Abstract
The nuclear pore complex (NPC) consists of multiple copies of ≈30 different proteins [nucleoporins (nups)], forming a channel in the nuclear envelope that mediates macromolecular transport between the cytosol and the nucleus. With <5% of the nup residues currently available in experimentally determined structures, little is known about the detailed structure of the NPC. Here, we use a combined computational and biochemical approach to assign folds for ≈95% of the residues in the yeast and vertebrate nups. These fold assignments suggest an underlying simplicity in the composition and modularity in the architecture of all eukaryotic NPCs. The simplicity in NPC composition is reflected in the presence of only eight fold types, with the three most frequent folds accounting for ≈85% of the residues. The modularity in NPC architecture is reflected in its hierarchical and symmetrical organization that partitions the predicted nup folds into three groups: the transmembrane group containing transmembrane helices and a cadherin fold, the central scaffold group containing β-propeller and α-solenoid folds, and the peripheral FG group containing predominantly the FG repeats and the coiled-coil fold. Moreover, similarities between structures in coated vesicles and those in the NPC support our prior hypothesis for their common evolutionary origin in a progenitor protocoatomer. The small number of predicted fold types in the NPC and their internal symmetries suggest that the bulk of the NPC structure has evolved through extensive motif and gene duplication from a simple precursor set of only a few proteins.
Keywords: coated vesicle, protocoatomer, evolution, fold assignment
The nuclear pore complex (NPC) is the only known selective gate for the passage of macromolecules across the nuclear envelope (NE) (1). The NPC is also one of the largest assemblies of defined structure in the cell, with a size of ≈50 MDa in yeast and up to 100 MDa in vertebrates. NPCs are common to all eukaryotes and are composed of a broadly conserved set of proteins termed nucleoporins (nups) (2) that have been fully cataloged for both yeast (3) and vertebrates (4).
Structural characterization of the whole NPC has proven challenging, because of its size and flexibility. A consensus low-resolution map of the NPC has emerged based largely on electron cryomicroscopy and tomography studies (5–8). The NPC is a ring of eight identical spokes. Each spoke can be divided into almost identical cytosolic and nuclear half-spokes that each consist of ≈25 different nups (1). At the center of the NPC is an aqueous channel serving as the conduit for the transport of macromolecules. Macromolecular transport is regulated by the filamentous FG repeat-containing nups that emanate from the NPC into the nucleoplasm and cytoplasm. A comparison of the vertebrate and yeast NPCs reveals that the main features of the complex are conserved (1, 2).
Experimentally determined atomic structures are currently available for only seven nup fragments: ≈20 residues of the Nsp1 FxFG repeat region (9), 38 residues of the C terminus of Nup1 (10), a 6-residue FG-repeat segment of the CAN nup (the human homolog of Nup159) (11), the autocatalytic fragment of the vertebrate Nup98 (12), the equivalent 147-residue NPC-targeting domain at the C terminus of Nup116 (13), and the N-terminal domains of human Nup133 (14) and yeast Nup159 (15). Together, these domains represent a mere 5% of the total number of nup amino acid residues. Moreover, only an additional 5% of the residues can be related to proteins of known structure via statistically significant sequence similarity (Results); hence, there is a paucity of high-resolution structural data on nups.
Despite the central role of the NPC in the cell biology of all modern eukaryotes, there has been until recently little information concerning its origin and evolution in protoeukaryotes. To address this question, it would be helpful to have structural characterizations of many nups because it is easier to recognize similarities in structure than in sequence, especially for distantly related proteins. We recently used bioinformatics tools supported by limited proteolysis data to assign folds to domains in seven nups comprising the Nup84 subcomplex in yeast (16). These assignments allowed us to propose an evolutionary relationship between the Nup84 subcomplex of the NPC and coated vesicles (16), and other relationships have subsequently been suggested (17). In this work, we extend our previous analysis and assign folds for domains in all known nups, resulting in a structural characterization of ≈95% of the nup residues. We discuss the implications of these fold assignments for the structural organization and evolution of the NPC.
Results
We applied a variety of bioinformatics methods to characterize the structures of the Saccharomyces cerevisiae nups (Fig. 1 and Table 1) (3) and their predicted vertebrate homologs (4). We first predicted secondary structure segments, transmembrane helices (TMHs), coiled coils, FG repeats, and disordered regions. Second, we detected nup homologs by sequence comparison methods, resulting in fold assignments for domains in some nups. Third, we also assigned folds to as many nup domains as possible using threading methods. And finally, for a given fold assignment, we applied an iterative process of sequence–structure alignment, comparative model building, and model evaluation to assess its accuracy. The fold assignments were also supported by protease-accessibility laddering experiments.
