Research ArticleCardiologyGenetics Free access | 10.1172/jci.insight.128799
Investigation of a dilated cardiomyopathy–associated variant in BAG3 using genome-edited iPSC-derived cardiomyocytes
Chris McDermott-Roe,1 Wenjian Lv,1 Tania Maximova,2 Shogo Wada,1 John Bukowy,3 Maribel Marquez,3 Shuping Lai,3 Amarda Shehu,2 Ivor Benjamin,3 Aron Geurts,3 and Kiran Musunuru1
1Division of Cardiology and Cardiovascular Institute, Perelman School of Medicine, University of Pennsylvania, Philadelphia, Pennsylvania, USA.
2Department of Computer Science, George Mason University, Fairfax, Virginia, USA.
3Cardiovascular Center & Department of Physiology, Medical College of Wisconsin, Milwaukee, Wisconsin, USA.
Address correspondence to: Chris McDermott-Roe or Kiran Musunuru, Smilow Center for Translational Research, University of Pennsylvania, 3400 Civic Center Boulevard, Philadelphia, Pennsylvania 19104, USA. Phone: 215.573.4717; Email: [email protected] (CMR). Phone: 215.573.4717; Email: [email protected] (KM).
Find articles by McDermott-Roe, C. in: JCI | PubMed | Google Scholar
1Division of Cardiology and Cardiovascular Institute, Perelman School of Medicine, University of Pennsylvania, Philadelphia, Pennsylvania, USA.
2Department of Computer Science, George Mason University, Fairfax, Virginia, USA.
3Cardiovascular Center & Department of Physiology, Medical College of Wisconsin, Milwaukee, Wisconsin, USA.
Address correspondence to: Chris McDermott-Roe or Kiran Musunuru, Smilow Center for Translational Research, University of Pennsylvania, 3400 Civic Center Boulevard, Philadelphia, Pennsylvania 19104, USA. Phone: 215.573.4717; Email: [email protected] (CMR). Phone: 215.573.4717; Email: [email protected] (KM).
Find articles by Lv, W. in: JCI | PubMed | Google Scholar
1Division of Cardiology and Cardiovascular Institute, Perelman School of Medicine, University of Pennsylvania, Philadelphia, Pennsylvania, USA.
2Department of Computer Science, George Mason University, Fairfax, Virginia, USA.
3Cardiovascular Center & Department of Physiology, Medical College of Wisconsin, Milwaukee, Wisconsin, USA.
Address correspondence to: Chris McDermott-Roe or Kiran Musunuru, Smilow Center for Translational Research, University of Pennsylvania, 3400 Civic Center Boulevard, Philadelphia, Pennsylvania 19104, USA. Phone: 215.573.4717; Email: [email protected] (CMR). Phone: 215.573.4717; Email: [email protected] (KM).
Find articles by Maximova, T. in: JCI | PubMed | Google Scholar
1Division of Cardiology and Cardiovascular Institute, Perelman School of Medicine, University of Pennsylvania, Philadelphia, Pennsylvania, USA.
2Department of Computer Science, George Mason University, Fairfax, Virginia, USA.
3Cardiovascular Center & Department of Physiology, Medical College of Wisconsin, Milwaukee, Wisconsin, USA.
Address correspondence to: Chris McDermott-Roe or Kiran Musunuru, Smilow Center for Translational Research, University of Pennsylvania, 3400 Civic Center Boulevard, Philadelphia, Pennsylvania 19104, USA. Phone: 215.573.4717; Email: [email protected] (CMR). Phone: 215.573.4717; Email: [email protected] (KM).
Find articles by
Wada, S.
in:
JCI
|
PubMed
|
Google Scholar
|

1Division of Cardiology and Cardiovascular Institute, Perelman School of Medicine, University of Pennsylvania, Philadelphia, Pennsylvania, USA.
2Department of Computer Science, George Mason University, Fairfax, Virginia, USA.
3Cardiovascular Center & Department of Physiology, Medical College of Wisconsin, Milwaukee, Wisconsin, USA.
Address correspondence to: Chris McDermott-Roe or Kiran Musunuru, Smilow Center for Translational Research, University of Pennsylvania, 3400 Civic Center Boulevard, Philadelphia, Pennsylvania 19104, USA. Phone: 215.573.4717; Email: [email protected] (CMR). Phone: 215.573.4717; Email: [email protected] (KM).
Find articles by Bukowy, J. in: JCI | PubMed | Google Scholar
1Division of Cardiology and Cardiovascular Institute, Perelman School of Medicine, University of Pennsylvania, Philadelphia, Pennsylvania, USA.
2Department of Computer Science, George Mason University, Fairfax, Virginia, USA.
3Cardiovascular Center & Department of Physiology, Medical College of Wisconsin, Milwaukee, Wisconsin, USA.
Address correspondence to: Chris McDermott-Roe or Kiran Musunuru, Smilow Center for Translational Research, University of Pennsylvania, 3400 Civic Center Boulevard, Philadelphia, Pennsylvania 19104, USA. Phone: 215.573.4717; Email: [email protected] (CMR). Phone: 215.573.4717; Email: [email protected] (KM).
Find articles by Marquez, M. in: JCI | PubMed | Google Scholar
1Division of Cardiology and Cardiovascular Institute, Perelman School of Medicine, University of Pennsylvania, Philadelphia, Pennsylvania, USA.
2Department of Computer Science, George Mason University, Fairfax, Virginia, USA.
3Cardiovascular Center & Department of Physiology, Medical College of Wisconsin, Milwaukee, Wisconsin, USA.
