Abstract
Resistance to fusidic acid in Staphylococcus aureus often results from acquisition of the fusB determinant or from mutations in the gene (fusA) that encodes the drug target (elongation factor G). We now report further studies on the genetic basis of resistance to this antibiotic in the staphylococci. Two staphylococcal genes that encode proteins exhibiting ca. 45% identity with FusB conferred resistance to fusidic acid in S. aureus. One of these genes (designated fusC) was subsequently detected in all fusidic acid-resistant clinical strains of S. aureus tested that did not carry fusB or mutations in fusA, and in strains of S. intermedius. The other gene (designated fusD) is carried by S. saprophyticus, explaining the inherent resistance of this species to fusidic acid. Fusidic acid-resistant strains of S. lugdunensis harbored fusB. Thus, resistance to fusidic acid in clinical isolates of S. aureus and other staphylococcal species frequently results from expression of FusB-type proteins.
Fusidic acid inhibits bacterial protein synthesis by preventing release of elongation factor G (EF-G) from the ribosome (4). It is used both topically and systemically for the treatment of staphylococcal disease (6).
Resistance to fusidic acid in Staphylococcus aureus occurs by horizontal acquisition of the fusB determinant, which encodes an EF-G-binding protein that protects the staphylococcal translation apparatus from inhibition by fusidic acid (13), or by spontaneous mutation in the gene encoding EF-G (fusA) (14). However, some fusidic acid-resistant S. aureus strains lack these mechanisms (14), and therefore additional, uncharacterized determinants of resistance to fusidic acid exist. In addition, little is known about the genetic basis of resistance to fusidic acid in staphylococci other than S. aureus.
Here we report on the basis of resistance to fusidic acid in strains of four different staphylococcal species. Two of these strains (S. saprophyticus ATCC 15305, S. aureus MSSA476) were chosen because their genomes encode homologues of the FusB protein (8, 10). Strains of S. lugdunensis and S. intermedius were examined since they exhibit phenotypic resistance to fusidic acid, the genetic basis of which is unknown. A collection of clinical S. aureus strains exhibiting resistance to fusidic acid, but not carrying fusB or resistance polymorphisms in fusA, was also characterized.
MATERIALS AND METHODS
Bacterial strains and culture.
Table 1 lists staphylococcal strains used and generated in the present study. S. lugdunensis strains were isolated from patients in Sweden and were identified on the basis of typical morphology and/or smell, resistance to desferrioxamine, and ornithine decarboxylase and pyrrolidonyl arylamidase activity (1). Escherichia coli XL-10 Gold and XL-1 Blue (Stratagene, Amsterdam, The Netherlands) were used as cloning hosts.
TABLE 1.
Staphylococcal strains used and generated in the present study and their susceptibilities to fusidic acid
| Strain | Fusidic acid MIC (μg/ml)a | Source or reference |
|---|---|---|
| S. aureus | ||
| RN4220 | 0.125 | 5 |
| CS957 | 3* | 14 |
| 74136 (EEFIC) | 4 | This study |
| MSSA476 | 8 | 8 |
| 649(pUB101) | 16 | 13 |
| RN4220(pCU1:fusB) | 16 | 13 |
| S. aureus clinical isolates with unknown mechanisms of resistance to fusidic acid | ||
| CS992 | 2 | 14 |
| CS730 | 3* | 14 |
| CS808 | 3* | 14 |
| CS866 | 3* | 14 |
| CS602 | 4 | This study |
| CS851 | 4 | This study |
| CS979 | 4 | This study |
| CS1083 | 4 | This study |
| CS1128 | 4 | This study |
| CS805 | 8 | This study |
| CS860 | 8 | This study |
| CS1076 | 8 | This study |
| S. aureus recombinant strains expressing FusB or FusB homologues | ||
| RN4220(pCU1) | 0.125 | This study |
| RN4220(pCU1:fusB) | 16 | This study |
| RN4220(pCU1:SAS0043) (fusC) | 16 | This study |
| RN4220(pCU1:SSP2165) (fusD) | 2 | This study |
| RN4220(pAJ96) | 0.125 | This study |
| RN4220(pAJ96:fusB) | 32 | This study |
| RN4220(pAJ96:SAS0043) (fusC) | 16 | This study |
| RN4220(pAJ96:SSP2165) (fusD) | 4 | This study |
| S. intermedius | ||
| CL1 | 4 | 7 |
| CL2 | 4 | 7 |
| NCTC 11048 | 0.064 | ATCC (no. 29663) |
| S. lugdunensis | ||
| 16641 | 4 | This study |
| 16496 | 4 | This study |
| 31440 | 4 | This study |
| 40869 | 0.064 | This study |
| S. saprophyticus | ||
| ATCC 15305 | 2 | 10 |
*, The MIC was determined by Etest in a previous study (14).
