Advertisement
Research ArticleEndocrinology Free access | 10.1172/JCI91761
Age-dependent human β cell proliferation induced by glucagon-like peptide 1 and calcineurin signaling
Chunhua Dai,1 Yan Hang,2 Alena Shostak,1 Greg Poffenberger,1 Nathaniel Hart,1 Nripesh Prasad,3 Neil Phillips,1 Shawn E. Levy,3 Dale L. Greiner,4 Leonard D. Shultz,5 Rita Bottino,6 Seung K. Kim,2,7 and Alvin C. Powers1,8,9
1Division of Diabetes, Endocrinology and Metabolism, Department of Medicine, Vanderbilt University Medical Center, Nashville, Tennessee, USA.
2Department of Developmental Biology, Stanford University School of Medicine, Stanford, California, USA.
3Hudson Alpha Institute for Biotechnology, Huntsville, Alabama, USA.
4Department of Molecular Medicine, Diabetes Center of Excellence, University of Massachusetts Medical School, Worcester, Massachusetts, USA.
5The Jackson Laboratory, Bar Harbor, Maine, USA.
6Institute of Cellular Therapeutics, Allegheny-Singer Research Institute, Allegheny Health Network, Pittsburgh, Pennsylvania, USA.
7Department of Medicine, Stanford University School of Medicine, Stanford California, USA.
8Department of Molecular Physiology and Biophysics, Vanderbilt University Medical Center, Nashville, Tennessee, USA.
9VA Tennessee Valley Healthcare System, Nashville, Tennessee, USA.
Address correspondence to: Alvin C. Powers, Departments of Medicine and Molecular Physiology and Biophysics, Vanderbilt University School of Medicine, 7465 Medical Research Building IV, 2215 Garland Avenue, Nashville, Tennessee 37232-0475, USA. Phone: 615.936.7678; Email: [email protected]. Or to: Seung K. Kim, Department of Developmental Biology and Medicine, Stanford University School of Medicine, 279 Campus Drive, Beckman Center B300, Stanford, California 94305-5329, USA. Phone: 650.723.6230; Email: [email protected].
Authorship note: C. Dai and Y. Hang contributed equally to this work.
Find articles by Dai, C. in: JCI | PubMed | Google Scholar
1Division of Diabetes, Endocrinology and Metabolism, Department of Medicine, Vanderbilt University Medical Center, Nashville, Tennessee, USA.
2Department of Developmental Biology, Stanford University School of Medicine, Stanford, California, USA.
3Hudson Alpha Institute for Biotechnology, Huntsville, Alabama, USA.
4Department of Molecular Medicine, Diabetes Center of Excellence, University of Massachusetts Medical School, Worcester, Massachusetts, USA.
5The Jackson Laboratory, Bar Harbor, Maine, USA.
6Institute of Cellular Therapeutics, Allegheny-Singer Research Institute, Allegheny Health Network, Pittsburgh, Pennsylvania, USA.
7Department of Medicine, Stanford University School of Medicine, Stanford California, USA.
8Department of Molecular Physiology and Biophysics, Vanderbilt University Medical Center, Nashville, Tennessee, USA.
9VA Tennessee Valley Healthcare System, Nashville, Tennessee, USA.
Address correspondence to: Alvin C. Powers, Departments of Medicine and Molecular Physiology and Biophysics, Vanderbilt University School of Medicine, 7465 Medical Research Building IV, 2215 Garland Avenue, Nashville, Tennessee 37232-0475, USA. Phone: 615.936.7678; Email: [email protected]. Or to: Seung K. Kim, Department of Developmental Biology and Medicine, Stanford University School of Medicine, 279 Campus Drive, Beckman Center B300, Stanford, California 94305-5329, USA. Phone: 650.723.6230; Email: [email protected].
Authorship note: C. Dai and Y. Hang contributed equally to this work.
Find articles by Hang, Y. in: JCI | PubMed | Google Scholar
1Division of Diabetes, Endocrinology and Metabolism, Department of Medicine, Vanderbilt University Medical Center, Nashville, Tennessee, USA.
2Department of Developmental Biology, Stanford University School of Medicine, Stanford, California, USA.
3Hudson Alpha Institute for Biotechnology, Huntsville, Alabama, USA.
4Department of Molecular Medicine, Diabetes Center of Excellence, University of Massachusetts Medical School, Worcester, Massachusetts, USA.
5The Jackson Laboratory, Bar Harbor, Maine, USA.
6Institute of Cellular Therapeutics, Allegheny-Singer Research Institute, Allegheny Health Network, Pittsburgh, Pennsylvania, USA.
7Department of Medicine, Stanford University School of Medicine, Stanford California, USA.
8Department of Molecular Physiology and Biophysics, Vanderbilt University Medical Center, Nashville, Tennessee, USA.
9VA Tennessee Valley Healthcare System, Nashville, Tennessee, USA.
Address correspondence to: Alvin C. Powers, Departments of Medicine and Molecular Physiology and Biophysics, Vanderbilt University School of Medicine, 7465 Medical Research Building IV, 2215 Garland Avenue, Nashville, Tennessee 37232-0475, USA. Phone: 615.936.7678; Email: [email protected]. Or to: Seung K. Kim, Department of Developmental Biology and Medicine, Stanford University School of Medicine, 279 Campus Drive, Beckman Center B300, Stanford, California 94305-5329, USA. Phone: 650.723.6230; Email: [email protected].
Authorship note: C. Dai and Y. Hang contributed equally to this work.
Find articles by Shostak, A. in: JCI | PubMed | Google Scholar
1Division of Diabetes, Endocrinology and Metabolism, Department of Medicine, Vanderbilt University Medical Center, Nashville, Tennessee, USA.
2Department of Developmental Biology, Stanford University School of Medicine, Stanford, California, USA.
3Hudson Alpha Institute for Biotechnology, Huntsville, Alabama, USA.
4Department of Molecular Medicine, Diabetes Center of Excellence, University of Massachusetts Medical School, Worcester, Massachusetts, USA.
5The Jackson Laboratory, Bar Harbor, Maine, USA.
6Institute of Cellular Therapeutics, Allegheny-Singer Research Institute, Allegheny Health Network, Pittsburgh, Pennsylvania, USA.
7Department of Medicine, Stanford University School of Medicine, Stanford California, USA.
8Department of Molecular Physiology and Biophysics, Vanderbilt University Medical Center, Nashville, Tennessee, USA.
9VA Tennessee Valley Healthcare System, Nashville, Tennessee, USA.
Address correspondence to: Alvin C. Powers, Departments of Medicine and Molecular Physiology and Biophysics, Vanderbilt University School of Medicine, 7465 Medical Research Building IV, 2215 Garland Avenue, Nashville, Tennessee 37232-0475, USA. Phone: 615.936.7678; Email: [email protected]. Or to: Seung K. Kim, Department of Developmental Biology and Medicine, Stanford University School of Medicine, 279 Campus Drive, Beckman Center B300, Stanford, California 94305-5329, USA. Phone: 650.723.6230; Email: [email protected].
Authorship note: C. Dai and Y. Hang contributed equally to this work.
Find articles by Poffenberger, G. in: JCI | PubMed | Google Scholar
1Division of Diabetes, Endocrinology and Metabolism, Department of Medicine, Vanderbilt University Medical Center, Nashville, Tennessee, USA.
2Department of Developmental Biology, Stanford University School of Medicine, Stanford, California, USA.
3Hudson Alpha Institute for Biotechnology, Huntsville, Alabama, USA.
4Department of Molecular Medicine, Diabetes Center of Excellence, University of Massachusetts Medical School, Worcester, Massachusetts, USA.
5The Jackson Laboratory, Bar Harbor, Maine, USA.
6Institute of Cellular Therapeutics, Allegheny-Singer Research Institute, Allegheny Health Network, Pittsburgh, Pennsylvania, USA.
7Department of Medicine, Stanford University School of Medicine, Stanford California, USA.
8Department of Molecular Physiology and Biophysics, Vanderbilt University Medical Center, Nashville, Tennessee, USA.
9VA Tennessee Valley Healthcare System, Nashville, Tennessee, USA.
Address correspondence to: Alvin C. Powers, Departments of Medicine and Molecular Physiology and Biophysics, Vanderbilt University School of Medicine, 7465 Medical Research Building IV, 2215 Garland Avenue, Nashville, Tennessee 37232-0475, USA. Phone: 615.936.7678; Email: [email protected]. Or to: Seung K. Kim, Department of Developmental Biology and Medicine, Stanford University School of Medicine, 279 Campus Drive, Beckman Center B300, Stanford, California 94305-5329, USA. Phone: 650.723.6230; Email: [email protected].
Authorship note: C. Dai and Y. Hang contributed equally to this work.
