Abstract
Multipotent Isl1+ heart progenitors give rise to three major cardiovascular cell types; cardiac, smooth muscle, and endothelial cells, and play a pivotal role in lineage diversification during cardiogenesis. A critical question is pinpointing when this cardiac-vascular lineage decision is made, and how this plasticity serves to coordinate cardiac chamber and vessel growth. The posterior domain of the Isl1-positive second heart field contributes to the SLN-positive atrial myocardium and myocardial sleeves in the cardiac inflow tract, where myocardial and vascular smooth muscle layers form anatomical and functional continuity. Herein, using a new atrial specific SLN-Cre knockin mouse line, we report that an Isl1+/SLN+ transient cell population contributes to cardiac as well as smooth muscle cells at the heart-vessel junction in cardiac inflow tract. The Isl1+/SLN+ cells are capable of giving rise to cardiac and smooth muscle cells until late gestational stages. These data suggest that the cardiac and smooth muscle cells in the cardiac inflow tract share a common developmental origin.
Keywords: cardiogenesis, myogenic progenitor, smooth muscle, great vessel, plasticity
1. Introduction
The cardiac, smooth muscle and endothelial cells arise from single, common multipotent precursor cells during embryogenesis [1-3]. Whereas three major cardiovascular lineages can arise from Flk1+/Bry+ mesodermal progenitors [1,4] and Flk1+/Isl1+/Nkx2.5+ cardiogenic colonies derived from mouse embryos around E8.0-8.5 [2], smooth muscle cell lineages can also arise from c-kit+/Nkx2.5+ cardiac progenitor cells isolated from a later stage [3]. These progenitor populations may represent discrete lineages that constitute the heart [5]. Alternatively, they may represent cells in the same lineage at different developmental stages. The comparison of these analyses suggests that the endothelial lineage separates from the myogenic lineage at an early stage, whereas cardiac and smooth muscle lineages are closely related until later stages of cardiogenesis [6,7]. Given the plasticity of progenitors becomes progressively restricted as embryogenesis proceeds, intriguing questions are which population displays cardiac-smooth muscle bipotency, how long the progenitors maintain their plasticity during cardiogenesis and what is the biological implication of smooth muscle differentiation capability of the cardiac progenitors.
The second heart field (SHF) is delineated by the expression of Isl1, a member of the LIM-homeodomain transcription factor family. This extra-crescent cardiac population is specified in the anterior lateral plate mesoderm adjacent to the first heart field (FHF) in the cardiac crescent [7-9]. After the primitive heart field forms a single straight heart tube, the SHF progenitors migrate from the splanchnic mesoderm towards the primitive heart tube and trigger its right-ward looping. These SHF progenitors continue to migrate and add additional myocardium to the primitive heart tube until midgestational stages. The SHF progenitors downregulate Isl1 expression immediately after completing their migration, which makes Isl1 a suitable marker of an immature state for cardiac progenitors. During this step, the Isl1+ cardiac progenitors in the anterior region of the SHF (aSHF, also known as anterior heart field; AHF) migrate to the arterial pole of the heart tube and form the outflow tract and right ventricle [10-12]. This anterior subpopulation is distinguishable from other subpopulations of Isl1-positive SHF progenitors by several molecular markers including specific enhancers of Mef2c [13] and FGF10 [10], suggesting that Isl1-positive cells are already heterogeneous at this stage. The Isl1+ progenitors in the aSHF eventually acquire a ventricular phenotype and contribute to the myocardium of the outflow tract and the right ventricle. Interestingly, this population also developmentally gives rise to smooth muscle cells in the root of the aorta [14]. On the other hand, Isl1-positive progenitors in posterior part of the SHF (pSHF) populate the venous pole of the primary heart tube slightly later, maintain high proliferative and migratory activity, and express Isl1 until midgestational stages [15,16]. This posterior subpopulation of late Isl1-positive progenitors eventually acquires an atrial phenotype and contributes to the myocardium in atrial chambers and myocardial sleeves in the inflow tract. Thus, at the linear heart tube stage, the Isl1+ cardiac progenitor population seems to be a pool of several different subsets of cardiac progenitors including aSHF and pSHF populations. Whereas Isl1-positive progenitors in the outflow tract/arterial pole/aSHF give rise to both cardiac and smooth muscle cells, it is not clear if myocardium and smooth muscle in the inflow tract share a common cellular origin in the venous pole/pSHF. This is in part because of the lack of the a specific lineage marker and in part because of the lack of single cell analysis.
Sarcolipin (SLN) is an inhibitor of sarco(endo)plasmic reticulum Ca2+-ATPase (SERCA) that is specifically expressed in atrial myocardium and skeletal muscle, and implicated in atrial specific contractile function [17-19]. SLN expression is gradually upregulated as the atrial myocytes maturate, and is a useful marker for the atrial muscle lineage.
Here, we report the generation of atrial specific SLN-Cre knockin mouse line, and the identification of Isl1+/SLN+ cells in the atrial lineage which contribute to atrial cardiomyocytes and smooth muscle cells in the cardiac inflow tract. To investigate the differentiation capability of Isl1+/SLN+ cells, we generated an atrial specific deleter mouse line by introducing Cre recombinase into the SLN locus. SLN-labeled atrial cells distribute in working and SA nodal atrial myocytes, as well as the smooth muscle cells in the cardiac inflow tract. In vitro assays indicate that Isl1+/SLN+ cells are capable of clonally differentiating into cardiomyocytes and smooth muscle cells from single progenitors. Interestingly, Isl1+/SLN+ cells retain smooth muscle competency until late gestational stages. These observations provide an insight into the origin of cardiac and smooth muscle cells at the boundary of the atrial chamber and inflow tract, and the mechanism underlying the formation of heart-vessel junctions.
2. Materials and methods
2.1. Generation of SLN-Cre mice
Exon 2 of the SLN locus including the 1st ATG was replaced with Cre cDNA. A correctly targeted R1 ES clone was screened by Southern blotting and genomic PCR. The recombination efficiency was 1/300 clones.