Address correspondence to: Chris McDermott-Roe or Kiran Musunuru, Smilow Center for Translational Research, University of Pennsylvania, 3400 Civic Center Boulevard, Philadelphia, Pennsylvania 19104, USA. Phone: 215.573.4717; Email: [email protected] (CMR). Phone: 215.573.4717; Email: [email protected] (KM).
Find articles by Lai, S. in: JCI | PubMed | Google Scholar
1Division of Cardiology and Cardiovascular Institute, Perelman School of Medicine, University of Pennsylvania, Philadelphia, Pennsylvania, USA.
2Department of Computer Science, George Mason University, Fairfax, Virginia, USA.
3Cardiovascular Center & Department of Physiology, Medical College of Wisconsin, Milwaukee, Wisconsin, USA.
Address correspondence to: Chris McDermott-Roe or Kiran Musunuru, Smilow Center for Translational Research, University of Pennsylvania, 3400 Civic Center Boulevard, Philadelphia, Pennsylvania 19104, USA. Phone: 215.573.4717; Email: [email protected] (CMR). Phone: 215.573.4717; Email: [email protected] (KM).
Find articles by Shehu, A. in: JCI | PubMed | Google Scholar
1Division of Cardiology and Cardiovascular Institute, Perelman School of Medicine, University of Pennsylvania, Philadelphia, Pennsylvania, USA.
2Department of Computer Science, George Mason University, Fairfax, Virginia, USA.
3Cardiovascular Center & Department of Physiology, Medical College of Wisconsin, Milwaukee, Wisconsin, USA.
Address correspondence to: Chris McDermott-Roe or Kiran Musunuru, Smilow Center for Translational Research, University of Pennsylvania, 3400 Civic Center Boulevard, Philadelphia, Pennsylvania 19104, USA. Phone: 215.573.4717; Email: [email protected] (CMR). Phone: 215.573.4717; Email: [email protected] (KM).
Find articles by Benjamin, I. in: JCI | PubMed | Google Scholar
1Division of Cardiology and Cardiovascular Institute, Perelman School of Medicine, University of Pennsylvania, Philadelphia, Pennsylvania, USA.
2Department of Computer Science, George Mason University, Fairfax, Virginia, USA.
3Cardiovascular Center & Department of Physiology, Medical College of Wisconsin, Milwaukee, Wisconsin, USA.
Address correspondence to: Chris McDermott-Roe or Kiran Musunuru, Smilow Center for Translational Research, University of Pennsylvania, 3400 Civic Center Boulevard, Philadelphia, Pennsylvania 19104, USA. Phone: 215.573.4717; Email: [email protected] (CMR). Phone: 215.573.4717; Email: [email protected] (KM).
Find articles by Geurts, A. in: JCI | PubMed | Google Scholar
1Division of Cardiology and Cardiovascular Institute, Perelman School of Medicine, University of Pennsylvania, Philadelphia, Pennsylvania, USA.
2Department of Computer Science, George Mason University, Fairfax, Virginia, USA.
3Cardiovascular Center & Department of Physiology, Medical College of Wisconsin, Milwaukee, Wisconsin, USA.
Address correspondence to: Chris McDermott-Roe or Kiran Musunuru, Smilow Center for Translational Research, University of Pennsylvania, 3400 Civic Center Boulevard, Philadelphia, Pennsylvania 19104, USA. Phone: 215.573.4717; Email: [email protected] (CMR). Phone: 215.573.4717; Email: [email protected] (KM).
Find articles by Musunuru, K. in: JCI | PubMed | Google Scholar
Published November 14, 2019 - More info
JCI Insight. 2019;4(22):e128799. https://doi.org/10.1172/jci.insight.128799.
© 2019 American Society for Clinical Investigation
Received: March 13, 2019; Accepted: October 10, 2019
-
Abstract
Mutations in B cell lymphoma 2–associated athanogene 3 (BAG3) are recurrently associated with dilated cardiomyopathy (DCM) and muscular dystrophy. Using isogenic genome-edited human induced pluripotent stem cell–derived cardiomyocytes (iPSC-CMs), we examined how a DCM-causing BAG3 mutation (R477H), as well as complete loss of BAG3 (KO), impacts myofibrillar organization and chaperone networks. Although unchanged at baseline, fiber length and alignment declined markedly in R477H and KO iPSC-CMs following proteasome inhibition. RNA sequencing revealed extensive baseline changes in chaperone- and stress response protein–encoding genes, and protein levels of key BAG3 binding partners were perturbed. Molecular dynamics simulations of the BAG3-HSC70 complex predicted a partial disengagement by the R477H mutation. In line with this, BAG3-R477H bound less HSC70 than BAG3-WT in coimmunoprecipitation assays. Finally, myofibrillar disarray triggered by proteasome inhibition in R477H cells was mitigated by overexpression of the stress response protein heat shock factor 1 (HSF1). These studies reveal the importance of BAG3 in coordinating protein quality control subsystem usage within the cardiomyocyte and suggest that augmenting HSF1 activity might be beneficial as a means to mitigate proteostatic stress in the context of BAG3-associated DCM.
-
Introduction
Dilated cardiomyopathy (DCM), characterized by enlarged ventricular dimensions and diminished systolic and diastolic function, represents the most common form of nonischemic cardiomyopathy and is a principal cause of heart failure (1). Exome sequencing studies and clinical testing have nominated nonsense, missense, and frameshift mutations in B cell lymphoma 2–associated athanogene 3 (BAG3) as potential causes of adult-onset, sporadic, and monogenic forms of DCM (2, 3). BAG3, identified via a yeast 2-hybrid screen based on its high-affinity interaction with HSC/HSP70 (4), is an evolutionarily conserved and multifunctional protein expressed in myriad cell types (5). In cardiac and skeletal muscle, BAG3 performs a vital protein quality control function by instigating removal of mechanically damaged proteins such as filamin C from Z-discs via the chaperone-assisted selective autophagy (CASA) pathway (6) and confers structural support by anchoring F-actin to α-actinin (7).