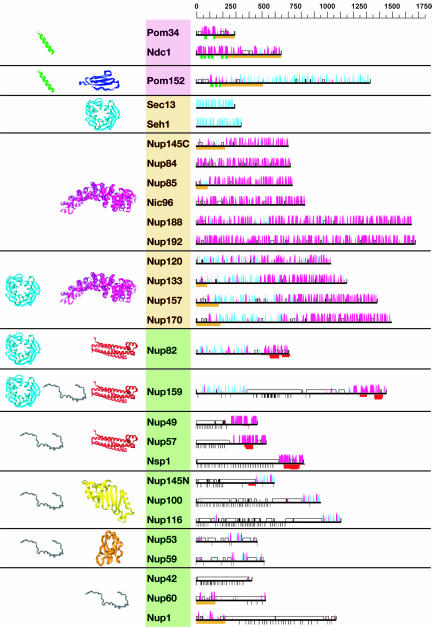
Fig. 1.
Predicted secondary structure and fold types of the nups. The names of the nups are boxed according to the group they define: transmembrane (pink), scaffold (orange), and FG repeat (green). The black horizontal lines on the right represent the sequence of each yeast nup. The predicted α-helices (magenta) and β-strands (cyan) are indicated by bars above each line. The height of the bars is proportional to the confidence of the prediction (39). Predicted transmembrane helices are shown in green, coiled coils are shown in red, FG repeats are shown in black, and unstructured regions are shown by an empty box. An orange block underlines regions of >50 residues to which a fold type could not be assigned. Representative models of the nup domains are colored according to the fold type and are shown on the left. Models are not to scale for visualization reasons. There are eight fold types. First, a TMH segment (green) is a hydrophobic 15- to 30-residue helical segment that spans the membrane. Second, cadherin domains (dark blue) have ≈110 residues that fold into a seven-stranded β-sandwich structure. Third, β-propellers (cyan) contain several blades arranged radially around a central axis, each blade consisting of a four-stranded antiparallel β-sheet. Fourth, α-solenoid domains are composed of numerous pairs of antiparallel α-helices stacked to form a solenoid. Fifth, coiled coils (red) generally display seven-residue repeats where the first and fourth residues of an α-helix are often hydrophobic. The coiled-coil structure is formed by helices (generally two) twisting together to bury their hydrophobic seams. Sixth, disordered FG-repeat segments are indicated schematically by a black curve. Seventh, the autoproteolytic domain of Nup98 (yellow) adopts a half-open, β-sandwich-like fold dominated by a large β-sheet with helices capping two of its ends. Finally, the RRM (orange) is a two-layer α/β sandwich typically found in proteins involved in RNA binding.
Table 1.
Nup domains are predicted to be clustered into eight fold types
| Name | Size | Domain range | Fold | Z score |
|---|---|---|---|---|
| POM34 | 299 | 64–150 (2)* | TMH | n.a. |
| NDC1 | 655 | 34–245 (6)* | TMH | n.a. |
| POM152 | 1,337 | 112–193 (3)* | TMH | n.a. |
| 750–1,337 | Cadherin | −4.06 (820–896) | ||
| SEC13† | 297 | 1–297 | β-Propeller | −4.8 (11–282) |
| SEH1† | 349 | 1–349 | β-Propeller | −5.7 (1–349) |
| NUP145C† | 712 | 200–512 | α-Solenoid | −10.4 (234–690) |
| NUP84† | 726 | 1–726 | α-Solenoid | −10.9 (301–726) |
| NUP85† | 744 | 100–744 | α-Solenoid | −11.8 (203–744) |
| NIC96 | 839 | 1–839 | α-Solenoid | −5.3 (622–742) |
| NUP188 | 1,655 | 1–1,655 | α-Solenoid | −6.3 (592–747) |
| NUP192 | 1,683 | 1–1,683 | α-Solenoid | −6.3 (420–716) |
| NUP120† | 1,037 | 1–450 | β-Propeller | −6.9 (1–398) |
| 500–1,037 | α-Solenoid | −8.6 (521–1,011) | ||
| NUP133† | 1,157 | 100–400 | β-Propeller | −8.0 (1–300) |
| 500–1,157 | α-Solenoid | −9.5 (167–371) | ||
| NUP157 | 1,391 | 150–600 | β-Propeller | −6.3 (167–371) |
| 750–1,391 | α-Solenoid | −6.5 (860–1,195) | ||
| NUP170 | 1,502 | 150–600 | β-Propeller | −4.0 (270–384) |
| 800–1,391 | α-Solenoid | −7.7 (879–1,345) | ||
| NUP82 | 713 | 1–500 | β-Propeller | −6.9 (27–198) |
| 550–713 | CC | n.a. | ||
| NUP159 | 1,460 | 1–400 | β-Propeller | −8.1 (1–400) |
| 400–1,150 | FG | n.a. | ||
| 1,250–1,460 | CC | n.a. | ||
| NUP49 | 472 | 1–250 | FG | n.a. |
| 350–400 | CC | n.a. | ||
| NUP57 | 541 | 1–250 | FG | n.a. |
| 350–450 | CC | n.a. | ||
| NSP1 | 823 | 1–600 | FG | n.a. |
| 650–823 | CC | n.a. | ||
| NUP145N | 605 | 1–450 | FG | n.a. |
| 455–605 | Nup98 | −8.8 (456–605) | ||
| NUP100 | 959 | 1–800 | FG | n.a. |
| 809–959 | Nup98 | −7.5 (808–955) | ||
| NUP116 | 1,113 | 1–950 | FG | n.a. |
| 963–1,113 | Nup98 | −9.2 (970–1,108) | ||
| NUP53 | 475 | 1–250, 356–475 | FG | n.a. |
| 251–355 | RRM | −4.8 (249–342) | ||
| NUP59 | 528 | 1–288, 391–528 | FG | n.a. |
| 290–390 | RRM | −4.5 (267–384) | ||
| NUP42 | 430 | 1–430 | FG | n.a. |
| NUP60 | 539 | 150–530 | FG | n.a. |
| NUP1 | 1,076 | 250–1,076 | FG | n.a. |
| Total | 24,117 | 22,876 | n.a. | n.a. |
Shown are the number of residues in the corresponding nup sequence (size), an estimate of the domain range, the fold type of the corresponding model (where CC is coiled coil, FG is FG repeat, and Nup98 is the autocatalytic domain of Nup98), and Z scores of the comparative models (46) for the residues indicated in the parentheses. The predicted TMH, FG repeat, and coiled coil folds were not modeled. n.a., not applicable.
*The value in parentheses is the count of TMHs.
†Indicates that the nup is characterized in ref. 16.
Fold Assignments.
