdulla M. A.et al.; HUGO Pan-Asian SNP Consortium; Indian Genome Variation Consortium , Mapping human genetic diversity in Asia. Science 326, 1541–1545 (2009). [DOI] [PubMed] [Google Scholar]
- 8.Hill C., et al., A mitochondrial stratigraphy for island southeast Asia. Am. J. Hum. Genet. 80, 29–43 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Capelli C., et al., A predominantly indigenous paternal heritage for the Austronesian-speaking peoples of insular Southeast Asia and Oceania. Am. J. Hum. Genet. 68, 432–443 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lipson M., et al., Reconstructing austronesian population history in Island Southeast Asia. Nat. Commun. 5, 4689 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mörseburg A., et al., Multi-layered population structure in Island Southeast Asians. Eur. J. Hum. Genet. 24, 1605–1611 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schlebusch C. M., et al., Genomic variation in seven Khoe-San groups reveals adaptation and complex African history. Science 338, 374–379 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wakeley J., Coalescent Theory (Roberts & Co., Greenwood Village, CO, 2008). [Google Scholar]
- 14.Lipson M., et al., Ancient genomes document multiple waves of migration in Southeast Asian prehistory. Science 361, 92–95 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Acabado S., The archaeology of pericolonialism: Responses of the “unconquered” to Spanish conquest and colonialism in Ifugao, Philippines. Int. J. Hist. Archaeol. 21, 1–26 (2017). [Google Scholar]
- 16.Skoglund P., et al., Genomic insights into the peopling of the southwest Pacific. Nature 538, 510–513 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Reid L. A., “Historical linguistics and Philippine hunter-gatherers” in The Language of Hunter-Gatherers, Güldemann T., McConvell P., Rhodes R. A., Eds. (Cambridge University Press, 2007), pp. 231–261. [Google Scholar]
- 18.Wang Chuan-Chao, et al., Genomic Insights into the Formation of Human Populations in East Asia. Nature, 10.1038/s41586-021-03336-2 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yang M. A., et al., Ancient DNA indicates human population shifts and admixture in northern and southern China. Science 369, 282–288 (2020). [DOI] [PubMed] [Google Scholar]
- 20.Ning C., et al., Ancient genomes from northern China suggest links between subsistence changes and human migration. Nat. Commun. 11, 2700 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bellwood P., Dizon E., The Batanes archaeological project and the “Out of Taiwan” hypothesis for Austronesian dispersal. J. Austronesian Stud. 1, 1–33 (2005). [Google Scholar]
- 22.Hung H., Prosperity and complexity without farming: The south China Coast, c. 5000–3000 BC. Antiquity 93, 325–341 (2019). [Google Scholar]
- 23.Bellwood P. S., Prehistory of the Indo-Malaysian Archipelago (ANU Press, Canberra, Australia, 2nd Ed., 1997). [Google Scholar]
- 24.Solheim W. G., Bulbeck D., Flavel A., Archaeology and culture in Southeast Asia: Unraveling the Nusantao (University of the Philippines Press, Diliman, Quezon City, 2006). [Google Scholar]
- 25.Soares P., et al., Climate change and postglacial human dispersals in southeast Asia. Mol. Biol. Evol. 25, 1209–1218 (2008). [DOI] [PubMed] [Google Scholar]
- 26.Blust R., The Austronesian languages (Asia-Pacific Linguistics, 2013). [Google Scholar]
- 27.Reid L. A., Possible non-Austronesian lexical elements in Philippine Negrito languages. Ocean. Linguist. 33, 37–72 (1994). [Google Scholar]
- 28.Blench R., Was there an austroasiatic presence in Island Southeast Asia prior to the austronesian expansion? Bulletin of the Indo-Pacific Prehistory Association 30, 133–144 (2011). [Google Scholar]
- 29.Adelaar K. A., “Borneo as a cross-roads for comparative Austronesian linguistics” in The Austronesians, Bellwood P., Fox J. J., Tryon D., Eds. (ANU Press, 1995), pp. 81–102. [Google Scholar]
- 30.Sjödin P.et al., Estimating divergence times from DNA sequences. Genetics, 10.1093/genetics/iyab008 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ma T., Rolett B. V., Zheng Z., Zong Y., Holocene coastal evolution preceded the expansion of paddy field rice farming. Proc. Natl. Acad. Sci. U.S.A. 117, 24138–24143 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gutaker R. M., et al., Genomic history and ecology of the geographic spread of rice. Nat. Plants 6, 492–502 (2020). [DOI] [PubMed] [Google Scholar]
- 33.Donohue M., et al., Farming and language in Island Southeast Asia: Reframing austronesian history. Curr. Anthropol. 51, 223–256 (2010). [Google Scholar]