Homozygous loss of Bag3 in mice results in severe muscle wasting and heart failure soon after birth (8). Similarly, Bag3 loss in zebrafish is coincident with cardiac dysfunction (3) and multisystem muscle failure in Drosophila (6). However, far less is known about the effect of naturally occurring mutations that occur in humans. This is crucial, since different BAG3 domains regulate distinct cellular functions (5), meaning different mutations could elicit distinct cellular defects and, hence, alter disease trajectories. Of > 250 human BAG3 mutations reported in clinical databases (e.g., ESP, ClinVar, and ExAC) as deleterious or potentially deleterious, just a handful have been phenotypically appraised (3, 9–11). Here, we employed genome-edited induced pluripotent stem cell–derived cardiomyocytes (iPSC-CMs) as a contextually accurate modality to examine the cell-autonomous effect of a BAG3 missense variant (c.1430G>A; R477H [RH]) linked to DCM (3). Our study represents the first exploration of a disease-linked BAG3 variant using engineered human iPSC-CMs. Underscoring BAG3’s crucial role in protein quality control, our data implicate the mutant allele as uncoupling the BAG3-HSC/HSP70 complex, dysregulating the chaperone system, and impairing myofiber maintenance. We also demonstrate that increasing expression of heat shock factor 1 (HSF1), a transcription factor that regulates expression of BAG3 and myriad stress-response genes, can lessen myofibrillar disarray in cardiomyocytes harboring BAG3 loss-of-function alleles.
-
Results
The RH mutation was introduced heterozygously into a healthy donor–derived iPSC line using the CRISPR-Cas9 system (Figure 1, A and B). A homozygous BAG3 KO line (BAG3-KO) was also established (Supplemental Figure 1; supplemental material available online with this article; https://doi.org/10.1172/jci.insight.128799DS1). BAG3-RH, BAG3-KO, and unedited isogenic control (BAG3-WT) lines were differentiated into cardiomyocytes (iPSC-CMs) via the Wnt/β-catenin modulation protocol (12) coupled with lactate selection (13), and purity was consistently ≥ 90% (Supplemental Figure 2). No obvious difference in BAG3 distribution was observed between BAG3-WT and BAG3-RH iPSC-CMs (Figure 1C). BAG3 promotes removal of damaged proteins from the sarcomeric Z-disc via the CASA pathway, and BAG3 loss-of-function causes myofibrillar disarray in mouse and fly (6, 8). However, based on gross examination of cardiac troponin T and α-actinin staining profiles, fiber formation and organization appeared unchanged across BAG3-WT, BAG3-RH, and BAG3-KO iPSC-CMs (Figure 1D).
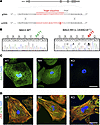 Figure 1
Figure 1Production and preliminary analysis of BAG3-R477H and BAG3-KO induced pluripotent stem cell (iPSC)-derived cardiomyocytes. (A) Schematic representation of the BAG3 gene, WT genomic DNA (gDNA) sequence, and the central part of the single-stranded oligonucleotide (ssODN) sequence used to introduce the c.1430G>A (R477H) mutation, which causes DCM in humans (3), into iPSCs. (B) Sanger sequence traces and corresponding amino acid sequences of an unedited iPSC line (BAG3-WT, left) and an iPSC line heterozygous for the c.1430G>A (BAG3-RH) mutation (right). In A and B, underlined/bolded and italicized nucleotides denote the variant of interest and synonymous Cas9-blocking mutations, respectively. (C) BAG3 localization in BAG3-WT (WT), BAG3-R477H (RH), and BAG3-KO (KO) iPSC–derived cardiomyocytes. Green, BAG3; blue, DAPI. Scale bar: 20 μm. (D) Visualization of myofibrillar organization in BAG3-WT (WT), BAG3-R477H (RH), and BAG3-KO (KO) iPSC–derived cardiomyocytes. Red, cardiac troponin T; green, α-actinin; blue, DAPI. Scale bar: 20 μm. Data are representative of 3 independent experiments.
Expression of BAG3 increased with age in mouse heart, as did the autophagy adapter protein P62 (Supplemental Figure 3), in line with increased autophagy in older cells (14). Since iPSC-CMs resemble mid- to late-embryonic cardiomyocytes (15) and favor the ubiquitin-proteasome system for proteolytic disposal, we posited that enforced reliance on autophagy would place increased stress on BAG3 and potentially lead to fiber disarray. Proteasome inhibition activates autophagy in various cell types, including cardiomyocytes (16, 17), and the 26s proteasome inhibitor MG132 caused accumulation of ubiquitinated proteins (Ub-proteins) and increased expression of BAG3 and P62 in healthy unedited iPSC-CMs (Supplemental Figure 3). MG132 also caused BAG3 to adopt a robust fiber distribution (Supplemental Figure 4). Using score-based classification and a quantitative approach, we firstly confirmed that in untreated iPSC-CMs, neither the BAG3-RH substitution nor BAG3 loss caused overt myofibrillar disorganization (Figure 2, A–D). We then analyzed fiber organization following proteasome inhibition and observed marked disarray in BAG3-RH and BAG3-KO iPSC-CMs relative to equivalently treated BAG3-WT iPSC-CMs (Figure 2, E–H). Proteasome inhibition also caused accumulation of Ub-proteins and enhanced expression of autophagy pathway proteins BAG3, P62, and LC3-II (Supplemental Figure 5). Ub-protein accumulation was most evident in MG132-treated BAG3-KO iPSC-CMs, and interestingly, induction of LC3-II by MG132 was greater in BAG3-RH iPSC-CMs and BAG3-KO iPSC-CMs than BAG3-WT iPSC-CMs (Supplemental Figure 5). Therefore, proteasome inhibition caused increased myofiber disarray in BAG3-RH and BAG3-KO iPSC-CMs and a pronounced, presumably adaptive induction of the macroautophagy pathway.