2.2. Preparation of cardiac mesenchymal feeder layer
Neonatal hearts were predigested with 0.5 mg/ml trypsin in HBSS at 4C overnight followed by strong digestion with collagenase at 37C for 1 hour (0.5 mg/ml in HBSS). Cardiac mesenchymal fibroblasts were separated from myocytes by differential plating for 1 hour twice. Fibroblasts from the first and the second differential plating were combined, grown until confluent and treated with 10μg/ml mitomycin C for 2 hours on the day before progenitors were seeded. The contamination of myocytes in the fibroblast fraction was less than 0.07% by cTnT staining.
2.3. Histology and immunostaining
Whole mount and section Xgal stainings were performed according to standard protocols. Double staining for Xgal and specific antibodies were performed as follows: 8μm frozen sections or cells were stained with Xgal followed by postfixation for 5min, 0.3% hydrogen peroxide treatment for 15min, blocking with 10% normal goat serum for 1 hour and antibody reaction in 3% normal goat serum at 4C overnight. Secondary antibody reaction was performed with Vectastain ABC kit (Vector lab) according to the manufacturer’s protocol. Section Xgal/Isl1 staining was performed as previously described [20]. The concentrations of the primary antibodies are; cTnT (1:200, Lab Vision Corp., Fremont, CA), smMHC (1:500, Biomedical Technologies Inc., Stoughton, MA), Isl1 (1:200, DSHB, Iowa City, IA), DsRed (1:500, Clontech, Mountain View, CA).
2.4. RT-PCR and qPCR
RNA was extracted with Trizol (Invitrogen, Carlsbad, CA) or Absolute nanoprep kit (Stratagene, Ceder Creek, TX) according to the manufacturer’s protocol, and cDNAs were synthesized with iScript kit (BioRad, Hercules, CA). Colony PCR was run for 35 cycles. Quantitative PCR was performed with the SYBR Green system and i-Cycler (BioRad, Hercules, CA).
2.5. Electron microscopic analysis
The SLNcre/+; R26R hearts were dissected and fixed in 1%PFA and 2.5% glutaraldehyde in PBS for 3 hours, and stained for 4 hours in Bluo-gal staining solution; 1 mg/ml Bluo-gal (Sigma), 10mM ferro/ferri cyanide, 2mM MgCl2, 0.02% NP40 and 0.01% NaDOC. Stained tissues were post-fixed for 30 min in a mixture of 1% osmium tetroxide and 2% glutaraldehyde in 0.15 M cacodylate buffer (on ice), washed several times in PBS, and dehydrated in graded ethanol and acetone (all steps on ice). Preparations were left overnight in a 1:1 mixture of Epon and acetone and then for 5–10 h in unpolymerized Epon. They were transferred to molds, oriented and placed at 60°C for 24 h to permit polymerization of the Epon. Sections were mounted on net grids (Ted Pella) and treated with uranyl acetate and lead citrate.
3. Results
3.1. SLN-Cre knockin strain is a sensitive and specific deleter line for the atrial lineage
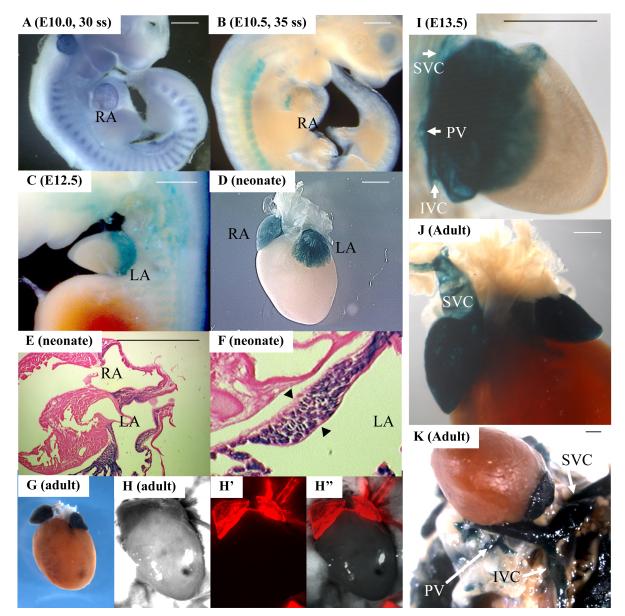
To generate an atrial specific Cre line driven by an internal promoter, we introduced Cre recombinase by homologous recombination into exon 2 of the SLN locus (Fig. 1A, B, C). SLNcre/+ heterozygotes displayed no morphological or fertility defects. While SLN mRNA is expressed at E10.0 (Fig. 2A), the βgal activity in SLNcre/+; R26R embryos was first detected in the atria at around E10.5, when Isl1 is still positive in the atrial lineage (Fig. 2B). After E12.5, the atrial myocardium was broadly and strongly labeled by R26R and CAG-DsRed reporter lines [21,22] (Fig. 2C-H”, and Fig. S1A, B). Section Xgal staining of the neonatal hearts revealed that the vast majority of the atrial myocytes were labeled (Fig. 2E, F), whereas none of the endocardial or epicardial cells were stained (Fig. 2F, arrowheads; Supplemental Table 1). Interestingly, HCN4-positive SA nodal cells were also labeled by SLN-Cre (Fig. S2). These data suggest that the SLN-Cre line is a sensitive and specific deleter line for the atrial lineage. To our knowledge, this mouse is the first deleter line in which Cre recombinase is driven by an internal atrial specific promoter.
Fig. 1. Generation of SLN-cre knock-in mouse line.
A. Schematic of SLN genomic locus, targeting vector design and recombinant alleles. Exon 2 which includes the start codon was replaced by Cre recombinase and neomycin resistant cassette by homologous recombination. FRT sites are indicated by filled triangles. DTA, diphtheria toxin A cassette.
B. Genomic Southern blot analysis of targeted ES cells and heterozygous mouse after removal of neo cassette using 5′ and 3′ probe shown in A.
C. Genomic PCR for mouse genotyping. Primer designs are shown in A.
Fig. 2. In vivo atrial lineage tracing.
