TraitBank: Practical semantics for organism attribute data
Abstract
Encyclopedia of Life (EOL) has developed TraitBank (http://eol.org/traitbank), a new repository for organism attribute (trait) data. TraitBank aggregates, manages and serves attribute data for organisms across the tree of life, including life history characteristics, habitats, distributions, ecological relationships and other data types. We describe how TraitBank ingests and manages these data in a way that leverages EOL’s existing infrastructure and semantic annotations to facilitate reasoning across the TraitBank corpus and interoperability with other resources. We also discuss TraitBank’s impact on users and collaborators and the challenges and benefits of our lightweight, scalable approach to the integration of biodiversity data.
1.Introduction
While human knowledge of life on Earth is vast, there is no easy way to query all the information accumulated in hundreds of years of biodiversity research and documentation. Even simple questions like “which plants have yellow flowers?” or “what do sharks eat?” are impossible to answer with confidence.
Biologists have captured and managed information about morphology, behavior, life history, and ecological interactions in many different ways. Most of this information survives in the form of free text or data tables in published papers, if it survives at all [20]. Lately communities have started to annotate those papers [3], extract information from text [28,40], and build special-purpose databases of trait data, for example, TRY11 for plants [24] and SeaLifeBase22 for marine organisms. In addition, modern researchers are more likely to archive and share data sets associated with their published studies in open data repositories such as Dryad33 [42], Ecological Archives44 and PANGAEA.55 While these are critical developments, there is still little standardization in how biologists talk about the characteristics of organisms, how they describe the context of their observations, and how they document the methods with which the data were collected. This means that the information in many data sets is not easily discovered, integrated or repurposed.
This lack of data standards impedes progress in the ecological, conservation, and phylogenetic research communities, who need effective ways to quickly discover and consume data in the coming era of data-intensive science [e.g., [17]–[19]]. For example, marine environmental modelers need high-quality inputs about large numbers of species in order to understand current and historical distributions of species; how these distributions are impacted by environmental changes such as climate change, overharvesting, or invasive species; how biological communities function to provide ecosystem services; and what could happen to these services under future scenarios that change the composition of these communities. Such large-scale data have also been identified by DIVERSITAS66 and the Group on Earth Observations Biodiversity Observation Network (GEO BON)77 as likely to be required by the Intergovernmental Platform on Biodiversity and Ecosystem Services (IPBES) [35]. Aggregating and standardizing these data, making them freely re-usable, and providing discovery mechanisms for them could facilitate rapid analyses for investigators interested in these urgent problems.
This paper describes TraitBank®, a system designed by the Encyclopedia of Life (EOL) to acquire, organize and serve biodiversity attribute data on a global scale across the entire tree of life88 – currently estimated at nearly two million species [9]. It describes our approaches to semantics, details TraitBank’s implementation, and evaluates the system with respect to implications for interoperability and impact on community and provider processes.
2.Approach
TraitBank mobilizes data from diverse sources including biodiversity databases (e.g., Global Biodiversity Information Facility (GBIF),99 Global Biotic Interactions (GloBI),1010 Ocean Biogeographic Information System (OBIS),1111 Paleobiology Database1212), literature repositories (e.g., Dryad, Ecological Archives, PANGAEA), natural history collections, and citizen science projects. Legacy or previously unpublished data are also represented, and some data are derived from text mining projects [28,40]. Access to the data is free and open. While some data sets are released under the Attribution Creative Commons License,1313 most TraitBank data can be used and redistributed without copyright restrictions.
In addition to traditional “trait data” like body size, flower color, and onset of fertility, TraitBank also features structured attributes like the number of sequences in GenBank, type specimen repository, and human population density within the geographic range of a taxon. TraitBank data include individual measurements (e.g., the wood density of a particular tree) as well as statistics (e.g., the mean body mass from a particular sample). In addition, there are facts derived from the literature (e.g., blue whales are known to prey on krill or dandelions have yellow flowers).
TraitBank leverages EOL’s existing network of content partners and Content Creation Community [39] and employs the EOL relational database frameworks (providing advanced taxonomic names resolution) in combination with existing data standards and domain ontologies. Rather than developing a comprehensive semantic framework for the integration of trait data, TraitBank simply links data records to relevant ontologies and controlled vocabularies. These links improve the discoverability and queriability of the data and provide interoperability with other semantic resources, but more principled inference is left to end users. This lightweight semantic approach allows for the efficient management of a large and diverse data store and ensures scalability as the system grows.
TraitBank is designed for use by a wide audience including biodiversity researchers, information and data scientists, but also teachers, students, and the public. It provides both human and machine accessible query interfaces, and trait data are displayed on EOL taxon pages making them readily accessible to the EOL user base of about 6 million unique users per year.1414
2.1.Data model
Fig. 1.
Data model and architecture for TraitBank/EOL. Elements are from Darwin Core except for the following extensions developed by EOL: Media (with Audubon Core), References (with BIBO), Associations (under development), and Agents. Only the most important properties are indicated. TraitBank elements may hold only pointers to elements managed in the EOL relational database management system (RDBMS), like taxon names and references.

To represent trait data, TraitBank uses and extends TDWG Darwin Core [43] (Fig. 1), the most widely used standard for exchange of biodiversity data. Darwin Core Archives are already the preferred method for sharing media, references, and taxonomic data with EOL. Other prominent initiatives like GBIF, OBIS, and the Atlas of Living Australia (ALA)1515 support Darwin Core, and it has gained wide acceptance in the natural history collection and citizen science communities. Adoption of this standard by an increasing number of projects will enable data providers to efficiently share their resources with multiple biodiversity information systems [2].
Each TraitBank record is associated with an Occurrence, which links to the taxon identifier. The Occurrence may also include the context in which the trait was recorded (e.g., geospatial information, dates, sex, life stage, individual count). LifeStage and Sex values are standardized whenever possible through links to terms from the Phenotypic Quality Ontology (PATO) [26] or the Uber Anatomy Ontology (UBERON) [27].