Find articles by Hart, N. in: JCI | PubMed | Google Scholar
1Division of Diabetes, Endocrinology and Metabolism, Department of Medicine, Vanderbilt University Medical Center, Nashville, Tennessee, USA.
2Department of Developmental Biology, Stanford University School of Medicine, Stanford, California, USA.
3Hudson Alpha Institute for Biotechnology, Huntsville, Alabama, USA.
4Department of Molecular Medicine, Diabetes Center of Excellence, University of Massachusetts Medical School, Worcester, Massachusetts, USA.
5The Jackson Laboratory, Bar Harbor, Maine, USA.
6Institute of Cellular Therapeutics, Allegheny-Singer Research Institute, Allegheny Health Network, Pittsburgh, Pennsylvania, USA.
7Department of Medicine, Stanford University School of Medicine, Stanford California, USA.
8Department of Molecular Physiology and Biophysics, Vanderbilt University Medical Center, Nashville, Tennessee, USA.
9VA Tennessee Valley Healthcare System, Nashville, Tennessee, USA.
Address correspondence to: Alvin C. Powers, Departments of Medicine and Molecular Physiology and Biophysics, Vanderbilt University School of Medicine, 7465 Medical Research Building IV, 2215 Garland Avenue, Nashville, Tennessee 37232-0475, USA. Phone: 615.936.7678; Email: [email protected]. Or to: Seung K. Kim, Department of Developmental Biology and Medicine, Stanford University School of Medicine, 279 Campus Drive, Beckman Center B300, Stanford, California 94305-5329, USA. Phone: 650.723.6230; Email: [email protected].
Authorship note: C. Dai and Y. Hang contributed equally to this work.
Find articles by Prasad, N. in: JCI | PubMed | Google Scholar
1Division of Diabetes, Endocrinology and Metabolism, Department of Medicine, Vanderbilt University Medical Center, Nashville, Tennessee, USA.
2Department of Developmental Biology, Stanford University School of Medicine, Stanford, California, USA.
3Hudson Alpha Institute for Biotechnology, Huntsville, Alabama, USA.
4Department of Molecular Medicine, Diabetes Center of Excellence, University of Massachusetts Medical School, Worcester, Massachusetts, USA.
5The Jackson Laboratory, Bar Harbor, Maine, USA.
6Institute of Cellular Therapeutics, Allegheny-Singer Research Institute, Allegheny Health Network, Pittsburgh, Pennsylvania, USA.
7Department of Medicine, Stanford University School of Medicine, Stanford California, USA.
8Department of Molecular Physiology and Biophysics, Vanderbilt University Medical Center, Nashville, Tennessee, USA.
9VA Tennessee Valley Healthcare System, Nashville, Tennessee, USA.
Address correspondence to: Alvin C. Powers, Departments of Medicine and Molecular Physiology and Biophysics, Vanderbilt University School of Medicine, 7465 Medical Research Building IV, 2215 Garland Avenue, Nashville, Tennessee 37232-0475, USA. Phone: 615.936.7678; Email: [email protected]. Or to: Seung K. Kim, Department of Developmental Biology and Medicine, Stanford University School of Medicine, 279 Campus Drive, Beckman Center B300, Stanford, California 94305-5329, USA. Phone: 650.723.6230; Email: [email protected].
Authorship note: C. Dai and Y. Hang contributed equally to this work.
Find articles by Phillips, N. in: JCI | PubMed | Google Scholar
1Division of Diabetes, Endocrinology and Metabolism, Department of Medicine, Vanderbilt University Medical Center, Nashville, Tennessee, USA.
2Department of Developmental Biology, Stanford University School of Medicine, Stanford, California, USA.
3Hudson Alpha Institute for Biotechnology, Huntsville, Alabama, USA.
4Department of Molecular Medicine, Diabetes Center of Excellence, University of Massachusetts Medical School, Worcester, Massachusetts, USA.
5The Jackson Laboratory, Bar Harbor, Maine, USA.
6Institute of Cellular Therapeutics, Allegheny-Singer Research Institute, Allegheny Health Network, Pittsburgh, Pennsylvania, USA.
7Department of Medicine, Stanford University School of Medicine, Stanford California, USA.
8Department of Molecular Physiology and Biophysics, Vanderbilt University Medical Center, Nashville, Tennessee, USA.
9VA Tennessee Valley Healthcare System, Nashville, Tennessee, USA.
Address correspondence to: Alvin C. Powers, Departments of Medicine and Molecular Physiology and Biophysics, Vanderbilt University School of Medicine, 7465 Medical Research Building IV, 2215 Garland Avenue, Nashville, Tennessee 37232-0475, USA. Phone: 615.936.7678; Email: [email protected]. Or to: Seung K. Kim, Department of Developmental Biology and Medicine, Stanford University School of Medicine, 279 Campus Drive, Beckman Center B300, Stanford, California 94305-5329, USA. Phone: 650.723.6230; Email: [email protected].
Authorship note: C. Dai and Y. Hang contributed equally to this work.
Find articles by Levy, S. in: JCI | PubMed | Google Scholar
1Division of Diabetes, Endocrinology and Metabolism, Department of Medicine, Vanderbilt University Medical Center, Nashville, Tennessee, USA.
2Department of Developmental Biology, Stanford University School of Medicine, Stanford, California, USA.
3Hudson Alpha Institute for Biotechnology, Huntsville, Alabama, USA.
4Department of Molecular Medicine, Diabetes Center of Excellence, University of Massachusetts Medical School, Worcester, Massachusetts, USA.
5The Jackson Laboratory, Bar Harbor, Maine, USA.
6Institute of Cellular Therapeutics, Allegheny-Singer Research Institute, Allegheny Health Network, Pittsburgh, Pennsylvania, USA.
7Department of Medicine, Stanford University School of Medicine, Stanford California, USA.
8Department of Molecular Physiology and Biophysics, Vanderbilt University Medical Center, Nashville, Tennessee, USA.
9VA Tennessee Valley Healthcare System, Nashville, Tennessee, USA.
Address correspondence to: Alvin C. Powers, Departments of Medicine and Molecular Physiology and Biophysics, Vanderbilt University School of Medicine, 7465 Medical Research Building IV, 2215 Garland Avenue, Nashville, Tennessee 37232-0475, USA. Phone: 615.936.7678; Email: [email protected]. Or to: Seung K. Kim, Department of Developmental Biology and Medicine, Stanford University School of Medicine, 279 Campus Drive, Beckman Center B300, Stanford, California 94305-5329, USA. Phone: 650.723.6230; Email: [email protected].
Authorship note: C. Dai and Y. Hang contributed equally to this work.
Find articles by Greiner, D. in: JCI | PubMed | Google Scholar
1Division of Diabetes, Endocrinology and Metabolism, Department of Medicine, Vanderbilt University Medical Center, Nashville, Tennessee, USA.
2Department of Developmental Biology, Stanford University School of Medicine, Stanford, California, USA.
3Hudson Alpha Institute for Biotechnology, Huntsville, Alabama, USA.
4Department of Molecular Medicine, Diabetes Center of Excellence, University of Massachusetts Medical School, Worcester, Massachusetts, USA.
5The Jackson Laboratory, Bar Harbor, Maine, USA.
6Institute of Cellular Therapeutics, Allegheny-Singer Research Institute, Allegheny Health Network, Pittsburgh, Pennsylvania, USA.
7Department of Medicine, Stanford University School of Medicine, Stanford California, USA.
8Department of Molecular Physiology and Biophysics, Vanderbilt University Medical Center, Nashville, Tennessee, USA.
9VA Tennessee Valley Healthcare System, Nashville, Tennessee, USA.
Address correspondence to: Alvin C. Powers, Departments of Medicine and Molecular Physiology and Biophysics, Vanderbilt University School of Medicine, 7465 Medical Research Building IV, 2215 Garland Avenue, Nashville, Tennessee 37232-0475, USA. Phone: 615.936.7678; Email: [email protected]. Or to: Seung K. Kim, Department of Developmental Biology and Medicine, Stanford University School of Medicine, 279 Campus Drive, Beckman Center B300, Stanford, California 94305-5329, USA. Phone: 650.723.6230; Email: [email protected].
Authorship note: C. Dai and Y. Hang contributed equally to this work.
Find articles by Shultz, L. in: JCI | PubMed | Google Scholar
1Division of Diabetes, Endocrinology and Metabolism, Department of Medicine, Vanderbilt University Medical Center, Nashville, Tennessee, USA.
2Department of Developmental Biology, Stanford University School of Medicine, Stanford, California, USA.
3Hudson Alpha Institute for Biotechnology, Huntsville, Alabama, USA.
4Department of Molecular Medicine, Diabetes Center of Excellence, University of Massachusetts Medical School, Worcester, Massachusetts, USA.