A. In situ hybridization for SLN at 30 somite stage. SLN is expressed in atria and myotomes.
B-H”. Contribution of SLN+ cells to atrial myocardium. Atrial specific labeling is shown by whole mount Xgal staining at E10.5 (B), E12.5 (C), neonatal (D) and adult (G) stages, and section Xgal staining of neonatal hearts (E, F) of SLN-cre; R26R mice. Note that the vast majority of the atrial myocytes are labeled. As indicated by black arrowheads, there were no Xgal-positive cells in endocardium or epicardium. H, H’ and H” show the hearts from adult SLN-cre; CAG-DsRed reporter mice. The expression of βgal and DsRed reporters is restricted to atrial myocytes throughout cardiogenesis and in the adult heart. Scale bar = 1mm.
I-K. Contribution of SLN+ cells to the cardiac inflow tract. Whole mount Xgal staining of the inflow tract of SLNcre/+; R26R embryo at E13.5 (I) and adult heart (J, K) shows that the proximal part of the SVC, IVC and PV are derived from SLN-positive cells (white arrows in I and K). Scale bar = 1mm.
IVC, inferior vena cava; LA, left atrium; PV, pulmonary vein; RA, right atrium; SVC, superior vena cava.
Xgal staining in the inflow region visualized the anatomical distribution of the myocardial sleeves of the venae cavae and pulmonary veins. The Xgal staining extended up to bifurcation of the internal jugular and subclavian veins in the cranial region, and down to the diaphragm in the thoracic cavity (Fig. 2I-K and S1C). The boundary of the right atrium and the venae cavae is demarcated by venous valves that also are derived from SLN-expressing cells (Fig. 3A, arrowhead). Whereas the muscular layer of the atrial chamber consists only of myocardial cells, the muscular wall of the venae cavae distal to the venous valves consist of two muscular layers – the outer myocardial layer derived from SLN-expressing cells and inner smooth muscle layer positive for smMHC, a definitive marker for vascular smooth muscle cells (Fig. 3A). The myocardial layer tapers off towards the periphery and generates myocardial sleeves in the great veins. Similar to the vena cavae, the proximal region of the pulmonary veins also showed a two layer-structure with outer myocardial sleeve and inner smooth muscle layer (Fig. S3).
Fig. 3. SLN-positive cells give rise to smooth muscle cells in the cardiac inflow tract.
A. Low magnification view of the junction between right atrium and vena cava of adult SLNcre/+; R26R mouse costained with Xgal and smMHC. The proximal region of the vena cava (VC) consists of two muscular layers; Xgal-positive outer myocardial layer (blue) and smMHC-positive inner smooth muscle layer (brown), and demarcated from the right atrium (RA) by venous valves (VV) which are also derived from SLN-positive cells.
B. Expression of a smooth muscle marker in βgal-labeled cells isolated from the cardiac inflow region of adult SLNcre/+; R26R mice. Tissues from the vena cava of adult SLNcre/+; R26R mouse were dissociated and plated onto a culture dish. Some cells express both βgal (blue) and smMHC (brown), suggesting that a fraction of SLN-positive cells gave rise to smooth muscle cells in the inflow tract.
C. Smooth muscle cells derived from SLN-expressing cells. Light microscopic analysis of 8μm sections shows the smMHC-positive cells in the inner layer costained with Xgal (scale bar = 50μm). Sections were cut along the line indicated in A.
D. 1μm sections show clear Xgal staining on smooth muscle cells (arrows) between a thin endothelial layer and thick myocardial layer (CM) (scale bar = 20μm). Sections were cut along the line indicated in A.
E, E’. Electron microscopic analysis (x49500) of the vena cava of adult SLNcre/+; R26R mouse stained with bluo-gal. Bluo-gal deposits (arrowheads) are found in smooth muscle cells (SMC) with non-striated myofilaments (black arrows). Note that the cardiomyocytes have striated myofilaments (white arrow).
CM, cardiomyocyte; col, collagen fiber; EC, endothelial cell; SMC, smooth muscle cell; MF, myofilament; RA, right atrium; VC, vena cava; VV, venous valve
3.2. SLN-positive cells give rise to smooth muscle cells in cardiac inflow tract
Further analysis of this two-layer structure revealed the close relationship between cardiac and smooth muscle lineages during cardiovascular development. Double staining of serial sections showed that about 5-10% of smMHC-positive cells in the inflow region are costained with Xgal (Fig. 3C, D, black arrows; Supplemental Table 1). To confirm this, the inflow region of the atrium from SLNcre/+; R26R adult mice was enzymatically dissociated and cultured on a fibronectin-coated dish. Consistent with section stainings, a fraction of βgal-labeled cells were costained for smMHC (Fig. 3B). Furthermore, electron microscopic analysis of Bluo-gal-stained SLNcre/+; R26R hearts revealed that the smooth muscle cells with non-striated myofilaments are labeled with Bluo-gal deposits on their membranes (Fig. 3E, E’, arrowheads). βgal-labeled smooth muscle cells were also found in myocardial sleeves in the pulmonary veins (Fig. S3). These data suggest the developmental contribution of SLN-positive cells to the smooth muscle cells in the cardiac inflow tract. It has been reported that SLN mRNA is expressed strongly in atria and esophageal muscle and least abundantly in skeletal muscle and bladder, but no expression has been detected in vascular smooth muscle [18]. Consistently, our SLN-cre lineage tracing experiments fail to detect any Xgal-positive smooth muscle cells in aortic or other major vasculature. Therefore, our findings raise the possibility that these two different cell types in the cardiac inflow region share a common cellular origin during cardiogenesis.