The Darwin Core MeasurementOrFact extension holds information about the trait measured and other metadata. MeasurementType describes the trait that was measured using a Uniform Resource Identifier, URI, from a domain ontology (e.g., Plant Trait Ontology [21] or Vertebrate Trait Ontology [29]). MeasurementValue holds either a number or a categorical value represented by a URI from an ontology, if possible (e.g., PATO or Environments Ontology (ENVO) [7]). Associated measurement metadata may include MeasurementUnit (mapped to the Units of Measurement Ontology, UO1616), MeasurementAccuracy and MeasurementMethod (not yet standardized), and StatisticalMethod, (e.g. mean or maximum), mostly mapped to the Semanticscience Integrated Ontology (SIO).1717 In addition to these frequently documented parameters, custom fields can be created to accommodate any metadata extracted from the source.
Interactions among species (e.g., predator-prey relationships) are handled using a new Associations Darwin Core extension which is still under development. This extension references two records in the Occurrence extension, with AssociationType indicating the type of relationship (e.g., X feeds on Y, A parasitizes B).
As with other content on EOL, provenance of TraitBank data is handled using rich attribution metadata via fields from Dublin Core1818 (e.g., bibliographicCitation, contributor, source) and Darwin Core (e.g., identifiedBy, recordedBy, measurementDeterminedBy), with structured references supported using an EOL extension based on the Bibliographic Ontology (BIBO).1919
2.2.Taxonomic semantics
Taxonomic names reconciliation is at the heart of any effort to integrate biodiversity information [33]. Since there is no comprehensive consensus classification for organism names, EOL maps each data record to names in multiple taxonomic hierarchies from several scientific providers. Synonyms, misspellings, ranks, and parent taxa are taken into account during the reconciliation process. Rather than attempt to fully capture these complex interactions semantically [15], TraitBank reflects data structures already developed to represent the multiple classifications managed in the EOL relational database [32].
Scientific names in TraitBank are designated with the Darwin Core property scientificName and are typically associated with Taxon URIs that have the rdf:type of Taxon. These in turn are associated to an EOL taxon page URL (e.g., http://eol.org/pages/328615) using the taxonConceptID predicate. These Taxon URIs associate a data point with a particular page and describe the parent/child relationships between the taxa. The parent/child relationships use the parentNameUsageID predicate.
3.Implementation
To ensure that TraitBank would meet the needs of the scientific community and to build a stakeholder base ready to use it, EOL convened workshops and an advisory panel early in the development process. Scientists who attended workshops sponsored by EOL’s Biodiversity Synthesis Center at the Field Museum over a period of four years provided high-level community requirements. A workshop in Washington, DC in September 2012 brought together more than twenty experts from biology and computer science, including semantics, to focus on the questions that could be addressed with a comprehensive, integrated trait repository and associated software and infrastructure requirements. Teleconferences with an 11-person panel of scientists and technologists drawn from the above workshops informed iterative design and development. Following the first production release of TraitBank in January 2014, further refinements to the technology were implemented on an as-needed basis, and the focus of the development team shifted to increasing the amount of content aggregated into TraitBank.
The initial data sets targeted for ingestion into TraitBank were chosen to quickly achieve broad taxonomic coverage for a number of commonly studied ecological and life history traits. In addition to iconic data sets like PanTHERIA [22], IUCN Redlist,2020 and the Global Wood Density Database [10], we looked especially for trait data that would be useful for marine biodiversity science, a focus of one of TraitBank’s sponsors. The TraitBank corpus has since grown to include more than 11 million data records sourced from over 50 data sets. They represent more than 300 attributes for over 1.7 million taxa (Table 1). Moving ahead, our strategy for data acquisition is guided by the needs of our audiences, sponsors, and partners.
Table 1
TraitBank contents as of 27 January 2015 as retrieved from http://eol.org/statistics. Trait types include both MeasurementTypes and AssociationTypes. An overview of TraitBank data sets is available at http://eol.org/collections/97700
| Data sets | 52 |
| Trait types | 331 |
| Individual data records | 11,063,667 |
| Taxa with at least one data record | 1,730,789 |
| Total triples | 218,893,457 |
3.1.Data import
Most TraitBank data are imported from other databases via PHP connectors or uploaded directly via Darwin Core Archive files.2121 A custom spreadsheet template is also available to support conversion of tabular data to a Darwin Core Archive.2222
If a data set introduces new concepts (attributes, values or metadata) to TraitBank, the new terms and their definitions must be added to the TraitBank URI registry before the data can be harvested [11]. Each attribute is mapped to broad subject categories (Distribution, Physical Description, Ecology, Life History and Behavior, Evolution and Systematics, Physiology and Cell Biology, Molecular Biology and Genetics, Conservation, Relevance to Humans and Ecosystems, Notes, Names and Taxonomy, Database and Repository Coverage), and basic semantic relationships are entered into the system (see below). Attributes are also ranked based on their putative audience appeal, so that attributes of greater interest to EOL audiences can be displayed more prominently in the EOL interface (see below).
3.2.Semantic annotation
Table 2
Some of the frequently referenced ontologies in TraitBank
| Subject Areas | Ontology | Example terms |
| Statistics | Semanticscience Integrated Ontology (SIO) [13] | mean, minimal value, standard deviation |
| Units of measure | Units of Measurement Ontology (UO) [16] | meter, years, degree Celsius |
| Habitat information | Environments Ontology (EnvO) [7] | wetland, desert, snow field |
| Attributes of organisms | Phenotype Quality Ontology (PATO) [26] | aerobic, conical, evergreen |
| Plant attributes | Plant Trait Ontology (TO) [21] | flower color, life cycle habit, salt tolerance |
| Animal attributes | Vertebrate Trait Ontology (VT) [29] | body mass, total life span, onset of fertility |
| Animal natural history | Animal Natural History and Life History Ontology (ETHAN) [31] | nocturnal, oviparous, scavenger |
If a provider supplies semantic annotations with their data, these mappings are preserved in TraitBank. However, only three TraitBank data partners, Environments-EOL [28], Global Biotic Interaction [36], and Polytraits [14] fall into this category. Most of the resources we aggregate are not “born semantic,” i.e., the data come to us with labels, some metadata, and sometimes an associated article explaining the rationale and methods of the study. In these cases, EOL staff analyze the meaning of each attribute and select formally-defined semantic terms to represent them. Terms from ontologies under active development by engaged communities are preferred. These include Open Biological and Biomedical Ontologies (OBO) Foundry ontologies such as Molecular Function (GO),2323 Plant Ontology (PO),2424 Phenotypic Quality (PATO)2525 and Chemical Entities of Biological Interest (CHEBI),2626 as well as OBO Foundry candidate ontologies such as Environment Ontology (ENVO),2727 Plant Trait Ontology (TO),2828 Uber Anatomy Ontology (UBERON)2929 and Ontology of Biological Attributes (OBA)3030 (Table 2).