5The Jackson Laboratory, Bar Harbor, Maine, USA.
6Institute of Cellular Therapeutics, Allegheny-Singer Research Institute, Allegheny Health Network, Pittsburgh, Pennsylvania, USA.
7Department of Medicine, Stanford University School of Medicine, Stanford California, USA.
8Department of Molecular Physiology and Biophysics, Vanderbilt University Medical Center, Nashville, Tennessee, USA.
9VA Tennessee Valley Healthcare System, Nashville, Tennessee, USA.
Address correspondence to: Alvin C. Powers, Departments of Medicine and Molecular Physiology and Biophysics, Vanderbilt University School of Medicine, 7465 Medical Research Building IV, 2215 Garland Avenue, Nashville, Tennessee 37232-0475, USA. Phone: 615.936.7678; Email: [email protected]. Or to: Seung K. Kim, Department of Developmental Biology and Medicine, Stanford University School of Medicine, 279 Campus Drive, Beckman Center B300, Stanford, California 94305-5329, USA. Phone: 650.723.6230; Email: [email protected].
Authorship note: C. Dai and Y. Hang contributed equally to this work.
Find articles by Bottino, R. in: JCI | PubMed | Google Scholar
1Division of Diabetes, Endocrinology and Metabolism, Department of Medicine, Vanderbilt University Medical Center, Nashville, Tennessee, USA.
2Department of Developmental Biology, Stanford University School of Medicine, Stanford, California, USA.
3Hudson Alpha Institute for Biotechnology, Huntsville, Alabama, USA.
4Department of Molecular Medicine, Diabetes Center of Excellence, University of Massachusetts Medical School, Worcester, Massachusetts, USA.
5The Jackson Laboratory, Bar Harbor, Maine, USA.
6Institute of Cellular Therapeutics, Allegheny-Singer Research Institute, Allegheny Health Network, Pittsburgh, Pennsylvania, USA.
7Department of Medicine, Stanford University School of Medicine, Stanford California, USA.
8Department of Molecular Physiology and Biophysics, Vanderbilt University Medical Center, Nashville, Tennessee, USA.
9VA Tennessee Valley Healthcare System, Nashville, Tennessee, USA.
Address correspondence to: Alvin C. Powers, Departments of Medicine and Molecular Physiology and Biophysics, Vanderbilt University School of Medicine, 7465 Medical Research Building IV, 2215 Garland Avenue, Nashville, Tennessee 37232-0475, USA. Phone: 615.936.7678; Email: [email protected]. Or to: Seung K. Kim, Department of Developmental Biology and Medicine, Stanford University School of Medicine, 279 Campus Drive, Beckman Center B300, Stanford, California 94305-5329, USA. Phone: 650.723.6230; Email: [email protected].
Authorship note: C. Dai and Y. Hang contributed equally to this work.
Find articles by Kim, S. in: JCI | PubMed | Google Scholar
1Division of Diabetes, Endocrinology and Metabolism, Department of Medicine, Vanderbilt University Medical Center, Nashville, Tennessee, USA.
2Department of Developmental Biology, Stanford University School of Medicine, Stanford, California, USA.
3Hudson Alpha Institute for Biotechnology, Huntsville, Alabama, USA.
4Department of Molecular Medicine, Diabetes Center of Excellence, University of Massachusetts Medical School, Worcester, Massachusetts, USA.
5The Jackson Laboratory, Bar Harbor, Maine, USA.
6Institute of Cellular Therapeutics, Allegheny-Singer Research Institute, Allegheny Health Network, Pittsburgh, Pennsylvania, USA.
7Department of Medicine, Stanford University School of Medicine, Stanford California, USA.
8Department of Molecular Physiology and Biophysics, Vanderbilt University Medical Center, Nashville, Tennessee, USA.
9VA Tennessee Valley Healthcare System, Nashville, Tennessee, USA.
Address correspondence to: Alvin C. Powers, Departments of Medicine and Molecular Physiology and Biophysics, Vanderbilt University School of Medicine, 7465 Medical Research Building IV, 2215 Garland Avenue, Nashville, Tennessee 37232-0475, USA. Phone: 615.936.7678; Email: [email protected]. Or to: Seung K. Kim, Department of Developmental Biology and Medicine, Stanford University School of Medicine, 279 Campus Drive, Beckman Center B300, Stanford, California 94305-5329, USA. Phone: 650.723.6230; Email: [email protected].
Authorship note: C. Dai and Y. Hang contributed equally to this work.
Find articles by Powers, A. in: JCI | PubMed | Google Scholar
Authorship note: C. Dai and Y. Hang contributed equally to this work.
Published September 18, 2017 - More info
J Clin Invest. 2017;127(10):3835–3844. https://doi.org/10.1172/JCI91761.
Copyright © 2017, American Society for Clinical Investigation
Received: December 8, 2016; Accepted: July 28, 2017
-
Abstract
Inadequate pancreatic β cell function underlies type 1 and type 2 diabetes mellitus. Strategies to expand functional cells have focused on discovering and controlling mechanisms that limit the proliferation of human β cells. Here, we developed an engraftment strategy to examine age-associated human islet cell replication competence and reveal mechanisms underlying age-dependent decline of β cell proliferation in human islets. We found that exendin-4 (Ex-4), an agonist of the glucagon-like peptide 1 receptor (GLP-1R), stimulates human β cell proliferation in juvenile but not adult islets. This age-dependent responsiveness does not reflect loss of GLP-1R signaling in adult islets, since Ex-4 treatment stimulated insulin secretion by both juvenile and adult human β cells. We show that the mitogenic effect of Ex-4 requires calcineurin/nuclear factor of activated T cells (NFAT) signaling. In juvenile islets, Ex-4 induced expression of calcineurin/NFAT signaling components as well as target genes for proliferation-promoting factors, including NFATC1, FOXM1, and CCNA1. By contrast, expression of these factors in adult islet β cells was not affected by Ex-4 exposure. These studies reveal age-dependent signaling mechanisms regulating human β cell proliferation, and identify elements that could be adapted for therapeutic expansion of human β cells.
-
Introduction
In type 1 or type 2 diabetes mellitus, an absolute or relative deficiency of functional pancreatic β cells underlies disease pathogenesis. Thus, investigations are focused on identifying native signaling pathways governing human pancreatic β cell proliferation, with the goal of expanding functional β cell mass. Islet β cells proliferate in neonatal humans and rodents, but the proliferation rate declines thereafter, and reaches its nadir by puberty (1–5). During postnatal β cell growth, studies in rodents suggest that hallmark β cell functions such as glucose sensing and insulin secretion mature (6–10). The mechanisms controlling age-dependent β cell proliferation and maturation are being intensely investigated, since deciphering these could accelerate development of β cell expansion strategies (3, 4, 8, 10–13).
Recent studies have identified age-dependent intrinsic and extrinsic regulators in mice and humans that influence or limit islet β cell proliferation. For example, with advancing age, increased expression of CDKN2A in β cells, which encodes the cell cycle inhibitor p16INK4a, limits β cell regeneration in mice and humans (3, 4, 11–13). Native extrinsic signals that regulate β cell proliferation include PDGF, prolactin (PRL), and glucagon-like peptide 1 (GLP-1). Recent studies have elucidated crucial signal transduction elements of these mitogens in β cells (4, 14). For example, work on mouse and human islets suggests that the mitogenic function of PDGF in β cells is age-dependent. While islet β cells from neonatal mice and human children express PDGF receptors (PDGFRs) and proliferate in response to PDGF-A, β cells from adult mice and humans lack PDGFR expression and are unresponsive to PDGF stimulation (4). Thus, attenuated receptor expression underlies one mechanism of age-dependent mitogenic restriction in β cells, underscored by the finding that expression of activated PDGFR protein in adult β cells led to β cell proliferation (4). PRL-stimulated β cell proliferation is also lacking in adult human islets and is accompanied by little or no PRL receptor expression in adult β cells (14). However, unlike the effects of PDGF signaling, ectopic expression of PRL receptor in adult β cells does not restore responsiveness to PRL (14), suggesting that restriction of β cell competence for PRL includes both attenuated receptor expression and reduced intracellular signal transduction. Thus, mechanisms limiting human β cell responses to PDGF and PRL appear distinct, although both involve age-dependent loss of cognate receptor expression.
