Advertisement
Review Free access | 10.1172/JCI84424
Targeting noncoding RNAs in disease
Brian D. Adams,1,2 Christine Parsons,3 Lisa Walker,4 Wen Cai Zhang,4 and Frank J. Slack4
1The RNA Institute, University at Albany, State University of New York, Albany, New York, USA.
2Investigative Medicine Program, Yale University Medical School, New Haven, Connecticut, USA.
3Department of Biochemistry, Bowdoin College, Brunswick, Maine, USA.
4Institute for RNA Medicine, Department of Pathology, Cancer Center, Beth Israel Deaconess Medical Center, Harvard Medical School, Boston, Massachusetts, USA.
Address correspondence to: Brian D. Adams, Research Faculty, The RNA Institute, University at Albany, State University of New York, 1400 Washington Avenue, Albany, New York 12222, USA. Or to: Brian D. Adams, Investigative Medicine Program, Yale University Medical School, 2 Church Street South, Suite 114, New Haven, Connecticut 06520, USA. Phone: 518.437.4447; E-mail: [email protected]. Or to: Frank J. Slack, Institute for RNA Medicine, Department of Pathology, Cancer Center, Beth Israel Deaconess Medical Center, Harvard Medical School, 330 Brookline Avenue, Boston, Massachusetts 02115, USA. Phone: 617.735.2601; E-mail: [email protected].
Authorship note: B.D. Adams and C. Parsons contributed equally to this work.
Find articles by
Adams, B.
in:
JCI
|
PubMed
|
Google Scholar
|

1The RNA Institute, University at Albany, State University of New York, Albany, New York, USA.
2Investigative Medicine Program, Yale University Medical School, New Haven, Connecticut, USA.
3Department of Biochemistry, Bowdoin College, Brunswick, Maine, USA.
4Institute for RNA Medicine, Department of Pathology, Cancer Center, Beth Israel Deaconess Medical Center, Harvard Medical School, Boston, Massachusetts, USA.
Address correspondence to: Brian D. Adams, Research Faculty, The RNA Institute, University at Albany, State University of New York, 1400 Washington Avenue, Albany, New York 12222, USA. Or to: Brian D. Adams, Investigative Medicine Program, Yale University Medical School, 2 Church Street South, Suite 114, New Haven, Connecticut 06520, USA. Phone: 518.437.4447; E-mail: [email protected]. Or to: Frank J. Slack, Institute for RNA Medicine, Department of Pathology, Cancer Center, Beth Israel Deaconess Medical Center, Harvard Medical School, 330 Brookline Avenue, Boston, Massachusetts 02115, USA. Phone: 617.735.2601; E-mail: [email protected].
Authorship note: B.D. Adams and C. Parsons contributed equally to this work.
Find articles by Parsons, C. in: JCI | PubMed | Google Scholar
1The RNA Institute, University at Albany, State University of New York, Albany, New York, USA.
2Investigative Medicine Program, Yale University Medical School, New Haven, Connecticut, USA.
3Department of Biochemistry, Bowdoin College, Brunswick, Maine, USA.
4Institute for RNA Medicine, Department of Pathology, Cancer Center, Beth Israel Deaconess Medical Center, Harvard Medical School, Boston, Massachusetts, USA.
Address correspondence to: Brian D. Adams, Research Faculty, The RNA Institute, University at Albany, State University of New York, 1400 Washington Avenue, Albany, New York 12222, USA. Or to: Brian D. Adams, Investigative Medicine Program, Yale University Medical School, 2 Church Street South, Suite 114, New Haven, Connecticut 06520, USA. Phone: 518.437.4447; E-mail: [email protected]. Or to: Frank J. Slack, Institute for RNA Medicine, Department of Pathology, Cancer Center, Beth Israel Deaconess Medical Center, Harvard Medical School, 330 Brookline Avenue, Boston, Massachusetts 02115, USA. Phone: 617.735.2601; E-mail: [email protected].
Authorship note: B.D. Adams and C. Parsons contributed equally to this work.
Find articles by Walker, L. in: JCI | PubMed | Google Scholar
1The RNA Institute, University at Albany, State University of New York, Albany, New York, USA.
2Investigative Medicine Program, Yale University Medical School, New Haven, Connecticut, USA.
3Department of Biochemistry, Bowdoin College, Brunswick, Maine, USA.
4Institute for RNA Medicine, Department of Pathology, Cancer Center, Beth Israel Deaconess Medical Center, Harvard Medical School, Boston, Massachusetts, USA.
Address correspondence to: Brian D. Adams, Research Faculty, The RNA Institute, University at Albany, State University of New York, 1400 Washington Avenue, Albany, New York 12222, USA. Or to: Brian D. Adams, Investigative Medicine Program, Yale University Medical School, 2 Church Street South, Suite 114, New Haven, Connecticut 06520, USA. Phone: 518.437.4447; E-mail: [email protected]. Or to: Frank J. Slack, Institute for RNA Medicine, Department of Pathology, Cancer Center, Beth Israel Deaconess Medical Center, Harvard Medical School, 330 Brookline Avenue, Boston, Massachusetts 02115, USA. Phone: 617.735.2601; E-mail: [email protected].
Authorship note: B.D. Adams and C. Parsons contributed equally to this work.
Find articles by Zhang, W. in: JCI | PubMed | Google Scholar
1The RNA Institute, University at Albany, State University of New York, Albany, New York, USA.
2Investigative Medicine Program, Yale University Medical School, New Haven, Connecticut, USA.
3Department of Biochemistry, Bowdoin College, Brunswick, Maine, USA.
4Institute for RNA Medicine, Department of Pathology, Cancer Center, Beth Israel Deaconess Medical Center, Harvard Medical School, Boston, Massachusetts, USA.
Address correspondence to: Brian D. Adams, Research Faculty, The RNA Institute, University at Albany, State University of New York, 1400 Washington Avenue, Albany, New York 12222, USA. Or to: Brian D. Adams, Investigative Medicine Program, Yale University Medical School, 2 Church Street South, Suite 114, New Haven, Connecticut 06520, USA. Phone: 518.437.4447; E-mail: [email protected]. Or to: Frank J. Slack, Institute for RNA Medicine, Department of Pathology, Cancer Center, Beth Israel Deaconess Medical Center, Harvard Medical School, 330 Brookline Avenue, Boston, Massachusetts 02115, USA. Phone: 617.735.2601; E-mail: [email protected].
Authorship note: B.D. Adams and C. Parsons contributed equally to this work.
Find articles by Slack, F. in: JCI | PubMed | Google Scholar
Authorship note: B.D. Adams and C. Parsons contributed equally to this work.
Published March 1, 2017 - More info
J Clin Invest. 2017;127(3):761–771. https://doi.org/10.1172/JCI84424.
Copyright © 2017, American Society for Clinical Investigation
-
Abstract
Many RNA species have been identified as important players in the development of chronic diseases, including cancer. Over the past decade, numerous studies have highlighted how regulatory RNAs such as microRNAs (miRNAs) and long noncoding RNAs (lncRNAs) play crucial roles in the development of a disease state. It is clear that the aberrant expression of miRNAs promotes tumor initiation and progression, is linked with cardiac dysfunction, allows for the improper physiological response in maintaining glucose and insulin levels, and can prevent the appropriate integration of neuronal networks, resulting in neurodegenerative disorders. Because of this, there has been a major effort to therapeutically target these noncoding RNAs. In just the past 5 years, over 100 antisense oligonucleotide–based therapies have been tested in phase I clinical trials, a quarter of which have reached phase II/III. Most notable are fomivirsen and mipomersen, which have received FDA approval to treat cytomegalovirus retinitis and high blood cholesterol, respectively. The continued improvement of innovative RNA modifications and delivery entities, such as nanoparticles, will aid in the development of future RNA-based therapeutics for a broader range of chronic diseases. Here we summarize the latest promises and challenges of targeting noncoding RNAs in disease.
-
Introduction
For decades, the central dogma that misregulated DNA, RNA, and proteins can result in disease states such as cancer has dominated our understanding of complex cellular processes. While many crucial, highly conserved metabolic and environmental protein-based feedback loops have been studied, a majority of these systems do not address the underlying genetic complexity associated with higher eukaryotic systems. This complexity is most likely derived from the noncoding RNA (ncRNA) portion of the genome, once thought of as “junk DNA.” In fact, approximately 98% of the human genome consists of noncoding DNA (1), and approximately 70% of the genome is actively transcribed, 2% of which codes for known protein-coding genes (2). Research over the last two decades revealed new classes of ncRNAs, including microRNAs (miRNAs), small nucleolar RNAs (snoRNAs), long ncRNAs (lncRNAs), and circular RNAs (circRNAs), all with different regulatory functions that effectively feed back into a larger RNA communication network that ultimately regulates the fundamental protein effectors of cellular function (Figure 1 and refs. 3–10).