Not all concepts encountered in TraitBank data sets can be matched to terms in current ontologies or controlled vocabularies. Especially in the life history and ecology domains ontology coverage is still sparse. EOL staff therefore regularly propose new terms for adoption into ontologies like PATO and CHEBI, and we are involved in efforts to extend the Relations Ontology (RO),3131 Population and Community Ontology (PCO),3232 and Biological Collections Ontology (BCO)3333 to improve coverage of the different dimensions of biotic interactions.
Many traits are highly complex and require referencing of more than one class, potentially from multiple ontologies. Some new terms are therefore created through Term Genie,3434 a post-composition tool that formally constructs composite attributes by combining classes from PATO and GO or UBERON. For example, secondary xylem volumetric density3535 (i.e., wood density) and cell shape3636 are attributes from TraitBank data sets that have been added to OBA.
The goal is for new TraitBank terms to become part of the most relevant ontologies so that they can be managed by domain experts and readily discovered by users and semantic web developers. Since adding new terms to ontologies can often take a considerable amount of time, EOL creates provisional URIs while term requests are under review.
TraitBank terms, their definitions, and URIs are listed in the TraitBank Data Glossary3737 which is populated automatically from the TraitBank URI registry. The entry for each attribute features a quick link to a data search for all relevant TraitBank records (see below). The URIs of EOL provisional terms resolve to relevant entries in this Data Glossary. As domain ontologies increase their coverage, fewer terms and definitions will have to be maintained in the TraitBank Data Glossary.
For terms imported from ontologies and controlled vocabularies, the Data Glossary entry can serve as a backup when the original resource is moved or temporarily unavailable. If the definition of a term changes in the source ontology, the Data Glossary entry also serves as a record of the definition implied in the TraitBank annotation. Links to individual glossary entries can be generated based on URIs (e.g., the OBA URI for cell shape is http://purl.obolibrary.org/obo/OBA_0000052, but the definition of this term can also be accessed in the EOL Data Glossary via this URL: http://eol.org/data_glossary#http___purl_obolibrary_org_obo_OBA_0000052).
3.3.Reasoning
Because of the complexity of semantic reasoning and the challenges of reasoning across highly heterogeneous or web-scale data sets [34,41] the availability of semantic reasoning capabilities was limited in the first release of TraitBank, with the goal to add additional reasoning later as the system matures and as demand requires. However, conversion relationships of units (e.g., from g to kg), logarithmic transformations, and some equivalent and inverse relationships (e.g., preysUpon and hasPredator) are already implemented. Eventually, reasoning can be expanded to infer values based on phylogeny, or to leverage semantic similarity for searches. As the corpus of data in TraitBank grows the value of this work will increase, and it is therefore a priority for the next phase of development.
3.4.Data quality
The quality of the data represented in TraitBank is highly variable. Early in the planning process, we made the decision to not only aggregate tightly curated data but to also recruit data in need of review (e.g., data from citizen science and text mining projects) and data of questionable provenance (e.g., summary statistics without original sources). Such provisional data can make important contributions to the biodiversity knowledge base in cases where no data from scientific studies are available, where such data cannot be shared and reused freely, or where the expert curated data are of limited scope. Feedback from stakeholders has since confirmed that, at least for some applications, provisional data are better than no data at all.
Data quality concerns may also extend to the accuracy of the semantic annotations in TraitBank. Most of these links are created by trained biologists, but not necessarily by domain experts. Also, when data sources provide only vague descriptions of attributes, values, and metadata there will be some conjecture involved in the selection of the appropriate semantic context.
Finally, taxonomic name reconciliation relies on algorithms that may yield suboptimal results if there are unresolved homonyms, unrecognized synonym relationships, contradictory taxonomic data from different providers or undocumented lexical variants of taxon or author names. As a result data records may sometimes not be associated with the most appropriate EOL taxon page.
TraitBank users in need of high quality data are advised to thoroughly check data sources, semantic annotations, and taxon mappings before employing the data in scientific analyses. The metadata needed to perform these assessments are provided alongside TraitBank records in all data delivery interfaces (see below).
3.5.Data search, download, and API
TraitBank data can be queried and downloaded through the EOL data search interface3838 which is accessible through numerous links on EOL web pages. A JSON-LD service is provided for machine access to the data, and relevant records are displayed on taxon pages throughout the EOL web site.
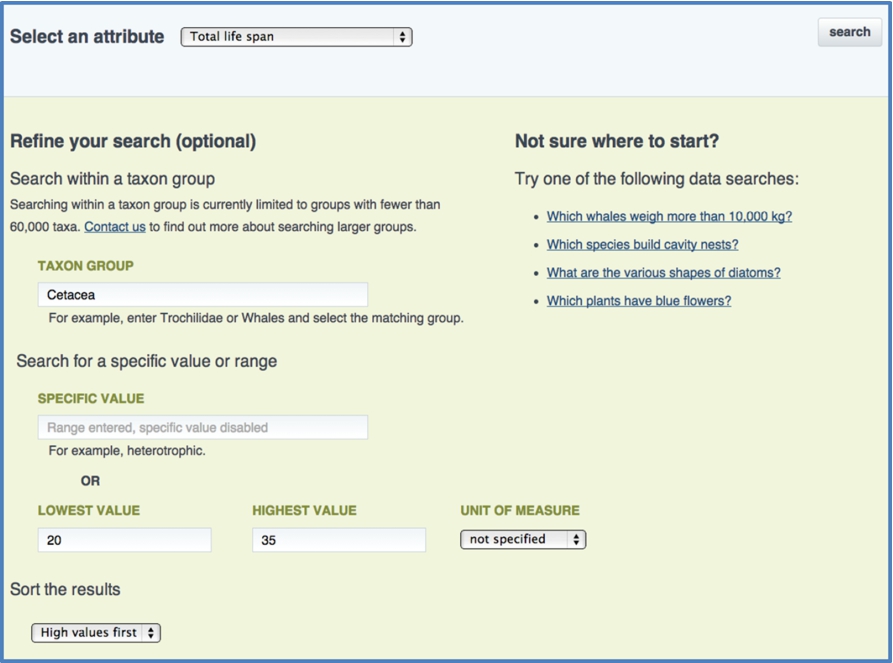
The EOL data search (Fig. 2) supports queries based on individual attributes. A generic search returns all TraitBank records for a given attribute like tail length or plant growth habit. Searches can be refined by specifying a value or range of values, and they can be restricted to a particular taxonomic group. Filtering by group currently relies on parent/child relationships in the National Center for Biotechnology Information (NCBI)3939 and Catalogue of Life [38] classifications, so only records for taxa that are featured in one or both of these hierarchies are returned for taxon-restriced queries.