The 28 nups were divided into 44 domains as follows. Using sequence analysis, we detected 12 FG repeats, 5 coiled-coil, and 3 TMH domains. An additional five domains could be assigned to a fold type by pp_scan based on statistically significant sequence similarity alone. Threading methods were then used to assign folds to the remaining 19 domains. The 44 domains together account for ≈95% of all nup residues. Each nup has at least one assigned domain. The only unassigned regions with >50 residues include segment 200–500 in Pom152, short segments at the N termini of Nup145C, Nup85, Nup133, Nup157, and Nup170, as well as the extreme C termini of Ndc1 and Pom34 (Fig. 1). However, most of these unassigned regions are predicted to contain little regular secondary structure.
The 44 nup domains were assigned to only eight fold types (Fig. 1 and Table 1): the α-solenoid fold covers the most residues (38%). The next most prevalent are the FG repeats (29%) and the β-propeller (16%) fold. Each of the other five fold types [the TMH fold, the cadherin fold, the coiled-coil fold, the autoproteolytic Nup98 domain, and the RNA recognition motif (RRM)] individually covers <5% of the total nup residues (Table 1).
In an effort to substantiate the yeast nup fold assignments and provide a complete structural coverage of the nups from yeast to vertebrate, we also assigned folds to the Rattus norvegicus nups (4) as well as their predicted Homo sapiens homologs. Although these orthologs generally share <40% sequence identity, their sequence similarity is statistically significant (i.e., the build_profile E value is <0.01). Fold assignments for nups from vertebrates matched those for their yeast homologs (Table 2, which is published as supporting information on the PNAS web site).
Assessment of Fold Assignments.
Our fold assignments are supported by seven considerations. First, for 16 of the 19 domains assigned by threading (Table 1), the assignment was statistically significant as defined by the authors of the threading methods (18–21) for at least three of the four methods. In addition, no other fold type occurred as frequently in the list of significant hits for any of the assigned domains. Moreover, although there are numerous sequence and structure variations among proteins of the same fold, the different servers often selected the same template structures. The folds for the remaining three domains are supported by other considerations (see below). Second, despite their low sequence similarity, the human orthologs of the yeast nups independently share their fold assignments without exception (Table 2). Third, assessment scores for comparative models based on each fold assignment were statistically significant when compared against the best models generated for random sequences of identical amino acid residue composition and length; all of the nup models were at least four standard deviations away from the mean score of the random models (Table 1). Fourth, secondary structure predictions from sequences largely matched the secondary structures in the corresponding comparative models. The agreement is as high as 87% of the residues for some of the models, when a three-state assignment (helix, strand, and other) is used. This agreement is the maximum possible level of consistency given the ≈75% accuracy of the secondary structure prediction methods (22). Fifth, protease accessibility laddering corroborates our fold assignments (Fig. 2; see also Table 3, which is published as supporting information on the PNAS web site). Limited proteolysis is expected to occur in regions that are exposed to the solvent and outside secondary structure segments, corresponding to domain boundaries and loops. Therefore, protease accessibility laddering can be used to test the predicted boundaries between domains and perhaps even regular secondary structure segments. For the multidomain nups, the strongest cleavage indeed occurs between the predicted domains (Fig. 2). Moreover, for the models in Table 1, 88% of the cuts occur within a residue of an exposed boundary of a regular secondary structure segment. In contrast, only 53% of the cuts are close to such boundaries in models of the same sequences based on randomly selected folds. This result is statistically significant with a P value of 0.05. Notably, proteolysis at equivalent sites is observed for Nup120 and Nup133 (16) as well as Nup157 and Nup170 (Fig. 2), which are all predicted to contain the same fold arrangement (Fig. 1 and Table 1). Sixth, circular dichroism and Fourier-transform infrared spectra of Nup85 are in agreement with our predictions, indicating a composition characteristic of α-solenoid domains (≈50% α-helix and 10% β-sheet) (23). Similarly, the circular dichroism spectrum for Sec13 is also in agreement with our model (<10% α-helix) (24). Finally, recent atomic structure determinations of the N-terminal domains of Nup133 (14) and Nup159 (15) as well as the NPC-targeting domain of Nup116 (13) directly confirmed our predictions for these proteins.
