Graphical abstract

Keywords: Audio, Image, Self-diagnosis, Deep learning, Reduced network complexity, Multimodal fusion
Abstract
The COVID-19 emerged at the end of 2019 and has become a global pandemic. There are many methods for COVID-19 prediction using a single modality. However, none of them predicts with 100% accuracy, as each individual exhibits varied symptoms for the disease. To decrease the rate of misdiagnosis, multiple modalities can be used for prediction. Besides, there is also a need for a self-diagnosis system to narrow down the risk of virus spread in testing centres. Therefore, we propose a robust IoT and deep learning-based multi-modal data classification method for the accurate prediction of COVID-19. Generally, highly accurate models require deep architectures. In this work, we introduce two lightweight models, namely CovParaNet for audio (cough, speech, breathing) classification and CovTinyNet for image (X-rays, CT scans) classification. These two models were identified as the best unimodal models after comparative analysis with the existing benchmark models. Finally, the obtained results of the five independently trained unimodal models are integrated by a novel dynamic multimodal Random Forest classifier. The lightweight CovParaNet and CovTinyNet models attain a maximum accuracy of 97.45% and 99.19% respectively even with a small dataset. The proposed dynamic multimodal fusion model predicts the final result with 100% accuracy, precision, and recall, and the online retraining mechanism enables it to extend its support even in a noisy environment. Furthermore, the computational complexity of all the unimodal models is minimized tremendously and the system functions effectively with 100% reliability even in the absence of any one of the input modalities during testing.
1. Introduction
The novel coronavirus disease 2019 (COVID-19) caused by the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection was first reported at the end of 2019 in China and swiftly spread across the globe [9]. The severity of the pandemic on the lives of the people can be reduced by early detection and treatment. There is also a shortage of health workers to take care of all the patients throughout the day. This makes it crucial to develop a preliminary remote self-testing procedure that provides immediate results and essentially allows testing at any place and any time. The symptoms such as cough, shortness of breath, voice breaks, lung infection are the most recorded among COVID-19 patients. Further, remote assistance is required in monitoring the patients’ symptoms. This can be provided by the Internet of Things (IoT) [15] and the retrieved data can be analyzed using machine learning techniques for diagnosis. Even if there are X-ray and CT scan centres established in remote villages, the availability of radiologists is still an issue. Hence, a lightweight remote diagnosis system that is easily accessible is necessary for immediate screening.
Earlier research has shown that vocal attributes of patients suffering from respiratory ailments have distinguishing features. These features can be extracted by suitable signal processing methods from symptomatic vocal traits, such as cough, speech, and breathing [13]. The extracted auditory input features can then be used to train a deep learning model for performing the preliminary screening of COVID-19[1]. Therefore, it is necessary to classify the specified three vocal traits of non-COVID-19 persons from those of COVID-19 patients [18]. Even though these symptoms may be absent in asymptomatic patients, it has been identified in a majority of the patients and is known to be a major cause of spreading in a social environment. Generally, temperature check is the precautionary method used in public places to identify potentially infected people. However, in addition to temperature checking, audio-based classification can prove to be significantly helpful in mitigating the spread of the disease in public places. Further, the coronavirus can inflict lung infections even on asymptomatic patients. Moreover, chest X-Rays and CT scans contain salient information about the damages caused in the lungs and can be used for accurate diagnosis of both symptomatic and asymptomatic individuals.
Nevertheless, using a single type of symptom may lead to declining prediction performance from a classification perspective. It would be preferable to integrate multiple input modalities and build multimodal fusion strategies to use complementary information from different types of symptoms. This allows us to gain better performance in terms of both accuracy and reliability [40]. Thus, in our work, we make use of 5 inputs such as cough, speech, breathing sounds, X-ray, and CT images to build the multimodal system. Multi-modal systems operate with multiple deep sub-models leading to an increase in system complexity. Hence, we propose lightweight architectures to construct the sub-models for each input modality. Multimodal fusion can generally be classified into early fusion, late fusion, and hybrid fusion. Early fusion method suffers from data scarcity and time synchronization difficulties [41]. The hybrid fusion method is a combination of early and late fusion and so faces the same problem as early fusion. Hence, we propose a machine learning-based Random Forest late fusion in our multimodal decision making, which is the best fusion strategy, especially with a smaller dataset. Furthermore, we introduce a dynamic retraining mechanism that allows the trained Random forest to update its parameters to support noisy environments.
The novelty and major contributions of our proposed work are as follows.
-
1.
To introduce an IoT-based cognitive framework that operates with multimodal data such as audios (cough, speech, breathing) and images (X-ray, CT Scan) for the effective diagnosis of COVID-19 with 100% accuracy.
-
2.
To propose two novel deep learning models with reduced network complexity namely CovParaNet and CovTinyNet for audio and image classification tasks respectively.
-
3.
To ensure 100% reliability even in noisy environments, a dynamic Random Forest-based late fusion method is used to fuse the predictions from lightweight unimodal models.
-
4.
To make the system modular and failproof even in the absence of one or more input data.
-
5.
To develop a lightweight dynamic multimodal framework that can easily be deployed in a real-time self-diagnosis/ remote monitoring system.
-
6.
To evaluate the performance of the proposed models with the existing benchmark models.
The remaining part of the work is organized as follows. Section 2 explains the literature survey. 3, 4 elaborates the proposed work and experimental details respectively. Performance metrics are discussed in Section 5. Section 6 gives the experimental evaluation. The final discussions are presented in Section 7, and Section 8 concludes the paper.
2. Literature survey
In this section, a review of some important literary works on COVID-19, audio, and image processing are presented. Abdulkareem et al. [28] developed an IoT-based COVID-19 prediction system with ML. In [9], Ahamad et al. used ML algorithms to identify COVID-19 with clinical data. However, the usage of clinical data requires a visit to the hospital. Previous studies show that cough can be used as a standalone symptom to diagnose a range of respiratory diseases [2], [5], [11]. In [2], Windmon et al. used cough as the primary symptom to predict pulmonary disease and heart failure. In [11] MFCC and CIF features on cough sounds were used to detect croup patients which yielded an accuracy of 86.09%. Jesus et al. used SVM in [7] and worked with KNN in [8] that was trained with MFCC, LPC, and spectral features to detect cough. However, the correlation with a particular disease was absent. Ali Imran[1] et al. developed a cough-based COVID-19 prediction app. Although the usage of 3 deep models reduces misdiagnosis, it increases the system complexity. In [3], the authors identified COVID-19 using MFCCs from cough. J. Laguarta et al. also proposed the use of MFCCs in [17] to diagnose COVID-19 with cough using 3 ResNet50s in parallel yielding an accuracy of 98.5%. Still, other input sounds can also be included to validate the model results.
Researchers have reported the feasibility of voice-based COVID-19 diagnosis in symptomatic and asymptomatic stages [18]. In [12], the authors compared the voice between healthy and COVID-19 patients by observing their pronunciation of vowel/a/. Cough sounds and speech (phonemes/ah/ and/z/, counting) were used in [13]. Z. Jiang et al. [14] introduced a non-contact method to screen the COVID-19 patients with respiratory characteristics. J. Acharya et al. [20] identified breathing sound anomalies for diagnosis of respiratory and pulmonary diseases. Nevertheless, the Hybrid CNN-RNN model used here only achieves a score of 66.31%. A public sentiment analysis in COVID-19 pandemic was performed in [33]. A new variant of the CNN model with parallel pooling structures was introduced in [4] to enhance the accuracy. As COVID-19 is a contagious disease, self-diagnosis methods are imperative. It lowers the number of individuals visiting healthcare facilities for getting tested and in turn mitigates the spread in these hotspots. IoT-based diagnosis for diseases namely asthma [6], obesity, high blood pressure, and diabetes [16] has been very popular in recent studies. IoT-based frameworks to identify COVID-19 were proposed in [10], [15], attaining a maximum accuracy of 92.95% with SVM in [10] and 74.7% in [15], providing more room for improvement.