Unless otherwise stated, strains were cultured in Luria-Bertani broth with aeration or on Luria-Bertani agar at 37°C. Susceptibility testing and determination of bacterial doubling times were performed as described previously (9).
DNA manipulation.
Recombinant DNA methods were standard (16). Routine PCR amplification was performed with Platinum Pfx (Invitrogen, Paisley, United Kingdom), while the FailSafe PCR System (Epicenter, Madison, WI) was used for long PCR. Southern hybridization for detection of fusB (14) was performed on EcoRI-digested DNA at 60°C. Detection of fusC was performed in an identical fashion but used a probe generated by PCR with the oligonucleotide primers fusCU and fusCL (Table 2).
TABLE 2.
Oligonucleotide primers used in the present study
| Designation | Sequence (5′-3′)a |
|---|---|
| fusU | TAAGCGGCCGCAAGATTCTTCAATATCGTCATCTA |
| groL | GCAGCGGCCGCAGCTCAAGCAATGATTCAAGAAGG |
| rpsU | ATGGCTGGTACCAACAAAGCATTTGCTCACTA |
| tufL | GCTGTGAGCTCTGTTTTACCATGGTCAACGTG |
| sas0043U | GTAGGATCCATTGGGAATGATAAATAGTGA |
| sas0043L | TTTGGATCCATCGATTAAGAGTGAGGTACA |
| ssp2165U | ATCGGATCCTGCTTTGTCTGTCACATCTAA |
| ssp2165L | ACGGATCCAGGTGGGGTTGTCTATA |
| fusBU | AATGGTACCACTTGTGAAAGGTTGAAAACAATGAAAACAATGATTTATCC |
| fusBL | AACAGAGCTCATTCCTTAATCTAGTTTATC |
| sapU | AAAATGGTACCACTTGTGAAAGGTTGAAAACAATGGAAAAACAACTTTAC |
| sapL | AATAGAGCTCGTTTTTGGTTTATTGAATCT |
| mssaU | AATGGTACCACTTGTGAAAGGTTGAAAACAATGAATAAAATAGAAGTGTA |
| mssaL | CAGGGAGCTCTGGATCTATTTTATTTTAAC |
| fusCU | GAGGAATATCATATGAATAAAATAGAAGTGTA |
| fusCL | AGAGTGGATCCCAAAATATAACAACCCTGATC |
Engineered regions are underlined.
Recombinant DNA constructs were routinely created by using plasmid pCU1 (2). Constructs for tetracycline-regulated gene expression were generated by using plasmid pAJ96, a derivative of pALC2073 (3) that carries a transcriptional terminator downstream of the cloning site. For the former, PCR amplicons were introduced into the BamHI restriction site of pCU1 by virtue of engineered BamHI restriction sites at the 5′ end of oligonucleotide primers (Table 2), while engineered KpnI/SacI restriction sites were used for ligation into pAJ96. Constructs were propagated in E. coli before recovery and introduction into S. aureus RN4220 by electroporation. Tetracycline (100 ng/ml) was used to induce expression from the xyl/tetO promoter on pAJ96.
GenBank accession numbers.
The fusA gene of S. intermedius (NCTC 11048) has been assigned GenBank accession number AY776250, and that from S. lugdunensis was assigned GenBank accession number DQ866810.
RESULTS
FusB homologues mediate resistance to fusidic acid in S. aureus and S. saprophyticus.