- 34.Brucato N., et al., Genomic admixture tracks pulses of economic activity over 2,000 years in the Indian Ocean trading network. Sci. Rep. 7, 2919 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hudjashov G., et al., Complex patterns of admixture across the Indonesian archipelago. Mol. Biol. Evol. 34, 2439–2452 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kusuma P., et al., Western Eurasian genetic influences in the Indonesian archipelago. Quat. Int. 416, 243–248 (2016). [Google Scholar]
- 37.Kusuma P., et al., The last sea nomads of the Indonesian archipelago: Genomic origins and dispersal. Eur. J. Hum. Genet. 25, 1004–1010 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.GenomeAsia100K Consortium , The GenomeAsia 100K Project enables genetic discoveries across Asia. Nature 576, 106–111 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Günther T., Jakobsson M., Genes mirror migrations and cultures in prehistoric Europe-a population genomic perspective. Curr. Opin. Genet. Dev. 41, 115–123 (2016). [DOI] [PubMed] [Google Scholar]
- 40.Purcell S., et al., PLINK: A tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 81, 559–575 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Price A. L., et al., Principal components analysis corrects for stratification in genome-wide association studies. Nat. Genet. 38, 904–909 (2006). [DOI] [PubMed] [Google Scholar]
- 42.Patterson N., Price A. L., Reich D., Population structure and eigenanalysis. PLoS Genet. 2, e190 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kumar S., Stecher G., Tamura K., MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Alexander D. H., Novembre J., Lange K., Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 19, 1655–1664 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jakobsson M., Rosenberg N. A., CLUMPP: A cluster matching and permutation program for dealing with label switching and multimodality in analysis of population structure. Bioinformatics 23, 1801–1806 (2007). [DOI] [PubMed] [Google Scholar]
- 46.Patterson N., et al., Ancient admixture in human history. Genetics 192, 1065–1093 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lipson M., Reich D., A working model of the deep relationships of diverse modern human genetic lineages outside of africa. Mol. Biol. Evol. 34, 889–902 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pickrell J. K., et al., Ancient west Eurasian ancestry in southern and eastern Africa. Proc. Natl. Acad. Sci. U.S.A. 111, 2632–2637 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Maples B. K., Gravel S., Kenny E. E., Bustamante C. D., RFMix: A discriminative modeling approach for rapid and robust local-ancestry inference. Am. J. Hum. Genet. 93, 278–288 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Browning B. L., Browning S. R., Improving the accuracy and efficiency of identity-by-descent detection in population data. Genetics 194, 459–471 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Browning S. R., Browning B. L., Accurate non-parametric estimation of recent effective population size from segments of identity by descent. Am. J. Hum. Genet. 97, 404–418 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Skoglund P., Götherström A., Jakobsson M., Estimation of population divergence times from non-overlapping genomic sequences: Examples from dogs and wolves. Mol. Biol. Evol. 28, 1505–1517 (2011). [DOI] [PubMed] [Google Scholar]
- 53.Li H., Durbin R., Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics 26, 589–595 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kircher M., “Analysis of high-throughput ancient DNA sequencing data” in Ancient DNA, Shapiro B., Hofreiter M., Eds. (SpringerHumana Press, 2012), pp. 197–228. [DOI] [PubMed] [Google Scholar]
- 55.Briggs A. W., et al., Patterns of damage in genomic DNA sequences from a Neandertal. Proc. Natl. Acad. Sci. U.S.A. 104, 14616–14621 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Green R. E., et al., A complete Neandertal mitochondrial genome sequence determined by high-throughput sequencing. Cell 134, 416–426 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Renaud G., Slon V., Duggan A. T., Kelso J., Schmutzi: Estimation of contamination and endogenous mitochondrial consensus calling for ancient DNA. Genome Biol. 16, 224 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Anonymized SNP genotype data have been deposited in the European Genome-Phenome Archive (EGAS00001005083), and will be made available upon request from the Data Access Committee, provided that the request is intended for academic purposes only and is in accordance with the consent provided by the study participants. Ancient DNA sequence data have been deposited as publically available in the European Nucleotide Archive (PRJEB43078).