Chest radiographs have been recently used in addition to the standard RT-PCR tests in clinical diagnosis. Yet, there aren’t adequate numbers of specialized radiologists in remote areas. Deep learning algorithms applied to radiography images can assist in providing an initial screening in remote areas. F. Shi et al. [34] analyzed the application of AI in COVID-19 predictions with X-ray and CT images. Mohammed et al. [30] made a benchmarking study and identified SVM as the best ML model for COVID-19 using X-Rays. Similarly, ResNet-50 followed by MobileNetV2 were identified as the best deep learning models [31]. Al-Waisy et al. proposed a COVID-CheXNet model [29] and COVID-DeepNet model [32] attaining 99.99% and 99.93% accuracy respectively. A lot of studies analyzed the use of CT scans for COVID-19 diagnosis [19], [21], [35]. A voting classifier called Guided WOA based on Particle Swarm Optimization (PSO) was introduced in [19]. Voulodimos in [35] proposed a few shot U-Net model that operates well in erroneous test conditions. However, their model’s performance in terms of metrics such as precision, recall, and f1-score are very low. The You Only Look Once (YOLO) models are primarily used for real-time object detection. YOLO uses Darknet as the backbone, which implements CNN to classify a detected object. In [24], the authors implemented YOLO to classify players and track ball movements in video clips of a basketball game. YOLOv3 tiny is used in [25] to detect broken corns, as the tiny variant has a good trade-off between memory, efficiency, and speed.
The existing works of literature mostly report COVID-19 prediction with a single modality of data. In [26], it is stated that significant knowledge can be extracted with multiple modalities, which is not possible with a single modality alone. A multimodal model was proposed in [27] that detects Alzheimer’s disease progression based on the early fusion of five types of multimodal time-series data. But, [40] states that early and hybrid fusion methods suffer from time synchronization problems. The authors also explain that each individual has a different response for a physiological signal, so there is a decline in performance if only a single modality is used. A multimodal late feature fusion strategy was used in [41] that makes use of a SoftMax classifier for emotion classification. Thus, utilizing multimodal data can pave the way for the development of an efficacious system for the prediction of COVID-19.
This review on various existing and reported methods on COVID-19 prediction using different modalities is summarized in Table 1 . (In Table 1, Acc. = accuracy, Sens. = sensitivity, Spec. = specificity, Rec. = recall.) Many of the aforementioned works concentrate on methods using clinical symptoms that need a visit to a clinic. Hence, there arises a need to develop a remote diagnosis system. Although a few studies make use of non-contact methods, they use a single modality of input, thereby making the system not completely reliable. To make the system accurate using a single modality, a few works have been proposed with a combination of two or more deep models. But, this may lead to additional complexity. Thus, it is critical to construct a system that operates on multiple modalities. At the same time, the sub-models considered must have a reduced network complexity. Furthermore, some of the late fusion strategies used for multimodal fusion in the existing works are max voting, softmax function, or weighted sum. However, these methods were not able to provide 100% accurate results. Hence, the use of a machine learning algorithm in the fusion task is necessary to attain highly accurate and reliable results. Also, to make the system robust to erroneous data in a noisy environment, it is necessary to introduce a dynamic mechanism that updates the learned parameters automatically.
Table 1.
Summary of previous methodologies on COVID-19 prediction.
| Literature/Year | Modality | Finding | Methods/Features | Result | Challenges/Research Gap |
|---|---|---|---|---|---|
| [3] (2020) | Cough | Classification: COVID-19/ Pneumonia/ Pertussis/ Others | LSTM, MFCC features, SVM | LSTM - 88% (Acc.), SVM- 94% (Acc.) | In [3], [17], [12], [14], [22], single modality of input was only used yielding only acceptable levels of confidence (<100%). Using additional modalities can boost the performance. |
| [17] (2020) | Cough | Classification: COVID-19/ Others | CNN, MFCC | 98.5% (Acc.), 94.2 (Spec.), 0.97 (AuC) | |
| [12] (2020) | Voice | Comparison of healthy and COVID-19 patients | Two-way ANOVA and Wilcoxon's rank-sum test | Significant differences observed between COVID-19 patients and the healthy participants. | |
| [14] (2020) | Respiratory characteristics | COVID-19 prediction | Bi-GRU | 83.69% (Acc.), 90.23% (Sens.) and 76.31% (Spec.) | |
| [22] (2020) | X-Rays | COVID-19 diagnosis | Patch-based CNN | 91.9% (Acc.) | |
| [9] (2020) | Clinical Symptoms - Age, fever, cough, etc. | To identify the highly correlated features in predicting COVID-19 | XGBoost | Significant symptoms identified: fever, cough, lung infection. | In [9], [28], diagnosis with clinical data requires a visit to the hospital. In addition, the performance also needs to be improved. |
| [28] (2021) | Laboratory findings | COVID diagnosis | Naive Bayes, Random Forest, and SVM | SVM - 95% (Acc.) | |
| [1] (2020) | Cough | Classification: COVID-19/ Bronchitis/ Pertussis/ Normal | Transfer learning based ML and DL models | 92.85%(Acc.) | In [1], [23], [29], [32], usage of 2 or 3 deep models with increased no. of layers for a single modality of input increases the system complexity. Thus, a lightweight architecture is required. |
| [23] (2020) | X-Rays | To identify normal, COVID-19, viral pneumonia | Fusion of ResNet-101 and ResNet-152 | 96.1%(Acc.) | |
| [29] (2020) | X-Rays | COVID-19 diagnosis | COVID-CheXNet (Score level fusion of ResNet-34 and HRNets) | 99.99%(Acc.) | |
| [32] (2021) | X-Rays | COVID-19 diagnosis | COVID-DeepNet model (Deep Belief Network (DBN) and Convolutional DBN) | 99.93%(Acc.) | |
| [35] (2021) | CT images | COVID-19 Infected Area Segmentation | UNet, dynamic retraining method | 95% (Confidence level) | Increased system overhead due to image retraining. |
| [21] (2020) | CT scans | COVID-19 prediction | Attention based Multiple instance learning | 97.9% (Acc.), 99.0% (AuC) | 3D imaging increases training and spatial complexity |
| [30] (2020) | X-Rays | Benchmarking study to identify the best ML model for COVID-19 | Entropy and TOPSIS methods | SVM was identified as the best model | In [30], [31], [19], a single modality of input was used and it cannot be relied upon due to varied symptoms among patients. Further, these models are not suitable for noisy environments. |
| [31] (2021) | X-Rays | Comparative study of deep learning models for COVID-19 diagnosis | Deep learning models | Resnet-50 (98.8%), MobileNetV2 (93.5%) (Acc.) | |
| [19] (2020) | CT scans | COVID-19 prediction | AlexNet | AuC of 0.995 | |
| [13] (2020) | Cough and Speech | COVID-19 prediction | RNN, Ensemble Stacking | 78% (Rec.) | In [13], [10], [15], in Spite of usage of multiple modalities or symptoms, the overall performance is not sufficient. |
| [10] (2020) | Fever, cough, shortness of breath | IoT based COVID diagnosis | ML models (SVM, KNN, Naive Bayes, etc.) | 92.95% (Acc.) | |
| [15] (2020) | Temperature, cough rate, respiratory rate, and blood oxygen saturation | IoT based COVID diagnosis | Fog based ML | SVM - 74.7% (Acc.) |
3. Proposed System
In this work, we propose a preliminary screening method for COVID-19 that can be done remotely by anyone without the need to visit a clinic. This work attempts to give a disease prediction result based on multimodal data including audio and image samples of the suspected patients. The existing prediction methods are based on a single mode of input which could be X-rays, CT scans, cough sounds, etc. There is a need for accuracy in prediction for this sensitive task which can be made possible by employing multimodal data consisting of audio data and image data such as cough, speech, breathing sounds, Chest X Rays, and CT images. The predictions made by each of the classifiers are fused by a machine learning model to determine the final result. In comparison with disease prediction made through a single mode of data, the proposed dynamic multimodal framework with an ensemble of trained models can nullify the number of false diagnoses as it combines the robustness of five individually trained lightweight and efficient models.