A whole-genome sequence analysis of S. aureus strain MSSA476 was recently reported (8). This strain is resistant to fusidic acid and carries a gene (SAS0043) encoding a homologue (YP_042173) of the FusB protein. The gene and the fusidic acid resistance phenotype may be associated (8), although this has not been confirmed. We also identified a staphylococcal gene (SSP2165) encoding a FusB homologue (YP_302255) in the genome of S. saprophyticus ATCC 15305 (10). Although most staphylococcal species are inherently susceptible to fusidic acid, S. saprophyticus is intrinsically resistant to the antibiotic (6). These two staphylococcal FusB homologues exhibit 44% (YP_042173) and 47% (YP_302255) identity to FusB and 41% identity to each other (Fig. 1).
FIG. 1.
Protein alignment between FusB, YP_042173 (FusC from S. aureus MSSA476), and YP_302255 (FusD from S. saprophyticus ATCC 15305). Gray shading indicates identity in two sequences; black shading indicates identity in all three sequences.
To establish whether these FusB-like proteins represent functional homologues of FusB (i.e., confer resistance to fusidic acid), the genes encoding them were PCR amplified, along with their upstream expression signals, using oligonucleotide primers SAS0043U/L and SSP2165U/L for SAS0043 and SSP2165, respectively (Table 2) and introduced into S. aureus RN4220 on plasmid pCU1. Both genes conferred resistance to fusidic acid (Table 1). Consequently, these proteins appear to form part of a FusB protein family that confer resistance to fusidic acid in staphylococci. Based on the precedent of ≤80% amino acid identity to represent the dividing line between one resistance determinant and a related one (12), we have designated these proteins FusC (YP_042173 from MSSA476) and FusD (YP_302255 from ATCC 15305), and their corresponding genes were designated fusC and fusD.
To establish the relative abilities of the FusB, FusC, and FusD proteins to confer resistance in S. aureus, constructs were generated enabling identical expression of the three genes. Thus, fusC and fusD were PCR amplified with oligonucleotide primers MSSAkpn/MSSAsac and SAPkpn/SAPsac (Table 2), resulting in upstream regions and ribosome binding sites identical to those of fusB. The fusB gene was amplified by using primers fusBkpn/fusBsac (Table 2). All three genes were expressed under identical induction conditions from the xyl/tetO promoter on plasmid pAJ96 in S. aureus RN4220. Under these conditions, FusB demonstrated a greater ability to protect S. aureus from fusidic acid compared to FusC and FusD (Table 1).
Carriage of antibiotic resistance determinants can impose a fitness cost in bacteria. Since constructs enabling equivalent expression of all three resistance genes had been constructed, it was possible to examine whether expression of fusB, fusC, or fusD was associated with a fitness cost in S. aureus. However, strains expressing these genes did not exhibit doubling times significantly different from RN4220 carrying pAJ96 (data not shown).
Resistance to fusidic acid mediated by FusB homologues in clinical strains of S. aureus.
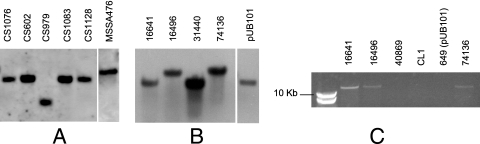
A proportion of fusidic acid-resistant S. aureus clinical strains carry neither fusB nor mutations in fusA (14). Such strains might be resistant to fusidic acid through expression of FusB homologues. Relevant strains (CS strains; Table 1) were screened for the presence of genes encoding FusB homologues by Southern hybridization. All strains tested positive for fusC (Fig. 2). To establish whether these strains harbored fusC or polymorphic variants cross-hybridizing with fusC, the fusC genes from these strains were PCR amplified and sequenced (primers SAS0043U and SAS0043L; Table 2). Since these primers correspond to the start and end of the fusC gene, this analysis could not provide DNA sequence information for the gene termini. However, a DNA sequence for >95% of the gene was obtained. The fusC genes encoded protein products identical (in the sequenced portion) to that encoded by fusC from MSSA476. However, fusC from strain CS979 carried a silent mutation (S162 was encoded by TCC rather than TCT).
FIG. 2.
Detection of fusB and fusC in staphylococci. (A) Detection of fusC by Southern hybridization in a representative set of fusidic acid-resistant clinical strains of S. aureus that harbor neither fusB nor resistance polymorphisms in fusA. MSSA476 is the positive control. (B) Detection of fusB by Southern hybridization in clinical strains of S. lugdunensis. pUB101 is the positive control, while S. aureus 74136 is an EEFIC strain (see the text). (C) PCR analysis maps fusB downstream of groEL in S. lugdunensis 16641 and 16496, the same location as that in EEFIC strain, S. aureus 74136. The other three strains are negative controls.