Search results can be explored in the EOL interface, or they can be downloaded as a CSV (comma-separated values) file. The CSV format is easily parsed and can be imported into common spreadsheet applications for manual or semi-automatic processing. The downloaded file features comprehensive information about each data record. It includes the unique EOL identifier for the associated taxon along with its scientific name and a common name if available. Each data row specifies the attribute label (e.g., egg size or leaf shape), the value (e.g., 38.5 or acicular), and units (e.g., mg or km) when appropriate. Most unit types are automatically normalized into comparable values. However, the raw value and units are also provided. In addition to attribute and value labels, all relevant URIs are provided. The metadata include the data provenance and context information such as life stage or geographical location.
To support data-driven web-applications, a JSON-LD application programming interface (API),4040 is available [25]. Based on EOL page identifiers (which are accessible through the EOL Search API4141) this service returns all TraitBank records for a given taxon; e.g., a URL of the form http://eol.org/api/traits/328067 will return all data for the kinkajou, Potus flavus, which has EOL page id #328067.
3.6.TraitBank data on EOL taxon pages
Fig. 2.
The EOL data search interface for TraitBank, accessible at http://eol.org/data_search.
TraitBank data are also displayed prominently on EOL taxon pages where they enrich the experience of millions of visitors each year. On many pages, these data fill important gaps by providing information that is not yet available in narrative form. Ubiquitous links to term definitions and data searches also encourage users to explore biodiversity data and give students and teachers easy access to sample data sets for instruction and projects.
The Overview tab, which is the information center of each EOL taxon page, features a sample of relevant data records. By default, these records are selected automatically based on global, dynamic attribute rankings. The principal criterion for these rankings is the relative level of interest expected in a general audience. For example, attributes like flower color or habitat are presumed to be of greater interest than things like outer ear length or germinative response to heat stimuli.
Fig. 3.
Part of a data tab of an EOL taxon page. Wood density is expanded to show rich metadata. Users can select info buttons (? icons) to access definitions of terms, URIs, and links to the glossary and data search interface.
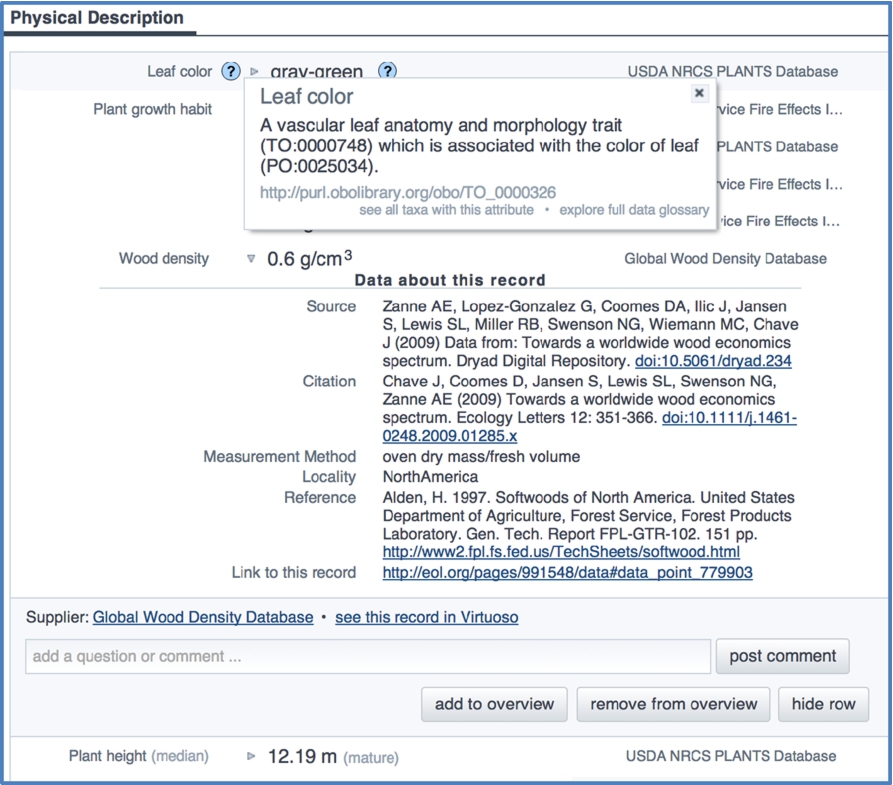
A comprehensive presentation of TraitBank data is provided in the Data tab of EOL taxon pages. The default view of this tab shows a simple list of attribute labels, values, and data providers, ordered by subject (Distribution, Physical Description, Ecology, etc.). A dynamic user interface (Fig. 3) gives access to the metadata for each record as well as URIs and definitions for attributes and categorical data values. Access to curation and commenting tools (see below), the data glossary, and data search interface are also provided.
Most TraitBank data are at the level of species or subspecies. For select physical, ecological, and life history attributes, the EOL Data tabs for higher taxa (genera, families, etc.) also feature summaries of the data represented among the taxonomic children of the group. Maximum and minimum values are displayed along with record and taxa counts and a quick link to a data search that yields relevant records.
3.7.Data curation
Any registered EOL member can review TraitBank content and report problems by adding comments to individual data records. EOL Curators – individuals with validated professional credentials – have the power to remove incorrect or suspect TraitBank records from public view. Flagged records remain visible to other curators and can be restored if flagged in error. Currently, TraitBank data providers do not receive notifications of comments and curator actions, but this feature will soon be available on an opt-in basis. This will allow data providers to benefit from the quality control activities of the EOL community.
EOL curators also participate in the selection of data for the Overview tabs of individual taxon pages. This activity is particularly important to ensure that the most interesting and informative records are highlighted for taxa of interest to a wide audience.