GLP-1 has a well-established role in stimulating β cell insulin secretion (the incretin effect), in addition to inducing insulin biosynthesis, and regulating β cell apoptosis (15–17). GLP-1 and its analogs have been previously reported to induce mouse β cell proliferation in an age-dependent manner (18). Prior studies investigating whether GLP-1 or exendin-4 (Ex-4) stimulates human β cell proliferation have yielded conflicting results (15, 17–22). Thus, it remains unclear whether GLP-1 can stimulate human β cell proliferation. GLP-1 stimulates β cell Ca2+ transients (23, 24) through the GLP-1 receptor (GLP-1R), and these are known to activate the calcium-dependent calcineurin/nuclear factor of activated T cells (NFAT) signaling pathway, a crucial regulator of β cell proliferation and function in neonatal and adult islets (25–28). However, the links between GLP-1R responses and downstream intrinsic regulators of human β cell proliferation like calcineurin/NFAT signaling have not yet been established.
To test the hypothesis that human β cell proliferative response to the GLP-1 analog Ex-4 is age-dependent, we used an in vivo transplantation strategy with human islets from juveniles and adults (3, 4, 10, 26). Here we report that Ex-4 stimulates β cell proliferation in transplanted juvenile, but not adult, human islets, and that this response requires intact calcineurin/NFAT signaling. Thus, these studies reveal age-dependent signaling pathways and mechanisms that stimulate human β cell proliferation.
-
Results
Age-dependent human islet cell proliferation profile after transplantation. To investigate the age-dependent proliferative potential of human islet cells in vivo, we transplanted juvenile (aged 0.5–9 years) or adult (20 years of age and older) human islets under the renal capsule of NOD.Cg-PrkdcscidIl2rgtm1Wjl/Sz (NSG) mice, an immunocompromised strain favorable for xenograft studies (ref. 29 and Figure 1A). Ki67 immunostaining of juvenile islet grafts 4 weeks after transplantation revealed a greater number of proliferating cells in juvenile insulin-positive β cells, glucagon-positive α cells, and somatostatin-positive δ cells than engrafted adult islets (Figure 1, B, C, F, G, J, and K). Compared with engrafted adult islets, juvenile islets had 8-fold more Ki67+ β cells (Figure 1, B–E, and Figure 1E, inset). We assessed phospho–histone H3 (pHH3), an independent marker of proliferation, and found that the number of pHH3+ β cells was greater in juvenile islets, confirming progression into G2 or M phases (Supplemental Figure 1; supplemental material available online with this article; https://doi.org/10.1172/JCI91761DS1). Thus, the greater proliferation of juvenile human β cells continued after transplantation, indicating that age-dependent β cell proliferation is maintained and independent of the native pancreatic environment.
 Figure 1
Figure 1Endocrine cell proliferation is greater in transplanted human juvenile islets than in adult islets. (A) Schematic of experimental design. Grafts were removed for analysis 6 weeks after transplantation. (B, C, F, G, J, and K) Images of juvenile (left panels) and adult (right panels) grafts labeled with insulin (INS, B and C), glucagon (GLU, F and G), or somatostatin (SOM, J and K) in green; Ki67 in red; DAPI in blue. Insets show proliferating Ki67+ cells. Scale bar: 50 μm (bar in K applies to all other images in this figure). (D, E, H, I, L, and M) Quantification of percentage Ki67+ β (D and E), α (H and I), and δ (L and M) cells of transplanted grafts from individual juvenile (Juv) and adult donors (n = 2–5 grafts per donor; age shown on x axis). The average number of β, α, and δ cells counted in each donor sample was approximately 6,000, 3,000, and 2,000, respectively. Insets are average percentage proliferating cells in each age group (β cells: data from D and E; α cells: data from H and I; δ cells: data from L and M). Error bars represent SEM. **P < 0.01; ***P < 0.001. An unpaired 2-tailed Student’s t test was used for statistical analysis. See also Supplemental Figure 1.
We also noted a higher percentage of Ki67+ α cells (Figure 1, F–I, and Figure 1I, inset) and Ki67+ δ cells (Figure 1, J–M, and Figure 1M, inset) in transplanted juvenile islets. To our knowledge, age-dependent proliferation of these islet cell subsets in humans has not been previously reported. In β and α cells, greater Ki67 labeling was noted in donors up to 3 years of age (Figure 1, D and H), while in δ cells, we observed a lower proliferation rate by that age (Figure 1L). Within the limits of our donor sampling size, these data suggest that the tempo of declining proliferation with age is faster in δ cells than in β or α cells.
Ex-4 signaling promotes proliferation in juvenile β cells. GLP-1 and GLP-1R agonists, like Ex-4, stimulate insulin secretion (the incretin effect, reviewed in refs. 15, 17, 30) and are used in the treatment of type 2 diabetes. However, prior studies (15, 17–19, 21, 22) are in conflict about the ability of GLP-1R agonists to stimulate human islet cell proliferation, possibly due to different in vitro treatments and assays used. Maintenance of age-dependent islet cell proliferation in our transplant system permitted in vivo studies of candidate mitogens such as GLP-1/Ex-4 in a physiological context. We hypothesized that human islet β cells have an age-dependent capacity to respond to Ex-4, and used our transplant system to investigate this possibility.
First, we examined whether expression of GLP-1R in human β cells was different with age. GLP1R mRNA levels were similar in juvenile and adult human islets (Figure 2A), suggesting that human β cell GLP-1R expression is not age-dependent. Moreover, in FACS-isolated islet cell subsets, GLP1R mRNA was principally expressed in β cells and was very low, or not detectable, in α cells (Supplemental Figure 2, A–C), consistent with prior findings (10, 31–35).
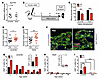 Figure 2
Figure 2Ex-4 promotes β cell proliferation only in juvenile islets. (A) Both juvenile and adult islets express a similar level of GLP1R mRNA measured by qPCR (juvenile: n = 6, 0.5–9 years old; adult: n = 6, 20–60 years old). (B) Experimental design. After a 2-week islet engraftment period, PBS or Ex-4 was delivered by osmotic pump. The grafts were removed and analyses were performed after 4 weeks of treatment. (C–E) Mouse random glucose (C) and human insulin (D and E) in mice with transplanted juvenile (D, n = 3 donors) or adult (E, n = 3 donors) human islets 48 hours after the implantation of pumps with PBS or Ex-4 (n = 13–14 samples). (F) Representative images of juvenile grafts labeled with insulin (green), Ki67 (red), and DAPI (blue). Insets show proliferating Ki67+ cells. Scale bar: 35 μm. (G and H) Percentage of β cell proliferation in grafts from individual juvenile and adult donors (n = 4–8 grafts per donor). Insets are average percentage in each age group. (I) Statistical analysis of data sets in G and H. Error bars represent SEM. *P < 0.05; **P < 0.01; ***P < 0.001. Unpaired 2-tailed Student’s t test or 1-way ANOVA followed by Newman-Keuls multiple-comparisons test (I) was used for statistical analysis. See also Supplemental Figures 2 and 3.
To investigate human islet responses to Ex-4, we infused Ex-4 in mice transplanted with human islets from juvenile or adult donors (Figure 2B); this infusion established pharmacologically relevant serum Ex-4 levels (Supplemental Figure 2D and ref. 36). As expected, blood glucose levels were reduced by Ex-4 within 24 hours and remained lower during the subsequent treatment period (Figure 2C and Supplemental Figure 2E). Circulating human insulin levels in NSG mice bearing human islets (Figure 2, D and E) were greater in mice infused with Ex-4. Moreover, this incretin effect was observed in mice with juvenile or adult islets. Thus, Ex-4 stimulation of insulin secretion in vivo by human islets is age-independent, suggesting that downstream signaling that regulates GLP-1–stimulated insulin secretion is similar in human juvenile and adult islets.
To investigate proliferative responses to Ex-4, we assessed β cell proliferation in transplanted juvenile and adult islets. Ex-4 stimulated β cell replication, marked by Ki67, in juvenile donor samples up to age 9 (Figure 2, F, G, and I), but not in islets from older donors (20 years and above; Figure 2, H and I). Unlike what was observed in β cells, we did not detect changes of α cell or δ cell proliferation in response to Ex-4 in juvenile or adult islets (Supplemental Figure 3, A–C). The lack of Ex-4 mitogenic effect on α cells is likely explained by our finding that human α cells did not express GLP1R mRNA (Supplemental Figure 2, A–C). Thus, these studies revealed age-dependent mitogenic effects of Ex-4 on β cells, while the insulin secretion effect was age-independent. This suggests that divergent downstream pathways regulate these two distinct effects of GLP-1R signaling in human β cells.
Ex-4 stimulates calcineurin/NFAT signaling in juvenile human islets. To understand the mechanisms underlying Ex-4 mitogenic effects in juvenile human islet grafts, we compared the transcriptomes of Ex-4–treated and control PBS-treated juvenile human islet grafts and found that 1,440 genes were differentially expressed (Figure 3A). Gene sets associated with signaling pathways governing cell proliferation and vesicle secretion were among those mostly upregulated in Ex-4–infused islet grafts. Gene sets comprising the NFAT pathway were at the top of this list (Figure 3B).