Fig. 2.
Mapping of domains of the yeast nups by protease accessibility laddering. The gels show immunoblots of limited proteolysis digests for Protein A-tagged versions of the nups. Each full-length nup is indicated by an arrow on the left side of the gel, as are the sizes of marker proteins (expressed in kDa). Secondary structure predictions are shown by using the conventions in Fig. 1. Proteolytic cleavage sites are identified by small, medium, and large arrows for weak, medium, and strong susceptibility sites, respectively. Where necessary, uncertainties in the precise cleavage positions are indicated by lines to the left of the sequence.
Discussion
We assigned folds to most domains in nups from S. cerevisiae, R. norvegicus and H. sapiens. These assignments pointed to a simplicity in the composition and modularity in the architecture of the NPC. The simplicity of the composition is reflected in the presence of only eight predicted fold types, most composed of multiple, simple repetitive units (Fig. 1 and Table 1). The modularity of the architecture is reflected in the partitioning of the nups according to their predicted folds into only three groups (Fig. 3). Such simplicity and modularity suggests that the evolution of the NPC occurred by many intragene and full-gene duplications followed by divergence.
Fig. 3.
Simplicity of the fold composition and modular architecture of the NPC. (a) The schematic structure and hierarchy of the NPC. Most of the nups consist of sequence repeats (step I). The nups assemble into multiple copies to form each half-spoke (step II) that dimerize to form the spokes (step III), which are themselves repeated eight times to form the complete NPC (step IV). (b) The architecture of the NPC ring, viewed in the plane of the NE, is segregated into the membrane (pink), scaffold (orange), and FG (green) groups. The domain fold types assigned to each group are indicated on the left side of the schema. The schema illustrates the coarse organization of the NPC and is not a precise map of the three-dimensional nup locations.
Simplicity of the NPC Composition.
The composition of the predicted domain folds in the yeast NPC is exceedingly simple, given that the NPC is an ≈50-MDa complex consisting of ≈480 proteins of ≈30 different types. The three most frequent domains (i.e., the α-solenoids, FG repeats, and β-propellers) account for 83% of the residues, whereas only five further fold types (i.e., the TMH, cadherin, coiled-coil, Nup98, and RRM folds) account for most of the remainder. The increased proteolytic susceptibility of the unassigned fragments suggests that they are enriched in unstructured and/or flexible regions. Therefore, with a possible exception of small undiscovered domains in the two membrane proteins Ndc1 and Pom34, it is likely that folds for all domains with defined structure have been detected. Thus, the conclusion that a small number of repeat elements is sufficient to build the NPC is unlikely to change.
Simplicity of the NPC Architecture.
The simplest possible architecture of the NPC must include membrane proteins for the anchoring of the NPC into the NE, the scaffold proteins that provide the framework for assembling the NPC, and the selective filter that presumably lines the central passage through which the transport occurs (1, 25). The fold assignments can be easily interpreted in terms of such a simple architecture as follows (Fig. 3).
The first group consists of the three membrane-spanning nup proteins, which were predicted to contain transmembrane α-helices and the cadherin fold (Fig. 1) and form the outermost group of nups (Fig. 3b) (3). The transmembrane α-helices of these pore membrane proteins likely help to anchor the NPC in the NE; in addition, they may help in stabilizing the curvature of the NE through the cadherin domains of Pom152. The cadherin domain has been observed to provide a bridge between two membranes (e.g., in desmosomes) (26). It is conceivable that it plays an equivalent role in stabilizing the interaction between the outer and inner nuclear membranes of the NE.