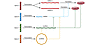 Figure 1
Figure 1The complexity of noncoding RNA gene networks. In this scenario, four genes are transcribed. However, only the splice variants mRNA-1 and mRNA-2 encoded by gene A are translated into protein products. These protein products can be regulated by noncoding RNAs (ncRNAs) that are encoded by genes B, C, and D, which interact with gene A at the RNA level in what is referred to as an RNA language or RNA network. Gene B encodes miRNA, which can interact with mRNAs at their 3′-UTR. Gene C encodes long noncoding RNA (lncRNA), which can interact with the protein products of gene A or serve as a decoy for certain miRNAs. Gene D encodes circular RNA (circRNA), which can sponge or serve as a decoy for any RNA binding event that indirectly regulates gene A protein products, such as lncRNA or miRNA interactions.
The first observation that RNA interference (RNAi) was possible occurred in transgenic plants, where an attempt was made to alter flower color in Petunia hybrida by reducing the expression of chalcone synthase by single-stranded antisense RNA (11). Soon after, Fire, Mello, and colleagues reported that injection of short dsRNA into Caenorhabditis elegans resulted in robust gene silencing (12). These early antisense technologies, however, used an endogenous RNA-induced silencing pathway. This concept was realized when miRNAs were discovered in C. elegans (8). Subsequent research identified miRNAs as regulators of biological processes, including development, proliferation, apoptosis, and cell metabolism (4, 13–18). These endogenous small RNAs are transcribed by RNA polymerase II, 5′-capped, and 3′-polyadenylated, and undergo sequential processing in the nucleus and cytoplasm by the RNase III enzymes DROSHA and DICER to generate an approximately 19- to 22-nt mature miRNA (19–21). The miRNA guides RNA-induced silencing complex (RISC) to the 3′-UTR of target mRNAs via sequence complementarity, resulting in degradation/destabilization or translational repression (22, 23). miRNAs regulate more than 60% of human protein-coding genes (24), and aberrant miRNA expression can alter the translational landscape within a cell, leading to deleterious phenotypic outcomes or disease states (18).
Another subset of ncRNAs transcribed by RNA polymerase II are lncRNAs (8). These transcripts are greater than 200 nt in length and are found in sense or antisense orientation to protein-coding genes, or within intergenic regions. lncRNAs regulate gene expression through a number of mechanisms, including epigenetic regulation via chromatin remodeling, transcriptional activation or repression, posttranscriptional modification of mRNA, or modulation of protein activity. lncRNAs can serve as scaffolds that recruit RNA-binding proteins (RBPs) into spatial proximity to each other, and/or guide RBPs to DNA. lncRNAs also function as decoys to compete for and disrupt protein-binding interactions, or to sponge miRNAs away from their natural mRNA targets (8, 25, 26). The X-inactive–specific transcript (XIST), which produces an approximately 20-kb RNA, was the first lncRNA discovered, and functions to balance X chromosome gene expression by recruiting polycomb repressive complex 2 (PRC2) to the inactive X chromosome. SHARP, SAF-A, and LBR are also required for XIST-mediated transcriptional silencing (27). Efforts to understand the epigenetic regulation mediated by lncRNAs have led to the finding that GWAS identify cancer risk loci outside of protein-coding regions, such as PTCSC3 in papillary thyroid carcinoma and ANRIL in type 2 diabetes, indicating that aberrant expression of lncRNAs associates with disease states (28–30).
As the biological significance of both small and long ncRNAs is becoming increasingly appreciated, numerous efforts have been initiated to target these ncRNAs in disease. Here we discuss development of therapeutic tools to target miRNA and lncRNAs in disease, the implications of these trials, and how continued elucidation of the underlying biology will serve as the basis for more effective rationales for ncRNA drug design and delivery.
-
General therapeutic strategies for ncRNA targeting and delivery
Essential RNA chemical modifications. As a class of molecules, miRNAs hold particular therapeutic potential because they regulate multiple gene targets within a particular signaling pathway, or a number of targets across several independent pathways (4, 18). Thus, the pleiotropic regulatory potential of miRNAs can alter cell growth and survival depending on the proper cell context, e.g., whether the miRNA targets functional pathways that are highly expressed in the specific cell or tissue of interest. Therefore, certain miRNAs may harbor stronger therapeutic effects when reintroduced or inhibited within a specific cell type. Efforts over the past decade have indicated that numerous miRNAs have the potential to become therapeutic targets, and the renewed interest in RNA therapeutics (31) has made it very likely that hundreds of novel RNA-based therapeutic agents may be developed within the next decade.
The current strategy to express or inhibit miRNA function involves delivering synthetic oligoribonucleotides (ORNs) that mimic the native miRNA duplex in cases in which miRNA overexpression is required, or generating a single-stranded antisense RNA to sequester the endogenous miRNA of interest for inhibition studies (Figure 2). As expected, use of unmodified ORNs encounters substantial stability and delivery barriers. To maintain RNA stability, modifications to protect ORNs against nucleases and enhance RNA binding affinity have been explored (32, 33). One of the first advancements was development of a 2′-O-methyl (2′-OMe) substitution in the sugar backbone of the RNA (34, 35), which significantly enhanced the stability of these RNAs in vitro. However, a variety of chemical modifications were developed over the past decade, including changes to the backbone structure of the ORN such as 2′-O-methoxyethyl (2′-MOE), 2′-fluoro, or locked nucleic acid (LNA) bases, along with oligomerization of the ORN with phosphorothioate linkages instead of a canonical phosphodiester bond (36–39). 2′-MOE– and LNA-based antisense oligonucleotide (ASO) strategies enhance RNA stability such that systemic delivery of these agents in vivo at doses of 10–20 mg/kg results in desired gene targeting within certain tissues (39, 40). Targeting moieties can also be directly conjugated to the ORN, such as N-acetylgalactosamine (GalNAc) for delivery to hepatocytes in liver disease. Regulus Therapeutics has developed an anti–miR-122 compound specifically for hepatitis C patients.
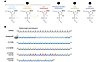 Figure 2
Figure 2Oligoribonucleotide modifications for therapeutic delivery. (A) Various oligoribonucleotide (ORN) modifications have been developed to improve RNA stability, including (i) substitutions of the 2′-OH of the ribose sugar, such as 2′-O-methyl (2′-OMe, light blue), 2′-O-methoxyethyl (2′-MOE, yellow), and 2′-fluoro (2′-F) substitutions (red); (ii) replacement of the phosphodiester linkage with a phosphorothioate (dark blue); (iii) locking of the conformation of the backbone with a methylene bridge using LNA modification (green); and (iv) DNA modification of the ribose sugar to a deoxyribose sugar (purple). (B) Examples of current modifications used in anti-miRNA ORN therapeutic strategies. Figure adapted with permission from Silence (ref. 33; Creative Commons user license available at http://creativecommons.org/licenses/by/2.0).
ORN modifications have been widely exploited for antisense approaches to sequester mature miRNAs. However, in thinking of methodologies to therapeutically reintroduce or re-express miRNAs, the available chemical modifications become rate-limiting. Synthetic miRNA duplexes must be stable and nuclease resistant, able to unwind to allow only mature guide strand loading into RISC, and have the guide strand properly recognize and bind its cognate mRNA target. Most chemical modifications involve addition of 2′-OMe groups across the passenger strand of the miRNA duplex, while 2′-OMe modifications can occur on the 3′ end of the mature miRNA strand (41). The 2′-OMe modification is sufficient to protect the ORN from degradation and immune detection and obviates the need for stable modifications, such as LNAs and phosphorothioate linkages, which might interfere with the required unwinding of the duplex while in RISC. Developing modified duplex RNAs that remain biologically active is a challenge, however. Attempts to deliver a single-stranded mature miRNA have failed (42), presumably because of its inability to load into RISC. Therefore, continued efforts are being undertaken to enhance the stability and cellular uptake of miRNA mimics for therapeutic applications.