3.1. Dataset Description
Three types of acoustic data which are considered are organized in 3 different datasets namely cough, speech, and breathing sound dataset. The cough audio dataset (289 audio samples in total) augmented from two different sources namely virufy [36] and coswara [37] GitHub repository is organized into two folders consisting of 200 COVID-19 negative cough samples and 89 positive cough samples after removing inconsistent (empty, defective) samples. To increase the dataset size, data augmentation was performed and the resulting dataset comprised 153 positive samples and 348 negative samples. The speech sound dataset collected consists of 76 positive samples and 200 negative samples. In the breathing sound dataset, the number of samples in the positive class is 193 and the negative class is 309 samples. Both speech and breathing datasets were collected from the coswara repository. In this study, raw X-ray and CT images were obtained from two different open online sources. The chest X-ray dataset [38] consists of 125 COVID-19 positive samples and 500 normal samples. The chest CT dataset [39] consists of 1252 COVID-19 positive samples and 1229 normal samples.
3.2. Deep Multimodal Learning for COVID-19 Prediction
The proposed architecture includes three major modules, namely diagnosis with audio data, image data, and multimodal fusion. Inputs collected from potential patients in real-time can be forwarded to the respective classifiers for obtaining predictions based on that particular data. The acoustic data including cough sounds, speech sounds (utterance of vowel ‘a’), and breathing sounds are used for training with the proposed CovParaNet model. Image data such as chest X Rays and CT scans are trained using our proposed CovTinyNet model. The data are also trained with the existing state-of-the-art models and the proposed models were identified as the best among them and is used in the multimodal fusion mechanism. We employ the method of late fusion here by a Machine Learning based dynamic Random Forest model on the predicted outcomes of the five separately trained models to get the final prediction. The proposed cognitive disease prediction system architecture is shown in Fig. 1 .
Fig. 1.
Cognitive Disease Prediction System Architecture.
3.2.1. Acoustic Data Classification
In general, audio classification consists of manual feature extraction, followed by a feature selection method, and finally, classification using machine learning algorithms. Another method is the usage of raw audio waveforms in classification using deep learning models. In our work, for the classification of audio data, we make use of a feature called MFCC that can be directly used as input for classification using a deep learning model. Here, the audio classification pipeline consists of dataset acquisition, pre-processing (feature extraction), and classification using CovParaNet (an enhanced CNN model).
3.2.1.1. Feature Extraction
The feature extraction stage involves extracting multi-dimensional MFCC feature vectors. Three types of audio samples with two classes each are transformed into the Mel scale for further processing. The Mel scale categorizes pitch where humans can interpret changes in pitch to be equal in length from each other along this scale. It is intended to make changes in frequency, such as with a spectrogram, more closely reflect audible changes. Mel scale provides a higher resolution in lower frequencies and vice versa. Since symptomatic acoustic data are known to have more energy in lower frequencies, the MFCC is a more suitable representation for these sounds.
There are multiple methods for transforming the frequency scale to the Mel scale. Here, the frequency f is converted into Mel scale m as:
| (1) |
The cepstral analysis is performed on the Mel spectrum of audio samples to compute their Cepstral coefficients, and these values are generally known as Mel Frequency Cepstral Coefficients (MFCCs).
3.2.1.2. Existing Predictive Models
Convolutional Neural Network (CNN) is a deep neural network widely used for image analysis. In recent studies, it was discovered that CNN can also be effectively used in sequential data analysis such as sound processing and natural language processing. The two primary operations in CNN are convolution and pooling. The convolution operation consists of various filters that extract features to build a feature map from the input data. Their corresponding spatial information is preserved using these learned feature maps. The pooling operations are used in dimensionality reduction of the feature maps obtained from the convolution operation. Activation functions like ReLU, leaky ReLU, etc., are used to transfer the gradient during training by backpropagation. However, sequential CNN may lead to the loss of information learned in the initial layers.
Recurrent Neural Network (RNN) is also a category of the artificial neural network, primarily used for temporal data by utilizing memory for processing inputs such as speech. All recurrent neural networks consist of repeating structures of neural networks which are generally a single tank layer. But, RNN networks are prone to gradient vanishing and exploding problems.
3.2.1.3. Cov-Para-Net Architecture
The CovParaNet architecture is used for training the three types of acoustic data separately. It comprises an input layer and three stages of convolution layers. In the first stage, three parallel convolution layers with different filter sizes of 8, 32, and 64 are used for feature learning. It is followed by a concatenation layer that merges the features extracted by the parallel convolution layers in the second stage and another convolution layer is introduced. The third stage is similar to the first stage with three parallel convolution layers. Then, a concatenation is performed and the features are flattened and 2 fully connected layers are used with the ReLU activation function in the first and SoftMax activation function in the second dense layer.
After every convolution layer, max pooling is performed to reduce the dimensional complexity. Batch normalization is also performed following max pooling to normalize the activations in the current layer before passing the values to the next layer and as a result speeding up the training process. The model is trained for 70 epochs with a batch size of 32. The optimizer used in the model is the Adam optimizer and sparse categorical cross-entropy is used as the loss function. The learning rate is fixed to a value of 0.0001. Finally, the output of the model gives the classification of the audio samples into COVID-19 positive or negative. The proposed parallel convolutions with different kernel sizes learn the important features in the initial stages that could easily be missed in the case of a sequential network. It also trains quickly due to reduced system complexity. The detailed algorithmic steps are depicted in Algorithm 1. The architectural specifications of the CovParaNet system are shown in Fig. 2 .
| Algorithm 1: COV-PARA-NET ALGORITHM |
|---|
| Input: A directory of audio samples X |
| Parameters: Classifier Model M, MFCC features array F, Sample rate R, Audio duration D, Number of mfcc arrays per segment N, Samples per segment S, Number of segments n, Hop length H, audio sample i, Class c, positive class , negative class , Time stamp l, r, Segment St, Frequency in Hertz f, Number of MFCCs p, Final MFCC features , Output from filter bank , Learning rate , Batch size BS, Loop variables j, k, Number of test samples t |
| Output: COVID-19 prediction result Y |
| 1: Initialize R = 44100, p = 13 |
| 2: S = R * D |
| 3: N = S/ H |
| 4: for each c in Xdo |
| 5: for each i and do |
| 6: Load i with R |
| 7: for j in 0 to n do |
| 8: St = Segment from l to r |
| 9: where, |
| 10: for k in 0 to N do |
| 11: f = Apply FFT on |
| 12: Convert to mel scale, ) |
| 13: ] |
| 14: end for |
| 15: end for |
| 16: end for |
| 17: end for |
| 18: Create M with specification given in Fig. 5 |
| 19: Compile M with Adam optimizer and = 0.0001 |
| 20: for j = 1 to 70 do |
| 21: Train M with , where BS = 32 |
| 22: end for |
| 23: for j = 1 to t do |
| 24: Y = Predict(M,i) |
| 25: end for |
| 26: return Y |
Fig. 2.