3.3. Isl1+/SLN+ cells represent a transient cell population in the venous pole
To examine the cellular origin of cardiac and smooth muscle cells in the inflow tract, we searched for Isl1-positive cells in the atrial lineage. As in situ hybridization and lineage tracing experiments indicated that SLN is expressed from E10.0 and on in the atria (Fig. 2A), and Isl1 expression continues until midgestation [15], we speculated that there is a spatial and temporal overlap of these two markers in the forming atria. Double staining for Xgal and Isl1 on SLNcre/+ × R26R embryos revealed Isl1/SLN double positive cells (Fig. 4). At E10.5, immature cardiac progenitors with strong Isl1 expression were found in the splanchnic mesoderm (Fig. 4A, arrowheads). Cells in the primary atrial septum are also positive for Isl1 (Fig. 4A, white arrow). A subset of cardiac cells in the atrial chamber and sinus venosus have already started to express SLN, and some are positive for both Isl1 and SLN (Fig. 4A, black arrows). At E13.5, Isl1+ cells in the splanchnic mesoderm are still negative for SLN (Fig. 4B, black arrowheads), although most of the atrial myocytes were already positive for βgal activity. Isl1+/SLN+cells were still found in dorsal mesenchymal protrusion (Fig. 4B, black arrows) [15,23-28]. These cells gradually lose Isl1 expression as they migrate towards the cushion and progressively acquire SLN (Fig. 4B, black arrows). These data suggest that a subset of Isl1+ cells give rise to SLN+ atrial myocytes, and that Isl1+/SLN+ cells represent a transient cell population that are committed to the mature atrial myocyte fate.
Fig. 4. Identification of Isl1+/SLN+ cells during cardiognesis.
Heart sections from SLNcre/+; R26R embryo at E10.5 (A) and E13.5 (B) were double-stained for Xgal and Isl1. Scale bar = 100μm.
A. At E10.5, most of the Isl1-positive cells in forming atria are negative for Xgal. Whereas cells in the splanchnic mesoderm (black arrowheads) and primary atrial septum (white arrow) strongly express Isl1, Xgal-positive cells with a weaker level of Isl1 are also seen in atrial free wall and sinus venosus (arrows).
B. At E13.5, most of the atrial myocytes have already started to express SLN. Isl1-positive immature cardiac progenitors in the splanchnic mesoderm (arrowhead) migrate into the septum and start to express SLN while maintaining Isl1 expression (black arrow). During the migration, atrial cells gradually lose Isl1 expression and gradually express SLN.
CA, common atrium; LA, left atrium; RA, right atrium; SV, sinus venosus
3.4. Single Isl1+/SLN+ cells give rise to both cardiac and smooth muscle cells
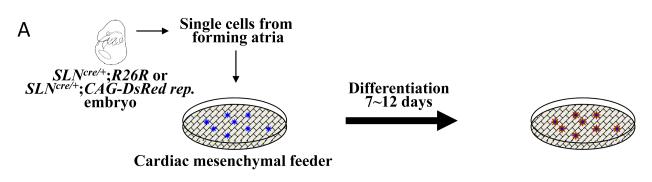
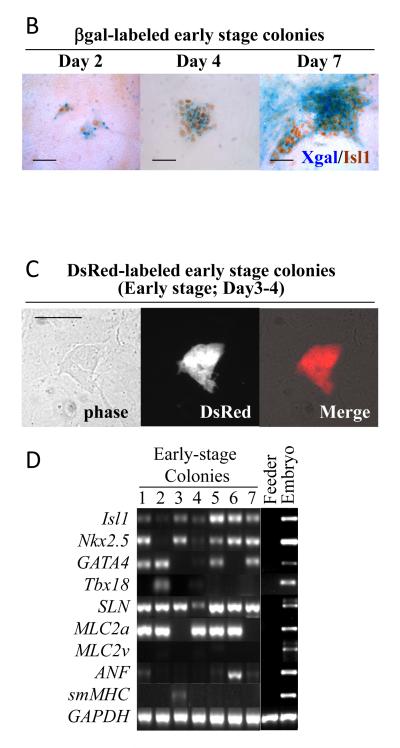
To examine whether cardiac and smooth muscle lineages can originate from a single common Isl1+/SLN+ cell, we dissected forming atrial tissue from E9.5 SLNcre/+; R26R embryos, dissociated them into single cells and cultured them at clonal density on cardiac mesenchymal feeder layers as previously described (Fig. 5A) [2,29,30]. βgal-labeled atrial cells grew to form colonies, and more than 90% of them were positive for Isl1 (Fig. 5B, D and Supplementary Table 2). Using a CAG-DsRed reporter line [22], the atrial progenitor colonies were visualized alive (Fig. 5C), picked up under the fluorescent microscope, and examined for their mRNA expression signature. After 3-4 days on feeders, the majority of the clones (early-stage colonies) were positive for early cardiac markers (Isl1, Nkx2.5, GATA4) and atrial markers (SLN, MLC2a) (Fig. 5D). However, the expression of ANF and smMHC were only occasionally found at this stage. Therefore, the feeder system is useful for propagating immature atrial cells in vitro.
Fig. 5. Isl1+/SLN+ cells clonally give rise to cardiac and smooth muscle cells.
A. The forming atria of E9.5 SLNcre/+× R26R or SLNcre/+× CAG-DsRed reporter embryos are dissected and dissociated with collagenase. βgal- or DsRed-labeled atrial progenitors are cultured on cardiac mesenchymal feeders at clonal density. The dissocated atrial tissues are grown for 7-12 days and then examined for cardiac and smooth muscle marker expression.
B. Colonies are stained for Isl1 at early stages (2, 4 and 7 days after isolation). βgal-labeled atrial cells maintain Isl1 expression up to 7 days on a cardiac mesenchymal feeder layer. Scale bar = 50μm.
C. DsRed-labeled atrial progenitor colonies grown for 4 days on feeders.
D. DsRed-labeled atrial progenitors are manually picked up 4 days after isolation, and examined for marker expression by RT-PCR. Shown are 7 representative colonies. Most of the colonies expressed early cardiac markers (Isl1, Nkx2.5 and GATA4) and atrial markers (SLN and MLC2a). None of them are positive for MLC2v. At this stage, only few colonies expressed smMHC.