RNA delivery vehicles. miRNA function is cell context dependent, and the ability to direct miRNA-based therapeutics to certain tissues remains a goal. Therefore, modifications are required to enhance cellular uptake of ORNs. ORNs with an electroneutral peptide-like backbone, such as peptide nucleic acids (PNAs), have been developed to silence miRNAs (43, 44). PNAs can be conjugated to cholesterol moieties to increase hydrophobicity, but also to cell-penetrating peptides to facilitate cellular uptake and enhanced tissue targeting. Adding lysine groups on PNAs can facilitate the endosomal release of PNA once taken up by target cells. Despite these advancements, a major impediment to RNA-based therapeutics is that ORN doses of more than 5 mg/kg are required before substantial therapeutic effects are observed in vivo, and these high doses pose risks of toxicity. Nanotechnology-based delivery strategies have significantly reduced the challenges of stability and target tissue specificity. These technologies allow RNA delivery at therapeutic doses of less than 2 mg/kg, reducing their toxicity profile (45). The most established nanodelivery agents used are lipid-based nanocarriers. These complexes are normally modified by PEGylation or through ionization to avoid unwanted aggregation in serum and bypass nonspecific tissue uptake. Cationic lipid complexes can also be decorated with tumor-targeting agents such as single-chain fragment variable (scFv) or transferrin (45, 46). Other lipid approaches involve neutral emulsion, where neutral phospholipids are used to package the miRNA, and therefore avoid issues of charge when attempting to deliver a highly polar RNA molecule across the cell membrane (47). This approach has proven successful for delivery of let-7 and miR-34a in preclinical KrasLSL-G12D lung cancer mouse models (47–49).
Numerous advances in delivery methodologies that do not rely on lipid-based approaches include polymer- and peptide-based systems. Poly(lactic-co-glycolic acid) (PLGA) nanoparticles can be used to deliver both siRNAs and charge-neutral PNA-modified miRNAs (50, 51). In addition to mitigating charge and electrostatic interactions that result in unfavorable miRNA delivery, PLGA nanoparticles can be decorated with surface peptides such as penetratin, to aid in the cellular uptake of the miRNA. Poly(amine-co-ester) terpolymer (PACE) nanoparticles are also attractive delivery platforms composed of a biodegradable and nontoxic polymer. They release therapeutic payloads gradually over time and can be coated with molecules such as polyE-mRGD to improve tumor targeting (52, 53). Additional delivery approaches include direct ORN conjugation using peptides such as pH low insertion peptide (pHLIP), which can deliver anti–miR-155 PNA to mice harboring B cell lymphomas in a pH-dependent manner (54, 55). Use of tumor-specific aptamers, small nucleic acids whose 3D structure binds to specific cell surface proteins identified by systematic evolution of ligands by exponential enrichment (SELEX), is also used to improve targeting of PNAs (56, 57). Importantly, many of these nanoparticle delivery methods are safe and biodegradable, indicating that they could rapidly be developed into “next-generation” miRNA therapeutics for clinical testing (Table 1 and Figure 3).
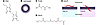 Figure 3
Figure 3Delivery vehicles/carriers for ncRNAs. The images depict the various delivery methodologies described in Table 1. Peptide nucleic acid (PNA) approaches (A) and lipid-based nanocarriers (B) are the most well-characterized delivery methods for RNA. Recently, new carriers have been developed, including poly(lactic-co-glycolic acid)–coated (PLGA-coated) nanoparticles (C), poly(amine-co-esters) such as PACE (D), and pH-sensitive peptides such as pHLIP (E).
New tools for lncRNA delivery. lncRNAs function as decoys, regulators of translation, and modular scaffolds that guide chromatin-modifying enzymes to specific genomic loci (8). lncRNAs are an attractive class of therapeutic target given the tight transcriptional regulation, tissue-specific expression, and dysregulation in disease. Associations of HOTAIR in breast cancer (58, 59) as well as MALAT1 in metastatic lung cancer (60, 61) are notable examples of this relationship. The first evidence that modulating lncRNAs or using their flanking regulatory regions could have therapeutic value originated from studies of the H19 gene, an lncRNA with oncogenic properties that is highly expressed in a wide range of tumors. Intratumoral injection of plasmids expressing diphtheria toxin under control of the H19 promoter led to tumor reduction in bladder cancer xenograft mouse models (62). From this work, several phase I/II clinical trials have been initiated to treat patients with bladder, ovarian, and pancreatic cancer. Therefore, there has been excitement over the prospect of developing therapeutic tools to modulate misregulated lncRNAs in disease.
While lncRNAs can be found in the cytoplasm, they are predominantly nuclear (63). Therefore, modified ASOs have been used in an effort to target the lncRNA in the nuclear compartment to initiate RNase H–dependent degradation of the particular target lncRNA. Continued advancement of this technology includes the use of 2′-OMe RNA and LNA modifications at both the 5′ and the 3′ end of the ASO to protect against nuclease activity (64, 65). Despite improvements, the highly structured nature of lncRNAs can preclude ASO binding to a predicted site within the lncRNA molecule. While one solution is to generate many siRNAs to a particular lncRNA, the cost of assessing off-target effects for each molecule attempted can be prohibitive.
Another approach to modulate lncRNA expression in cells is to target promoter regions of genes. A majority of gene loci contain promoter-associated RNA (pRNA) transcripts that are transcribed in the sense orientation just upstream of an mRNA or lncRNA transcriptional start site (66, 67). pRNAs function as cis-regulatory elements by keeping the surrounding chromatin in an epigenetically active state. The use of ASOs or LNAs directed toward these pRNAs induces the recruitment of the nuclear RISC complex and a subsequent remodeling of the local chromatin (59). This involves recruitment of HDAC1, EZH2, and DNMT3A, which promotes enrichment of silent-state chromatin modifications, heterochromatin formation, and ultimately transcriptional gene silencing.
Although lncRNAs represent appealing pharmacological and therapeutic targets, inhibiting lncRNAs in vivo remains a challenge. Many limitations of delivering ASOs for lncRNAs are similar for all RNAi-based therapies. However, one tractable approach has been the use of small molecules that disrupt lncRNA-chromatin–modifying complexes to alter the epigenetic state of the target cell. It is well accepted that lncRNAs can regulate JARID2 and EZH2 (68), and if small molecules can be developed to target the binding cleft between the lncRNA and its protein-binding partner, these interactions may be disrupted. Similar approaches have been used for targeting miRNA function or disrupting the miRNA processing machinery (69–71). Taken together, these delivery efforts, along with further elucidation of lncRNA regulatory mechanisms, will ultimately lead to the development of effective therapeutic strategies that target lncRNAs in vivo.
-
The role of ncRNAs in chronic disease
Development of ncRNA-based therapies could be considered a novel tool in treating a number of chronic diseases. For instance, protein products that are known culprits of the disease (e.g., MYC in cancer) have proven difficult to target (72). The development of RNAi-based drugs enabled highly specific targeting of genes of interest at the mRNA level. An extension of RNAi-based therapies is the ability to target the endogenous ncRNA transcriptome within a particular disease. Indeed, aberrant miRNA and lncRNA expression has been linked to a number of chronic illnesses, from heart disease to diabetes to cancer. Given the cell-specific context in which most ncRNAs operate, identifying the key misregulated ncRNAs within a particular disease context will lead to identification of ncRNA candidates for therapeutic targeting (Table 2).
ncRNAs support development of diabetes. Investigators have elucidated that ncRNAs are key players in metabolic pathways that support development of both type 1 and type 2 diabetes. Mice with an inducible β cell deletion of Dicer harbor a diabetic phenotype due to a reduction in insulin production, despite normal islet architecture (73). Loss of miR-26a, miR-148, miR-182, and miR-200, among other miRNAs, may be responsible for the reduced insulin phenotype, as these miRNAs positively regulate insulin transcription. A more actionable therapeutic target is miR-375, given its high abundance in pancreatic β cells. Modulation of miR-375 activity by 2′-OMe, ORN knockdown, or recombinant adenovirus overexpression in MIN6 cells, an insulin-secreting glucose-responsive pancreatic β cell line, alters glucose-stimulated insulin secretion and exocytosis through a novel pathway involving myotrophin (encoded by Mtpn) (74). miR-375 KO mice are hyperglycemic owing to decreased pancreatic β cell mass secondary to reduced proliferation, while in leptin KO mice, miR-375 levels are elevated and correlate with increased β cell mass (75). Clinically, miR-375 shows potential as a biomarker, as it is overexpressed in the serum of type 2 diabetic patients (76), and circulating miR-375 levels dramatically increase in mice before the onset of hyperglycemia (77). Therefore, while miR-375 is itself essential for normal glucose homeostasis and β cell turnover, dampening the effects of aberrant miR-375 expression in hyperglycemic patients represents a viable therapeutic strategy.