 Figure 2
Figure 2Proteasome inhibition causes myofibrillar disarray in BAG3-R477H and BAG3-KO induced pluripotent stem cell–derived cardiomyocytes (iPSC-CMs). (A) Visualization of myofibrillar organization in unedited (WT), BAG3-R477H knock-in (RH), and BAG3-KO (KO) iPSC-CMs, maintained under standard culture conditions. Red, cardiac troponin T; green, α-actinin. (B) Quantification of myofibrillar disorganization/organization in BAG3-WT, BAG3-RH, and BAG3-KO iPSC-CMs based on manual classification (n > 150 cells per genotype). (C) Quantification of myofibrillar disorganization/organization in BAG3-WT, BAG3-RH, and BAG3-KO iPSC-CMs from corresponding cells in A. Left panels show individual myofiber boundaries (identified from cardiac troponin channel) defined using an edge detection algorithm in MATLAB (see Methods) and are displayed as red or blue if aligned or unaligned, respectively, with the predicted long axis of the parent cell, indicated in the top right corner. Right panels display each myofiber from the corresponding parent cell (left panels) as a single point and are plotted according to their length (y axis) and angle relative to the predicted long axis (x axis) of the parent cell. The dashed box represents ±15°of the predicted long axis of the cell. (D) Relative myofiber alignment based on edge detection measurements in BAG3-WT, BAG3-RH, and BAG3-KO iPSC-CMs. E–H are as described in A–D, respectively, but with the addition of MG132 (25 μM, 15 hours). In A and E, scale bar: 20 μm. In D and H, each data point represents average myofiber alignment from a single cell. For each genotype, 15 randomly selected cells were analyzed. Boxplots show median and interquartile range (IQR), whiskers extend 1.5 times the IQR, and individual data points are displayed as red points. *P < 0.05 (unpaired 2-tailed t test) following Bonferroni correction for multiple comparisons. Images in A and E display representative cells from 1 experiment, and data are representative of 3 independent experiments.
RNA sequencing (RNA-Seq) revealed marked transcriptional differences between BAG3-WT, BAG3-RH, and BAG3-KO iPSC-CMs (Figure 3A). The majority of chaperone- and heat shock protein–encoding genes detected (48 of 80) were differentially expressed between BAG3-RH and BAG3-WT iPSC-CMs (P < 0.01, FDR < 0.1) (Figure 3B). In BAG3-RH vs. BAG3-WT iPSC-CMs, BAG1 — whose activity is reciprocally linked to that of BAG3 (14) — was the most highly upregulated, while BAG3 binding partner HSPB6 (18) was the most repressed. At the protein level, increased expression of HSPA8 (HSC70), a generic marker of cell stress and chief BAG3 binding partner, was evident, while HSPB6 and HSPB8 levels diminished (Figure 3C). Hence, the BAG3-RH mutation disequilibrated expression and stability of key mediators of protein quality control and the stress response.
 Figure 3
Figure 3Dysregulated expression and stability of chaperones and heat shock genes/proteins in BAG3-R477H and BAG3-KO induced pluripotent stem cell–derived cardiomyocytes (iPSC-CMs). (A) Principal components analysis of RNA-Seq gene expression profiles from unedited (WT), BAG3-R477H (RH), and BAG3-KO (KO) iPSC-CM. PC1 and PC2 denote principal components 1 and 2, respectively. (B) Relative transcript abundance of all chaperone and heat shock protein (HSP) genes in BAG3-RH vs. BAG3-WT iPSC-CMs (expressed as log2 fold change). Red and gray points denote transcripts at P < 0.01 and FDR < 0.1 calculated by quasi-likelihood F-test (QLF test) in edgeR (28) and have been adjusted for multiple testing. (C) Western blot for BAG3, key binding partners, and GAPDH (loading control) in BAG3-WT (WT), BAG3-RH (RH), and BAG3-KO (KO) iPSC-CMs.
We next tested if the RH mutation affects interaction between BAG3 and HSC70. Using a theoretically derived construct of the BAG3-HSC70 interface, we conducted molecular dynamics (MD) simulations, assessing protein backbone trajectories and amino acid side-chain interactions. Simulations predicted that the RH mutation causes a loss of H-bonding between BAG3 and HSC70 (Figure 4A), as well as a mild but obvious increase in BAG3 protein backbone flexibility (Figure 4B), which in sum could be envisaged to destabilize the complex. To explore this experimentally, coimmunoprecipitation assays were conducted on FLAG-tagged forms of BAG3-WT, BAG3-RH, and a BAG domain–deficient form of BAG3 (BAG3-BAGΔ) following stable expression in HL-1 cardiomyocytes. As expected, BAG3-WT bound robustly to HSC70, while BAG3-BAGΔ did not (Supplemental Figure 6), and supporting our in silico studies, BAG3-RH bound less (~40%) HSC70 than BAG3-WT (Figure 4, C and D).