3.8.Architecture and technology
TraitBank is built on the RDF triple store integrated into the open source edition of the OpenLink Virtuoso Universal Server.4242 This datastore is accessed by EOL’s application servers and backend data harvesting engine [32]. Virtuoso was selected over other candidate technologies such as Neo4j4343 because using an RDF triple store made it easier to import and blend standard URI-based ontologies, URIs provided by content partners, and when necessary newly minted EOL URIs. The SPARQL4444 query language works well to efficiently query complex chains of relationships including recursive queries needed for traversing taxonomic hierarchies.
All code is available under an MIT open source license and is published to the EOL project on GitHub.4545
4.Evaluation and conclusions
The amount of available biodiversity information has transcended our ability to process and analyze it. TraitBank addresses this impediment with an efficient, pragmatic approach to trait data integration that bridges taxon-specific and technology-specific systems. By organizing distributed knowledge from diverse sources into a lightweight, scalable framework, we facilitate its retrieval and reuse for a variety of applications, ranging from large-scale synthetic analyses of biodiversity to linked data products like the Knowledge Graph4646 and hands-on data science in the classroom.
4.1.Feedback from stakeholders
TraitBank was released in January 2014 after private (September 2013) and public (October 2013) beta test releases, with each test followed by a survey. Informal demonstrations to communities at several conferences have also been used to gather feedback. Some of the most valuable insights about the needs of TraitBank users were gained during the EOL-NESCent-BHL research sprint [30]. This event, scheduled only a week after TraitBank’s public launch, brought together a diverse group of biologists and informaticians to tackle large-scale ecological and evolutionary questions with the aid of resources provided by EOL and the Biodiversity Heritage Library (BHL).4747 During the four-day meeting, members of the TraitBank team had the opportunity to interact with users while they explored the TraitBank corpus and used it to assemble their own data sets.
Based on user feedback and observations of user behavior, new features were added to TraitBank (e.g., JSON-LD access on a taxon by taxon basis) and the data search and download functions have been revised. In addition, new data sets were imported to TraitBank in response to specific user requests.
Several improvements suggested by users are still in the planning stages. These include support for more complex data queries, with multiple facets across traits, metadata, values, and taxa, improved presentation of results including visualizations, an R-interface for access to TraitBank data, and better performance of searches filtered by taxonomic group. Also, TraitBank’s geographic keyword vocabulary is not yet standardized. Most locations are currently stored as text strings, preventing reasoning on geographic distribution data. These records need to be mapped to gazetteers like GAZ,4848 Geonames4949 and MarineRegions.org.
4.2.Implications for interoperability
TraitBank fosters semantic interoperability both within and across domains by using URIs from ontologies that are also used in other systems. As the use of semantic technologies is already prevalent in genomics, morphology, ecology, and developmental biology communities, it makes sense to link newly exposed and annotated biodiversity trait information to these efforts. On the other hand, where existing ontologies do not yet capture knowledge adequately (e.g., missing terms, missing relations, missing definitions, complex taxonomic and nomenclatural semantics), our approach still allows progress in knowledge management and sharing in the most practical sense, even if not all elements of the system are interoperable.
Recent efforts to automate the description and measurement of organisms [3,6,23] accelerate the pace of data generation. While semantic annotation and open access publishing are likely to become an integral part of modern scientific workflows, standardization across data sets and domains remains in its infancy [12]. We expect that the semantic annotation of TraitBank resources will long remain a work in progress. The rapid growth and diversification of the corpus of data frequently requires the exploration of new subject areas. Even the annotation of existing data sets is often an iterative process as best practices develop in response to evolving needs for integration, new ontology resources, and feedback from domain and knowledge representation experts.
4.3.Impact on semantic community, data providers and research community
TraitBank is a starting point for the untangling of the vast riches compiled through centuries of biodiversity exploration. It will take time for it to mature into a comprehensive, consistent knowledge management platform that can supply highly curated, analysis-ready data products. Based on our experience so far, domain ontologies will have to become much more detailed if they are to be applied to the backlog of biodiversity data. Achieving the desired level of complexity without sacrificing interoperability will be an ongoing challenge. Because of its broad scope, TraitBank is in an ideal position to provide the stewards of many relevant domain ontologies with use cases that can help to optimize the development of their resources. We also anticipate that the prominent use of semantics in TraitBank will result in increased usage of ontologies in research applications.
TraitBank complements taxon or subject-specific trait databases by filling gaps (both in taxonomic and attribute space), by recruiting new types of data (e.g., from text-mining, citizen-science, and specimen data digitization efforts) and by integrating knowledge across the tree of life and multiple scientific domains. To promote progress in the aggregation of comprehensive data sets of particular interest to scientists and the public, EOL has funded projects like GloBi (Global Biotic Interactions) [36] and Environments-EOL [28]. For these communities and other ongoing projects like Polytraits and OBIS, TraitBank provides a live platform for distribution and re-use that exposes their data to broader audiences and promotes significant community curation. For legacy data providers, such as the authors of literature-derived data sets, TraitBank improves discoverability of data that otherwise would not be exposed to the Linked Open Data (LOD) community [5]. Once provisioned to TraitBank, data can be discovered and re-used for a wide range of use cases, from simple fact-finding to “big data” modeling studies. Through its association with the Encyclopedia of Life web site, TraitBank also brings awareness of data science and interoperability efforts to novel audiences. Some of these new data users may themselves become data providers, e.g., through participation in citizen science5050 or transcription crowdsourcing projects.5151
With TraitBank only a year old, it is somewhat premature to assess its impact on scientific research. The TraitBank data search interface has so far been accessed over 5,000 times, and more than 1,500 data packages have been downloaded. Also, papers citing TraitBank as a data source are starting to appear in the literature (e.g., [1,4,8,28,36,37,44]). Future development efforts will focus on improving TraitBank’s utility for research by improving the search interface, exposing the data in more advanced machine-readable formats, employing standardized data quality descriptors, replacing provisional EOL terms with community-managed terms, and exploring the best use of reasoning within the EOL-TraitBank framework.
Notes
14 Data from 1 October 2013 to 30 September 2014.
Acknowledgements
Support for TraitBank was provided by the Alfred P. Sloan Foundation, the Smithsonian Institution, the Marine Biological Laboratory, and the John D. and Catherine T. MacArthur Foundation. The production hardware infrastructure for the EOL website was supported by the Harvard Faculty of Arts and Sciences (FAS) Sciences Division Research Computing Group and the Smithsonian Institution. The TraitBank development team wishes to specifically thank Dr. Jesse Ausubel for his support and for his commitment to the entire Encyclopedia of Life initiative.