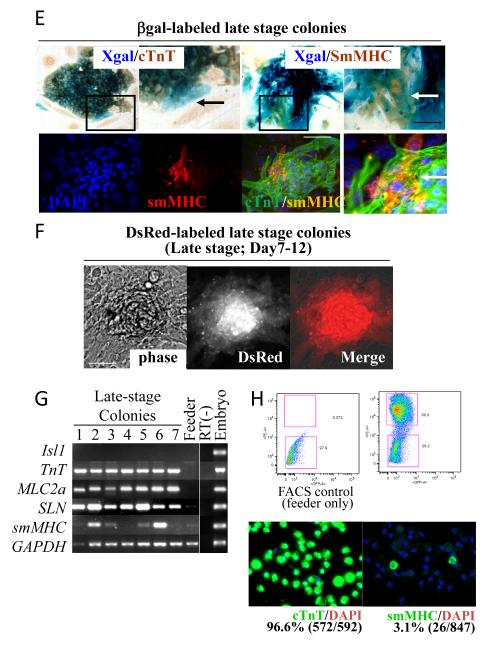
E. βgal-labeled atrial progenitors are differentiated for 7-12 days and stained for Xgal, cTnT and/or smMHC. Whereas most of the cells in Xgal-positive colonies were positive for cTnT, some in the periphery were negative (black arrow). These peripheral cells are co-stained with smMHC (white arrows). Lower panels show cTnT/smMHC immunofluorescent staining of a representative colony, indicating that these two cell types differentiate from a single cell (white arrow).
F. DsRed-labeled atrial colonies grown for 12 days on feeders.
G. Expression profile of atrial colonies. After 7-12 days (late stage) on feeders, colonies were picked up and examined for marker expression. RT-PCR revealed that Isl1 is already downregulated at this stage but SLN and MLC2a are still positive. Importantly, smMHC becomes positive in more than half of the colonies.
H. Ex vivo quantification of the percentage of smooth muscle cells derived from atrial lineage. After 7-12 days (late stage) on feeders, DsRed-labeled atrial colonies were FACS-sorted, attached on glass slides by cytospin, and stained for cTnT (left bottom panel) or smMHC (right bottom panel). Note that 3.1% of the progeny of atrial colonies were smooth muscle cells, while 96.6% were cardiomyocytes.
After differentiation for 7~12 days in culture, cTnT-negative cells were found in the periphery of the βgal-labeled colonies (Fig. 5E, black arrow). These peripheral cells were co-stained with Xgal/smMHC (Fig. 5E, white arrows). RT-PCR revealed that no colony was positive for Isl1 at this stage (Fig. 5G). Whereas all the clones examined gave rise to cTnT-positive cardiomyocytes, 51.3% of them were capable of giving rise to smMHC-positive cells (Fig. 5G and Supplementary Table 2). Given that 90.3% of the colonies express both Isl1 and SLN, these data suggest that more than half of the Isl1+/SLN+ cells at E9.5 are capable of giving rise to both cardiac and smooth muscle cells. While more than a half of Isl1+/SLN+ cells are smooth muscle competent, not all of them give rise to smooth muscle cells. To examine the destination of Isl1+/SLN+ cells, DsRed-labeled cells were propagated for 7~12 days on feeders, and FACS-sorted onto glass slides by cytospin for smooth muscle staining. As shown in Fig. 5H, 3.1% of the progeny of atrial cells gave rise to smMHC-positive smooth muscle cells ex vivo consistent with in vivo lineage tracing data (Supplemental Table 1).
Interestingly, atrial colonies derived from later stage (E12.5 and 15.5) showed a decrease in smooth muscle differentiation capability (Supplementary Table 2), suggesting that smooth muscle competency is gradually restricted as the atrial myocytes mature.
Taken together, these data suggest that Isl1-positive cells in the atrial lineage contribute to cardiac as well as smooth muscle cells in the cardiac inflow tract during migration from splanchnic mesoderm.
In conclusion, we reported (1) a novel Cre mouse line which specifically and sensitively labels the atrial cardiac lineage, (2) the developmental contribution of the atrial cells into the smooth muscle cells in cardiac inflow tract, (3) the smooth muscle competency of immature atrial cells, and (4) the gradual loss of smooth muscle competency during cardiogenesis. Together, these findings provide an insight into the mechanism underlying the formation of the boundary between atrial myocardium and vascular smooth muscle layer. The myocardial-smooth muscle junction in the vena cava is often affected in some types of congenital heart disease. Moreover, the myocardial sleeve in pulmonary vein is highly arrhythmogenic in diseased and/or aged hearts. Elucidating how the plasticity of heart progenitors serves the cardiovascular morphogenesis will provide a better understanding of the pathogenesis of these diseases.
4. Discussion
The heart is the first functional organ that develops during embryogenesis, and it is unique in that it has to be operational while it is still forming. Therefore, the embryonic heart needs to strike a delicate balance between its formation and its functionality, which accounts for the fact that 1% of human live births are complicated by congenital heart diseases. A major question in cardiogenesis research is how this balance is regulated at the cellular level.
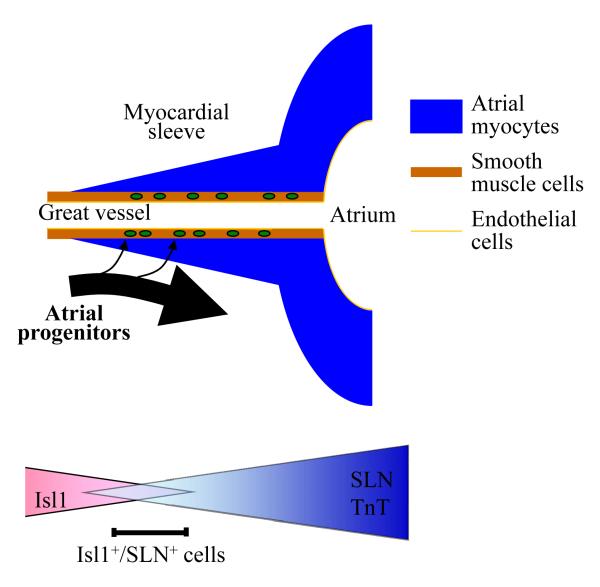
Using a newly generated atrial specific SLN-Cre knockin mouse line, we have demonstrated that the atrial subset of Isl1-positive SHF progenitors are capable of giving rise to both cardiac and smooth muscle cells until unexpectedly later stages. During the migration from splanchnic mesoderm, a majority of Isl1+/SLN+ cells contribute to the atrial myocardium, while a subset of them are incorporated into the inner layer of the inflow tract and give rise to smooth muscle cells (Fig. 6). Instead of making binary cell fate decision at certain developmental stages, our data suggest that the atrial cells gradually lose their plasticity during cardiogenesis.