Despite preclinical evidence for use of certain miRNAs in the treatment of diabetes, Regulus Therapeutics has developed lead compounds such as RG-125(AZD4076), a GalNAc-conjugated anti–miR-103/107 ORN that can improve insulin sensitivity in preclinical mouse models. Use of anti–miR-103/107 and transient adenovirus-mediated expression of miR-103/107 in Mir103-transgenic mice was sufficient to induce an impairment of glucose homeostasis and alter insulin sensitivity (78, 79). Use of RG-125(AZD4076) in a diet-induced obesity model promoted reduced fasting blood glucose and plasma insulin levels as well as enhanced glucose tolerance, suggesting that miR-103/107 plays a central role in insulin sensitivity. AstraZeneca purchased RG-125(AZD4076) from Regulus, and together these companies are developing the compound to treat nonalcoholic steatohepatitis in patients with type 2 diabetes (80).
ncRNAs and the aging heart. ncRNAs control important processes in cardiac development and physiology, and aberrant expression of these ncRNAs have been linked to cardiovascular disease. α-MHC-Cre–mediated deletion of Dicer results in postnatal lethality due to cardiomyopathy (81), and a series of miRNAs abundantly expressed in striated muscle (myomiRs) are thought to be crucial regulators of cardiomyocyte proliferation, morphogenesis, and remodeling (82, 83). Inhibition of one of these myomiRs, miR-208a, by systemic delivery of antisense ORNs can improve cardiac function and survival during hypertension-induced heart failure in Dahl hypertensive rats (84). Importantly, approximately 80% of mice harboring a homozygous deletion of Mir208a had significant disruption of normal electrical conductivity resulting in atrial fibrillation (85). Therefore, proper dosing strategies will be required in nonhuman primates before therapeutic approaches are implemented.
miRNA profiling also indicated that members of the miR-15 family, including miR-195, miR-15a, miR-16, and miR-497, were important regulators of postnatal cardiomyocyte mitotic arrest by controlling cell cycle regulators, including Chek1. miR-15a is also upregulated during cardiac ischemia and heart failure (84). rAAV9-mediated delivery of anti–miR-15a, which has been used to deliver siRNAs to cardiac tissue in rats (85) and is a safe and effective delivery method widely used in clinical trials, may be an approach to alleviate the pathological hypertrophy and fibrosis associated with reduced cardiac function and failure. However, miRagen Therapeutics is currently in the preclinical stage of development of MGN-1374, an LNA-based anti–miR-15/195 compound (86). MGN-1374 was shown to enhance heart function and promote cardiomyocyte proliferation in murine and porcine models (87), indicating that this compound has potential to move forward for phase I trials.
lncRNAs are also associated with cardiac disease and could be targeted by standard ORN approaches. The lncRNA ANRIL was identified in large-scale GWAS for coronary artery disease (88) and is expressed in vascular endothelial and coronary smooth muscle cells (89). ANRIL may regulate the INK4a/b locus by recruiting PRC2 to impose a repressive chromatin state, thereby altering the senescent state of cells (90). It is not clear whether overexpression or knockdown of ANRIL would affect cardiomyocytes; therefore, the therapeutic augmentation of cardiac function by targeting of ANRIL during ischemic events is unclear. It will be important to elucidate the mechanisms of lncRNAs in other cardiac injury models, such as chemotherapy-induced cardiotoxicity, or inflammation-mediated ischemia, before fully pursuing lncRNA-based therapeutic strategies.
ncRNAs in neurogenesis and Alzheimer’s disease. Within the CNS, ncRNAs are expressed in specific spatiotemporal patterns and play a crucial role in buffering gene products along neuronal processes and synaptic junctions (91, 92). Improper integration of this information, perhaps through loss of the regulatory networks imparted by ncRNAs, can result in disease pathogenesis. For instance, ATXN8OS, an antisense RNA that encodes a CUG repeat, has been implicated in spinocerebellar ataxia type 8 (SCA8). SCA8 is an autosomal dominant disorder thought to be caused by a polyglutamine repeat expansion within the ATXN8 gene (93, 94). However, transgenic mice expressing ATXN8OS harbor dysregulated muscleblind-like splicing regulator 1–mediated pre-mRNA splicing. It would be interesting if a siRNA-based therapeutic strategy could be developed to target either ATXN8OS or the repeat expansion tracts within the regulatory regions of the ATXN8 or the ATXN8OS gene, or to correct the splicing defects of downstream effector genes such as Gabt4.
ncRNAs have also been reported to be dysregulated in Alzheimer’s disease (AD). For instance, miR-9 controls neuronal stem cell differentiation, is enriched within synapses, and is downregulated in AD. miR-9 appears to regulate the amyloid-β (Aβ) induction of the CAMKK2/AMPK pathway, and is itself controlled by Aβ42 (95). In primary neuronal cultures, miR-9 overexpression inhibited CAMKK2, attenuated Aβ42-induced synaptotoxicity, and regulated the hyperphosphorylation of tau proteins, which combine to form toxic intracellular aggregations. Therefore, ncRNAs are actionable targets for treating neurological disorders. However, a more robust panel of ncRNA targets to correct the phenotypes associated with AD without causing unexpected toxicity to normal brain or spinal function is needed. Once these candidates are identified, delivering ORNs across the blood-brain barrier will present another challenge. The antisense strategies originally used to target miRNAs in vivo, termed antagomiRs, are delivered to many tissues, with the notable exception of the brain (96). This suggests that certain chemical modifications of a single-stranded RNA are not sufficient for robust delivery and uptake into the CNS without an additional carrier. While certain delivery agents are being developed, companies including miRagen Therapeutics have an anti–miR-155 compound using LNA-based chemistries that may enable investigational new drugs for patients with ALS (97). Relying on newer chemically modified antisense ORNs may prove effective, given that modulation of certain miRNAs can promote tau phosphorylation in rat primary hippocampal neurons and impaired learning when injected directly into the hippocampal dentate gyrus in 3-month-old C57BL/6 WT mice (98).
-
ncRNAs in cancer
Hanahan and Weinberg’s landmark review describes six molecular hallmarks that are shared among all cancer types (99). In particular, successful initiation of tumorigenesis requires that cells develop the ability to sustain proliferative signaling, evade growth suppressors, enable replicative immortality, resist cell death, induce angiogenesis, and activate invasion and metastasis. It is clear that ncRNAs are dysregulated in cancer and have been implicated in regulating fundamental tumorigenic processes (Table 3). This is explained by reciprocal or double-negative transcription factor–miRNA feedback loops that result in the fine-tuning of gene expression during normal cellular function (100–102). During tumorigenesis, these regulatory loops become disrupted. Similarly, lncRNAs such as MALAT-1, HOTAIR, and ANRIL are elevated in cancer while MEG3, lincRNA-p21, and PTENP1 appear to be downregulated (103). While the mechanisms by which lncRNAs contribute to cancer pathogenesis are still unclear, accumulating evidence suggests that lncRNAs regulate chromatin-modifying proteins such as PRC2 and coREST (104, 105). Given that hypermethylation of gene promoter regions occurs often in cancer, lncRNAs may harbor tumor-suppressive or oncogenic functions depending on how dysregulation of the respective lncRNA affects the activity of the aforementioned chromatin-modifying complexes.
Oncogenic ncRNAs can inhibit growth suppressors. For instance, a key regulator of CDKN1B is the miR-221/222 family of miRNAs, which are overexpressed in multiple cancer types. Targeting of CDKN1B by this miRNA family promotes enhanced cell cycle progression from G1 to S phase resulting in sustained proliferation (100). miR-Zip knockdown of miR-221 in an MDA-MB-231 triple-negative breast cancer mouse model inhibited tumor growth, confirming the pro-proliferative effect of miR-221 in vivo (101, 102). The targeting of another oncogenic miRNA, miR-155, is being pursued given that miR-155 supports lymphoma development in mouse models in the absence of Myc (50). miRagen Therapeutics has announced a phase I clinical trial for use of an unmodified LNA anti–miR-155 molecule, MRG-106, against cutaneous T cell lymphoma (CTCL). Since delivery of MRG-106 will be via direct injection into skin lesions, this compound’s delivery efficacy should be high, and it will be interesting to know how CTCL lesions respond to MRG-106 treatment.