The second group is formed by the nups predicted to contain primarily the FG repeat and coiled-coil domains, in addition to the infrequent Nup98 and RRM folds (Fig. 1). The FG repeats were shown to provide low-affinity, high-specificity interactions with transport factors involved in active transport through the NPC (27) and appear overall to be localized toward the inside of the NPC (3). Therefore, it is reasonable to suggest that this second group of nups coats the central pore surface (Fig. 3b), providing an interaction region for transport factors around the central channel of the NPC. Moreover, the frequent mediation of protein interactions through coiled coils (28) suggests that the coiled-coil domains anchor the FG nups into the bulk of the NPC.
The third group consists of the nups predicted to contain only α-solenoid and β-propeller folds (Fig. 1). Every nup without a transmembrane helix, FG repeat, or coiled-coil domain contains the α-solenoid or β-propeller or both folds. Given the proposed outer and inner groups of the membrane and FG nups, respectively, we suggest that the third group of nups forms the structural scaffold of the NPC located mainly between the outer and inner groups, thus anchoring the two groups to form a complete NPC (Fig. 3b). This proposal is further supported by our previous observation relating the seven nups in the Nup84 subcomplex to the proteins covering the clathrin, COPI, and COPII coated vesicles (16). In coated vesicles, the clathrin-like and adaptor proteins constitute the structural scaffold of the protein coat surrounding the membrane of the vesicle in a lattice fashion (29), perhaps similarly to the role of the scaffold nups in the NPC. We extend the original proposal [the “protocoatomer hypothesis” (16)] and suggest that most if not all of the scaffold nups and the coated vesicle proteins originated from a common precursor complex (i.e., the protocoatomer). Both α-solenoid and β-propeller folds provide extensive solvent-accessible surfaces that appear well suited for binding other proteins. Moreover, the α-solenoid and β-propeller folds seem to be sufficiently robust for significant variation in sequence while retaining the core structure, thus allowing optimization of interactions with a multitude of other partners.
The fold assignments for the nups, therefore, (i) reinforce the protocoatomer hypothesis (16), (ii) extend the hypothesis to all of the nups in the scaffold group, (iii) imply that the scaffold nups form the structural core of the NPC, and (iv) further indicate that same aspects of the organization of the NPC are similar to those of coated vesicles.
Evolution of the NPC.
The simplicity of the putative NPC composition and architecture also suggests how the NPC evolved. The three major predicted domain types in nups (α-solenoid, FG repeat, and β-propeller) are composed of repeat motifs. This observation indicates extensive intragenic duplication in the evolution of the NPC and is consistent with a prior observation that repeat proteins play an important role in eukaryotic evolution in general (30). In folds with repeats, the number of repeats can vary even between orthologs, indicating that rapid loss and gain of repeats occur frequently. Indeed, such variation in repeat number is seen for the FG repeats of Nsp1, even in different strains of the same species (31). Sequence similarity among repeats may erode quickly, which makes the reconstruction of the evolutionary trajectory difficult despite common features in sequence and structure.
The eight predicted fold types of the 44 assigned domains comprising the 28 yeast nups indicate extensive gene duplication and structural redundancy in the building blocks of the NPC; all of the nups presumably originated from a minimal set of precursor proteins by extensive intragenic and intergenic duplication events. In fact, the same eight fold types are also found in the nups from H. sapiens. Interestingly, additional domains are found in higher eukaryotic nups. These predicted domains include zinc finger, Ran binding, Ran GAP (a GTPase activating domain for Ran), and cyclophilin domains. Many of these domains occur within a single protein, Nup358, a 3,224-residue vertebrate protein (32, 33). These domains constitute additional elaborations to the core NPC that occurred during the diversification of eukaryotes.
Karyopherins.
Karyopherins mediate the specific transfer of cargo macromolecules through the NPC via their interactions with the FG nups (27). The crystallographic structures of karyopherins show that they assume an α-solenoid fold (34). We have proposed here that approximately one-third of the nups also contain α-solenoid domains. Moreover, these α-solenoid-containing nups, like other nups, can dynamically associate and dissociate from NPC (35). This mobility is linked with the targeting of the α-solenoid-containing nups to more than one kind of structure in the cell; for example, a complex of several α-solenoid-containing nups localizes to kinetochores during mitosis (36), perhaps reflecting an ancient association between kinetochores and the NE that is still seen in dinoflagellates (37). Given these considerations, it is tempting to suggest that the karyopherins and the α-solenoid nups may share the same ancient precursor. According to this proposal, karyopherins evolved from the nups that were an integral structural part of an early NPC but diverged so that the interaction became transient as required by a developing cargo transport function.
Evolution and Emergence.