 Figure 4
Figure 4Molecular dynamics simulations and coimmunoprecipitation assays implicate the BAG3-R477H mutation as weakening the BAG3-HSC70 interaction. (A) Ribbon representations of theoretically derived models (see Methods) of WT and R477H BAG3-HSC70 complexes generated using I-TASSER (30). The BAG domain 3-helix bundle is shown in blue, and the HSC70 ATPase domain is shown in gray. Molecular dynamics simulations predict that ARG477 (green asterisk) hydrogen bonds to GLU283 in HSC70 but HIS477 (red asterisk) does not. (B) Superimposed first normal mode motions (see Methods) of BAG3-WT (dark blue, opaque) and BAG3-R477H (light blue, transparent) BAG domain trajectories over a 20-ns molecular dynamics simulation. Ball-and-stick representation displays position of ARG477 (blue) and HIS477 (green). Note increased motion of the RH protein predictive of weaker interaction with HSC70. (C) Coimmunoprecipitation analysis of BAG3-HSC70 binding. Left panel shows Western blot detection of stably expressed FLAG-tagged BAG3-WT and BAG3-RH in HL-1 cardiomyocytes. Right panel shows coimmunoprecipitation (co-IP) analysis of how R477H substitution affects BAG3-HSC70 interaction. Each lane corresponds to a distinct sample. (D) Quantification of band densities in C. Individual data points are displayed as red points, and boxplot shows median and interquartile range; whiskers extend 1.5 times the IQR. *P < 0.05 (unpaired 2-tailed t test).
Finally, we tested if stimulating the stress response system would reduce myofibrillar disarray induced by proteasome inhibition in mutant BAG3 iPSC-CMs. The stress-inducible transcription factor HSF1 drives expression of BAG3 and various chaperones, including HSP70, and regulates proteasome function (19). In line with a protective effect, MG132-induced fiber disarray was less severe in BAG3-RH iPSC-CMs transduced with an HSF1 lentivirus than in BAG3-RH iPSC-CMs transduced with a control lentivirus (Figure 5). Hence, promoting the heat shock pathway lessens proteasome inhibition–mediated myofibrillar disarray in the context of BAG3 loss-of-function and could hold potential as a therapeutic entry point for treatment of BAG3-associated DCM.
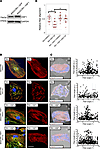 Figure 5
Figure 5Overexpression of HSF1 mitigates proteasome inhibition–induced myofibrillar disarray in induced pluripotent stem cell–derived cardiomyocytes (iPCS-CMs) with the BAG3-R477H (RH) mutation. (A) Western blot for HSF1 and GAPDH (loading control) in BAG3-RH iPCS-CMs transduced with HSF1 and GFP lentivirus. (B) Visualization of myofibrillar organization in untransduced and HSF1-transduced BAG3-RH iPSC-CMs following treatment with MG132 (25 μM, 15 hours) or DMSO. Red, cardiac troponin T; green, α-actinin; blue, DAPI. Images display representative cells. Right panels show cardiac troponin channel only for clarity. Scale bar: 20 μm. (C) Quantification of myofibrillar disorganization/disorganization in cells in B. Left panels show individual myofiber boundaries (identified from cardiac troponin channel) defined using an edge detection algorithm in MATLAB (see Methods) and are displayed as red or blue if aligned or unaligned, respectively, with the predicted long axis of the parent cell, indicated in the top right corner. Right panels display each myofiber from the corresponding parent cell (left panels) as a single point and are plotted according to their length (y axis) and angle relative to the predicted long axis (x axis) of the parent cell. The dashed box represents ±15° of the predicted long axis of the cell. (D) Relative myofiber alignment based on edge detection measurements in untransduced BAG3-RH iPSC-CMs and BAG3-RH iPSC-CMs overexpressing HSF1 following addition of MG132 (25 μM, 15 hours) or DMSO. Each data point represents average myofiber alignment from a single cell. For each condition, 17 randomly selected cells per analyzed. Boxplots show median and interquartile range (IQR); whiskers extend 1.5 times the IQR. *P < 0.05 (unpaired 2-tailed t test) following Bonferroni correction for multiple comparisons. Data is representative of 3 independent experiments.
-
Discussion
BAG3 plays a critical role in maintaining myofibrillar organization, and loss causes severe muscle dysfunction in several model organisms. In humans, missense variants in BAG3 recurrently associate with dilated cardiomyopathy (DCM) and muscular dystrophy. As in the case of the RH variant, symptom onset often occurs in mid-life to later life. How such mutations initiate disease remains largely unexplored. To our knowledge, this represents the first use of genome-edited iPSC-CMs to interrogate a DCM-associated BAG3 mutation. In brief, our study revealed the following: (a) Basal myofibrillar organization in iPSC-CMs is unaffected by the DCM-associated BAG3-RH variant or total loss of BAG3; (b) proteasome inhibition causes marked fiber disarray in both BAG3-RH and BAG3-KO iPSC-CMs; (c) hundreds of genes, including those with chaperoning and stress response functions, are dysregulated in BAG3-RH and BAG3-KO iPSC-CMs at baseline; (d) the ordinarily high-affinity interaction between BAG3 and HSC/HSP70 is weakened by the RH variant; and (e) stimulating the HSF1-driven stress response pathway reduces proteasome inhibition–mediated fiber disarray in BAG3-RH iPSC-CMs.