Additionally, tumor-suppressive ncRNAs can target growth promoters. While numerous miRNAs are implicated in this process, recently, miR-34a has made headlines as the first miRNA mimic–based therapy to enter human clinical trials (ClinicalTrials.gov, NCT01829971). In preclinical models, systemic administration of miR-34a, packaged as a neutral lipid emulsion, either alone or in combination with let-7 (48, 49), reduced lung tumor volumes in KrasLSL-G12D p53loxP/loxP mice. The therapeutic effectiveness of tumor regression induced by miR-34a is unsurprising, given that miR-34a harbors strong tumor-suppressive properties across a broad spectrum of tumor types. A recent approach developed by the Asbestos Diseases Research Institute and EnGeneIC was instead used to re-express miR-16 in patients with malignant pleural mesothelioma and advanced non–small cell lung cancer (NSCLC) failing standard therapy (NCT02369198). These TargomiRs are hoped to deliver miR-16 in NSCLC as effectively as MRX34 in liver cancer. Interestingly, recent work indicates that the growth-suppressive effect of miR-16 is enhanced when it is coupled with miR-34a (105), highlighting the importance of combinatorial miRNA therapeutic approaches.
ncRNAs can also alter the senescent state of cancer cells. The miRNA regulation of sirtuins (e.g., miR-34a and SIRT1; ref. 47) is a classic example, given that sirtuins regulate the activity of gene products involved in cellular metabolism, inflammation, transcription, and cell survival. ncRNAs also regulate telomeric regions of chromosomes, which progressively shorten during cell division, eventually triggering senescence. Cancer cells circumvent this by expressing telomerase reverse transcriptase (TERT), which adds telomeric repeats to the 3′ end of chromosomes. The lncRNA TERRA is transcribed from the same locus and binds to telomerase, inhibiting its activity in vitro (106). Subsequently, tumor cells actively downregulate TERRA, extending the longevity of cancer cells through telomerase-mediated lengthening of chromosomal ends. An additional senescence-promoting ncRNA is GAS5, which induces downregulation of CDK6 in pancreatic, bladder, and gastric cancer and in some cases is anticorrelated with tumor size and advanced clinical stage (107, 108). Inhibition of GAS5 is associated with an increased frequency of cells in S phase, increased CDK6 protein levels, and altered p21/WAF1 levels, such that the incidence of cellular senescence is reduced (108). In an in vivo colorectal cancer model, tumors ectopically expressing GAS5 harbored significantly reduced tumor volumes and tumor burden, suggesting that targeting GAS5 is a potential therapeutic (109).
ncRNAs can also alter the cellular survival response to uncontrolled proliferation and DNA damage (110). miR-221/222 can abrogate programmed cell death in numerous cancers, and inhibition of these miRNAs increases apoptosis through activation of SIRT1 and/or downregulation of PUMA (111, 112). The oncogenic lncRNA ANRIL was also implicated in resistance to cell death. ANRIL was discovered by mapping of a germline deletion in a family with melanoma–neural system tumor syndrome. ANRIL functions by epigenetically silencing its target genes via recruitment of PRC2 to gene regions, including the P15 (also known as CDKN2B) gene loci (113). Therefore, under a normal physiological state, ANRIL functions as a balancer of p15 expression, and this homeostatic relationship becomes disrupted in cancer (114). siRNA studies in xenograft mouse models indicated that knockdown of ANRIL results in slower tumor growth and lower tumor weight than in controls (115). Overall, this suggests that ANRIL is a regulator of the key genetic networks controlling cell death in cancer, and could be a potential therapeutic target.
ncRNAs can also regulate neovascularization, a key feature of the tumorigenic process described by Judah Folkman nearly 5 decades ago (116). As tumors increase in size, associated capillary outgrowth occurs from the tumor to surrounding stroma and host blood vessels. This dependence on vessel formation for tumor growth indicated that tumor angiogenesis could be targeted therapeutically (117). ncRNAs play a role in tumor angiogenesis, given that miRNA-deficient tumors generated from Dicer KO mice are poorly vascularized and hypoxic (118, 119). Numerous miRNAs have been shown to modulate angiogenesis in mice (120–122). miR-7, for example, was delivered as a biodegradable polymeric nanoparticle to inhibit angiogenesis in a chick chorioallantoic membrane assay, as well as in a human glioblastoma xenograft model (123). A growing number of lncRNAs, such as HULC and HOXD-AS1, have also been identified as angiogenesis regulators (124). However, the lncRNA MEG3, part of the TP53 regulatory feedback loop, appears to play a more predominant role in angiogenesis (125). Enhanced expression of proangiogenic genes coincides with low MEG3 levels in human tumors, and in mice maternal deletion of Meg3 results in a significant increase in microvessel formation in the brain. These studies highlight the growing list of angiogenic factors that could serve as targets for cancer treatment.
While the molecular underpinning of metastasis has yet to be elucidated, it is clear that ncRNAs are fundamental regulators of this epithelial-to-mesenchymal transition (EMT). miR-21 is a classic example of an ncRNA that promotes the migration and invasion of cells in a broad range of cancers, primarily through downregulation of phosphatase and tensin homolog (PTEN) and programmed cell death protein 4 (PDCD4). Inhibition of miR-21 in chicken embryo metastasis assays is associated with less intravasation of colorectal tumor cells (126, 127). miR-21–mediated regulation of PTEN results in increased IL-6 signaling, subsequently inducing key characteristics of EMT, including PI3K activation and enhanced levels of phosphorylated STAT3 and NF-κB in HER2-positive breast cancer cells (128). In a different in vivo study using MDA-MB-231 breast cancer cells, delivery of anti–miR-10b in a nanoparticle formulation prevented lymph node metastasis (129). Additionally, a number of antimetastatic targets are regulated by miR-10b, including HOXD10, TBX5, and PTEN, and blocking miR-10b activity inhibits the expression of prometastatic factors such as RhoC (130, 131). Other groups have targeted the cell-extrinsic composition of the metastatic microenvironment. Le et al. found that extracellular vesicles derived from highly metastatic 4T1E breast cancer cells promoted the metastasis of weakly metastatic 4T07 cells in vivo (132). miR-200 was implicated in this process as a prometastatic factor despite the canonical role of the miR-200 family in suppressing EMT (133–135). Similarly, Zetter and colleagues found that metastatic cells from the lymph nodes of mice with prostate tumors exhibited an increasingly epithelial phenotype and an increased level of miR-200 (136). These studies raise the prospect of targeting exosomes to block tumor metastasis as a new and innovative therapeutic approach.
-
Conclusions
The number of ncRNAs that have the potential to become therapeutic targets is steadily growing. ncRNAs can regulate numerous aspects of cellular biology. Canonical miRNAs such as miR-34, miR-155, and miR-200b were developed or are in development into therapeutic drugs to treat specific patient populations with chronic or rare diseases. Over the past 5 to 10 years, new ncRNA targets have emerged on the basis of strong in vitro and in vivo data indicating that these ncRNAs regulate fundamental cellular processes. Many novel lncRNAs are being discovered, and the mechanism of action for each lncRNA varies depending on the cellular context. In this Review, we have focused on key ncRNAs that may eventually be developed into novel therapies in the respective disease setting. Currently, over 100 antisense-based oligonucleotide therapies are in clinical trials. While the stability of these small yet powerful RNAs has been vastly improved, the issues associated with targeting these RNAs to the correct tissues of interest still remain. Biomedical engineering and nanoparticle developmental efforts have begun to develop the tools that allow for this tissue targeting. These second-generation miRNA-based therapeutics offer the potential for a greater delivery payload to the tissue site while reducing RNA-mediated toxicity. Overall, the continued development of innovative RNA modifications and delivery entities such as nanoparticles will aid in the development of future RNA-based therapeutics for a broader range of chronic diseases.
-
Acknowledgments
This work was supported by grants to FJS from the NIH (P50 CA196530, R01 CA157749, and R01 CA131301). BDA was supported by a Career Development Award from P50 CA196530. CP was supported by a Sackler/National Science Foundation REU grant and by the Bowdoin College Alumni Council Internship Fund. We thank Eleni Anastasiadou for critical reading of the manuscript.
Address correspondence to: Brian D. Adams, Research Faculty, The RNA Institute, University at Albany, State University of New York, 1400 Washington Avenue, Albany, New York 12222, USA. Or to: Brian D. Adams, Investigative Medicine Program, Yale University Medical School, 2 Church Street South, Suite 114, New Haven, Connecticut 06520, USA. Phone: 518.437.4447; E-mail: [email protected]. Or to: Frank J. Slack, Institute for RNA Medicine, Department of Pathology, Cancer Center, Beth Israel Deaconess Medical Center, Harvard Medical School, 330 Brookline Avenue, Boston, Massachusetts 02115, USA. Phone: 617.735.2601; E-mail: [email protected].
-
Footnotes
CP’s present address is: Department of Medicine, University of Connecticut Health Center, Farmington, Connecticut, USA.
Conflict of interest: F.J. Slack has ownership interest (including patents; US 7,893,034) in, and is a consultant and advisory board member for, Mirna Therapeutics. B.D. Adams holds patent interests with, and consults with, AUM LifeTech.