CovParaNet Architecture.
3.2.2. Visual Data Classification
Radiology imaging techniques can be coupled with deep learning for automated and accurate diagnosis of COVID-19. Both X Rays and CT scan images have been known to contain notable information regarding the virus. The two datasets are trained by our lightweight and efficient CovTinyNet model.
3.2.2.1. Existing Predictive Models
The emergence of deep learning techniques has revolutionized the field of artificial intelligence. Deep learning means, an increase in the number of layers, and as a consequence, the network size and complexity are also increased. Generally, an increase in network size increases the performance of the model. But real-time classification tasks require an efficient model with reduced network complexity.
Deep learning models such as Inception, DenseNet, ResNet, MobileNet, U-Net, RCNN, and YOLO are widely used in recent times for image analysis tasks. Inception is a 27 layer deep CNN architecture generally used for image classification. Resnet architecture uses residual and skip connections. Here the sum of output in the early layers is used as the input for the later layers. This avoids the loss or abstraction of information learned in the initial stages. In DenseNet, the output obtained from the previous layer is concatenated instead of addition. The major limitation of these is the increased complexity owing to its huge number of layers and skip connections. Region-based Convolutional Neural Network (RCNN) and You Only Look Once (YOLO) models are generally used for object detection. RCNN uses a set of boxes called regions in the image to check if the object is present in one of the boxes, which leads to the increased time taken for convergence.
U-Net is a type of CNN architecture mainly used for image segmentation. It consists of an encoder and decoder path which are concatenated with skip connections. These skip connections are known to provide local information to the global information during upsampling. The YOLO architecture is mainly used for object detection, classification, object localization, and segmentation. The variants such as YOLOv1, YOLOv2, and YOLOv3 performed better than the respective previous versions. YOLOv2 uses Darknet19 and YOLOv3 has Darknet53. These models are very efficient but have higher complexity and occupy larger memory spaces. Nevertheless, the tiny variant of YOLOv3 executes much faster in real-time and occupies much less memory. Hence it is the most desirable model for a real-time system.
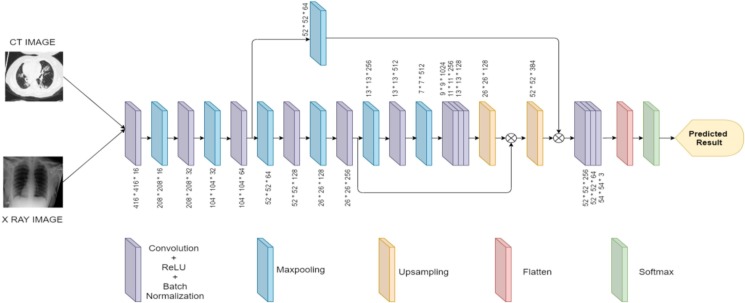
3.2.2.2. Cov-Tiny-Net Architecture Description
The architecture of YOLOv3 tiny is used as a basis for the construction of the CovTinyNet model. As YOLOv3 tiny has been proven to be a state-of-the-art model for real-time object detection, our model has been designed by enhancing the network structure of the existing yolov3 tiny. The number of layers used in the architecture has been decreased to increase the speed and accuracy for real-time diagnosis. We have also introduced additional skip connections as in U-Net architecture to retain local information. This helps the model to easily localize the abnormality in the medical images and correctly classify them. The reduction in the size of the model also makes it easier to be deployed on any remote device. Our CovTinyNet model consists of 12 convolution layers for feature learning and 7 max-pooling layers for dimensionality reduction. Each convolution is followed by a ReLU activation function and batch normalization is performed to normalize the activations. The feature maps are flattened into a feature vector and the final prediction is given by a softmax layer. The detailed steps followed are depicted in Algorithm 2. The architectural specification of the CovTinyNet is depicted in Fig. 3 .
| Algorithm 2: COV-TINY-NET ALGORITHM |
|---|
| Input: A directory of image samples X |
| Parameters: Classifier Model M, Class c, positive class , negative class , Learning rate , Batch size BS, epoch T, Loop variable i, j, Feature map F, Feature vector V, Weights W, Class probabilities P, Number of test samples t |
| Output: COVID-19 prediction result Y |
| 1: Initialize parameters T = 50, = 0.0003, BS = 32 |
| 2: for i = 1 to T do |
| 3: preprocess images , where X and |
| 4: F = generate |
| 5: = reshape feature maps F |
| 6: feature vector V = flatten |
| 7: produce class probabilities P |
| 8: W = update(W) |
| 9: end for |
| 10: Load the trained model M with W |
| 11: for j = 1 to t do |
| 12: Y = Predict(M,i) |
| 13: end for |
| 14: return Y |
Fig. 3.
CovTinyNet Architecture.
3.3. The Proposed Dynamic Multimodal Fusion Strategy
Random Forest (RF) is a type of machine learning model that follows the ensemble learning method. The multimodal fusion strategy is based on Random Forest. A Random Forest is an ensemble of decision trees constructed with a randomized subset of samples and features. The randomness in both instances and features allows the diversity of the base learners and avoids overfitting. The predictions made by the unimodal models are stacked to the RF which gives the final prediction result.
Furthermore, RF is known for handling missing data effectively. This allows the system to function even in the absence of any of the inputs. The prediction scores of the incorrectly classified samples are further fed to the trained RF to dynamically adjust its learned parameters through retraining. Feeding the entire dataset for retraining all the unimodal models increases the system complexity. In contrast, in our proposed system, only the RF fusion model undergoes retraining with the misclassified unimodal prediction scores. This makes the system dynamic without increasing its complexity. The detailed algorithm is depicted in Algorithm 3.
| Algorithm 3: Dynamic multimodal fusion algorithm |
|---|
| Input: Validation dataset X |
| Parameters: Validation Dataset X = {}, Test sample = {}, Cough sample , Speech sample , Breathing sample , X-ray sample , CT sample , Number of test subject n, Number of models k, Loop variables , Prediction score vector P, Prediction Score Train set , Prediction Score Test set , Random Forest Classifier model RFC, No. of decision trees n_estimators, No. of features n_features, No. of features for best split max_features, Minimum samples to split internal node min_samples_split, Misclassification scores vector F |
| Output: COVID-19 final prediction result Y |
| 1: Initialize n = 200, k = 5, n_estimators = 10, min_samples_split = 2 |
| 2: for i = 1 to n do |
| 3: for j = 1 to k do |
| 4: |
| 5: end for |
| 6: end for |
| 7: Split P into train set and |
| 8: Build the RFC model with max_features = sqrt(n_features) |
| 9: Train the RFC with |
| 10: Y = Test with trained RFC |
| 11: for i = 1 to 25 do |
| 12: if is an incorrect prediction then |
| 13: Append to F |
| 14: end if |
| 15: end for |
| 16: Retrain RFC with F to update learned parameters |
| 17: return Y |
4. Experiments
The experiments were first performed for unimodal acoustic models, followed by visual models. It is followed by experimentation with multimodal models. The detailed explanations are given in the following sections. Table 2 summarizes the experimental values of hyperparameters considered for optimization and the final tuned values for the parameters.
Table 2.