Cardiac fiber proteins experience significant mechanical stress and undergo continual replacement. Multicomponent chaperone complexes comprising BAG3, HSC70, CHIP, and other small HSPs play a key role by promoting refolding of fiber proteins, as well as targeting irreversibly damaged proteins for degradation via the proteasome and chaperone-assisted selective autophagy (CASA), which is distinct from the canonical macroautophagy pathway. In our studies, myofibrillar organization was not adversely affected by either the BAG3-RH variant or wholesale loss of BAG3 at baseline. However, in the setting of proteasome inhibition, which, as anticipated, caused accumulation of Ub-proteins and activated the autophagy pathway, fiber organization worsened in BAG3-RH and BAG3-KO iPSC-CMs. These observations indicate an adaptive effort to maintain proteostasis in response to diminished proteasome function and signify that, in such a setting, cells harboring BAG3 mutations fail to compensate sufficiently. Conversion rate of cytosol-localized LC3-I to the autophagosome-associated LC3-II moiety reflects macroautophagic flux. Relative to WT cells, proteasome inhibition–mediated LC3-II induction was greater in BAG3-RH and BAG3-KO cells. Hence, rather than impairing autophagy, the RH variant potentiated its activation in the context of proteasome stress. Interestingly, BAG3 loss-of-function has an opposing effect on autophagy in the setting of nutrient stress (9). Cell type, age, and maturity may explain these differences, especially since BAG3 has been proposed to underlie an aging-dependent switch from ubiquitin-proteasome– to autophagy-lysosome–based degradative pathways (14). Alternatively, it is possible that proteasome stress activates cardiomyocyte autophagy via a noncanonical pathway, as proposed elsewhere (20).
Key to BAG3’s role in protein quality control is its interaction with HSC/HSP70. The RH residue is in BAG3’s BAG domain, an ~80 amino acid–long 3-helix bundle motif located at the C-terminus of the protein, which mediates binding with HSC/HSP70 and subsequent assembly of multimeric chaperone complexes. Exploiting high-sequence similarity across the BAG family, as well as preexisting structural data, we modeled the BAG3-HSC70 binding interface by protein threading and examined how the RH mutation affects the interaction using MD simulations. In the context of the RH mutation, partial uncoupling of the BAG3-HSC70 complex was observed. This was supported by coimmunoprecipitation experiments in which a subtle reduction in binding between BAG3-RH and HSC70 was measured. It will be informative to determine if this model can successfully predict the uncoupling potential of other, novel BAG domain mutations suspected to cause disease.
Our findings tally with a previous study in which knockdown of Bag3 in rat neonatal cardiomyocytes had no obvious impact on myofibril length or Z-disc periodicity in the absence of mechanical stress (7). Contrary to our observations, overt myofibrillar disarray was reported in BAG3-deficient iPSC-CMs at baseline (21). This disparity could originate from the use of different iPSC lines and suggests that genetic heterogeneity — within and beyond BAG3 — modifies phenotypic severity. Interestingly, a recent study identified a panel of BAG3 variants enriched in African American DCM patients, which — while not disease causing — were associated with worse outcomes, such as increased hospitalizations and heart transplant (22).
Case-by-case genome editing in iPSCs is laborious, and a limitation of our study is that a single RH line and a single KO line were generated. We recently employed a bacterial recombinase system (23) to introduce a panel of variants of uncertain significance (VUS) into the TNNT2 gene (24). Such an approach would be well suited to appraise the growing list of VUS in BAG3.
In conclusion, missense variants in BAG3’s BAG domain are recurrently encountered in DCM patients. In this study, we aimed to elucidate molecular-level consequences of the RH substitution. Our data imply a pathologic mechanism in which BAG3-RH improperly engages HSC/HSP70. We propose that this impairs formation of larger multimeric chaperone complexes required for essential protein quality control duties, including but likely not limited to myofibrillar maintenance. The observation that fiber disorganization was only apparent when cells were forced to use autophagy — regarded as a key degradative mechanism for damaged proteins in older cells — suggests that BAG3 variant expressivity is influenced by age-related dynamics in protein quality control subsystem usage and provides a potential explanation for the delayed onset of BAG3-associated DCM and heart failure observed in many patients. The severity of fiber disarray was mitigated by overexpression of HSF1. This could be because HSF1 stimulates the stress response program, increasing expression of chaperones and cochaperones; bolstering the holdase and foldase functions, which appear to be lost when BAG3 is mutated and ultimately reducing the proteastatic deficit associated with BAG3 loss of function.
-
Methods
Genome editing. Genome editing was conducted as previously described (25) using an sgRNA targeting the GGGACGAGCCGATGTGCGTC sequence, a presynthesized (IDT) single-stranded oligodeoxynucleotide (5′-CCTGATGATCGAAGAGTATTTGACCAAAGAGCTGCTGGCCCTGGATTCAGTGGACCCCGAGGGGCGGGCAGACGTCCATCAGGCCAGGAGAGACGGTGTCAGGAAGGTTCAGACCATCTTGGAAAAACTTGAACAGAAAG-3′, variant underlined and italicized), and AAVS1 targeting reagents. BAG3-KO cells were established by transfecting pX330 expressing an sgRNA targeting the GGGACGAGCCGATGTGCGTC sequence. Transfected cells were maintained in puromycin-containing media for 1 week. Distinct colonies were expanded, PCR-amplified, and genotyped via Sanger sequencing. Potential off-target loci, identified using ZiFiT Targeter Version 4.2 (26), were also sequenced. Isolated knock-in iPSC lines were expanded for pluripotency immunocytochemistry, karyotyping, and cryopreservation.
Edited iPSCs were differentiated into cardiomyocytes using the Palacek lab’s Wnt/β-catenin modulation protocol (12) with metabolic selection (13). Typically, beating sheets were evident after 10 days, and cardiac troponin T positivity as measured by flow cytometry was typically ~90%. Phenotypic characterization studies were conducted at day 30–40.
Lentivirus-mediated overexpression of HSF1. HSF1 CDS was amplified from iPSC-CM cDNA and cloned into pLenti-Puro. Lentivirus was generated in HEK293-Lx cells using pMD2.G and psPAX2 plasmids. For transduction, 5–10 μL concentrated lentivirus was added to cardiomyocytes grown in 12-well plates for protein studies or on coverslips in 24-well plates for immunocytochemistry studies. HSF1 protein expression was confirmed by Western blotting and immunofluorescence microscopy.