Resistance to fusidic acid in S. intermedius and S. lugdunensis.
Resistant strains of S. intermedius (7) and S. lugdunensis were ∼64-fold less susceptible to fusidic acid than wild-type, sensitive strains of the same species (Table 1). These resistant strains were screened initially for nucleotide substitutions in fusA. Since the DNA sequence of fusA from these species was not available, oligonucleotide primers for PCR amplification and sequencing of this locus were designed against regions of the upstream (rpsG) and downstream genes (tufA) that are conserved between S. aureus and Bacillus subtilis. Thus, the primers rpsU and tufL (Table 2) were used to PCR amplify the entire fusA gene from both fusidic acid-sensitive and resistant strains, which were then sequenced in toto.
The fusA genes of S. lugdunensis and S. intermedius encode proteins with 95 and 92% identity, respectively, to EF-G from S. aureus N315. No nucleotide polymorphisms were identified in the fusA genes of the fusidic acid-resistant S. lugdunensis strains relative to the sensitive strains. However, six coding polymorphisms in fusA from S. intermedius CL1 and CL2 were identified compared to S. intermedius NCTC 11048 (NCTC 11048 residue shown first); N187T, V197T, D231E, N294D, S368T, and T488S. To examine whether these polymorphisms were responsible for the observed fusidic acid-resistance phenotype in CL1/CL2, the fusA genes of CL1, CL2, NCTC 11048 (negative control) and S. aureus CS957 (positive control; Table 1) were expressed in trans from pAJ96 in S. aureus RN4220. Expression of fusA from CS957 conferred resistance to fusidic acid, although expression of the S. intermedius fusA genes did not (data not shown).
Subsequently, representative fusidic acid-sensitive and -resistant strains of S. lugdunensis and S. intermedius were tested for the presence of fusB and fusC by Southern hybridization. Fusidic acid-resistant S. intermedius carried fusC (data not shown). The sequences of the fusC genes from S. intermedius CL1 and CL2 were identical to those found in MSSA476, with the exception of the same silent mutation identified in strain CS979.
Fusidic acid-resistant S. lugdunensis strains carried fusB (Fig. 2), a finding that was confirmed by PCR amplification and sequencing. We recently determined that fusB in the epidemic European fusidic acid-resistant impetigo clone (EEFIC) of S. aureus is located on a genomic island called SaRIfusB, integrated into the chromosome downstream of the groEL gene (14a). Since fusB is present on a similar-sized EcoRI fragment in fusidic acid-resistant S. lugdunensis and in representatives of the EEFIC such as S. aureus 74136 (Fig. 2), we postulated that fusB may reside at the same chromosomal locus in S. lugdunensis. Southern hybridization established that the fusB-positive S. lugdunensis strains carried this resistance gene on the chromosome, since no hybridization of the fusB probe was detected with plasmid preparations (data not shown). Furthermore, using oligonucleotide primers specific for fusB (fusU; Table 2) and conserved regions of staphylococcal groEL (groL; Table 2), a PCR product (∼15 kb) was generated from S. aureus 74136 and the fusidic acid-resistant S. lugdunensis strains but not from fusidic acid-sensitive strains or from those that carry plasmid-borne fusB (Fig. 2).
DISCUSSION
Early work on fusidic acid resistance in S. aureus (11) suggested that horizontally acquired (FusB-type) resistance was the most prevalent mechanism among clinical strains. This suggestion has recently been strengthened by the finding that the major fusidic acid-resistant clone of S. aureus (i.e., EEFIC), which is present in several European countries, carries the fusB determinant (14). The present study further underlines the importance of this resistance determinant in staphylococci, both for the prototypical fusB gene and for genes encoding functional homologues.
Two staphylococcal genes encoding functional homologues of FusB were identified in the present study: one from S. aureus MSSA476 (fusC) and the other from S. saprophyticus ATCC 15305 (fusD). In the latter species, fusD appears to be a normal component of the genome, explaining the inherent resistance of this species to fusidic acid (6). Maintenance of a fusidic acid resistance determinant by S. saprophyticus, which provides protection against an antibiotic the organism is unlikely to encounter, supports previous arguments that the FusB family of proteins originally evolved to provide a housekeeping function unrelated to fusidic acid resistance (13). Furthermore, expression of FusB and the homologues did not impose a fitness cost, suggesting that maintenance of these determinants in the absence of antibiotic selection pressure does not disadvantage the organisms in which they occur naturally.