References
[1] | N.F. Angeli, J. Otegui, M. Wood and E.P. Gomez-Ruiz, A process to support species conservation planning and climate change readiness in protected areas, PeerJ PrePrints 2: ((2014) ), e492v2. doi:10.7287/peerj.preprints.492v2. |
[2] | E. Baker, S. Rycroft and V.S. Smith, Linking multiple biodiversity informatics platforms with Darwin Core archives, Biodiversity Data Journal 2: ((2014) ), e1039. doi:10.3897/BDJ.2.e1039. |
[3] | J.P. Balhoff, W.M. Dahdul, C.R. Kothari, H. Lapp, J.G. Lundberg, P. Mabee, P.E. Midford, M. Westerfield and T.J. Vision, Phenex: Ontological annotation of phenotypic diversity, PLoS ONE 5: (5) ((2010) ), 10. doi:10.1371/journal.pone.0010500. |
[4] | J.-Y. Barnagaud, J. Papaïx, O. Gimenez and J.-C. Svenning, Dynamic spatial interactions between the native invader brown-headed cowbird and its hosts, Diversity and Distributions 21: (5) ((2014) ), 511–522. doi:10.1111/ddi.12275. |
[5] | C. Bizer, T. Heath and T. Berners-Lee, Linked data – the story so far, International Journal on Semantic Web and Information Systems 5: (3) ((2009) ), 1–22. doi:10.4018/jswis.2009081901. |
[6] | J.G. Burleigh, K. Alphonse, A.J. Alverson, H.M. Bik, C. Blank, A.L. Cirranello, H. Cui, M. Daly, T.G. Dietterich, G. Gasparich, J. Irvine, M. Julius, S. Kaufman, E. Law, J. Liu, L. Moore, M.A. O’Leary, M. Passarotti, S. Ranade, N.B. Simmons, D.W. Stevenson, R.W. Thacker, C. Theriot E, S. Todorovic, P.M. Velazco, R.L. Walls, J.M. Wolfe and M. Yu, Next-generation phenomics for the Tree of Life, PLoS Currents Tree of Life (2013 Jun 26), Edition 1, 2013. doi:10.1371/currents.tol.085c713acafc8711b2ff7010a4b03733. |
[7] | P.L. Buttigieg, N. Morrison, B. Smith, C.J. Mungall, S.E. Lewis and the ENVO Consortium, The environment ontology: Contextualising biological and biomedical entities, Journal of Biomedical Semantics 4: ((2013) ), 43. doi:10.1186/2041-1480-4-43. |
[8] | I.R. Caldwell and E.M. Hart, Using encyclopedia of life’s TraitBank to identify plant traits associated with vulnerability, PeerJ PrePrints 2: ((2014) ), e491v1. doi:10.7287/peerj.preprints.491v1. |
[9] | A.D. Chapman, Numbers of Living Species in Australia and the World Report, 2nd edn, Commonwealth of Australia, Department of the Environment and Water Resources, (2009) , http://www.environment.gov.au/node/13876. |
[10] | J. Chave, D. Coomes, S. Jansen, S.L. Lewis, N.G. Swenson and A.E. Zanne, Towards a worldwide wood economics spectrum, Ecology Letters 12: ((2009) ), 351–366. doi:10.1111/j.1461-0248.2009.01285.x. |
[11] | M. Courtot, F. Gibson, A.L. Lister, J. Malone, D. Schober, R.R. Brinkman and A. Ruttenberg, MIREOT: The minimum information to reference an external ontology term, Applied Ontology 6: ((2011) ), 23–33. doi:10.1038/npre.2009.3576.1. |
[12] | A.R. Deans, S.E. Lewis, E. Huala, S.S. Anzaldo, M. Ashburner, J.P. Balhoff, D.C. Blackburn, J.A. Blake, J.G. Burleigh, B. Chanet, L.D. Cooper, M. Courtot, S. Csösz, H. Cui, W. Dahdul, S. Das, T.A. Dececchi, A. Dettai, R. Diogo, R.E. Druzinsky, M. Dumontier, N.M. Franz, F. Friedrich, G.V. Gkoutos, M. Haendel, L.J. Harmon, T.F. Hayamizu, Y. He, H.M. Hines, N. Ibrahim, L.M. Jackson, P. Jaiswal, C. James-Zorn, S. Köhler, G. Lecointre, H. Lapp, C.J. Lawrence, N. Le Novère, J.G. Lundberg, J. Macklin, A.R. Mast, P.E. Midford, I. Mikó, C.J. Mungall, A. Oellrich, D. Osumi-Sutherland, H. Parkinson, M.J. Ramírez, S. Richter, P.N. Robinson, A. Ruttenberg, K.S. Schulz, E. Segerdell, K.C. Seltmann, M.J. Sharkey, A.D. Smith, B. Smith, C.D. Specht, R.B. Squires, R.W. Thacker, A. Thessen, J. Fernandez-Triana, M. Vihinen, P.D. Vize, L. Vogt, C.E. Wall, R.L. Walls, M. Westerfeld, R.A. Wharton, C.S. Wirkner, J.B. Woolley, M.J. Yoder, A.M. Zorn and P. Mabee, Finding our way through phenotypes, PLOS Biology 3: (1) ((2015) ), e1002033. doi:10.1371/journal.pbio.1002033. |
[13] | M. Dumontier, C.J. Baker, J. Baran, A. Callahan, L. Chepelev, J. Cruz-Toledo, N.R. Del Rio, G. Duck, L.I. Furlong, N. Keath, D. Klassen, J.P. McCusker, N. Queralt-Rosinach, M. Samwald, N. Villanueva-Rosales, M.D. Wilkinson and R. Hoehndorf, The Semanticscience Integrated Ontology (SIO) for biomedical research and knowledge discovery, Journal of Biomedical Semantics 5: (1) ((2014) ), 14. doi:10.1186/2041-1480-5-14. |
[14] | S. Faulwetter, V. Markantonatou, C. Pavloudi, N. Papageorgiou, K. Keklikoglou, E. Chatzinikolaou, E. Pafilis, G. Chatzigeorgiou, K. Vasileiadou, T. Dailianis, L. Fanini, P. Koulouri, C. Arvanitidis and Polytraits, A database on biological traits of marine polychaetes, Biodiversity Data Journal 2: ((2014) ), e1024. doi:10.3897/BDJ.2.e1024. |