Fig. 6. A model for the migration of atrial progenitors.
During the migration from the splanchnic mesoderm, Isl1-progentiors in the pSHF gradually lose Isl1 expression and start to express atrial specific markers including SLN. The Isl1+/SLN+ cells represent a transient cell population that is already committed to the atrial lineage, but still maintain proliferative and migratory activities. Isl1+/SLN+ cells mainly contribute to atrial myocardium, but some contribute to smooth muscle cells in the cardiac inflow tract. This smooth muscle competency is gradually lost during the migration and maturation.
Diversification of atrial cell populations
The SHF has been characterized by several different techniques; dye injection, detection of specific markers and retrospective single cell fate tracing experiments. Different methods and different markers have determined slightly different populations of the cardiac lineages [8,10-12,31]. Atrial cells arise from the posterior part of Isl1-positive heart field [16,27,28]. The Isl1+/SLN+progenitors represent components of the posterior region of the secondary heart field [32], and retain proliferative activity [33] and bipotency until later stages of cardiogenesis, which apparently relates to their generation of the myocardial/smooth muscle sleeves that serve as the junctional boundary to connect the great vessels and the cardiac chambers into a functional syncytium. Interestingly, there are several diseases of the atrium and inflow tract, including a common form of congenital heart disease where the pulmonary venous inflow tract is ectopic or absent, and atrial fibrillation that relates to a reemergence of ectopic electrical activity in the pulmonary veins. The finding of bipotent Isl1+/SLN+ cells suggests the possibility that some of atrial diseases might arise from dys-regulation of the bipotency in atrial lineages.
The Isl1+/SLN+atrial progenitor population includes at least three populations; working atrial myocytes, Nkx2.5−/Tbx18+ cardiac progenitors in the sinus venosus and mediastinal myocardium in the dorsal mesocardium [16,23,24,33-35]. These three populations derive from Isl1+ posterior SHF progenitors and eventually acquire SLN expression, although our experiments cannot tell which population display smooth muscle competency. Discovery of earlier and later markers will help establish the lineage tree of the posterior SHF/venous pole population.
Lineage proximity of cardiac and smooth muscle cells
Smooth muscle cells appear to be recruited locally from a wide range of mesodermal tissues and differentiate in response to specific local signals. Our data suggest that during the migration from the posterior part of the SHF, Isl1+/SLN+ cells mainly contribute to mature atrial myocytes, but some cells escape from the cardiac lineage and give rise to the smooth muscle cells in the inflow tract (Fig. 6).
Given that SLN is relatively late marker of the atrial lineage, it is not unexpected that SLN+ cells contribute only to 5-10% of the inflow smooth muscle cells. It is rather significant in that atrial cells give rise to smooth muscle cells in vivo even after the expression of SLN gene, which is involved in atrial specific contractile function [17, 18].
It has been known that cardiac cells can transdifferentiate into smooth muscle cells even at the postnatal stage at low frequency upon certain culture conditions. However, little is known as to whether it is a dedifferentiation-redifferentiation process or a direct transition from differentiated cardiomyocyte to differentiated smooth muscle cell. Wu, et al. reported that Nkx2.5-positive cardiac progenitors downregulate Nkx2.5 before committing to smooth muscle lineage [3]. Since Nkx2.5 is also expressed in atrial lineage, it is likely that atrial cells shut of cardiac transcriptional program in order to give rise to smooth muscle cells. Although our study cannot give a clear answer to this question, it raises the possibility that the smooth muscle competency of the postnatal myocytes may be a trace of the developmental potential of the embryonic cardiac progenitors and myocytes.
A fundamental question relates to how the smooth muscle differentiation of cardiac progenitors serves to coordinate the morphogenesis of the atrial chamber and great vessels. An interesting speculation is that the smooth muscle competency of the atrial cells is required to anchor the great veins to the atrial chambers. Mal-connection of myocardium and vascular walls causes various types of congenital and adult heart diseases including anomalous pulmonary venous return and cor triatriatum [36]. In the aging and/or diseased hearts, the junction between pulmonary vein and left atria is highly arrhythmogenic and can be the foci of atrial fibrillation, one of the major cause of thromboembolic stroke [37-41]. Likewise, the heart-vessel junction at the arterial pole of the heart is often affected by various congenital malformations and acquired diseases including DeBakey Type I and II aortic dissection. It would be of great interest to analyze the pathogenesis of these congenital and acquired heart diseases from the viewpoint of progenitor cell plasticity.
Supplementary Material
Acknowledgements
This work is supported by NIH, Jean Le Ducq foundation, Harvard Stem Cell Institute, Massachusetts General Hospital, UCLA, and Eli and Edythe Broad Center of Regenerative Medicine and Stem Cell Research. We thank Marianne Cilluffo at the Electron Microscopic Lab in Brain Research Institute, UCLA.