 Figure 3
Figure 3Gene expression in human islets and juvenile grafts after treatment with Ex-4 and/or FK506. (A and B) RNA-Seq analysis of PBS- or Ex-4–treated juvenile grafts (n = 3 grafts per treatment, 6-year-old donor). (A) Number of genes regulated by Ex-4 (≥1.5-fold) in juvenile grafts (Ex-4 vs. PBS); 1,115 genes are upregulated and 325 are downregulated by Ex-4 treatment. (B) Signaling pathways upregulated by Ex-4. (C) Genes of the NFATC family are expressed more highly in juvenile islets (n = 5 donors, age: 0.5, 1.2, 1.7, 3, and 4 years) than adult (n = 5 donors, age: 29, 43, 50, 53, and 60 years). The average expression level in juvenile islets is defined as 1.00 for each gene. (D) Schematic of transplantation and treatment with Ex-4 plus FK506. Two weeks after transplantation, a pump with PBS or Ex-4 was implanted, and a second pump with saline or FK506 was implanted in the last 2 weeks. Grafts were removed at 4 weeks. (E–G) Juvenile graft gene expression measured by qPCR. Ex-4 upregulates genes of the NFAT family (E), cell cycle regulators (F), and transcription factors (G), and the effects were diminished by FK506 (n = 5 graft samples per treatment from 0.2- and 6-year-old donors). The average expression level in PBS+saline–treated islets is defined as 1.00 for each gene. Error bars represent SEM. *P < 0.05, **P < 0.01, ***P < 0.001 vs. PBS+saline; †P < 0.05, ††P < 0.01, †††P < 0.001 vs. Ex-4+saline; no significant differences between PBS+FK506 and Ex-4+FK506. Unpaired 2-tailed Student’s t test or 1-way ANOVA followed by Newman-Keuls multiple-comparisons test (E–G) was used for analysis of statistical significance. See also Supplemental Figures 4 and 5.
A prior study reported that signaling through the calcineurin/NFAT pathway is critical for juvenile β cell proliferation (26). For example, this pathway stimulates expression of multiple β cell transcription factors and cell cycle activators, including NFATC1, FOXM1, cyclin-dependent kinases (CDKs), and the A-type cyclins (26, 27, 37, 38). To unravel signaling links between Ex-4/GLP-1R and calcineurin/NFAT pathways in human islets, we first measured human islet mRNAs encoding all NFAT members and found that NFATC2 and NFATC3 were most abundant (Supplemental Figure 4, A and B). In human islets, levels of mRNAs encoding NFATC1, NFATC2, and NFATC4, but not NFATC3, declined with advancing age (Figure 3C), similar to a prior observation that levels of NFATC transcription factors are lower in older mouse islets (26). We also found that important cell cycle regulators (CCNA1, CCNA2, CDK1, and CDK4) and key transcription factors such as FOXM1 are expressed more highly in juvenile human islets (Supplemental Figure 4, C–F). By contrast, mRNA levels of CDKN2A and CDKN2B, which respectively encode the β cell cycle inhibitors p16INK4a and p15INK4b, were greater in adult human islets (Supplemental Figure 4C and refs. 4, 10, 11, 13).
After Ex-4 treatment, we observed an increase in mRNAs encoding NFATC1, FOXM1, CCNA1, and CDK1 in juvenile human islet grafts (Figure 3, E–G) but not in adult islet grafts (Supplemental Figure 5A), suggesting that calcineurin/NFAT signaling was stimulated by Ex-4 only in juvenile human islets. We also found increased mRNAs encoding other possible targets of calcineurin/NFAT signaling (Figure 3, E–G) thought to promote cell proliferation, including NFATC3, NFATC4, CDK1 (39–41), and EGR3 (42). However, mRNA levels of NFATC2, CDKN2A, CDKN2B, or EZH2, a regulator of CDKN2A expression and proliferation in β cells (3, 4), were not detectably altered in juvenile islets after Ex-4 infusion. Together these data suggest that Ex-4 stimulates calcineurin/NFAT signaling and thus enhances expression of key transcriptional and cell cycle regulators that promote human juvenile β cell proliferation. These studies also reveal that the expression of other calcineurin/NFAT factors (e.g., NFATC2) and intrinsic inhibitors of β cell proliferation (e.g., CDKN2A) was not changed by Ex-4 exposure.
Ex-4 mitogenic effect on human β cells prevented by the calcineurin inhibitor FK506. To test whether the mitogenic effects of Ex-4 on juvenile human islets are mediated by calcineurin/NFAT signaling activation, we infused Ex-4 in mice engrafted with juvenile human islets and simultaneously infused FK506, a potent specific calcineurin inhibitor (Figure 3D). In mice coinfused with Ex-4 and saline, we observed increased nuclear localization of β cell NFATC4, and increased proliferation of transplanted human islet β cells quantified by Ki67 labeling (Figure 4, A–C). In contrast, simultaneous infusion of FK506 with Ex-4 prevented increases of β cell NFATC4 nuclear localization and proliferation (Figure 4, A–C). In control mice receiving PBS plus saline or PBS plus FK506, we did not detect differences in human islet β cell NFATC4 nuclear localization or proliferation (Figure 4, A–C), indicating that basal proliferation in transplanted juvenile human β cells was insensitive to FK506. Furthermore, little apoptosis was observed in human islet grafts after either treatment (Figure 4D and Table 1). Collectively, these data suggest that calcineurin/NFAT signaling is required for Ex-4–stimulated human juvenile islet β cell proliferation. Supporting this view, we observed that increased expression of calcineurin/NFAT signaling targets induced by Ex-4 was reduced to basal levels in human islet grafts recovered from mice simultaneously infused with Ex-4 and FK506, including NFATC1, NFATC3, NFATC4, FOXM1, CCNA1, and CDK1 (Figure 3, E–G). Together, these data demonstrate that calcineurin signaling in human juvenile islets is required for β cell proliferation.
 Figure 4
Figure 4Calcineurin/NFAT signaling mediates the mitogenic effect of Ex-4 in juvenile β cells. (A) Ex-4 stimulated NFAT translocation. Insulin (green), NFATC4 (NFAT3) (red), DAPI (blue). Scale bar: 15 μm (applies to the other images in A). Arrows point to NFATC4+ β cells, and insets show NFATC4+ β cells. (B) Representative images of β cell proliferation in juvenile grafts. Insulin (green), Ki67 (red), DAPI (blue). Arrows point to proliferating Ki67+ cells (also showed in insets). Scale bar: 35 μm. (C) FK506 blocked the β cell proliferation stimulated by Ex-4 in juvenile grafts (n = 4 donors, 0.2, 1.8, 6, and 9 years old). See also Supplemental Figure 6. (D) Representative images of TUNEL assay including positive control. Insulin (green), TUNEL (red), DAPI (blue). Scale bar: 50 μm (applies to all images in D). See Table 1 for the quantification of TUNEL+ β cells in PBS+saline–treated, Ex-4+saline–treated, PBS+FK506–treated, and Ex-4+FK506–treated graft samples from 3 juvenile donors. Error bars represent SEM. **P < 0.01. Unpaired 2-tailed Student’s t test or 1-way ANOVA followed by Newman-Keuls multiple-comparisons test (C) was used for statistical analysis. See also Supplemental Figure 6.
-
Discussion
Understanding the basis of declining proliferation observed with advancing age in human pancreatic islet β cells (1–3, 5) would reveal potential strategies for therapeutic human β cell expansion in diabetes. Studies with rodent islets have suggested that GLP-1 stimulates cell proliferation, but most studies on human islets have relied on in vitro assays and reported variable effects of GLP-1 on proliferation (21, 22, 43). To test the hypothesis that Ex-4, a GLP-1 analog, stimulates human islet cell proliferation in an age-dependent fashion, we developed an in vivo engraftment system that maintained the proliferative competence of juvenile human islet cells and an infrastructure to systematically compare human juvenile and adult islets. We found that Ex-4 stimulates human β cell proliferation in human juvenile, but not adult, islets. The unresponsiveness to Ex-4 in adult human β cells did not reflect absence of GLP-1R expression or signal transduction, since Ex-4 stimulated insulin secretion in both juvenile and adult human β cells. We show that this β cell proliferative response to Ex-4 requires calcineurin/NFAT signaling and also suggest that β cell mitogenic responses to Ex-4 may be limited in adult islets by calcineurin/NFAT–independent factors, including potent inhibitors of β cell proliferation like the cyclin inhibitor CDKN2A (p16INK4A). Thus, this work reveals an age-dependent pathway for β cell proliferation in human juvenile islets (Figure 5).