Reference information: J Clin Invest. 2017;127(3):761–771.
https://doi.org/10.1172/JCI84424.
-
References
- Mattick JS. Non-coding RNAs: the architects of eukaryotic complexity. EMBO Rep. 2001;2(11):986–991.
- Djebali S, et al. Landscape of transcription in human cells. Nature. 2012;489(7414):101–108.
- Mattick JS, Makunin IV. Non-coding RNA. Hum Mol Genet. 2006;15(suppl 1):R17–R29.View this article via: PubMed Google Scholar
- Esquela-Kerscher A, Slack FJ. Oncomirs — microRNAs with a role in cancer. Nat Rev. 2006;6(4):259–269.
- Memczak S, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495(7441):333–338.
- Volinia S, et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci U S A. 2006;103(7):2257–2261.
- Lee RC, Feinbaum RL, Ambros V. The C.elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75(5):843–854.
- Rinn JL, Chang HY. Genome regulation by long noncoding RNAs. Annu Rev Biochem. 2012;81:145–166.
- Kiss T. Small nucleolar RNA-guided post-transcriptional modification of cellular RNAs. EMBO J. 2001;20(14):3617–3622.
- Kiss T. Small nucleolar RNAs: an abundant group of noncoding RNAs with diverse cellular functions. Cell. 2002;109(2):145–148.
- Napoli C, Lemieux C, Jorgensen R. Introduction of a chimeric chalcone synthase gene into petunia results in reversible co-suppression of homologous genes in trans. Plant Cell. 1990;2(4):279–289.View this article via: PubMed Google Scholar
- Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391(6669):806–811.
- Kato M, Slack FJ. microRNAs: small molecules with big roles — C.elegans to human cancer. Biol Cell. 2008;100(2):71–81.
- Zhou J, Ng SB, Chng WJ. LIN28/LIN28B: an emerging oncogenic driver in cancer stem cells. Int J Biochem Cell Biol. 2013;45(5):973–978.
- Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136(2):215–233.
- Mendell JT, Olson EN. MicroRNAs in stress signaling and human disease. Cell. 2012;148(6):1172–1187.
- Finnerty JR, Wang WX, Hébert SS, Wilfred BR, Mao G, Nelson PT. The miR-15/107 group of microRNA genes: evolutionary biology, cellular functions, and roles in human diseases. J Mol Biol. 2010;402(3):491–509.
- Adams BD, Kasinski AL, Slack FJ. Aberrant regulation and function of microRNAs in cancer. Curr Biol. 2014;24(16):R762–R776.
- Macfarlane LA, Murphy PR. MicroRNA: biogenesis, function and role in cancer. Curr Genomics. 2010;11(7):537–561.
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116(2):281–297.
- Lee Y, et al. MicroRNA genes are transcribed by RNA polymerase II. EMBO J. 2004;23(20):4051–4060.
- Winter J, Jung S, Keller S, Gregory RI, Diederichs S. Many roads to maturity: microRNA biogenesis pathways and their regulation. Nat Cell Biol. 2009;11(3):228–234.
- Fabian MR, Sonenberg N, Filipowicz W. Regulation of mRNA translation and stability by microRNAs. Annu Rev Biochem. 2010;79:351–379.
- Friedman RC, Farh KK, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19(1):92–105.View this article via: PubMed Google Scholar
- Lee JT. Epigenetic regulation by long noncoding RNAs. Science. 2012;338(6113):1435–1439.
- Ponting CP, Oliver PL, Reik W. Evolution and functions of long noncoding RNAs. Cell. 2009;136(4):629–641.
- McHugh CA, et al. The Xist lncRNA interacts directly with SHARP to silence transcription through HDAC3. Nature. 2015;521(7551):232–236.
- Cheetham SW, Gruhl F, Mattick JS, Dinger ME. Long noncoding RNAs and the genetics of cancer. Br J Cancer. 2013;108(12):2419–2425.
- Pasmant E, Sabbagh A, Vidaud M, Bièche I. ANRIL, a long, noncoding RNA, is an unexpected major hotspot in GWAS. FASEB J. 2011;25(2):444–448.
- Jendrzejewski J, et al. The polymorphism rs944289 predisposes to papillary thyroid carcinoma through a large intergenic noncoding RNA gene of tumor suppressor type. Proc Natl Acad Sci U S A. 2012;109(22):8646–8651.
- Zeliadt N. Big pharma shows signs of renewed interest in RNAi drugs. Nat Med. 2014;20(2):109.
- Van Rooij E, Purcell AL, Levin AA. Developing MicroRNA therapeutics. Circ Res. 2012;110(3):496–507.
- Stenvang J, Petri A, Lindow M, Obad S, Kauppinen S. Inhibition of microRNA function by antimiR oligonucleotides. Silence. 2012;3(1):1.
- Hutvágner G, Simard MJ, Mello CC, Zamore PD. Sequence-specific inhibition of small RNA function. PLoS Biol. 2004;2(4):E98.
- Meister G, Landthaler M, Dorsett Y, Tuschl T. Sequence-specific inhibition of microRNA-and siRNA-induced RNA silencing. RNA. 2004;10(3):544–550.
- Elmén J, et al. LNA-mediated microRNA silencing in non-human primates. Nature. 2008;452(7189):896–899.
- Ørom UA, Kauppinen S, Lund AH. LNA-modified oligonucleotides mediate specific inhibition of microRNA function. Gene. 2006;372:137–141.
- Davis S, Lollo B, Freier S, Esau C. Improved targeting of miRNA with antisense oligonucleotides. Nucleic Acids Res. 2006;34(8):2294–2304.
- Esau C, et al. miR-122 regulation of lipid metabolism revealed by in vivo antisense targeting. Cell Metab. 2006;3(2):87–98.
- Obad S, et al. Silencing of microRNA families by seed-targeting tiny LNAs. Nat Genet. 2011;43(4):371–378.
- Henry JC, Azevedo-Pouly ACP, Schmittgen TD. MicroRNA replacement therapy for cancer. Pharm Res. 2011;28(12):3030–3042.
- Henry JC, et al. miR-199a-3p targets CD44 and reduces proliferation of CD44 positive hepatocellular carcinoma cell lines. Biochem Biophys Res Commun. 2010;403(1):120–125.
- Fabani MM, Gait MJ. miR-122 targeting with LNA/2′-O-methyl oligonucleotide mixmers, peptide nucleic acids (PNA), and PNA-peptide conjugates. RNA. 2008;14(2):336–346.View this article via: PubMed Google Scholar
- Nielsen PE, Egholm M, Buchardt O. Peptide nucleic acid (PNA). A DNA mimic with a peptide backbone. Bioconjug Chem. 1994;5(1):3–7.
- Misso G, et al. Mir-34: a new weapon against cancer? Mol Ther Nucleic Acids. 2014;3:e194. View this article via: PubMed Google Scholar
- Pirollo KF, et al. Tumor-targeting nanocomplex delivery of novel tumor suppressor RB94 chemosensitizes bladder carcinoma cells in vitro and in vivo. Clin Cancer Res. 2008;14(7):2190–2198.
- Trang P, et al. Systemic delivery of tumor suppressor microRNA mimics using a neutral lipid emulsion inhibits lung tumors in mice. Mol Ther. 2011;19(6):1116–1122.
- Kasinski AL, Slack FJ. miRNA-34 prevents cancer initiation and progression in a therapeutically resistant K-ras and p53-induced mouse model of lung adenocarcinoma. Cancer Res. 2012;72(21):5576–5587.
- Kasinski AL, et al. A combinatorial microRNA therapeutics approach to suppressing non-small cell lung cancer. Oncogene. 2015;34(27):3547–3555.
- Babar IA, et al. Nanoparticle-based therapy in an in vivo microRNA-155 (miR-155)-dependent mouse model of lymphoma. Proc Natl Acad Sci U S A. 2012;109(26):E1695–E1704.
- Woodrow KA, Cu Y, Booth CJ, Saucier-Sawyer JK, Wood MJ, Saltzman WM. Intravaginal gene silencing using biodegradable polymer nanoparticles densely loaded with small-interfering RNA. Nat Mater. 2009;8(6):526–533.
- Zhou J, et al. Biodegradable poly(amine-co-ester) terpolymers for targeted gene delivery. Nat Mater. 2011;11(1):82–90.
- Adams BD, et al. miR-34a silences c-SRC to attenuate tumor growth in triple-negative breast cancer. Cancer Res. 2016;76(4):927–939.