Immunocytochemistry studies. iPSC-CMs were stained, as appropriate, with antibodies against BAG3 (Proteintech, 10599-1-AP), cardiac troponin T (Abcam, ab45932), and α-actinin (MilliporeSigma, A7811) overnight at 4°C. Cells were then incubated with Alexa Fluor 555–conjugated goat anti-rabbit (Thermo Fisher Scientific, A27039), Alexa Fluor 488–conjugated goat anti-mouse (Thermo Fisher Scientific, A-11001), Alexa Fluor 647–conjugated goat anti-chicken (Thermo Fisher Scientific, A-21449) secondary antibodies as appropriate for 1 hour at room temperature. Cells were mounted (Abcam) and viewed at ×100 magnification (oil immersion) using a Nikon A1+ confocal microscope. Myofibrillar organization was assessed before and after treatment with MG132 (25 μM for 15 hours). For qualitative analyses, cells were stained with cardiac troponin T and α-actinin antibodies, and ×20 magnification images (~20 field of view and ~200 cells per genotype) were acquired with a Nikon Eclipse 80i fluorescence microscope for user-based classification. For quantitative analysis, thin filament fibers were defined by edge detection (MATLAB, MathWorks). Area and angle relative to cell long axis was determined for each fiber object. For presentation purposes, each fiber object is plotted based on its length and angle relative to the predefined long axis of the cell.
RNA-Seq. Poly-A mRNA was prepared from day-30 iPSC-CM cultures (3 independent wells per genotype) with RNeasy columns (Qiagen), followed by oligo-dT selection. Sample quality was assessed using the Agilent Bioanalyzer 2100. Library preparation was conducted with TruSeq RNA Library Prep Kit v2 chemistry, and sequencing was carried out on an Illumina HiSeq 2500. FASTQ reads were aligned to human genome (hg19) with STAR 2.5.2a (27). Transcript counts per million files were normalized by the Cox-Reid method and compared via PCA (FactoMineR) and quasi-likelihood F-tests in edgeR (28). Data can be accessed at Gene Expression Omnibus (accession number GSE138633)
BAG3 homology modeling. The BAG1/HSC70 structure (PDB 1HX1; ref. 29) served as a template for homology modeling using I-TASSER (30). The RH mutant was generated by substituting Arginine at position 477 for histidine. Side-chain conformations were predicted using SCWRL4, and tertiary structure was refined with ENCoM Normal Mode Analysis. MD simulations of the WT and Arg447His mutant (20-ns long) were performed using the AMBER package (31) with the ff14sb force field. Both models were simulated without additional restraints.
Coimmunoprecipitation studies. Protein lysates from HL-1 cells (MilliporeSigma, SCC065) expressing FLAG-tagged WT, RH, and BAG3-BAGΔ were incubated with Protein A–conjugated Dynabeads (Thermo Fisher Scientific) precomplexed with anti-FLAG antibody (Cell Signaling Technology, 14793S) overnight at 4°C. Coimmunoprecipitates were subjected to SDS-PAGE and Western blotting as standard. Blots were probed with a BAG3 antibody (Abcam, ab47124) to ensure capture and HSC70 (Cell Signaling Technology, 8444S) to assess BAG3-HSC70 binding.
Statistics. The MATLAB code used for analysis of myofibers is available upon request. For all statistical tests other than those involving RNA-Seq data (see above), P values were calculated with an unpaired 2-tailed Student’s t test with Bonferroni correction for multiple comparisons and were considered significant when P < 0.05.
Study approval. All mouse experiments were performed according to procedures approved by the University of Pennsylvania IACUC.
-
Author contributions
CMR designed the study. CMR, WL, MM, SW, and SL conducted the experiments. CMR, TM, JB, MM, and AS acquired data. CMR wrote the manuscript. AG, IB, and KM funded the study.
-
Acknowledgments
This study was partially funded by the Steven Cullen Healthy Heart Award (CMR and AMG), made possible by the Research and Education Program Fund, a component of the Advancing a Healthier Wisconsin endowment at the Medical College of Wisconsin.
Address correspondence to: Chris McDermott-Roe or Kiran Musunuru, Smilow Center for Translational Research, University of Pennsylvania, 3400 Civic Center Boulevard, Philadelphia, Pennsylvania 19104, USA. Phone: 215.573.4717; Email: [email protected] (CMR). Phone: 215.573.4717; Email: [email protected] (KM).
-
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Copyright: © 2019, American Society for Clinical Investigation.
Reference information: JCI Insight. 2019;4(22):e128799.https://doi.org/10.1172/jci.insight.128799.
-
References
- McNally EM, Golbus JR, Puckelwartz MJ. Genetic mutations and mechanisms in dilated cardiomyopathy. J Clin Invest. 2013;123(1):19–26.
- Villard E, et al. A genome-wide association study identifies two loci associated with heart failure due to dilated cardiomyopathy. Eur Heart J. 2011;32(9):1065–1076.View this article via: CrossRef Google Scholar
- Norton N, et al. Genome-wide studies of copy number variation and exome sequencing identify rare variants in BAG3 as a cause of dilated cardiomyopathy. Am J Hum Genet. 2011;88(3):273–282.View this article via: CrossRef Google Scholar
- Takayama S, Xie Z, Reed JC. An evolutionarily conserved family of Hsp70/Hsc70 molecular chaperone regulators. J Biol Chem. 1999;274(2):781–786.