The concept of emergence suggests that, in a structured system, at each level, units associate to form a unit of the level above (38). Each new unit formed by the integration of subunits from the level below has “emerged” characteristics and capacities not present at any lower level of integration. This emergence can be observed in the NPC model, in which individual sequence motifs multimerize to form much larger proteins (Fig. 3a, step I) that assemble into multiple copies to form each half-spoke (Fig. 3a, step II) that eventually dimerize to form the spokes (Fig. 3a, step III), which are themselves repeated eight times to form the complete NPC (Fig. 3a, step IV). Thus, the entire NPC appears to be based on the hierarchical repetition of simple structural motifs to form an elaborate structure. In addition, it is reasonable to suggest that this hierarchical structure evolved through a series of gene duplications and divergencies.
Conclusion
We find simplicity at several levels in the NPC architecture. There are only eight predicted fold types assumed by the domains of 28 yeast nups. Except for the two most infrequent folds, they all consist of repeats of regular secondary structure segments. This composition indicates the central role of intragene and gene duplication in the evolution of the NPC. Thus, the entire NPC appears to be modular to a much greater extent than has previously been realized. Moreover, the eight fold types are partitioned between three groups of the NPC structure: the membrane, scaffold, and FG groups (Fig. 3b). It is conceivable that even karyopherins, which transiently interact with the NPC during their mediation of the transport through the pore, evolved via duplication and divergence from the ancestors common to both karyopherins and scaffold nups. It remains to be seen whether the three-dimensional structure of the NPC, once solved, reflects the simplicity of the architecture suggested by the proposed fold assignments.
Materials and Methods
Predicting Sequence Features of Nups.
The secondary structure, TMH, disordered, and coiled-coil regions were predicted from sequence by the psi-pred (39), phobius v2.0 (40), and disopred servers (41) and the coils program (28), respectively. FG repeat domains were identified as the disordered regions containing repeats of any of the following sequence motifs: FG, FxF, FxFG, GLFG, SAFG, PSFG, and SAFGxPSFG, where we allowed x to be any residue type.
Sequence-Based Searching for Nup Homologs.
Disordered segments were excluded from sequence comparisons, because they tend to fail the assessment of statistical significance of a sequence match. The nups were divided into domains by an iterative manual process relying on predicted secondary structure, gaps in multiple sequence alignments, and sequence–structure alignments from threading. For each nup domain, homologous sequences in the nonredundant uniprot90 database (42) were detected by the build_profile command of modeller-8 (43). build_profile is an iterative database-searching tool that relies on local dynamic programming to generate alignments and a robust estimate of their statistical significance (44); as a result, build_profile detects ≈50% more homologs than psi-blast (45). Each of the resulting nup profiles was scanned against our database of profiles for the domains of known structure, using the pp_scan command of modeller-8. pp_scan compares pairs of sequence profiles by a local dynamic programming algorithm using the correlation coefficient between profile columns as the scoring function.
Fold Assignment by Threading and Model Assessment.
Nup segments unmatched to a known structure by the sequence-based searches were submitted to the mgenthreader (18), fugue (20), agape (19), and hhsearch (21) fold assignment web servers. We combined these results into consensus fold assignments and evaluated the fold assignments by an iterative process of alignment, comparative model building, and model assessment (16).
Protease Accessibility Laddering.
We performed proteolytic domain mapping (16, 47) for nine yeast nups. Together with the previous mapping of the seven nups in the Nup84 subcomplex (16), we thus cover all nups without FG repeats. The protease accessibility laddering data were used to assess the fold assignments. Quantification of the agreement between the observed proteolytic cuts and the model is based on the assumption that the proteases cut only next to specific residues (i.e., Lys, Arg, and Asp) that are exposed to the solvent and located between secondary structure elements (i.e., exposed loops). Each model was assessed as follows. We compared the number of experimentally observed cleavage locations occurring in exposed regions between secondary structure segments of the model with the corresponding number expected by chance. This latter number was obtained by modeling the sequence using randomly selected folds as templates.
Supplementary Material
Acknowledgments
We thank the members of the A.S. and M.P.R. laboratories for discussions about the NPC, especially Maya Topf and Fred Davis. We also thank Joe Fernandez and the Proteomic Resource Center of The Rockefeller University for protein sequence analysis. This work was supported by The Sandler Family Supporting Foundation, Sun Microsystems, IBM, Intel, and National Institutes of Health Grants GM62529 and GM54762 (to A.S.); by an Irma T. Hirschl Career Scientist Award, a Sinsheimer Scholar Award, a grant from the Rita Allen Foundation, and National Institutes of Health Grants GM062427 and RR022220 (to M.P.R.); and by National Institutes of Health Grants RR00862 (to B.T.C.) and CA89810 (to B.T.C. and M.P.R.).
Glossary
Abbreviations:
- NPC
nuclear pore complex
- NE
nuclear envelope
- nup
nucleoporin
- TMH
transmembrane helix
- RRM
RNA recognition motif.