Footnotes
Disclosures None declared.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Kattman SJ, Huber TL, Keller GM. Multipotent flk-1+ cardiovascular progenitor cells give rise to the cardiomyocyte, endothelial, and vascular smooth muscle lineages. Dev Cell. 2006;11:723–32. doi: 10.1016/j.devcel.2006.10.002. [DOI] [PubMed] [Google Scholar]
- [2].Moretti A, Caron L, Nakano A, Lam JT, Bernshausen A, Chen Y, et al. Multipotent embryonic isl1+ progenitor cells lead to cardiac, smooth muscle, and endothelial cell diversification. Cell. 2006;127:1151–65. doi: 10.1016/j.cell.2006.10.029. [DOI] [PubMed] [Google Scholar]
- [3].Wu SM, Fujiwara Y, Cibulsky SM, Clapham DE, Lien CL, Schultheiss TM, et al. Developmental origin of a bipotential myocardial and smooth muscle cell precursor in the mammalian heart. Cell. 2006;127:1137–50. doi: 10.1016/j.cell.2006.10.028. [DOI] [PubMed] [Google Scholar]
- [4].Yang L, Soonpaa MH, Adler ED, Roepke TK, Kattman SJ, Kennedy M, Henckaerts E, Bonham K, Abbott GW, Linden RM, et al. Human cardiovascular progenitor cells develop from a KDR+ embryonic-stem-cell-derived population. Nature. 2008;453:524–28. doi: 10.1038/nature06894. [DOI] [PubMed] [Google Scholar]
- [5].Garry DJ, Olson EN. A common progenitor at the heart of development. Cell. 2006;127:1101–4. doi: 10.1016/j.cell.2006.11.031. [DOI] [PubMed] [Google Scholar]
- [6].Kattman SJ, Adler ED, Keller GM. Specification of multipotential cardiovascular progenitor cells during embryonic stem cell differentiation and embryonic development. Trends Cardiovasc Med. 2007;17:240–6. doi: 10.1016/j.tcm.2007.08.004. [DOI] [PubMed] [Google Scholar]
- [7].Martin-Puig S, Wang Z, Chien KR. Lives of a heart cell: tracing the origins of cardiac progenitors. Cell Stem Cell. 2008;2:320–31. doi: 10.1016/j.stem.2008.03.010. [DOI] [PubMed] [Google Scholar]
- [8].Cai CL, Liang X, Shi Y, Chu PH, Pfaff SL, Chen J, et al. Isl1 identifies a cardiac progenitor population that proliferates prior to differentiation and contributes a majority of cells to the heart. Dev Cell. 2003;5:877–89. doi: 10.1016/s1534-5807(03)00363-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Laugwitz KL, Moretti A, Caron L, Nakano A, Chien KR. Islet1 cardiovascular progenitors: a single source for heart lineages? Development. 2008;135:193–205. doi: 10.1242/dev.001883. [DOI] [PubMed] [Google Scholar]
- [10].Kelly RG, Brown NA, Buckingham ME. The arterial pole of the mouse heart forms from Fgf10-expressing cells in pharyngeal mesoderm. Dev Cell. 2001;1:435–40. doi: 10.1016/s1534-5807(01)00040-5. [DOI] [PubMed] [Google Scholar]
- [11].Mjaatvedt CH, Nakaoka T, Moreno-Rodriguez R, Norris RA, Kern MJ, Eisenberg CA, et al. The outflow tract of the heart is recruited from a novel heart-forming field. Dev Biol. 2001;238:97–109. doi: 10.1006/dbio.2001.0409. [DOI] [PubMed] [Google Scholar]
- [12].Waldo KL, Kumiski DH, Wallis KT, Stadt HA, Hutson MR, Platt DH, et al. Conotruncal myocardium arises from a secondary heart field. Development. 2001;128:3179–88. doi: 10.1242/dev.128.16.3179. [DOI] [PubMed] [Google Scholar]
- [13].Dodou E, Verzi MP, Anderson JP, Xu SM, Black BL. Mef2c is a direct transcriptional target of ISL1 and GATA factors in the anterior heart field during mouse embryonic development. Development. 2004;131:3931–42. doi: 10.1242/dev.01256. [DOI] [PubMed] [Google Scholar]
- [14].Waldo KL, Hutson MR, Ward CC, Zdanowicz M, Stadt HA, Kumiski D, et al. Secondary heart field contributes myocardium and smooth muscle to the arterial pole of the developing heart. Dev Biol. 2005;281:78–90. doi: 10.1016/j.ydbio.2005.02.012. [DOI] [PubMed] [Google Scholar]
- [15].Sun Y, Liang X, Najafi N, Cass M, Lin L, Cai CL, et al. Islet 1 is expressed in distinct cardiovascular lineages, including pacemaker and coronary vascular cells. Dev Biol. 2007;304:286–96. doi: 10.1016/j.ydbio.2006.12.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Galli D, Dominguez JN, Zaffran S, Munk A, Brown NA, Buckingham ME. Atrial myocardium derives from the posterior region of the second heart field, which acquires left-right identity as Pitx2c is expressed. Development. 2008;135:1157–67. doi: 10.1242/dev.014563. [DOI] [PubMed] [Google Scholar]
- [17].Odermatt A, Taschner PE, Scherer SW, Beatty B, Khanna VK, Cornblath DR, et al. Characterization of the gene encoding human sarcolipin (SLN), a proteolipid associated with SERCA1: absence of structural mutations in five patients with Brody disease. Genomics. 1997;45:541–53. doi: 10.1006/geno.1997.4967. [DOI] [PubMed] [Google Scholar]
- [18].Minamisawa S, Wang Y, Chen J, Ishikawa Y, Chien KR, Matsuoka R. Atrial chamber-specific expression of sarcolipin is regulated during development and hypertrophic remodeling. J Biol Chem. 2003;278:9570–75. doi: 10.1074/jbc.m213132200. [DOI] [PubMed] [Google Scholar]
- [19].Babu GJ, Bhupathy P, Carnes CA, Billman GE, Periasamy M. Differential expression of sarcolipin protein during muscle development and cardiac pathophysiology. J Mol Cell Cardiol. 2007;43:215–22. doi: 10.1016/j.yjmcc.2007.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Xu H, Chen L, Baldini A. In vivo genetic ablation of the periotic mesoderm affects cell proliferation survival and differentiation in the cochlea. Dev Biol. 2007;310:329–40. doi: 10.1016/j.ydbio.2007.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Soriano P. Generalized lacZ expression with the ROSA26 Cre reporter strain. Nat Genet. 1999;21:70–1. doi: 10.1038/5007. [DOI] [PubMed] [Google Scholar]