The FusB-like proteins of the staphylococci are more closely related to each other than to FusB homologues found in other gram-positive bacteria (13; data not shown). This suggests that these proteins are all descended from a single, ancestral, staphylococcal FusB. Based on the substantial sequence divergence of the three staphylococcal proteins (Fig. 1), the original recruitment of the ancestral fusB to the staphylococci probably occurred a long time ago and not as a consequence of the clinical introduction of fusidic acid in the 1960s.
Of the three determinants, fusD, found in S. saprophyticus, is the only gene encoding a FusB-like protein for which there is currently no evidence for spread between staphylococcal species. In contrast, we detected fusC in fusidic acid-resistant strains of S. intermedius and in 12 fusidic acid-resistant strains of S. aureus. Indeed, fusC is responsible for resistance to fusidic acid in all S. aureus strains that we have examined that do not carry fusB or resistance mutations in fusA, including four previously described strains (14). Thus, to date, all examples of fusidic acid resistance in clinical strains of S. aureus are the result of expression of a FusB-type protein (encoded by fusB or fusC) or mutations in fusA. For those seeking to detect fusB and fusC in clinical strains, it is important to note that the level of nucleotide sequence homology between these genes is ∼60%, which explains why we did not previously observe hybridization of a fusB probe to DNA from strains we now know to contain fusC (14).
Fusidic acid-resistant strains of S. lugdunensis carried the fusB determinant on the chromosome, downstream of groEL (Fig. 2), the locus occupied by the SaRIfusB genomic island in the EEFIC strain of S. aureus. The emergence of fusidic acid resistance in strains of S. lugdunensis in Sweden has occurred after clonal expansion of the EEFIC in Sweden (15) and in a geographical area with a high prevalence of EEFIC strains. This suggests the possibility that fusB in S. lugdunensis has been acquired from S. aureus, although further studies will be required to support this hypothesis.
The fusB gene has also recently been reported in bovine isolates of coagulase negative staphylococci (17). That report, together with our own studies, therefore illustrates the importance of FusB and FusB homologues for resistance to fusidic acid in several staphylococcal species.
Acknowledgments
This study was supported by a research grant to I.C. from LEO Pharma and a BBSRC-CASE PhD studentship in conjunction with LEO.
Footnotes
Published ahead of print on 26 February 2007.
REFERENCES
- 1.American Society for Microbiology. 1992. Clinical microbiology procedures handbook. American Society for Microbiology, Washington, DC.
- 2.Augustin, J., R. Rosenstein, B. Wieland, U. Schneider, N. Schnell, G. Engelke, K. D. Entian, and F. Gotz. 1992. Genetic analysis of epidermin biosynthetic genes and epidermin-negative mutants of Staphylococcus epidermidis. Eur. J. Biochem. 2041149-1154. [DOI] [PubMed] [Google Scholar]
- 3.Bateman, B. T., N. P. Donegan, T. M. Jarry, M. Palma, and A. L. Cheung. 2001. Evaluation of a tetracycline-inducible promoter in Staphylococcus aureus in vitro and in vivo and its application in demonstrating the role of sigB in microcolony formation. Infect. Immun. 697851-7857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bodley, J. W., F. J. Zieve, L. Lin, and S. T. Zieve. 1969. Formation of the ribosome-G factor-GDP complex in the presence of fusidic acid. Biochem. Biophys. Res. Commun. 37437-443. [DOI] [PubMed] [Google Scholar]
- 5.Fairweather, N., S. Kennedy, T. J. Foster, M. Kehoe, and G. Dougan. 1983. Expression of a cloned Staphylococcus aureus alpha-hemolysin determinant in Bacillus subtilis and Staphylococcus aureus. Infect. Immun. 411112-1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Greenwood, D. 2003. Fusidanes, p. 297-299. In R. G. Finch (ed.), Antibiotic and chemotherapy, 8th ed. Churchill Livingstone, London, England.