Footnotes
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Rout M. P., Aitchison J. D. J. Biol. Chem. 2001;276:16593–16596. doi: 10.1074/jbc.R100015200. [DOI] [PubMed] [Google Scholar]
- 2.Suntharalingam M., Wente S. R. Dev. Cell. 2003;4:775–789. doi: 10.1016/s1534-5807(03)00162-x. [DOI] [PubMed] [Google Scholar]
- 3.Rout M. P., Aitchison J. D., Suprapto A., Hjertaas K., Zhao Y., Chait B. T. J. Cell Biol. 2000;148:635–651. doi: 10.1083/jcb.148.4.635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cronshaw J. M., Krutchinsky A. N., Zhang W., Chait B. T., Matunis M. J. J. Cell Biol. 2002;158:915–927. doi: 10.1083/jcb.200206106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beck M., Forster F., Ecke M., Plitzko J. M., Melchior F., Gerisch G., Baumeister W., Medalia O. Science. 2004;306:1387–1390. doi: 10.1126/science.1104808. [DOI] [PubMed] [Google Scholar]
- 6.Yang Q., Rout M. P., Akey C. W. Mol. Cell. 1998;1:223–234. doi: 10.1016/s1097-2765(00)80023-4. [DOI] [PubMed] [Google Scholar]
- 7.Jarnik M., Aebi U. J. Struct. Biol. 1991;107:291–308. doi: 10.1016/1047-8477(91)90054-z. [DOI] [PubMed] [Google Scholar]
- 8.Stoffler D., Fahrenkrog B., Aebi U. Curr. Opin. Struct. Biol. 1999;11:391–401. doi: 10.1016/S0955-0674(99)80055-6. [DOI] [PubMed] [Google Scholar]
- 9.Bayliss R., Littlewood T., Stewart M. Cell. 2000;102:99–108. doi: 10.1016/s0092-8674(00)00014-3. [DOI] [PubMed] [Google Scholar]
- 10.Liu S. M., Stewart M. J. Mol. Biol. 2005;349:515–525. doi: 10.1016/j.jmb.2005.04.003. [DOI] [PubMed] [Google Scholar]
- 11.Fribourg S., Braun I. C., Izaurralde E., Conti E. Mol. Cell. 2001;8:645–656. doi: 10.1016/s1097-2765(01)00348-3. [DOI] [PubMed] [Google Scholar]
- 12.Hodel A. E., Hodel M. R., Griffis E. R., Hennig K. A., Ratner G. A., Xu S., Powers M. A. Mol. Cell. 2002;10:347–358. doi: 10.1016/s1097-2765(02)00589-0. [DOI] [PubMed] [Google Scholar]
- 13.Robinson M. A., Park S., Sun Z.-Y. J., Silver P. A., Wagner G., Hogle J. M. J. Biol. Chem. 2005;280:35723–35732. doi: 10.1074/jbc.M505068200. [DOI] [PubMed] [Google Scholar]
- 14.Berke I. C., Boehmer T., Blobel G., Schwartz T. U. J. Cell Biol. 2004;167:591–599. doi: 10.1083/jcb.200408109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Weirich C. S., Erzberger J. P., Berger J. M., Weis K. Mol. Cell. 2004;16:749–760. doi: 10.1016/j.molcel.2004.10.032. [DOI] [PubMed] [Google Scholar]
- 16.Devos D., Dokudovskaya S., Alber F., Williams R., Chait B. T., Sali A., Rout M. P. PLoS Biol. 2004;2:e380. doi: 10.1371/journal.pbio.0020380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mans B. J., Anantharaman V., Aravind L., Koonin E. V. Cell Cycle. 2004;3:1612–1637. doi: 10.4161/cc.3.12.1316. [DOI] [PubMed] [Google Scholar]
- 18.McGuffin L. J., Jones D. T. Bioinformatics. 2003;19:874–881. doi: 10.1093/bioinformatics/btg097. [DOI] [PubMed] [Google Scholar]
- 19.Przybylski D., Rost B. J. Mol. Biol. 2004;341:255–269. doi: 10.1016/j.jmb.2004.05.041. [DOI] [PubMed] [Google Scholar]
- 20.Shi J., Blundell T. L., Mizuguchi K. J. Mol. Biol. 2001;310:243–257. doi: 10.1006/jmbi.2001.4762. [DOI] [PubMed] [Google Scholar]
- 21.Soding J. Bioinformatics. 2005;21:951–960. doi: 10.1093/bioinformatics/bti125. [DOI] [PubMed] [Google Scholar]
- 22.Koh I. Y., Eyrich V. A., Marti-Renom M. A., Przybylski D., Madhusudhan M. S., Eswar N., Grana O., Pazos F., Valencia A., Sali A., Rost B. Nucleic Acids Res. 2003;31:3311–3315. doi: 10.1093/nar/gkg619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Denning D. P., Patel S. S., Uversky V., Fink A. L., Rexach M. Proc. Natl. Acad. Sci. USA. 2003;100:2450–2455. doi: 10.1073/pnas.0437902100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Saxena K., Gaitatzes C., Walsh M. T., Eck M., Neer E. J., Smith T. F. Biochemistry. 1996;35:15215–15221. doi: 10.1021/bi961616x. [DOI] [PubMed] [Google Scholar]