 Figure 5
Figure 5Model of GLP-1 mitogenic effect mediated by calcineurin/NFAT signaling in juvenile human cells. GLP-1 (or Ex-4) binds and activates GLP-1R, which increases intracellular calcium concentration (56), Ca2+-sensitive phosphatase, and calcineurin (Cn). Calcineurin dephosphorylates NFATc factors to expose their nuclear localization sequences, which triggers rapid entry into the nucleus. In the nucleus, NFATc proteins assemble on DNA with partner proteins (termed NFATn) to activate transcription of target genes, including cell cycle activators (e.g., cyclins, CDKs) and proliferation-promoting transcription factors (TFs) such as FOXM1, which are all produced at a low level in adult islets. GLP-1/calcineurin/NFAT signaling does not appear to regulate the expression of age-associated cell cycle inhibitors such as CDKN2A (normally very low level in juvenile islets or its repressor EZH2 in juvenile human β cells, which may explain the absence of proliferative response to GLP-1 or Ex-4 in adult islets.
We used transplantation of human juvenile and adult islets into immunodeficient NSG mice (44, 45) to investigate mechanisms controlling human islet cell proliferation. Compared with retrospective studies of cadaveric pancreata or in vitro islet culture–based assays, this in vivo transplantation strategy afforded experimental flexibility, including relatively prolonged observation times, prospective exposure of islets to sequential or concurrent schedules of drugs, and serial measures of insulin secretion in response to Ex-4. In our transplantation model, the maintenance of greater levels of β cell proliferation after transplantation indicates that the proliferation resulted from intrinsic human islet cell signals, and was not dependent on the pancreatic environment or circulating human factors.
Recent studies suggest that insulin secretion by human islets may mature in an age-dependent manner (10), similar to findings in rodents (6–9). Using our transplant-based system, however, we found that Ex-4 stimulated insulin secretion similarly in both juvenile and adult islet grafts, consistent with similar levels of GLP-1R expression in these islets. By contrast, Ex-4 stimulated β cell proliferation only in islets from juvenile donors. This suggests that age-dependent factors in adult β cells may suppress proliferation or that adult β cells lack factors mediating the proliferative effect of Ex-4/GLP-1R signaling, without affecting the incretin effects of this signaling pathway (reviewed in ref. 46). These findings are also supported by studies here (Figure 2A) and elsewhere (10, 33) revealing that GLP1R expression did not detectably change with age in human islet β cells. An additional finding from our work is that human α cell and δ cell proliferation also appears to decline with age, corroborating a recent mass cytometry–based study (47). However, unlike in juvenile human β cells, Ex-4 did not stimulate proliferation of these other islet cell subsets. This is consistent with findings here and from prior studies that GLP1R expression in human islet cells is restricted to β cells (Supplemental Figure 2, A–C, and refs. 10, 31–35). The circulating Ex-4 level in our system (~4 ng/ml) was much higher than the basal plasma GLP-1 level in mice (48, 49) or healthy humans (50) but close to the therapeutic concentration target for GLP-1R agonists in clinical use (36). The duration of Ex-4 infusion in our study was shorter (28 days) in comparison with a recent study (16) in which continuous exposure (>250 days) of transplanted islets to the GLP-1R agonist liraglutide was found to impair β cell function. Our findings raise the possibility that relatively brief exposure to pharmacological levels of Ex-4 may be sufficient to stimulate human β cell proliferation, and should motivate further studies of Ex-4 dosing and schedule effects on β cell growth and function.
Another principal finding here is that Ex-4 effects on human β cell proliferation and gene expression require calcineurin/NFAT signaling. Calcineurin/NFAT signaling is a calcium-responsive pathway required for postnatal β cell expansion in mice and humans (26, 27, 51). Prior observations that Ex-4/GLP-1R signaling may provoke Ca2+ transients in β cells (19, 23, 24, 52) suggested a connection between GLP-1R and calcineurin/NFAT signaling pathways. Our findings here provide direct evidence for stimulation of calcineurin/NFAT signaling by Ex-4 in juvenile human islets. FK506 is a potent calcineurin inhibitor used in multiple clinical settings, and a role for calcineurin/NFAT in human β cell function has also been inferred from the striking observation that 10%–30% of patients requiring immunosuppression with FK506 develop diabetes mellitus (28). Here we showed that infusion of FK506 eliminated the increased juvenile β cell proliferation and gene expression alteration evoked by Ex-4. Together, these findings suggest that the calcineurin/NFAT signaling pathway is crucial for mediating the mitogenic effect of GLP-1R in human juvenile β cells. In addition to NFAT, calcineurin signaling regulates other factors, including CTRC2, RCAN1, and IRS2 (53–55). Our study did not detect an effect of Ex-4 or FK506 on the transcript levels of these factors (Supplemental Figure 5). However, we cannot exclude roles of these or additional factors in the Ex-4 effects we observed in human β cells.
Age-dependent decline in human β cell responsiveness to mitogens was found previously to reflect loss of mitogen receptor expression in β cells. For example, age-dependent reduction of receptors for PDGF and PRL in human β cells has been noted (4, 14). Similarly to PDGFR, GLP-1R signal transduction can stimulate Ca2+ transients to activate NFATC transcription factors (47). Thus, we speculate that PDGFR and GLP-1R may signal in parallel to regulate β cell proliferation, a possibility requiring further studies. In contrast to age-dependent loss of PDGFR expression and signaling competence observed previously (4), our current work indicates that loss of mitogenic responses by adult human β cells to Ex-4 is not the result of reduced GLP-1R expression (Figure 2A) or failure to activate GLP-1R signaling in cells (Figures 2, C–E). Instead, other age-dependent mechanisms constraining β cell proliferation appear to limit adult β cell responses to Ex-4. For example, expression of intrinsic β cell growth regulators like CDKN2A, CDKN2B, NFATC2, and EZH2 (3, 4, 12, 56) was not detectably affected by Ex-4 exposure, suggesting a cell-autonomous basis for age-dependent mitogenic responses by human islets to Ex-4. Thus, findings here provide motivation for future investigations aiming to rejuvenate human adult β cell mitogenic responses to GLP-1 agonists.
In summary, use of a system for investigating age-dependent phenotypes in human pancreatic islets revealed that human β cell mitogenic responses to GLP-1 are age-dependent and require intact calcineurin/NFAT signal transduction. Identification of direct and indirect targets of calcineurin/NFAT activation in human islet cells could suggest steps for achieving therapeutic aims like functional β cell expansion.
-
Methods
Human islet procurement, quality assessment, and transplantation. Deidentified human pancreatic islets were obtained from previously healthy, nondiabetic organ donors. Islets from 14 juvenile donors (ages 0.2, 0.5, 1.2, 1.7, 1.8, 3, 3, 4, 5, 5, 6, 7, 9, and 9 years) were procured through the National Disease Research Interchange (http://ndriresource.org) and the International Institute for the Advancement of Medicine (http://www.iiam.org). Juvenile islets were isolated at the Institute of Cellular Therapeutics of the Allegheny Health Network (Pittsburgh, Pennsylvania, USA) as previously described (3, 4, 26). Islets from 12 adult donors (ages 20, 21, 29, 30, 40, 43, 43, 48, 49, 53, 60, and 60 years, BMI ranges 24.1–29.6) were obtained from the Integrated Islet Distribution Program (http://iidp.coh.org). Islet function (i.e., glucose-induced insulin secretion) was assessed by islet perifusion assay on the day of arrival, as previously described (57, 58). Each human islet preparation was handpicked on the day of arrival for gene expression studies (57). Male NOD.Cg-PrkdcscidIl2rgtm1Wjl/Sz (NSG) mice (44, 45), aged 12–18 weeks, were used for transplantation. Human islets were transplanted under the kidney capsule as previously described (29, 59, 60).
Ex-4 and FK506 administration. Two weeks after human islet transplantation, Ex-4 (24 nmol/kg/d; California Peptide Research Inc.) or 1× PBS was delivered by micro-osmotic pumps (Alzet 1004) implanted in recipient NSG mice (Figure 2B). For FK506 (tacrolimus) experiments, a second pump (Alzet 1002) loaded with FK506 (0.25 mg/kg/d; Astellas Ireland Co.) or saline was implanted 2 weeks after the Ex-4 pump was placed (Figure 4A). Serum Ex-4 levels in recipient mice were measured using the Exendin-4 EIA kit (Phoenix Pharmaceuticals Inc.).
Immunohistochemistry of human islet grafts. Recovered human islet grafts were fixed in 4% paraformaldehyde in PBS and cryopreserved. Five-micrometer-thick frozen sections were cut and stained as previously described (60). Primary antibodies used were rabbit anti–human Ki67 (Abcam, ab15580), guinea pig anti–human insulin (DAKO, A0564), mouse anti-glucagon (Abcam, ab10988), goat anti-somatostatin (Santa Cruz Biotechnology, sc7819), and rabbit anti–histone H3 (phospho S10) (Abcam, ab5176). Images were obtained using an Olympus BX41 fluorescence microscope. Proliferating islet cells were quantified by counting of Ki67+ and hormone-positive cells as described previously (57, 60, 61). Apoptosis was assessed by TUNEL (Millipore, S7165) following the manufacturer’s instructions.