[15] | N.M. Franz and D. Thau, Biological taxonomy and ontology development: Scope and limitations, Biodiversity Informatics 7: ((2010) ), 45–66. |
[16] | G.V. Gkoutos, P.N. Schofield and R. Hoehndorf, The Units Ontology: A tool for integrating units of measurement in science, Database: The Journal of Biological Databases and Curation 2012: ((2012) ), bas033. doi:10.1093/database/bas033. |
[17] | A. Guisan, Biodiversity: Predictive traits to the rescue, Nature Climate Change 4: (3) ((2014) ), 175–176. doi:10.1038/nclimate2157. |
[18] | M. Harfoot and D. Roberts Taxonomy: Call for ecosystem modelling data, Nature 505: (7482) ((2014) ), 160. doi:10.1038/505160a. |
[19] | L.J. Harmon, J. Baumes, C. Hughes, J. Soberon, C.D. Specht, W. Turner, C. Lisle and R.W. Thacker, Comparative analysis workflows for the Tree of Life, PLOS Currents Tree of Life (2013 Jun 26), Edition 1, 2013. doi:10.1371/currents.tol.099161de5eabdee073fd3d21a44518dc. |
[20] | P. Heidorn, Shedding light on the dark data in the long tail of science, Library Trends 57: ((2008) ), 280–299, Institutional Repositories: Current State and Future, S. Sheeves and M. Cragin, eds, http://hdl.handle.net/2142/9127. |
[21] | P. Jaiswal, D. Ware, J. Ni, K. Chang, W. Zhao, S. Schmidt, X. Pan, K. Clark, L. Teytelman, S. Cartinhour, L. Stein and S. McCouch, Gramene: Development and integration of trait and gene ontologies for rice, Comparative and Functional Genomics 3: (2) ((2002) ), 132–136. doi:10.1002/cfg.156. |
[22] | K.E. Jones, J. Bielby, M. Cardillo, S.A. Fritz, J. O’Dell, C.D.L. Orme, K. Safi, W. Sechrest, E.H. Boakes, C. Carbone, C. Connolly, M.J. Cutts, J.K. Foster, R. Grenyer, M. Habib, C.A. Plaster, S.A. Price, E.A. Rigby, J. Rist, A. Teacher, O.R.P. Bininda-Emonds, J.L. Gittleman, G.M. Mace and A. Purvis, PanTHERIA: A species-level database of life history, ecology, and geography of extant and recently extinct mammals, Ecology 90: ((2009) ), 2648. doi:10.1890/08-1494.1. |
[23] | R.H. Kao, C.M. Gibson, R.E. Gallery, C.L. Meier, D.T. Barnett, K.M. Docherty, K.K. Blevins, P.D. Travers, E. Azuaje, Y.P. Springer, K.M. Thibault, V.J. McKenzie, M. Keller, L.F. Alves, E.-L.S. Hinckley, J. Parnell and D. Schimel, NEON terrestrial field observations: Designing continental-scale, standardized sampling, Ecosphere 3: (12) ((2012) ), 1–17. doi:10.1890/ES12-00196.1. |
[24] | J. Kattge, S. Díaz, S. Lavorel, I.C. Prentice, P. Leadley, G. Bönisch, E. Garnier, M. Westoby, P.B. Reich, I.J. Wright, J.H.C. Cornelissen, C. Violle, S.P. Harrison, P.M. Van Bodegom, M. Reichstein, B.J. Enquist, N.A. Soudzilovskaia, D.D. Ackerly, M. Anand, O. Atkin, M. Bahn, T.R. Baker, D. Baldocchi, R. Bekker, C.C. Blanco, B. Blonder, W.J. Bond, R. Bradstock, D.E. Bunker, F. Casanoves, J. Cavender-Bares, J.Q. Chambers, F.S. Chapin III, J. Chave, D. Coomes, W.K. Cornwell, J.M. Craine, B.H. Dobrin, L. Duarte, W. Durka, J. Elser, G. Esser, M. Estiarte, W.F. Fagan, J. Fang, F. Fernández-Méndez, A. Fidelis, B. Finegan, O. Flores, H. Ford, D. Frank, G.T. Freschet, N.M. Fyllas, R.V. Gallagher, W.A. Green, A.G. Gutierrez, T. Hickler, S.I. Higgins, J.G. Hodgson, A. Jalili, S. Jansen, C.A. Joly, A.J. Kerkhoff, D. Kirkup, K. Kitajima, M. Kleyer, S. Klotz, J.M.H. Knops, K. Kramer, I. Kühn, H. Kurokawa, D. Laughlin, T.D. Lee, M. Leishman, F. Lens, T. Lenz, S.L. Lewis, J. Lloyd, J. Llusià, F. Louault, S. Ma, M.D. Mahecha, P. Manning, T. Massad, B.E. Medlyn, J. Messier, A.T. Moles, S.C. Müller, K. Nadrowski, S. Naeem, Ü. Niinemets, S. Nöllert, A. Nüske, R. Ogaya, J. Oleksyn, V.G. Onipchenko, Y. Onoda, J. Ordoñez, G. Overbeck, W.A. Ozinga, S. Patiño, S. Paula, J.G. Pausas, J. Peñuelas, O.L. Phillips, V. Pillar, H. Poorter, L. Poorter, P. Poschlod, A. Prinzing, R. Proulx, A. Rammig, S. Reinsch, B. Reu, L. Sack, B. Salgado-Negret, J. Sardans, S. Shiodera, B. Shipley, A. Siefert, E. Sosinski, J.-F. Soussana, E. Swaine, N. Swenson, K. Thompson, P. Thornton, M. Waldram, E. Weiher, M. White, S. White, S.J. Wright, B. Yguel, S. Zaehle, A.E. Zanne and C. Wirth, TRY – a global database of plant traits, Global Change Biology 17: (9) ((2011) ), 2905–2935. doi:10.1111/j.1365-2486.2011.02451.x. |
[25] | M. Lanthaler and C. Gütl, On using JSON-LD to create evolvable RESTful services, in: Proc. of the 3rd International Workshop on RESTful Design WSREST 2012 at WWW2012, R. Alarcon, C. Pautasso, E. Wilde, eds, ACM Press, (2012) , pp. 25–32. doi:10.1145/2307819.2307827. |
[26] | P.M. Mabee, M. Ashburner, Q. Cronk, G.V. Gkoutos, M. Haendel, E. Segerdell, C. Mungall and M. Westerfield, Phenotype ontologies: The bridge between genomics and evolution, Trends in Ecology & Evolution 22: (7) ((2007) ), 345–350. doi:10.1016/j.tree.2007.03.013. |