Hyperparameters considered for optimization of proposed models.
| Model | Hyperparameter | Range Considered | Optimal Value |
|---|---|---|---|
| CovParaNet | Learning rate | [0.0003, 0.0001, 0.01] | 0.0001 |
| Batch Size | [32, 64] | 32 | |
| Epochs | [30, 50, 70, 100] | 70 | |
| Sampling rate | [22050, 44100] | 44100 Hz | |
| No. of MFCCs | [13, 26] | 13 | |
| Convolution Layers | [3, 6, 7, 9, 13] | 7 | |
| Activation function | Fixed | ReLU | |
| Optimizer | Fixed | Adam | |
| Loss Function | Fixed | Sparse Categorical Cross Entropy Loss | |
| Train test split | Fixed | 75% - 25% | |
| CovTinyNet | Learning rate | [0.0003, 0.0001, 0.01] | 0.0003 |
| Batch Size | [32, 64] | 32 | |
| Epochs | [30, 50, 70, 100] | 50 | |
| Convolution Layers | [11, 12, 13] | 12 | |
| Activation function | Fixed | ReLU | |
| Optimizer | Fixed | Adam | |
| Loss Function | Fixed | Cross Entropy Loss | |
| Train test split | Fixed | 80% - 20% | |
| Dynamic Multimodal Fusion | Number of decision trees | Fixed | 10 |
| Minimum samples to split internal node | Fixed | 2 | |
| Train test split | Fixed | 75% - 25% |
4.1. Acoustic unimodal experiments
The audio files present in the three datasets are preprocessed by extracting Mel Frequency Cepstral Coefficients (MFCCs). The number of audio samples per recording is determined by the product of the sample rate and the duration of the audio recording. Segmentation of the audio files is performed to increase the training data. After the segmentation of each audio, each segment is further divided into multiple samples and finally, the count of MFCC arrays will be the quotient obtained by dividing the samples per segment by the hop length. These MFCC arrays are extracted and stored as a JSON file which would be used to train the CovParaNet model. An audio library called librosa aids in the extraction of the MFCC features from the audio files. The implementation of the CovParaNet is performed in google colab workspace with Keras which uses TensorFlow as the backend engine. The JSON file with the extracted features organized in a 2D format is fed as an input into the models used for classifying the audios as COVID-19 positive and negative. 75% of the input dataset was used for training by CovParaNet and 25% was used for testing. The model is trained for 70 epochs and is optimized using Adam optimizer. The trained CovParaNet model is exported as a Keras h5 model which can be used later for prediction. Finally, the trained model is used for testing the unseen data to evaluate the performance.
4.2. Visual unimodal experiments
The implementation of the CovTinyNet model is performed using Pytorch and the fastai library in the google colab workspace. The constructed model with the said specifications is trained with 80% of the image dataset. The model is trained for 50 epochs with a learning rate of 0.0003 and Adam is used as the optimizer function. The trained CovTinyNet model is exported as a pth model and can be used later for prediction. The remaining 20% of the dataset is used as the testing set and is used for performance evaluation. In addition, some of the state-of-the-art models such as ResNet-18, ResNet-50, MobileNet-v2, DenseNet-121, U-Net are also trained and tested for comparison with the proposed CovTinyNet model.
4.3. Dynamic multimodal experiments
The best-performing acoustic and visual models are loaded. The testing dataset with 100 positive and 100 negative samples containing the symptoms such as cough, speech, breathing sound, X-ray, and CT image of the patients are constructed. The speech dataset only has 76 positive samples. So, data augmentation was performed to increase its size to 100. All the samples in the test set are fed into the respective loaded trained models to obtain the unimodal predictions. The prediction scores given by the respective models are split into 75%-25% to be used as training and testing data respectively by the Random Forest fusion model to give the final result. The Random Forest model is initialized with parameters such as n_estimators = 10 and min_samples_split = 2. The performance of the proposed dynamic Random Forest-based fusion strategy is compared with the traditional max voting fusion technique. The incorrectly predicted scores are then appended to a misclassification scores vector which is then used for dynamic retraining by the learned Random Forest.
5. Performance metrics
Accuracy is the percentage of correctly classified normal along with abnormal samples out of total samples, and it is given by the ratio of the sum of the count of true positives and the count of true negative samples to the total number of samples.
| (2) |
where, TP is True positive, TN is True Negative, FP is False Positive, FN is False Negative.
Precision is obtained by dividing the number of correctly classified positive samples by the total number of predicted samples that are positive.
| (3) |
The recall is the ratio of the number of positive samples that are correctly classified to the total number of positive examples.
| (4) |
The f1 score is calculated using the obtained values of precision and recall and will be closer to the lesser value of precision or recall.
| (5) |
In a classification problem, a confusion matrix is used to give an overview of the predicted results. The class-wise count of correct and incorrect predictions is given. It also reports the types of errors made by the chosen classifier.
In a 2 class classification problem, a 2-by-2 matrix is used where the row represents the samples in the predicted class and the column represents the samples in the actual class.
The four values in the confusion matrix are True Positive (TP) is the number of truly positive instances that were classified as positive using the classifier model, False Positive (FP) is the number of truly negative instances that were classified as positive using the classifier model, False Negative (FN) is the number of truly positive instances that were classified as negative using the classifier model and True Negative (TN) is the number of truly negative instances that were classified as negative using the classifier model.
The Receiver Operating Characteristic curve (ROC) estimates the performance of the classification model using True Positive Rate (TPR) and False Positive Rate (FPR).
| (6) |
| (7) |
ROC plots the difference between TPR and FPR with different classification thresholds.
6. Experimental evaluation
The performance evaluation of the proposed models is based on accuracy, precision, recall, f1-score, and area under the ROC curve. The training and validation graphs for accuracy and loss for the CovParaNet models are given in Fig. 4 and Fig. 5 . The validation accuracy increases and the loss decreases exponentially with progress in the number of epochs. On reaching 70 epochs, the graph provides stable convergence by attaining the highest accuracy and minimized loss. Furthermore, the confusion matrices obtained for the respective classifiers are shown in Fig. 8 and Fig. 10. The average number of false-positive predictions given by CovParaNet is 4 and CovTinyNet is 0. The average number of false-negative predictions given by CovParaNet is 27 and CovTinyNet is 2. From this, it is obvious that the number of false predictions given by both models is very less compared to the correctly predicted ones.
Fig. 4.
Training and validation accuracy of CovParaNet.
Fig. 5.
Training and validation loss of CovParaNet.
Fig. 8.
Confusion Matrices of CovParaNet.
Fig. 10.
Confusion Matrix of CovTinyNet.
The hyperparameters of the proposed model have been optimized to attain stable convergence. The range of parameters and the optimal value identified with experiments for the three proposed models are reported in Table 2. The ReLU activation function gives a non-linear way of feature extraction. The usage of max-pooling layers in our architecture significantly reduces the overfitting problem.
When the learning rate was too low, there was an escalation in the training time as the process stuck at local minima failing to converge over an optimal solution. On the contrary, when it was too high, it converged quickly providing non-optimal results. A range of learning rate values was experimented with and an optimal value for the respective models was obtained. It was observed after experimentation that a batch size of 32 provided a stable convergence. The number of epochs was set to an ideal value after a series of trials with the value set to 30, 50, 70, and 100. The accuracy of CovParaNet and CovTinyNet reached a maximum of 70 and 50 epochs respectively and started to overfit on further increase in the number of epochs.