RNA isolation, cDNA synthesis, and quantitative reverse transcriptase PCR. Total RNA was extracted from isolated human islets or recovered human islet grafts using an RNAqueous RNA isolation kit (Ambion). RNA quality control and quantity assessment (QC/QA) was performed using a Bioanalyzer instrument in the Vanderbilt Technologies for Advanced Genomics (VANTAGE) core laboratory. Only samples with a 28S/18S ratio greater than 1.2 and an RNA integrity number greater than 8.1 were used for subsequent analysis. cDNA was synthesized using a High Capacity cDNA Reverse Transcription Kit (Applied Biosystems, 4368814) according to the manufacturer’s instructions. Quantitative PCR (qPCR) was performed using TaqMan assays (Supplemental Table 1) and reagents from Applied Biosystems as previously described (57). ACTB and TFRC were used as endogenous controls in islet gene measurement, while SYPL1, SV2A, and CHDA were used for control genes in graft gene assay. Relative changes in mRNA expression were calculated by the comparative ΔCt method using the Applied Biosystems StepOne Plus System.
RNA-Seq library preparation, sequencing, and data normalization. The PolyA-enriched RNA sequencing library was prepared as previously described (62). Paired-end sequencing (100 million 100-bp paired-end reads) was performed on an Illumina HiSeq2500 sequencer (Illumina Inc.). The quality checks on raw sequence data for each sample were performed using FastQC (Babraham Bioinformatics). Raw reads were mapped to the reference hg19 using TopHat version 2.0. The alignment metrics of the mapped reads were estimated using SAMtools. Aligned reads were imported to the commercial data analysis platform AvadisNGS (Strand Scientifics). Samples were grouped and transcript abundance was quantified for this final read list using trimmed means of M-values (TMM) as the normalization method (63). RNA sequencing data have been deposited in the NCBI’s Gene Expression Omnibus (GEO) with ID code GSE100660.
Statistics. Statistical significance of differences was determined by 2-tailed Student’s t test or 1-way ANOVA followed by Newman-Keuls multiple-comparisons tests. A P value less than 0.05 was considered statistically significant. Values represent mean ± SEM. In RNA-Seq analysis, differential expression of genes was calculated on the basis of fold changes (using the default cutoff ≥ ±1.5) observed in comparisons between defined conditions, and the P value of the differentially expressed gene list was estimated by ANOVA using Benjamini-Hochberg corrections of 0.05 for FDR. Differentially expressed genes were used for the functional pathway analysis using Ingenuity Pathway Analysis (IPA; Qiagen Inc.).
Study approval. All human islet studies were approved by the Vanderbilt Institutional Review Board. All animal studies were approved by the Vanderbilt Institutional Animal Care and Use Committee.
-
Author contributions
CD, YH, SKK, and ACP designed the experiments. YH, CD, SKK, and ACP wrote the original manuscript draft, and YH, CD, SEL, DLG, LDS, RB, SKK, and ACP reviewed and edited the manuscript. CD, AS, GP, NH, N Prasad, N Phillips, and RB performed the experiments and analyzed the data.
-
Acknowledgments
We gratefully acknowledge organ donors and their families. We thank R. Banerjee, W. Goodyer, S. Kundu, and H. Chen for advice or help with tissue procurement, X. Gu for assistance with tissue processing and morphometry, and members of the Powers and Kim laboratories for comments on the manuscript. We thank David D’Alessio (Duke University) for helpful discussions. We thank Peng Wang and Andrew Stewart for providing the NFATC4 antibody. YH is supported by a Juvenile Diabetes Research Foundation (JDRF) postdoctoral fellowship (3-PDF-2014-196-A-N). Joint work in the Powers and Kim groups was supported by the NIH Beta Cell Biology Consortium (UO1DK89532) and the NIH Human Islet Research Network (UC4DK104211). Work in the Shultz and Greiner groups was supported by grants from the NIH Human Islet Research Network (UC4DK104218) and the Helmsley Charitable Trust. Work in the Kim group was also supported by grants from the Helmsley Charitable Trust, the H.L. Snyder Foundation, the Elser Foundation, the Doolittle Trust, the JDRF, the Howard Hughes Medical Institute, and the NIH (DK102612). Work in the Powers group was supported by grants from the NIH (DK66636, DK72473, DK89572, DK104211, DK108120, DK106755, DK094199, DK097829), the JDRF, the Department of Veterans Affairs, and the Cell Imaging Shared Resource and the Islet Procurement and Analysis Core of the Vanderbilt Diabetes Research and Training Center (DK20593).
Address correspondence to: Alvin C. Powers, Departments of Medicine and Molecular Physiology and Biophysics, Vanderbilt University School of Medicine, 7465 Medical Research Building IV, 2215 Garland Avenue, Nashville, Tennessee 37232-0475, USA. Phone: 615.936.7678; Email: [email protected]. Or to: Seung K. Kim, Department of Developmental Biology and Medicine, Stanford University School of Medicine, 279 Campus Drive, Beckman Center B300, Stanford, California 94305-5329, USA. Phone: 650.723.6230; Email: [email protected].
-
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Reference information: J Clin Invest. 2017;127(10):3835–3844.https://doi.org/10.1172/JCI91761.
-
References
- Teta M, Long SY, Wartschow LM, Rankin MM, Kushner JA. Very slow turnover of β-cells in aged adult mice. Diabetes. 2005;54(9):2557–2567.
- Meier JJ, et al. β-Cell replication is the primary mechanism subserving the postnatal expansion of β-cell mass in humans. Diabetes. 2008;57(6):1584–1594.
- Chen H, et al. Polycomb protein Ezh2 regulates pancreatic β-cell Ink4a/Arf expression and regeneration in diabetes mellitus. Genes Dev. 2009;23(8):975–985.
- Chen H, et al. PDGF signalling controls age-dependent proliferation in pancreatic β-cells. Nature. 2011;478(7369):349–355.
- Gregg BE, et al. Formation of a human β-cell population within pancreatic islets is set early in life. J Clin Endocrinol Metab. 2012;97(9):3197–3206.
- Avrahami D, et al. Aging-dependent demethylation of regulatory elements correlates with chromatin state and improved β cell function. Cell Metab. 2015;22(4):619–632.
- Jermendy A, et al. Rat neonatal β cells lack the specialised metabolic phenotype of mature β cells. Diabetologia. 2011;54(3):594–604.
- Aguayo-Mazzucato C, et al. Mafa expression enhances glucose-responsive insulin secretion in neonatal rat β cells. Diabetologia. 2011;54(3):583–593.
- Blum B, Hrvatin SS, Schuetz C, Bonal C, Rezania A, Melton DA. Functional β-cell maturation is marked by an increased glucose threshold and by expression of urocortin 3. Nat Biotechnol. 2012;30(3):261–264.
- Arda HE, et al. Age-dependent pancreatic gene regulation reveals mechanisms governing human β cell function. Cell Metab. 2016;23(5):909–920.
- Krishnamurthy J, et al. p16INK4a induces an age-dependent decline in islet regenerative potential. Nature. 2006;443(7110):453–457.
- Zhou JX, et al. Combined modulation of polycomb and trithorax genes rejuvenates β cell replication. J Clin Invest. 2013;123(11):4849–4858.
- Helman A, et al. p16(Ink4a)-induced senescence of pancreatic beta cells enhances insulin secretion. Nat Med. 2016;22(4):412–420.
- Chen H, et al. Augmented Stat5 signaling bypasses multiple impediments to lactogen-mediated proliferation in human β-cells. Diabetes. 2015;64(11):3784–3797.
- Campbell JE, Drucker DJ. Pharmacology, physiology, and mechanisms of incretin hormone action. Cell Metab. 2013;17(6):819–837.
- Abdulreda MH, Rodriguez-Diaz R, Caicedo A, Berggren PO. Liraglutide compromises pancreatic β cell function in a humanized mouse model. Cell Metab. 2016;23(3):541–546.
- Yang C, et al. Lixisenatide accelerates restoration of normoglycemia and improves human β-cell function and survival in diabetic immunodeficient NOD-scid IL-2rg(null) RIP-DTR mice engrafted with human islets. Diabetes Metab Syndr Obes. 2015;8:387–398.View this article via: PubMed Google Scholar
- Tschen SI, Dhawan S, Gurlo T, Bhushan A. Age-dependent decline in β-cell proliferation restricts the capacity of β-cell regeneration in mice. Diabetes. 2009;58(6):1312–1320.