- 25.Fabre E., Hurt E. C. Annu. Rev. Genet. 1997;31:277–313. doi: 10.1146/annurev.genet.31.1.277. [DOI] [PubMed] [Google Scholar]
- 26.Wheelock M. J., Johnson K. R. Annu. Rev. Cell Dev. Biol. 2003;19:207–235. doi: 10.1146/annurev.cellbio.19.011102.111135. [DOI] [PubMed] [Google Scholar]
- 27.Strawn L. A., Shen T., Shulga N., Goldfarb D. S., Wente S. R. Nat. Cell Biol. 2004;6:197–206. doi: 10.1038/ncb1097. [DOI] [PubMed] [Google Scholar]
- 28.Lupas A. Methods Enzymol. 1996;266:513–525. doi: 10.1016/s0076-6879(96)66032-7. [DOI] [PubMed] [Google Scholar]
- 29.Fotin A., Cheng Y., Sliz P., Grigorieff N., Harrison S. C., Kirchhausen T., Walz T. Nature. 2004;432:573–579. doi: 10.1038/nature03079. [DOI] [PubMed] [Google Scholar]
- 30.Marcotte E. M., Pellegrini M., Yeates T. O., Eisenberg D. J. Mol. Biol. 1999;293:151–160. doi: 10.1006/jmbi.1999.3136. [DOI] [PubMed] [Google Scholar]
- 31.Wimmer C., Doye V., Grandi P., Nehrbass U., Hurt E. C. EMBO J. 1992;11:5051–5061. doi: 10.1002/j.1460-2075.1992.tb05612.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wu J., Matunis M. J., Kraemer D., Blobel G., Coutavas E. J. Biol. Chem. 1995;270:14209–14213. doi: 10.1074/jbc.270.23.14209. [DOI] [PubMed] [Google Scholar]
- 33.Reverter D., Lima C. D. Nature. 2005;435:687–692. doi: 10.1038/nature03588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Conti E., Izaurralde E. Curr. Opin. Cell Biol. 2001;13:310–319. doi: 10.1016/s0955-0674(00)00213-1. [DOI] [PubMed] [Google Scholar]
- 35.Rabut G., Doye V., Ellenberg J. Nat. Cell Biol. 2004;6:1114–1121. doi: 10.1038/ncb1184. [DOI] [PubMed] [Google Scholar]
- 36.Loiodice I., Alves A., Rabut G., Van Overbeek M., Ellenberg J., Sibarita J. B., Doye V. Mol. Biol. Cell. 2004;15:3333–3344. doi: 10.1091/mbc.E03-12-0878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Costas E., Goyanes V. Cytogenet. Genome Res. 2005;109:268–275. doi: 10.1159/000082409. [DOI] [PubMed] [Google Scholar]
- 38.Jacob F. Science. 1977;196:1161–1166. doi: 10.1126/science.860134. [DOI] [PubMed] [Google Scholar]
- 39.McGuffin L. J., Bryson K., Jones D. T. Bioinformatics. 2000;16:404–405. doi: 10.1093/bioinformatics/16.4.404. [DOI] [PubMed] [Google Scholar]
- 40.Kall L., Krogh A., Sonnhammer E. L. J. Mol. Biol. 2004;338:1027–1036. doi: 10.1016/j.jmb.2004.03.016. [DOI] [PubMed] [Google Scholar]
- 41.Ward J. J., McGuffin L. J., Bryson K., Buxton B. F., Jones D. T. Bioinformatics. 2004;20:2138–2139. doi: 10.1093/bioinformatics/bth195. [DOI] [PubMed] [Google Scholar]
- 42.Apweiler R., Bairoch A., Wu C. H., Barker W. C., Boeckmann B., Ferro S., Gasteiger E., Huang H., Lopez R., Magrane M., et al. Nucleic Acids Res. 2004;32:D115–D119. doi: 10.1093/nar/gkh131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sali A., Blundell T. L. J. Mol. Biol. 1993;234:779–815. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- 44.Pearson W. R. J. Mol. Biol. 1998;276:71–84. doi: 10.1006/jmbi.1997.1525. [DOI] [PubMed] [Google Scholar]
- 45.Altschul S. F., Madden T. L., Schäffer A. A., Zhang J., Zhang Z., Miller W., Lipman D. J. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Melo F., Sanchez R., Sali A. Protein Sci. 2002;11:430–448. doi: 10.1002/pro.110430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dokudovskaya S. S., Williams R., Dews D., Sali A., Chait B. T., Rout M. P. Structure (London) 2006. in press. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.