The training accuracy and loss graphs of the CovTinyNet model trained with CT images and X-ray images are illustrated in Fig. 6 and Fig. 7 respectively. The model attains convergence after training for 50 epochs. The training time taken by the CovTinyNet is 5 h. The Receiver Operating Characteristic curve obtained for the CovTinyNet model is depicted in Fig. 9 . Fig. 9(a) shows that the area under the ROC curve obtained for CT was 100% and Fig. 9(b) shows AuROC for X-ray was 99%. This manifests the diagnostic capability of the proposed classifiers. The obtained values for the considered performance metrics are presented in Table 3, Table 4 . From Table 3, Table 4, it can be observed that all the proposed unimodal models perform robustly.
Fig. 6.
Accuracy Graph of CovTinyNet.
Fig. 7.
Loss Graph of CovTinyNet.
Fig. 9.
ROC Curve of CovTinyNet.
Table 3.
Comparison of CovParaNet with existing models for acoustic dataset.
| Modality | Model | Accuracy | Precision | Recall | F1Score |
|---|---|---|---|---|---|
| Cough | CovParaNet | 97.12% | 96.96% | 94.11% | 96% |
| CNN | 92.57% | 86.45% | 88.12% | 87.28% | |
| RNN | 91.84% | 90.90% | 82.05% | 86.25% | |
| Speech | CovParaNet | 96.08% | 100% | 82.87% | 91% |
| CNN | 94.63% | 100% | 82.54% | 90.43% | |
| RNN | 93.91% | 100% | 77.65% | 87.42% | |
| Breathing | CovParaNet | 97.45% | 100% | 93.53% | 97% |
| CNN | 96.33% | 95.65% | 94.86% | 95.25% | |
| RNN | 96.73% | 98.44% | 92.88% | 95.58% |
Table 4.
Comparsion of CovTinyNet with state-of-the-arts for visual dataset.
| Modality | Model | No. of Layers | Accuracy | Precision | Recall | F1Score | AUROC |
|---|---|---|---|---|---|---|---|
| Chest X-ray | CovTinyNet | 12 | 98.40% | 100% | 89% | 94% | 99.45% |
| ResNet-18 | 18 | 97.60% | 100% | 84.21% | 91.43% | 99.90% | |
| ResNet-50 | 50 | 98.33% | 100% | 96.67% | 98.31% | 99.75% | |
| MobileNetV2 | 53 | 96.80% | 100% | 86.67% | 92.86% | 99.64% | |
| DenseNet-121 | 121 | 96.80% | 100% | 78.95% | 88.24% | 98.51% | |
| UNet | 23 | 97.38% | 97.90% | 96.68% | 97.29% | 98.80% | |
| Chest CT | CovTinyNet | 12 | 99.19% | 100% | 98.32% | 99.15% | 99.98% |
| ResNet-18 | 18 | 97.38% | 98.08% | 96.96% | 97.51% | 99.84% | |
| ResNet-50 | 50 | 96.17% | 98.03% | 94.68% | 96.32% | 99.52% | |
| MobileNetV2 | 53 | 93.55% | 92.62% | 95.44% | 94.01% | 98.91% | |
| DenseNet-121 | 121 | 97.38% | 98.45% | 96.58% | 97.50% | 99.67% | |
| UNet | 23 | 96.00% | 85.00% | 89.47% | 87.18% | 95.60% |
Further, the performance of CovParaNet was compared with that of CNN and RNN which are widely used for sound classification tasks in the existing works. Table 3 provides the comparison of the performance of CovParaNet with CNN and RNN for cough, speech, and breathing sound datasets. From the table, it can be observed that the CovParaNet gives the highest performance in terms of accuracy, precision, recall, and f1-score compared to the other models for the cough dataset. Though all the models, including CovParaNet, gives 100% precision for the speech dataset, the CovParaNet gives the highest performance for other metrics such as accuracy, recall, and f1-score. For the breathing dataset, the accuracy, precision, and f1-score values of the CovParaNet are very high compared to the other models.
From Table 4, it can be seen that the CovTinyNet gives the highest accuracy and 100% precision for the X-ray dataset. Even though the recall and f1-score of the ResNet-50 are higher compared to the CovTinyNet model, the number of layers in ResNet-50 is 4 times higher compared to that of the CovTinyNet. This proves that the CovTinyNet gives the best performance with reduced complexity. Table 4 indicates that the CovTinyNet gives the best performance in terms of all the metrics such as accuracy, precision, recall, f1-score, and AuROC for the CT images dataset.
Table 5 provides a comparison of the performance of the proposed Random Forest-based dynamic multimodal fusion model with the widely used MaxVoting fusion method. Using a machine learning model to train the prediction score enables us to build the best model that predicts the test set with 100% accuracy, precision, recall, and f1-score. When a rare test case with incorrect predictions given by 3 unimodal models was encountered, the MaxVoting classifier gave incorrect predictions. But, the Random Forest Classifier handled the said case efficiently giving accurate predictions. There was no case where 4 or more unimodal models gave incorrect predictions at the same time due to the highest accuracy of individual models. When a highly improbable case where 4 or more unimodal models give inaccurate predictions, the dynamic retraining method updates the learned parameters of the RF model making it 100% accurate.
Table 5.
Comparison of Dynamic Multimodal fusion with MaxVoting fusion
| Model | Accuracy | Precision | Recall | F1Score |
|---|---|---|---|---|
| Dynamic Multimodal Fusion | 100% | 100% | 100% | 100% |
| MaxVoting Fusion | 98% | 96.15% | 100% | 98.04% |
7. Discussions
The optimized CovParaNet model makes the training process much quicker due to the reduction in network complexity. In addition, the parallel convolution layers reduce the loss of information in the initial stages resulting in better performance. The experimental evaluation also proves this as the accuracy attained by CovParaNet for all three acoustic modes is above 96%. Furthermore, the precision for the speech model and the breathing sounds model reaches 100%. The lightweight CovTinyNet model with comparatively very few convolution layers for an image classification task shows a great performance of 98.4% accuracy and 100% precision in X-ray image classification, and 99.19% accuracy and 100% precision in CT image classification.
The multimodal Random Forest Fusion model gives the final fusion result for the test set with 100% accuracy, precision, recall, and f1-score making it highly accurate. This also proves that late fusion strategy is more efficacious than early or hybrid fusion as they suffer from time synchronization and data scarcity problems, especially in the case of a limited dataset. And also, one of the main advantages of using the Random Forest is that it handles the problem of missing data appreciably. This acts as a failover mechanism allowing the diagnosis system to provide results even with incomplete inputs. The online retraining method used here updates the RF multimodal fusion model when it misclassifies an input due to a noisy environment. The respective prediction scores are sent for retraining instead of the entire dataset. This method makes the system dynamic, at the same time reduces overhead that can be caused if the entire input data is sent for retraining in an online environment.
The limitation of our proposed work is the use of limited data size, especially for the audio classification tasks. The experimental results of our work can be more accurate when a dataset with a larger size is used. Also, our system predicts a single disease ie., the COVID-19. The possible comorbidities or misclassification with other diseases like pneumonia and other lung disorders will be considered in our future works.