[27] | C.J. Mungall, C. Torniai, G.V. Gkoutos, S.E. Lewis and M.A. Haendel, Uberon, an integrative multi-species anatomy ontology, Genome Biology 13: ((2012) ), R5. doi:10.1186/gb-2012-13-1-r5. |
[28] | E. Pafilis, S.P. Frankild, J. Schnetzer, L. Fanini, S. Faulwetter, C. Pavloudi, A. Vasileiadou, P. Leary, J. Hammock, K. Schulz, C.S. Parr, C. Arvanitidis and L.J. Jensen, ENVIRONMENTS and EOL: Identification of environment ontology terms in text and the annotation of the Encyclopedia of Life, Bioinformatics 31: (11) ((2015) ), 1872–1874. doi:10.1093/bioinformatics/btv045. |
[29] | C.A. Park, S.M. Bello, C.L. Smith, Z.-L. Hu, D.H. Munzenmaier, R. Nigam, J.R. Smith, M. Shimoyama, J.T. Eppig and J.M. Reecy, The vertebrate trait ontology: A controlled vocabulary for the annotation of trait data across species, Journal of Biomedical Semantics 4: (1) ((2013) ), 13. doi:10.1186/2041-1480-4-13. |
[30] | C.S. Parr and C.R. McClain, EOL-BHL-NESCent Research Sprint Report, PeerJ PrePrints 2: ((2014) ), e503v1. doi:10.7287/peerj.preprints.503v1. |
[31] | C.S. Parr, J. Sachs, A. Parafiynyk, T. Wang, R. Espinosa and T. Finin, ETHAN: The Evolutionary Trees and Natural History Ontology, Tech report, University of Maryland, Baltimore County, 2006, http://aisl.umbc.edu/get/softcopy/id/1025/1025.pdf. |
[32] | C.S. Parr, N. Wilson, P. Leary, K.S. Schulz, K. Lans, L. Walley, J.A. Hammock, A. Goddard, J. Rice, M. Studer, J.T.G. Holmes and R.J. Corrigan Jr., The Encyclopedia of Life v2: Providing global access to knowledge about life on Earth, Biodiversity Data Journal 2: ((2014) ), e1079. doi:10.3897/BDJ.2.e1079. |
[33] | D.J. Patterson, S. Faulwetter and A. Shipunov, Principles for a names-based cyberinfrastructure to serve all of biology, Zootaxa 1950: ((2008) ), 153–163. |
[34] | P.R.O. Payne, Chapter 1: Biomedical knowledge integration, PLoS Computational Biology 8: (12) ((2012) ), e1002826. doi:10.1371/journal.pcbi.1002826. |
[35] | H.M. Pereira, S. Ferrier, M. Walters, G.N. Geller, R.H.G. Jongman, R.J. Scholes, M.W. Bruford, N. Brummitt, S.H.M. Butchart, A.C. Cardoso, N.C. Coops, E. Dulloo, D.P. Faith, J. Freyhof, R.D. Gregory, C. Heip, R. Höft, G. Hurtt, W. Jetz, D.S. Karp, M.A. McGeoch, D. Obura, Y. Onoda, N. Pettorelli, B. Reyers, R. Sayre, J.P.W. Scharlemann, S.N. Stuart, E. Turak, M. Walpole and M. Wegmann, Essential biodiversity variables, Science 339: (6117) ((2013) ), 277–278. doi:10.1126/science.1229931. |
[36] | J.H. Poelen, J.D. Simons and C.J. Mungall, Global biotic interactions: An open infrastructure to share and analyze species-interaction datasets, Ecological Informatics 24: ((2014) ), 148–159. doi:10.1016/j.ecoinf.2014.08.005. |
[37] | E. Quintero, A.E. Thessen, P. Arias-Caballero and B. Ayala-Orozco, A statistical assessment of population trends for data deficient mexican amphibians, PeerJ 2: ((2014) ), e703. doi:10.7717/peerj.703. |
[38] | Y. Roskov, T. Kunze, T. Orrell, L. Abucay, L. Paglinawan, A. Culham, N. Bailly, P. Kirk, T. Bourgoin, G. Baillargeon, W. Decock, A. De Wever and V. Didžiulis (eds), in: Species 2000 & ITIS Catalogue of Life, 2013 Annual Checklist, Species 2000: Naturalis, Leiden, the Netherlands, 2013. Digital resource at http://www.catalogueoflife.org/annual-checklist/2013/. |
[39] | D. Rotman, K. Procita, D. Hansen, C.S. Parr and J. Preece, Supporting content curation communities: The case of the Encyclopedia of Life, Journal of the American Society for Information Science and Technology 63: (6) ((2012) ), 1–29. doi:10.1002/asi.22633. |
[40] | A.E. Thessen and C.S. Parr, Knowledge extraction and semantic annotation of text from the Encyclopedia of Life, PLoS ONE 9: (3) ((2014) ), e89550. doi:10.1371/journal.pone.0089550. |
[41] | J. Urbani, Three laws learned from web-scale reasoning, in: 2013 AAAI Fall Symposium Series, Semantics for Big Data, F. van Harmelen, J.A. Hendler, P. Hitzler, K. Janowicz and Program Cochairs, eds, (FS-13-04), (2013) , https://www.aaai.org/ocs/index.php/FSS/FSS13/paper/view/7585. |
[42] | T. Vision, The Dryad digital repository: Published evolutionary data as a part of the greater data ecosystem, Nature Precedings 713: ((2010) ), 1, http://hdl.handle.net/10101/npre.2010.4595.1. |
[43] | J. Wieczorek, D. Bloom, R. Guralnick, S. Blum, M. Döring, R. Giovanni, T. Robertsib and D. Vieglais, Darwin Core: An evolving community-developed biodiversity data standard, PLoS ONE 7: (1) ((2012) ), e29715. doi:10.1371/journal.pone.0029715. |
[44] | C. Wright and K. Seltmann, Usage patterns of blue flower color representation by Encyclopedia of Life content providers, Biodiversity Data Journal 2: ((2014) ), e1143. doi:10.3897/BDJ.2.e1143. |