- Perfetti R, Zhou J, Doyle ME, Egan JM. Glucagon-like peptide-1 induces cell proliferation and pancreatic-duodenum homeobox-1 expression and increases endocrine cell mass in the pancreas of old, glucose-intolerant rats. Endocrinology. 2000;141(12):4600–4605.
- Rankin MM, Kushner JA. Adaptive beta-cell proliferation is severely restricted with advanced age. Diabetes. 2009;58(6):1365–1372.
- Tschen SI, Georgia S, Dhawan S, Bhushan A. Skp2 is required for incretin hormone-mediated β-cell proliferation. Mol Endocrinol. 2011;25(12):2134–2143.
- Rutti S, Sauter NS, Bouzakri K, Prazak R, Halban PA, Donath MY. In vitro proliferation of adult human β-cells. PLoS One. 2012;7(4):e35801.
- Gromada J, Bokvist K, Ding WG, Holst JJ, Nielsen JH, Rorsman P. Glucagon-like peptide 1 (7-36) amide stimulates exocytosis in human pancreatic β-cells by both proximal and distal regulatory steps in stimulus-secretion coupling. Diabetes. 1998;47(1):57–65.View this article via: PubMed Google Scholar
- Shigeto M, et al. GLP-1 stimulates insulin secretion by PKC-dependent TRPM4 and TRPM5 activation. J Clin Invest. 2015;125(12):4714–4728.
- Crabtree GR, Olson EN. NFAT signaling: choreographing the social lives of cells. Cell. 2002;109(suppl):S67–S79.View this article via: PubMed Google Scholar
- Goodyer WR, Gu X, Liu Y, Bottino R, Crabtree GR, Kim SK. Neonatal β cell development in mice and humans is regulated by calcineurin/NFAT. Dev Cell. 2012;23(1):21–34.
- Heit JJ, et al. Calcineurin/NFAT signalling regulates pancreatic β-cell growth and function. Nature. 2006;443(7109):345–349.
- Oetjen E, et al. Inhibition of human insulin gene transcription by the immunosuppressive drugs cyclosporin A and tacrolimus in primary, mature islets of transgenic mice. Mol Pharmacol. 2003;63(6):1289–1295.
- Dai C, et al. Stress-impaired transcription factor expression and insulin secretion in transplanted human islets. J Clin Invest. 2016;126(5):1857–1870.
- Kjems LL, Holst JJ, Vølund A, Madsbad S. The influence of GLP-1 on glucose-stimulated insulin secretion: effects on β-cell sensitivity in type 2 and nondiabetic subjects. Diabetes. 2003;52(2):380–386.
- Kedees MH, Grigoryan M, Guz Y, Teitelman G. Differential expression of glucagon and glucagon-like peptide 1 receptors in mouse pancreatic α and β cells in two models of alpha cell hyperplasia. Mol Cell Endocrinol. 2009;311(1-2):69–76.
- Tornehave D, Kristensen P, Rømer J, Knudsen LB, Heller RS. Expression of the GLP-1 receptor in mouse, rat, and human pancreas. J Histochem Cytochem. 2008;56(9):841–851.
- Blodgett DM, et al. Novel observations from next-generation RNA sequencing of highly purified human adult and fetal islet cell subsets. Diabetes. 2015;64(9):3172–3181.
- Wang YJ, et al. Single-cell transcriptomics of the human endocrine pancreas. Diabetes. 2016;65(10):3028–3038.
- Segerstolpe Å, et al. Single-cell transcriptome profiling of human pancreatic islets in health and type 2 diabetes. Cell Metab. 2016;24(4):593–607.
- Fineman M, et al. Pharmacokinetics and pharmacodynamics of exenatide extended-release after single and multiple dosing. Clin Pharmacokinet. 2011;50(1):65–74.
- Bugliani M, et al. The direct effects of tacrolimus and cyclosporin A on isolated human islets: a functional, survival and gene expression study. Islets. 2009;1(2):106–110.
- Zhang H, et al. The FoxM1 transcription factor is required to maintain pancreatic beta-cell mass. Mol Endocrinol. 2006;20(8):1853–1866.
- Fiaschi-Taesch NM, et al. Human pancreatic β-cell G1/S molecule cell cycle atlas. Diabetes. 2013;62(7):2450–2459.
- Tiwari S, et al. Early and late G1/S cyclins and Cdks act complementarily to enhance authentic human β-cell proliferation and expansion. Diabetes. 2015;64(10):3485–3498.
- Fiaschi-Taesch N, et al. Survey of the human pancreatic β-cell G1/S proteome reveals a potential therapeutic role for cdk-6 and cyclin D1 in enhancing human β-cell replication and function in vivo. Diabetes. 2009;58(4):882–893.
- Li S, et al. The transcription factors Egr2 and Egr3 are essential for the control of inflammation and antigen-induced proliferation of B and T cells. Immunity. 2012;37(4):685–696.
- Parnaud G, et al. Proliferation of sorted human and rat β cells. Diabetologia. 2008;51(1):91–100.View this article via: PubMed Google Scholar
- Ishikawa F, et al. Development of functional human blood and immune systems in NOD/SCID/IL2 receptor {gamma} chain(null) mice. Blood. 2005;106(5):1565–1573.
- Shultz LD, et al. Human lymphoid and myeloid cell development in NOD/LtSz-scid IL2Rγ null mice engrafted with mobilized human hemopoietic stem cells. J Immunol. 2005;174(10):6477–6489.
- Butler PC, Meier JJ, Butler AE, Bhushan A. The replication of β cells in normal physiology, in disease and for therapy. Nat Clin Pract Endocrinol Metab. 2007;3(11):758–768.
- Wang YJ, et al. Single-cell mass cytometry analysis of the human endocrine pancreas. Cell Metab. 2016;24(4):616–626.
- Mulvihill EE, et al. Cellular sites and mechanisms linking reduction of dipeptidyl peptidase-4 activity to control of incretin hormone action and glucose homeostasis. Cell Metab. 2017;25(1):152–165.
- Smith EP, et al. The role of β cell glucagon-like peptide-1 signaling in glucose regulation and response to diabetes drugs. Cell Metab. 2014;19(6):1050–1057.
- Kuhre RE, et al. Fructose stimulates GLP-1 but not GIP secretion in mice, rats, and humans. Am J Physiol Gastrointest Liver Physiol. 2014;306(7):G622–G630.
- Soleimanpour SA, et al. Calcineurin signaling regulates human islet β-cell survival. J Biol Chem. 2010;285(51):40050–40059.
- Leech CA, et al. Molecular physiology of glucagon-like peptide-1 insulin secretagogue action in pancreatic β cells. Prog Biophys Mol Biol. 2011;107(2):236–247.
- Peiris H, et al. RCAN1 regulates mitochondrial function and increases susceptibility to oxidative stress in mammalian cells. Oxid Med Cell Longev. 2014;2014:520316. View this article via: PubMed Google Scholar
- Peiris H, et al. Increased expression of the glucose-responsive gene, RCAN1, causes hypoinsulinemia, β-cell dysfunction, and diabetes. Endocrinology. 2012;153(11):5212–5221.
- Screaton RA, et al. The CREB coactivator TORC2 functions as a calcium- and cAMP-sensitive coincidence detector. Cell. 2004;119(1):61–74.
- Kushner JA. The role of aging upon β cell turnover. J Clin Invest. 2013;123(3):990–995.
- Dai C, et al. Islet-enriched gene expression and glucose-induced insulin secretion in human and mouse islets. Diabetologia. 2012;55(3):707–718.
- Kayton NS, et al. Human islet preparations distributed for research exhibit a variety of insulin-secretory profiles. Am J Physiol Endocrinol Metab. 2015;308(7):E592–E602.
- Brissova M, et al. Intraislet endothelial cells contribute to revascularization of transplanted pancreatic islets. Diabetes. 2004;53(5):1318–1325.
- Brissova M, et al. Islet microenvironment, modulated by vascular endothelial growth factor-A signaling, promotes β cell regeneration. Cell Metab. 2014;19(3):498–511.
- Dai C, et al. Pancreatic islet vasculature adapts to insulin resistance through dilation and not angiogenesis. Diabetes. 2013;62(12):4144–4153.
- Alam SG, et al. The mammalian LINC complex regulates genome transcriptional responses to substrate rigidity. Sci Rep. 2016;6:38063. View this article via: PubMed Google Scholar
- Robinson MD, Oshlack A. A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol. 2010;11(3):R25.
-
Version history
- Version 1 (September 18, 2017): Electronic publication
- Version 2 (October 2, 2017): Print issue publication



Copyright © 2024 American Society for Clinical Investigation
ISSN: 0021-9738 (print), 1558-8238 (online)