8. Conclusion
In this study, a cognitive system for COVID-19 prediction using multimodal data was proposed. Existing works majorly use only a single modality of input for the identification of coronavirus. The proposed system considers multiple modalities such as cough sounds, speech sounds, breathing sounds, X-ray images, and CT images for the diagnosis of the disease. The system is driven by two of our proposed models namely CovParaNet and CovTinyNet for unimodal classification. The CovParaNet and the CovTinyNet models attain 100% precision even with a smaller dataset. Further, the AuROC value obtained for the CovTinyNet exceeds 99% for both X-ray and CT datasets. Though 5 deep learning models (for 5 different modalities) were used, the size of the models was optimized with only 7 and 12 layer deep architectures making it easier to be deployed in a real-time diagnosis system to yield instant accurate predictions. In addition to the individual robustness, the minute false predictions are further avoided by the machine learning-based multimodal fusion method. The prediction scores given by the unimodal models are processed by a Random Forest-based late fusion strategy to compute the final result. Furthermore, to overcome the noisy environment, the online retraining method was introduced. This makes the system dynamic with minimized overhead. The system can also function in the absence of any one of the input modalities due to the minimized dependency between the models. In comparison with the existing systems that widely use a max voting classifier or a softmax function, the Random Forest fusion model gives highly convincing results with 100% accuracy, 100% precision, and 100% recall. The developed framework can be effortlessly coupled with IoT for providing a self-diagnosis system, bringing forth a method of non-contact diagnosis that can be accessed with ease from anywhere.
CRediT authorship contribution statement
Jayachitra V P: Conceptualization, Methodology, Validation, Formal analysis, Investigation, Project administration, Writing - original draft, Supervision, Software. Nivetha S: Methodology, Validation, Visualization, Formal analysis, Investigation, Software, Writing - original draft. Nivetha R: Validation, Visualization, Investigation, Software, Writing - original draft. Harini R: Software, Data curation, Investigation, Visualization.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- 1.Ali Imran, Iryna Posokhova, Haneya N. Qureshi, Usama Masood, Muhammad Sajid Riaz, Kamran Ali, Charles N. John, MD Iftikhar Hussain, Muhammad Nabeel, AI4COVID-19: AI enabled preliminary diagnosis for COVID-19 from cough samples Via an app, Informatics in Medicine Unlocked, Elsevier, Volume 20, 100378, ISSN 2352–9148, 2020. doi: 10.1016/j.imu.2020.100378. [DOI] [PMC free article] [PubMed]
- 2.Windmon Anthony, Minakshi Mona, Bharti Pratool, Chellappan Sriram, Johansson Marcia, Jenkins Bradlee A., Athilingam Ponrathi R. TussisWatch: A Smart-phone System to Identify Cough Episodes as Early Symptoms of Chronic Obstructive Pulmonary Disease and Congestive Heart Failure. IEEE J. Biomed. Health Inform. July 2019;23(4):1566–1573. doi: 10.1109/JBHI.2018.2872038. [DOI] [PubMed] [Google Scholar]
- 3.D.Sudaroli Vijayakumar, Monica Sneha, Low Cost Covid-19 preliminary Diagnosis utilizing cough samples and keenly intellective deep learning approaches, Alexandria Engineering Journal, Elsevier, Volume 60, Pages 549–557, ISSN 1110–0168, 2020. doi: 10.1016/j.aej.2020.09.032.
- 4.F. Demir, A.M. Ismael and A. Sengur, Classification of Lung Sounds With CNN Model Using Parallel Pooling Structure, in IEEE Access, vol. 8, pp. 105376-105383, 2020, https://doi.10.1109/ACCESS.2020.3000111.
- 5.Gowri Sree Rudraraju, Shubha Deepti Palreddy, Baswaraj Mamidgi, Narayana Rao Sripada, Y. Padma Sai, Naveen Kumar Vodnala, Sai Praveen Haranath, Cough sound analysis and objective correlation with spirometry and clinical diagnosis, Informatics in Medicine Unlocked, Elsevier, Volume 19, 100319, ISSN 2352–9148, 2020. doi: 10.1016/j.imu.2020.100319.
- 6.H. Fouad, Azza S. Hassanein, Ahmed M. Soliman, Haytham Al-Feel, Analyzing patient health information based on IoT sensor with AI for improving patient assistance in the future direction, Measurement, Elsevier, Volume 159, 107757, ISSN 0263–2241, 2020. doi: 10.1016/j.measurement.2020.107757.
- 7.Monge-Alvarez J., Hoyos-Barcelo C., San-Jose-Revuelta L.M., Casaseca-de-la-Higuera P. A Machine Hearing System for Robust Cough Detection Based on a High-Level Representation of Band-Specific Audio Features. IEEE Trans. Biomed. Eng. Aug. 2019;66(8):2319–2330. doi: 10.1109/TBME.2018.2888998. [DOI] [PubMed] [Google Scholar]
- 8.Monge-Alvarez J., Hoyos-Barcelo C., Lesso P., Casaseca-de-la-Higuera P. Robust Detection of Audio-Cough Events Using Local Hu Moments. IEEE J. Biomed. Health Inform. Jan. 2019;23(1):184–196. doi: 10.1109/JBHI.2018.2800741. [DOI] [PubMed] [Google Scholar]
- 9.Md. Martuza Ahamad, Sakifa Aktar, Md. Rashed-Al-Mahfuz, Shahadat Uddin, Pietro Lió, Haoming Xu, Matthew A. Summers, Julian M.W. Quinn, Mohammad Ali Mon, A machine learning model to identify early stage symptoms of SARS-Cov-2 infected patients, Expert Systems with Applications, Elsevier, Volume 160, 113661, ISSN 0957–4174, 2020. doi: 10.1016/j.eswa.2020.113661. [DOI] [PMC free article] [PubMed]
- 10.Mwaffaq Otoom, Nesreen Otoum, Mohammad A. Alzubaidi, Yousef Etoom, Rudaina Banihani, An IoT-based framework for early identification and monitoring of COVID-19 cases, Biomedical Signal Processing and Control, Elsevier, Volume 62, 2020, 102149, ISSN 1746-8094. https://doi.org/10.1016/j.bspc.2020.102149. [DOI] [PMC free article] [PubMed]
- 11.Sharan R.V., Abeyratne U.R., Swarnkar V.R., Porter P. Automatic Croup Diagnosis Using Cough Sound Recognition. IEEE Trans. Biomed. Eng. Feb. 2019;66(2):485–495. doi: 10.1109/TBME.2018.2849502. [DOI] [PubMed] [Google Scholar]
- 12.Maral Asiaee, Amir Vahedian-azimi, Seyed Shahab Atashi, Abdalsamad Keramatfar, Mandana Nourbakhsh, Voice Quality Evaluation in Patients With COVID-19: An Acoustic Analysis, Journal of Voice, Elsevier, ISSN 0892–1997, 2020. doi: 10.1016/j.jvoice.2020.09.024. [DOI] [PMC free article] [PubMed]
- 13.Pinkas G., Karny Y., Malachi A., Barkai G., Bachar G., Aharonson V. SARS-CoV-2 Detection From Voice. IEEE Open J. Eng. Med. Biol. 2020;1:268–274. doi: 10.1109/OJEMB.2020.3026468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Z. Jiang et al., Detection of Respiratory Infections Using RGB-Infrared Sensors on Portable Device, in IEEE Sensors Journal, vol. 20, no. 22, pp. 13674–13681, 15 Nov. 15, 2020, doi: 10.1109/JSEN.2020.3004568. [DOI] [PMC free article] [PubMed]