- 7.Guardabassi, L., M. E. Loeber, and A. Jacobson. 2004. Transmission of multiple antimicrobial-resistant Staphylococcus intermedius between dogs affected by deep pyoderma and their owners. Vet. Microbiol. 9823-27. [DOI] [PubMed] [Google Scholar]
- 8.Holden, M. T., E. J. Feil, J. A. Lindsay, S. J. Peacock, N. P. Day, M. C. Enright, T. J. Foster, C. E. Moore, L. Hurst, R. Atkin, A. Barron, N. Bason, S. D. Bentley, C. Chillingworth, T. Chillingworth, C. Churcher, L. Clark, C. Corton, A. Cronin, J. Doggett, L. Dowd, T. Feltwell, Z. Hance, B. Harris, H. Hauser, S. Holroyd, K. Jagels, K. D. James, N. Lennard, A. Line, R. Mayes, S. Moule, K. Mungall, D. Ormond, M. A. Quail, E. Rabbinowitsch, K. Rutherford, M. Sanders, S. Sharp, M. Simmonds, K. Stevens, S. Whitehead, B. G. Barrell, B. G. Spratt, and J. Parkhill. 2004. Complete genomes of two clinical Staphylococcus aureus strains: evidence for the rapid evolution of virulence and drug resistance. Proc. Natl. Acad. Sci. USA 1019786-9791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hurdle, J. G., A. J. O'Neill, and I. Chopra. 2004. The isoleucyl-tRNA synthetase mutation V588F conferring mupirocin resistance in glycopeptide-intermediate Staphylococcus aureus is not associated with a significant fitness burden. J. Antimicrob. Chemother. 53102-104. [DOI] [PubMed] [Google Scholar]
- 10.Kuroda, M., A. Yamashita, H. Hirakawa, M. Kumano, K. Morikawa, M. Higashide, A. Maruyama, Y. Inose, K. Matoba, H. Toh, S. Kuhara, M. Hattori, and T. Ohta. 2005. Whole genome sequence of Staphylococcus saprophyticus reveals the pathogenesis of uncomplicated urinary tract infection. Proc. Natl. Acad. Sci. USA 10213272-13277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lacey, R. W., and V. T. Rosdahl. 1974. An unusual “penicillinase plasmid” in Staphylococcus aureus: evidence for its transfer under natural conditions. J. Med. Microbiol. 71-9. [DOI] [PubMed] [Google Scholar]
- 12.Levy, S. B., L. M. McMurry, T. M. Barbosa, V. Burdett, P. Courvalin, W. Hillen, M. C. Roberts, J. I. Rood, and D. E. Taylor. 1999. Nomenclature for new tetracycline resistance determinants. Antimicrob. Agents Chemother. 431523-1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.O'Neill, A. J., and I. Chopra. 2006. Molecular basis of fusB-mediated resistance to fusidic acid in Staphylococcus aureus. Mol. Microbiol. 59664-676. [DOI] [PubMed] [Google Scholar]
- 14.O'Neill, A. J., A. R. Larsen, A. S. Henriksen, and I. Chopra. 2004. A fusidic acid-resistant epidemic strain of Staphylococcus aureus carries the fusB determinant, whereas fusA mutations are prevalent in other resistant isolates. Antimicrob. Agents Chemother. 483594-3597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14a.O'Neill, A. J., A. R. Larsen, R. Skov, A. S. Henriksen, and I. Chopra. 7 March 2007. Characterization of the epidemic European fusidic acid-resistant impetigo clone (EEFIC) of Staphylococcus aureus. J. Clin. Microbiol. doi: 10.1128/JCM.01984-06. [DOI] [PMC free article] [PubMed]
- 15.Osterlund, A., T. Eden, B. Olsson-Liljequist, S. Haeggman, and G. Kahlmeter. 2002. Clonal spread among Swedish children of a Staphylococcus aureus strain resistant to fusidic acid. Scand. J. Infect. Dis. 34729-734. [DOI] [PubMed] [Google Scholar]
- 16.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, New York, NY.
- 17.Yazdankhah, S. P., A. W. Asli, H. Sorum, H. Oppegaard, and M. Sunde. 2006. Fusidic acid resistance, mediated by fusB, in bovine coagulase-negative staphylococci. J. Antimicrob. Chemother. 2323. [DOI] [PubMed] [Google Scholar]