- Cheng CJ, et al. MicroRNA silencing for cancer therapy targeted to the tumour microenvironment. Nature. 2015;518(7537):107–110.View this article via: PubMed Google Scholar
- Yao L, et al. pHLIP peptide targets nanogold particles to tumors. Proc Natl Acad Sci U S A. 2013;110(2):465–470.
- Catuogno S, Rienzo A, Di Vito A, Esposito CL, de Franciscis V. Selective delivery of therapeutic single strand antimiRs by aptamer-based conjugates. J Control Release. 2015;210:147–159.
- Pofahl M, Wengel J, Mayer G. Multifunctional nucleic acids for tumor cell treatment. Nucleic Acid Ther. 2014;24(2):171–177.
- Gupta RA, et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464(7291):1071–1076.
- Roberts TC, Wood MJ. Therapeutic targeting of non-coding RNAs. Essays Biochem. 2013;54(1):127–145.View this article via: PubMed Google Scholar
- Tano K, et al. MALAT-1 enhances cell motility of lung adenocarcinoma cells by influencing the expression of motility-related genes. FEBS Lett. 2010;584(22):4575–4580.
- Ji P, et al. MALAT-1, a novel noncoding RNA, and thymosin beta4 predict metastasis and survival in early-stage non-small cell lung cancer. Oncogene. 2003;22(39):8031–8041.
- Smaldone MC, Davies BJ. BC-819, a plasmid comprising the H19 gene regulatory sequences and diphtheria toxin A, for the potential targeted therapy of cancers. Curr Opin Mol Ther. 2010;12(5):607–616.View this article via: PubMed Google Scholar
- Fatemi RP, Velmeshev D, Faghihi MA. De-repressing LncRNA-targeted genes to upregulate gene expression: focus on small molecule therapeutics. Mol Ther Nucleic Acids. 2014;3:e196. View this article via: PubMed Google Scholar
- Rettig GR, Behlke MA. Progress toward in vivo use of siRNAs-II. Mol Ther. 2012;20(3):483–512.
- Kurreck J, Wyszko E, Gillen C, Erdmann VA. Design of antisense oligonucleotides stabilized by locked nucleic acids. Nucleic Acids Res. 2002;30(9):1911–1918.
- Bonasio R, Shiekhattar R. Regulation of transcription by long noncoding RNAs. Annu Rev Genet. 2014;48:433–455.
- Villegas VE, Zaphiropoulos PG. Neighboring gene regulation by antisense long non-coding RNAs. Int J Mol Sci. 2015;16(2):3251–3266.
- Kaneko S, et al. Interactions between JARID2 and noncoding RNAs regulate PRC2 recruitment to chromatin. Mol Cell. 2014;53(2):290–300.
- Paris O, et al. Direct regulation of microRNA biogenesis and expression by estrogen receptor beta in hormone-responsive breast cancer. Oncogene. 2012;31(38):4196–4206.
- Davis BN, Hata A. Regulation of MicroRNA Biogenesis: A miRiad of mechanisms. Cell Commun Signal. 2009;7:18. View this article via: PubMed Google Scholar
- Xiao Z, Chen Y. Small molecule targeting miR-34a for cancer therapy. Mol Cell Oncol. 2015;2(1):e977160.
- Ferrarelli LK. Targeting the “undruggable” MYCN. Sci Signal. 2014;7(344):ec260. View this article via: CrossRef Google Scholar
- Melkman-Zehavi T, et al. miRNAs control insulin content in pancreatic β-cells via downregulation of transcriptional repressors. EMBO J. 2011;30(5):835–845.
- Poy MN, et al. A pancreatic islet-specific microRNA regulates insulin secretion. Nature. 2004;432(7014):226–230.
- Poy MN, et al. miR-375 maintains normal pancreatic alpha- and beta-cell mass. Proc Natl Acad Sci U S A. 2009;106(14):5813–5818.
- Higuchi C, et al. Identification of circulating miR-101, miR-375 and miR-802 as biomarkers for type 2 diabetes. Metab Clin Exp. 2015;64(4):489–497.
- Erener S, Mojibian M, Fox JK, Denroche HC, Kieffer TJ. Circulating miR-375 as a biomarker of β-cell death and diabetes in mice. Endocrinology. 2013;154(2):603–608.
- Zhao E, et al. Obesity and genetics regulate microRNAs in islets, liver, and adipose of diabetic mice. Mamm Genome. 2009;20(8):476–485.
- Lovis P, et al. Alterations in microRNA expression contribute to fatty acid-induced pancreatic β-cell dysfunction. Diabetes. 2008;57(10):2728–2736.
- Ohno M, et al. The flavonoid apigenin improves glucose tolerance through inhibition of microRNA maturation in miRNA103 transgenic mice. Sci Rep. 2013;3:2553. View this article via: PubMed Google Scholar
- Trajkovski M, et al. MicroRNAs 103 and 107 regulate insulin sensitivity. Nature. 2011;474(7353):649–653.
- Christoph. A microRNA therapeutic targeting microRNA-103/107 for the treatment of NASH in patients with type 2 diabetes/pre-diabetes, selected as clinical candidate by AstraZeneca. miRNA Blog, miRNA Res Ind News. April 7, 2015. http://mirnablog.com/a-microrna-therapeutic-targeting-microrna-103107-for-the-treatment-of-nash-in-patients-with-type-2-diabetespre-diabetes-selected-as-clinical-candidate-by-astrazeneca/ Accessed February 10, 2017.
- Chen JF, et al. Targeted deletion of Dicer in the heart leads to dilated cardiomyopathy and heart failure. Proc Natl Acad Sci U S A. 2008;105(6):2111–2116.
- van Rooij E, Sutherland LB, Qi X, Richardson JA, Hill J, Olson EN. Control of stress-dependent cardiac growth and gene expression by a microRNA. Science. 2007;316(5824):575–579.
- Zhao Y, Samal E, Srivastava D. Serum response factor regulates a muscle-specific microRNA that targets Hand2 during cardiogenesis. Nature. 2005;436(7048):214–220.
- Montgomery RL, et al. Therapeutic inhibition of miR-208a improves cardiac function and survival during heart failure. Circulation. 2011;124(14):1537–1547.
- Callis TE, et al. MicroRNA-208a is a regulator of cardiac hypertrophy and conduction in mice. J Clin Invest. 2009;119(9):2772–2786.
- Hydbring P, Badalian-Very G. Clinical applications of microRNAs. F1000Res. 2013;2:136. View this article via: CrossRef Google Scholar
- Hullinger TG, et al. Inhibition of miR-15 protects against cardiac ischemic injury. Circ Res. 2012;110(1):71–81.
- Yap KL, et al. Molecular interplay of the noncoding RNA ANRIL and methylated histone H3 lysine 27 by polycomb CBX7 in transcriptional silencing of INK4a. Mol Cell. 2010;38(5):662–674.
- Mercer TR, Dinger ME, Sunkin SM, Mehler MF, Mattick JS. Specific expression of long noncoding RNAs in the mouse brain. Proc Natl Acad Sci U S A. 2008;105(2):716–721.
- Bak M, et al. MicroRNA expression in the adult mouse central nervous system. RNA. 2008;14(3):432–444.
- Daughters RS, et al. RNA gain-of-function in spinocerebellar ataxia type 8. PLoS Genet. 2009;5(8):e1000600.
- Smith P, Al Hashimi A, Girard J, Delay C, Hébert SS. In vivo regulation of amyloid precursor protein neuronal splicing by microRNAs. J Neurochem. 2011;116(2):240–247.
- Chang F, Zhang LH, Xu WP, Jing P, Zhan PY. microRNA-9 attenuates amyloidβ-induced synaptotoxicity by targeting calcium/calmodulin-dependent protein kinase kinase 2. Mol Med Rep. 2014;9(5):1917–1922.View this article via: PubMed Google Scholar
- Krützfeldt J, et al. Silencing of microRNAs in vivo with ‘antagomirs’. Nature. 2005;438(7068):685–689.
- Ali SS, et al. Pathological microRNAs in acute cardiovascular diseases and microRNA therapeutics. J Acute Dis. 2015;5(1):9–15.
- Banzhaf-Strathmann J, et al. MicroRNA-125b induces tau hyperphosphorylation and cognitive deficits in Alzheimer’s disease. EMBO J. 2014;33(15):1667–1680.
- Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100(1):57–70.
- Herranz H, Cohen SM. MicroRNAs and gene regulatory networks: managing the impact of noise in biological systems. Genes Dev. 2010;24(13):1339–1344.
- Tsang J, Zhu J, van Oudenaarden A. MicroRNA-mediated feedback and feedforward loops are recurrent network motifs in mammals. Mol Cell. 2007;26(5):753–767.