- [22].Vintersten K, Monetti C, Gertsenstein M, Zhang P, Laszlo L, Biechele S, et al. Mouse in red: red fluorescent protein expression in mouse ES cells, embryos, and adult animals. Genesis. 2004;40:241–6. doi: 10.1002/gene.20095. [DOI] [PubMed] [Google Scholar]
- [23].Soufan AT, van den Hoff MJ, Ruijter JM, de Boer PA, Hagoort J, Webb S, et al. Reconstruction of the patterns of gene expression in the developing mouse heart reveals an architectural arrangement that facilitates the understanding of atrial malformations and arrhythmias. Circ Res. 2004;95:1207–15. doi: 10.1161/01.RES.0000150852.04747.e1. [DOI] [PubMed] [Google Scholar]
- [24].Anderson RH, Brown NA, Moorman AF. Development and structures of the venous pole of the heart. Dev Dyn. 2006;235:2–9. doi: 10.1002/dvdy.20578. [DOI] [PubMed] [Google Scholar]
- [25].Mommersteeg MT, Soufan AT, de Lange FJ, van den Hoff MJ, Anderson RH, Christoffels VM, et al. Two distinct pools of mesenchyme contribute to the development of the atrial septum. Circ Res. 2006;99:351–3. doi: 10.1161/01.RES.0000238360.33284.a0. [DOI] [PubMed] [Google Scholar]
- [26].Moorman AF, Christoffels VM, Anderson RH, van den Hoff MJ. The heart-forming fields: one or multiple? Philos Trans R Soc Lond B Biol Sci. 2007;362:1257–65. doi: 10.1098/rstb.2007.2113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Snarr BS, O’Neal JL, Chintalapudi MR, Wirrig EE, Phelps AL, Kubalak SW, et al. Isl1 expression at the venous pole identifies a novel role for the second heart field in cardiac development. Circ Res. 2007;101:971–4. doi: 10.1161/CIRCRESAHA.107.162206. [DOI] [PubMed] [Google Scholar]
- [28].Snarr BS, Wirrig EE, Phelps AL, Trusk TC, Wessels A. A spatiotemporal evaluation of the contribution of the dorsal mesenchymal protrusion to cardiac development. Dev Dyn. 2007;236:1287–94. doi: 10.1002/dvdy.21074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Kruithof BP, van den Hoff MJ, Wessels A, Moorman AF. Cardiac muscle cell formation after development of the linear heart tube. Dev Dyn. 2003;227:1–13. doi: 10.1002/dvdy.10269. [DOI] [PubMed] [Google Scholar]
- [30].Bu L, Jiang X, Martin-Puig S, Caron L, Zhu S, Shao Y, et al. Human ISL1 heart progenitors generate diverse multipotent cardiovascular cell lineages. Nature. 2009;460:113–7. doi: 10.1038/nature08191. [DOI] [PubMed] [Google Scholar]
- [31].Dyer LA, Kirby ML. The role of secondary heart field in cardiac development. Dev Biol. 2009;336:137–44. doi: 10.1016/j.ydbio.2009.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Buckingham M, Meilhac S, Zaffran S. Building the mammalian heart from two sources of myocardial cells. Nat Rev Genet. 2005;6:826–35. doi: 10.1038/nrg1710. [DOI] [PubMed] [Google Scholar]
- [33].Soufan AT, van den Berg G, Ruijter JM, de Boer PA, van den Hoff MJ, Moorman AF. Regionalized sequence of myocardial cell growth and proliferation characterizes early chamber formation. Circ Res. 2006;99:545–52. doi: 10.1161/01.RES.0000239407.45137.97. [DOI] [PubMed] [Google Scholar]
- [34].Christoffels VM, Mommersteeg MT, Trowe MO, Prall OW, de Gier-de Vries C, Soufan AT, et al. Formation of the venous pole of the heart from an Nkx2-5-negative precursor population requires Tbx18. Circ Res. 2006;98:1555–63. doi: 10.1161/01.RES.0000227571.84189.65. [DOI] [PubMed] [Google Scholar]
- [35].Mommersteeg MT, Brown NA, Prall OW, de Gier-de Vries C, Harvey RP, Moorman AF, et al. Pitx2c and Nkx2-5 are required for the formation and identity of the pulmonary myocardium. Circ Res. 2007;101:902–9. doi: 10.1161/CIRCRESAHA.107.161182. [DOI] [PubMed] [Google Scholar]
- [36].Nakano A, Nakano H, Chien KR. Multipotent islet-1 cardiovascular progenitors in development and disease. Cold Spring Harb Symp Quant Biol. 2008;73:297–306. doi: 10.1101/sqb.2008.73.055. [DOI] [PubMed] [Google Scholar]
- [37].Jais P, Haissaguerre M, Shah DC, Chouairi S, Gencel L, Hocini M, et al. A focal source of atrial fibrillation treated by discrete radiofrequency ablation. Circulation. 1997;95:572–6. doi: 10.1161/01.cir.95.3.572. [DOI] [PubMed] [Google Scholar]
- [38].Haissaguerre M, Jais P, Shah DC, Takahashi A, Hocini M, Quiniou G, et al. Spontaneous initiation of atrial fibrillation by ectopic beats originating in the pulmonary veins. N Engl J Med. 1998;339:659–66. doi: 10.1056/NEJM199809033391003. [DOI] [PubMed] [Google Scholar]
- [39].Haissaguerre M, Shah DC, Jais P, Hocini M, Yamane T, Deisenhofer I, et al. Electrophysiological breakthroughs from the left atrium to the pulmonary veins. Circulation. 2000;102:2463–5. doi: 10.1161/01.cir.102.20.2463. [DOI] [PubMed] [Google Scholar]
- [40].Chen PS, Chou CC, Tan AY, Zhou S, Fishbein MC, Hwang C, et al. The mechanisms of atrial fibrillation. J Cardiovasc Electrophysiol. 2006;17(Suppl 3):S2–7. doi: 10.1111/j.1540-8167.2006.00626.x. [DOI] [PubMed] [Google Scholar]
- [41].Ocorr K, Reeves NL, Wessells RJ, Fink M, Chen HS, Akasaka T, et al. KCNQ potassium channel mutations cause cardiac arrhythmias in Drosophila that mimic the effects of aging. Proc Natl Acad Sci U S A. 2007;104:3943–8. doi: 10.1073/pnas.0609278104. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.