- 15.Vedaei S.S., et al. COVID-SAFE: An IoT-Based System for Automated Health Monitoring and Surveillance in Post-Pandemic Life. IEEE Access. 2020;8:188538–188551. doi: 10.1109/ACCESS.2020.3030194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.W.N. Ismail, M.M. Hassan, H.A. Alsalamah and G. Fortino, CNN-Based Health. Laguarta, F. Hueto and B. Subirana, COVID-19 Artificial Intelligence Diagnosis Using Only Cough Recordings, IEEE Open Journal of Engineering in Medicine and Biology, vol. 1, pp. 275–281, 2020. Model for Regular Health Factors Analysis in Internet-of-Medical Things Environment, in IEEE Access, vol. 8, pp. 52541–52549, 2020, doi: 10.1109/ACCESS.2020.2980938. [DOI] [PMC free article] [PubMed]
- 17.Laguarta J., Hueto F., Subirana B. COVID-19 Artificial Intelligence Diagnosis Using Only Cough Recordings. IEEE Open J. Eng. Med. Biol. 2020;1:275–281. doi: 10.1109/OJEMB.2020.3026928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Quatieri T.F., Talkar T., Palmer J.S. A Framework for Biomarkers of COVID-19 Based on Coordination of Speech-Production Subsystems. IEEE Open J. Eng. Med. Biol. 2020;1:203–206. doi: 10.1109/OJEMB.2020.2998051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.El-Kenawy E.-S.M., Ibrahim A., Mirjalili S., Eid M.M., Hussein S.E. Novel Feature Selection and Voting Classifier Algorithms for COVID-19 Classification in CT Images. IEEE Access. 2020;8:179317–179335. doi: 10.1109/ACCESS.2020.3028012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Acharya J., Basu A. Deep Neural Network for Respiratory Sound Classification in Wearable Devices Enabled by Patient Specific Model Tuning. IEEE Trans. Biomed. Circuits Syst. June 2020;14(3):535–544. doi: 10.1109/TBCAS.2020.2981172. [DOI] [PubMed] [Google Scholar]
- 21.Han Z., et al. Accurate Screening of COVID-19 Using Attention-Based Deep 3D Multiple Instance Learning. IEEE Trans. Med. Imaging. Aug. 2020;39(8):2584–2594. doi: 10.1109/TMI.2020.2996256. [DOI] [PubMed] [Google Scholar]
- 22.Oh Y., Park S., Ye J.C. Deep Learning COVID-19 Features on CXR Using Limited Training Data Sets. IEEE Trans. Med. Imaging. Aug. 2020;39(8):2688–2700. doi: 10.1109/TMI.2020.2993291. [DOI] [PubMed] [Google Scholar]
- 23.Wang N., Liu H., Xu C. 2020 IEEE 10th International Conference on Electronics Information and Emergency Communication (ICEIEC) 2020. Deep Learning for The Detection of COVID-19 Using Transfer Learning and Model Integration; pp. 281–284. [DOI] [Google Scholar]
- 24.Yoon Y., et al. Analyzing Basketball Movements and Pass Relationships Using Realtime Object Tracking Techniques Based on Deep Learning. IEEE Access. 2019;7:56564–56576. doi: 10.1109/ACCESS.2019.2913953. [DOI] [Google Scholar]
- 25.Liu Z., Wang S. Broken Corn Detection Based on an Adjusted YOLO With Focal Loss. IEEE Access. 2019;7:68281–68289. doi: 10.1109/ACCESS.2019.2916842. [DOI] [Google Scholar]
- 26.Ali Pournemat, Peyman Adibi, Jocelyn Chanussot, Semisupervised charting for spectral multimodal manifold learning and alignment, Pattern Recognition, Elsevier, Volume 111, 107645, ISSN 0031–3203, 2021. doi: 10.1016/j.patcog.2020.107645.
- 27.Shaker El-Sappagh, Tamer Abuhmed, S.M. Riazul Islam, Kyung Sup Kwak, Multimodal multitask deep learning model for Alzheimer’s disease progression detection based on time series data, Neurocomputing, Elsevier, Volume 412, Pages 197–215, ISSN 0925–2312, 2020. doi: 10.1016/j.neucom.2020.05.087.
- 28.K.H. Abdulkareem et al., Realizing an Effective COVID-19 Diagnosis System Based on Machine Learning and IOT in Smart Hospital Environment, in IEEE Internet of Things Journal, doi: 10.1109/JIOT.2021.3050775. [DOI] [PMC free article] [PubMed]
- 29.Al-Waisy A.S., Al-Fahdawi S., Mohammed M.A., et al. COVID-CheXNet: hybrid deep learning framework for identifying COVID-19 virus in chest X-rays images. Soft Comput. 2020 doi: 10.1007/s00500-020-05424-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mohammed M.A., et al. Benchmarking Methodology for Selection of Optimal COVID-19 Diagnostic Model Based on Entropy and TOPSIS Methods. IEEE Access. 2020;8:99115–99131. doi: 10.1109/ACCESS.2020.2995597. [DOI] [Google Scholar]
- 31.Mohammed M.A., Abdulkareem K.H., Garcia-Zapirain B., Mostafa S.A., Maashi M.S., et al. A comprehensive investigation of machine learning feature extraction and classification methods for automated diagnosis of covid-19 based on x-ray images. Computers, Mater. Continua. 2021;66(3):3289–3310. doi: 10.32604/cmc.2021.012874. [DOI] [Google Scholar]
- 32.Al-Waisy A.S., Mohammed M.A., Al-Fahdawi S., Maashi M.S., Garcia-Zapirain B., et al. Covid-deepnet: hybrid multimodal deep learning system for improving covid-19 pneumonia detection in chest x-ray images. Computers, Mater. Continua. 2021;67(2):2409–2429. doi: 10.32604/CMC.2021.012955. [DOI] [Google Scholar]
- 33.Albahli S., Algsham A., Aeraj S., Alsaeed M., Alrashed M., et al. Covid-19 public sentiment insights: a text mining approach to the gulf countries. Computers, Mater. Continua. 2021;67(2):1613–1627. doi: 10.32604/CMC.2021.014265. [DOI] [Google Scholar]
- 34.Shi F., et al. Review of Artificial Intelligence Techniques in Imaging Data Acquisition, Segmentation, and Diagnosis for COVID-19. IEEE Rev. Biomed. Eng. 2021;14:4–15. doi: 10.1109/RBME.2020.2987975. [DOI] [PubMed] [Google Scholar]
- 35.Voulodimos Athanasios, Protopapadakis Eftychios, Katsamenis Iason, Doulamis Anastasios, Doulamis Nikolaos. A Few-Shot U-Net Deep Learning Model for COVID-19 Infected Area Segmentation in CT Images. Sensors. 2021;21(6):2215. doi: 10.3390/s21062215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Virufy COVID-19 Open Cough Dataset, https://github.com/virufy/ virufy-data.
- 37.Coswara-Data, https://github.com/iiscleap/Coswara-Data.
- 38.Cohen JP, Morrison P, Dao L (2020) COVID-19 image data collection. https://github.com/ieee8023/covid-chestxray-dataset, https://arxiv.org/abs/ 2003.11597.
- 39.Soares, Eduardo, Angelov, Plamen, Biaso, Sarah, Higa Froes, Michele, and Kanda Abe, Daniel. SARS-CoV-2 CT-scan dataset: A large dataset of real patients CT scans for SARS-CoV-2 identification. medRxiv (2020), doi: 10.1101/2020.04.24.20078584.
- 40.X. Zhang et al., Emotion Recognition From Multimodal Physiological Signals Using a Regularized Deep Fusion of Kernel Machine, in IEEE Transactions on Cybernetics, doi: 10.1109/TCYB.2020.2987575. [DOI] [PubMed]
- 41.Pandeya Y.R., Lee J. Deep learning-based late fusion of multimodal information for emotion classification of music video. Multimed Tools Appl. 2021;80:2887–2905. doi: 10.1007/s11042-020-08836-3. [DOI] [Google Scholar]