- Martinez NJ, Walhout AJ. The interplay between transcription factors and microRNAs in genome-scale regulatory networks. Bioessays. 2009;31(4):435–445.
- Dong Q, et al. MicroRNA let-7a inhibits proliferation of human prostate cancer cells in vitro and in vivo by targeting E2F2 and CCND2. PLoS One. 2010;5(4):e10147.
- Trang P, et al. Regression of murine lung tumors by the let-7 microRNA. Oncogene. 2010;29(11):1580–1587.
- Bandi N, Vassella E. miR-34a and miR-15a/16 are co-regulated in non-small cell lung cancer and control cell cycle progression in a synergistic and Rb-dependent manner. Mol Cancer. 2011;10:55. View this article via: PubMed Google Scholar
- Redon S, Reichenbach P, Lingner J. The noncoding RNA TERRA is a natural ligand and direct inhibitor of human telomerase. Nucleic Acids Res. 2010;38(17):5797–5806.
- Liu Z, et al. Downregulation of GAS5 promotes bladder cancer cell proliferation, partly by regulating CDK6. PLoS One. 2013;8(9):e73991.
- Guo X, et al. GAS5 inhibits gastric cancer cell proliferation partly by modulating CDK6. Oncol Res Treat. 2015;38(7–8):362–366.View this article via: PubMed Google Scholar
- Yin D, He X, Zhang E, Kong R, De W, Zhang Z. Long noncoding RNA GAS5 affects cell proliferation and predicts a poor prognosis in patients with colorectal cancer. Med Oncol. 2014;31(11):253.
- Jovanovic M, Hengartner MO. miRNAs and apoptosis: RNAs to die for. Oncogene. 2006;25(46):6176–6187.
- Wang L, et al. Effects of microRNA-221/222 on cell proliferation and apoptosis in prostate cancer cells. Gene. 2015;572(2):252–258.
- Yang X, et al. Down-regulation of mir-221 and mir-222 restrain prostate cancer cell proliferation and migration that is partly mediated by activation of SIRT1. PLoS One. 2014;9(6):e98833.
- Naemura M, Murasaki C, Inoue Y, Okamoto H, Kotake Y. Long noncoding RNA ANRIL regulates proliferation of non-small cell lung cancer and cervical cancer cells. Anticancer Res. 2015;35(10):5377–5382.View this article via: PubMed Google Scholar
- Nie FQ, et al. Long noncoding RNA ANRIL promotes non-small cell lung cancer cell proliferation and inhibits apoptosis by silencing KLF2 and P21 expression. Mol Cancer Ther. 2015;14(1):268–277.
- Zhu H, Li X, Song Y, Zhang P, Xiao Y, Xing Y. Long non-coding RNA ANRIL is up-regulated in bladder cancer and regulates bladder cancer cell proliferation and apoptosis through the intrinsic pathway. Biochem Biophys Res Commun. 2015;467(2):223–228.
- Folkman J. Tumor angiogenesis: therapeutic implications. N Engl J Med. 1971;285(21):1182–1186.
- Klagsbrun M, Soker S. VEGF/VPF: the angiogenesis factor found? Curr Biol. 1993;3(10):699–702.
- Folkman J, Klagsbrun M. Angiogenic factors. Science. 1987;235(4787):442–447.
- Chen S, et al. Global microRNA depletion suppresses tumor angiogenesis. Genes Dev. 2014;28(10):1054–1067.
- Cimmino A, et al. miR-15 and miR-16 induce apoptosis by targeting BCL2. Proc Natl Acad Sci U S A. 2005;102(39):13944–13949.
- Kuehbacher A, Urbich C, Zeiher AM, Dimmeler S. Role of Dicer and Drosha for endothelial microRNA expression and angiogenesis. Circ Res. 2007;101(1):59–68.
- Stahlhut C, Suárez Y, Lu J, Mishima Y, Giraldez AJ. miR-1 and miR-206 regulate angiogenesis by modulating VegfA expression in zebrafish. Development. 2012;139(23):4356–4364.
- Babae N, et al. Systemic miRNA-7 delivery inhibits tumor angiogenesis and growth in murine xenograft glioblastoma. Oncotarget. 2014;5(16):6687–6700.
- Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: insights into functions. Nat Rev Genet. 2009;10(3):155–159.
- Zhou Y, Zhang X, Klibanski A. MEG3 noncoding RNA: a tumor suppressor. J Mol Endocrinol. 2012;48(3):R45–R53.
- Li X, et al. MicroRNA-21 (miR-21) post-transcriptionally downregulates tumor suppressor PDCD4 and promotes cell transformation, proliferation, and metastasis in renal cell carcinoma. Cell Physiol Biochem. 2014;33(6):1631–1642.
- Meng F, Henson R, Wehbe-Janek H, Ghoshal K, Jacob ST, Patel T. MicroRNA-21 regulates expression of the PTEN tumor suppressor gene in human hepatocellular cancer. Gastroenterology. 2007;133(2):647–658.
- De Mattos-Arruda L, et al. MicroRNA-21 links epithelial-to-mesenchymal transition and inflammatory signals to confer resistance to neoadjuvant trastuzumab and chemotherapy in HER2-positive breast cancer patients. Oncotarget. 2015;6(35):37269–37280.View this article via: PubMed Google Scholar
- Yigit MV, et al. Context-dependent differences in miR-10b breast oncogenesis can be targeted for the prevention and arrest of lymph node metastasis. Oncogene. 2013;32(12):1530–1538.
- Ma L, et al. Therapeutic silencing of miR-10b inhibits metastasis in a mouse mammary tumor model. Nat Biotechnol. 2010;28(4):341–347.
- Liu Z, Zhu J, Cao H, Ren H, Fang X. miR-10b promotes cell invasion through RhoC-AKT signaling pathway by targeting HOXD10 in gastric cancer. Int J Oncol. 2012;40(5):1553–1560.View this article via: PubMed Google Scholar
- Le MT, et al. miR-200-containing extracellular vesicles promote breast cancer cell metastasis. J Clin Invest. 2014;124(12):5109–5128.
- Perdigão-Henriques R, et al. miR-200 promotes the mesenchymal to epithelial transition by suppressing multiple members of the Zeb2 and Snail1 transcriptional repressor complexes. Oncogene. 2016;35(2):158–172.
- Burk U, et al. A reciprocal repression between ZEB1 and members of the miR-200 family promotes EMT and invasion in cancer cells. EMBO Rep. 2008;9(6):582–589.
- Sundararajan V, Gengenbacher N, Stemmler MP, Kleemann JA, Brabletz T, Brabletz S. The ZEB1/miR-200c feedback loop regulates invasion via actin interacting proteins MYLK and TKS5. Oncotarget. 2015;6(29):27083–27096.
- Banyard J, et al. Regulation of epithelial plasticity by miR-424 and miR-200 in a new prostate cancer metastasis model. Sci Rep. 2013;3:3151. View this article via: PubMed Google Scholar
- Edmonds MD, et al. MicroRNA-31 initiates lung tumorigenesis and promotes mutant KRAS-driven lung cancer. J Clin Invest. 2016;126(1):349–364.
- Zhang WC, et al. Tumour-initiating cell-specific miR-1246 and miR-1290 expression converge to promote non-small cell lung cancer progression. Nat Commun. 2016;7:11702. View this article via: PubMed Google Scholar
- Ge Y, Zhang L, Nikolova M, Reva B, Fuchs E. Strand-specific in vivo screen of cancer-associated miRNAs unveils a role for miR-21(*) in SCC progression. Nat Cell Biol. 2016;18(1):111–121.View this article via: PubMed Google Scholar
- Mitra RN, et al. Nanoparticle-mediated miR200-b delivery for the treatment of diabetic retinopathy. J Control Release. 2016;236:31–37.
- Arun G, et al. Differentiation of mammary tumors and reduction in metastasis upon Malat1 lncRNA loss. Genes Dev. 2016;30(1):34–51.
- Fu WM, et al. Long noncoding RNA Hotair mediated angiogenesis in nasopharyngeal carcinoma by direct and indirect signaling pathways. Oncotarget. 2016;7(4):4712–4723.View this article via: PubMed Google Scholar
- Yin DD, Liu ZJ, Zhang E, Kong R, Zhang ZH, Guo RH. Decreased expression of long noncoding RNA MEG3 affects cell proliferation and predicts a poor prognosis in patients with colorectal cancer. Tumour Biol. 2015;36(6):4851–4859.
-
Version history
- Version 1 (March 1, 2017): Print issue publication



Copyright © 2024 American Society for Clinical Investigation
ISSN: 0021-9738 (print), 1558-8238 (online)