- Rosati A, Graziano V, De Laurenzi V, Pascale M, Turco MC. BAG3: a multifaceted protein that regulates major cell pathways. Cell Death Dis. 2011;2:e141. View this article via: CrossRef Google Scholar
- Arndt V, et al. Chaperone-assisted selective autophagy is essential for muscle maintenance. Curr Biol. 2010;20(2):143–148.View this article via: CrossRef Google Scholar
- Hishiya A, Kitazawa T, Takayama S. BAG3 and Hsc70 interact with actin capping protein CapZ to maintain myofibrillar integrity under mechanical stress. Circ Res. 2010;107(10):1220–1231.View this article via: CrossRef Google Scholar
- Homma S, Iwasaki M, Shelton GD, Engvall E, Reed JC, Takayama S. BAG3 deficiency results in fulminant myopathy and early lethality. Am J Pathol. 2006;169(3):761–773.View this article via: CrossRef Google Scholar
- Fang X, et al. Loss-of-function mutations in co-chaperone BAG3 destabilize small HSPs and cause cardiomyopathy. J Clin Invest. 2017;127(8):3189–3200.
- Arimura T, Ishikawa T, Nunoda S, Kawai S, Kimura A. Dilated cardiomyopathy-associated BAG3 mutations impair Z-disc assembly and enhance sensitivity to apoptosis in cardiomyocytes. Hum Mutat. 2011;32(12):1481–1491.
- Myers VD, et al. Association of Variants in BAG3 With Cardiomyopathy Outcomes in African American Individuals. JAMA Cardiol. 2018;3(10):929–938.
- Lian X, et al. Directed cardiomyocyte differentiation from human pluripotent stem cells by modulating Wnt/β-catenin signaling under fully defined conditions. Nat Protoc. 2013;8(1):162–175.
- Tohyama S, et al. Distinct metabolic flow enables large-scale purification of mouse and human pluripotent stem cell-derived cardiomyocytes. Cell Stem Cell. 2013;12(1):127–137.View this article via: CrossRef Google Scholar
- Gamerdinger M, Hajieva P, Kaya AM, Wolfrum U, Hartl FU, Behl C. Protein quality control during aging involves recruitment of the macroautophagy pathway by BAG3. EMBO J. 2009;28(7):889–901.View this article via: CrossRef Google Scholar
- Uosaki H, et al. Transcriptional Landscape of Cardiomyocyte Maturation. Cell Rep. 2015;13(8):1705–1716.View this article via: CrossRef Google Scholar
- Ding WX, et al. Linking of autophagy to ubiquitin-proteasome system is important for the regulation of endoplasmic reticulum stress and cell viability. Am J Pathol. 2007;171(2):513–524.View this article via: CrossRef Google Scholar
- Zhu K, Dunner K, McConkey DJ. Proteasome inhibitors activate autophagy as a cytoprotective response in human prostate cancer cells. Oncogene. 2010;29(3):451–462.View this article via: CrossRef Google Scholar
- Chen Y, et al. Bcl2-associated athanogene 3 interactome analysis reveals a new role in modulating proteasome activity. Mol Cell Proteomics. 2013;12(10):2804–2819.
- Willis MS, Patterson C. Hold me tight: Role of the heat shock protein family of chaperones in cardiac disease. Circulation. 2010;122(17):1740–1751.View this article via: CrossRef Google Scholar
- Liu BQ, et al. BAG3-dependent noncanonical autophagy induced by proteasome inhibition in HepG2 cells. Autophagy. 2013;9(6):905–916.
- Judge LM, et al. A BAG3 chaperone complex maintains cardiomyocyte function during proteotoxic stress. JCI Insight. 2017;2(14):94623.
- Myers VD, et al. Association of Variants in BAG3 With Cardiomyopathy Outcomes in African American Individuals. JAMA Cardiol. 2018;3(10):929–938.
- Zhu F, et al. DICE, an efficient system for iterative genomic editing in human pluripotent stem cells. Nucleic Acids Res. 2014;42(5):e34. View this article via: CrossRef Google Scholar
- Lv W, et al. Functional Annotation of TNNT2 Variants of Uncertain Significance With Genome-Edited Cardiomyocytes. Circulation. 2018;138(24):2852–2854.
- Mitzelfelt KA, et al. Efficient Precision Genome Editing in iPSCs via Genetic Co-targeting with Selection. Stem Cell Reports. 2017;8(3):491–499.View this article via: CrossRef Google Scholar
- Sander JD, Maeder ML, Reyon D, Voytas DF, Joung JK, Dobbs D. ZiFiT (Zinc Finger Targeter): an updated zinc finger engineering tool. Nucleic Acids Res. 2010;38(Web Server issue):W462–W468.View this article via: PubMed Google Scholar
- Dobin A, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29(1):15–21.View this article via: CrossRef Google Scholar
- Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26(1):139–140.
- Sondermann H, Scheufler C, Schneider C, Hohfeld J, Hartl FU, Moarefi I. Structure of a Bag/Hsc70 complex: convergent functional evolution of Hsp70 nucleotide exchange factors. Science. 2001;291(5508):1553–1557.View this article via: CrossRef Google Scholar
- Yang J, Yan R, Roy A, Xu D, Poisson J, Zhang Y. The I-TASSER Suite: protein structure and function prediction. Nat Methods. 2015;12(1):7–8.
- Maier JA, Martinez C, Kasavajhala K, Wickstrom L, Hauser KE, Simmerling C. ff14SB: Improving the Accuracy of Protein Side Chain and Backbone Parameters from ff99SB. J Chem Theory Comput. 2015;11(8):3696–3713.
-
Version history
- Version 1 (November 14, 2019): Electronic publication








