Advertisement
Research ArticleHematology Free access | 10.1172/JCI81749
Arginine methyltransferase PRMT5 is essential for sustaining normal adult hematopoiesis
Fan Liu,1,2 Guoyan Cheng,1,2 Pierre-Jacques Hamard,1,2 Sarah Greenblatt,1,2 Lan Wang,1,2 Na Man,1,2 Fabiana Perna,3 Haiming Xu,3 Madhavi Tadi,1,2 Luisa Luciani,1,2 and Stephen D. Nimer1,2,4
1Department of Biochemistry and Molecular Biology and
2Sylvester Comprehensive Cancer Center, University of Miami, Miller School of Medicine, Miami, Florida, USA.
3Molecular Pharmacology and Chemistry Program, Sloan Kettering Institute, Memorial Sloan Kettering Cancer Center, New York, New York, USA.
4Department of Internal Medicine, University of Miami, Miller School of Medicine, Miami, Florida, USA.
Address correspondence to: Stephen D. Nimer, 1120 NW 14th Street, CRB 660A (C241), Miami, Florida 33136, USA. Phone: 305.243.1775; E-mail: [email protected].
Find articles by Liu, F. in: JCI | PubMed | Google Scholar
1Department of Biochemistry and Molecular Biology and
2Sylvester Comprehensive Cancer Center, University of Miami, Miller School of Medicine, Miami, Florida, USA.
3Molecular Pharmacology and Chemistry Program, Sloan Kettering Institute, Memorial Sloan Kettering Cancer Center, New York, New York, USA.
4Department of Internal Medicine, University of Miami, Miller School of Medicine, Miami, Florida, USA.
Address correspondence to: Stephen D. Nimer, 1120 NW 14th Street, CRB 660A (C241), Miami, Florida 33136, USA. Phone: 305.243.1775; E-mail: [email protected].
Find articles by Cheng, G. in: JCI | PubMed | Google Scholar
1Department of Biochemistry and Molecular Biology and
2Sylvester Comprehensive Cancer Center, University of Miami, Miller School of Medicine, Miami, Florida, USA.
3Molecular Pharmacology and Chemistry Program, Sloan Kettering Institute, Memorial Sloan Kettering Cancer Center, New York, New York, USA.
4Department of Internal Medicine, University of Miami, Miller School of Medicine, Miami, Florida, USA.
Address correspondence to: Stephen D. Nimer, 1120 NW 14th Street, CRB 660A (C241), Miami, Florida 33136, USA. Phone: 305.243.1775; E-mail: [email protected].
Find articles by Hamard, P. in: JCI | PubMed | Google Scholar
1Department of Biochemistry and Molecular Biology and
2Sylvester Comprehensive Cancer Center, University of Miami, Miller School of Medicine, Miami, Florida, USA.
3Molecular Pharmacology and Chemistry Program, Sloan Kettering Institute, Memorial Sloan Kettering Cancer Center, New York, New York, USA.
4Department of Internal Medicine, University of Miami, Miller School of Medicine, Miami, Florida, USA.
Address correspondence to: Stephen D. Nimer, 1120 NW 14th Street, CRB 660A (C241), Miami, Florida 33136, USA. Phone: 305.243.1775; E-mail: [email protected].
Find articles by Greenblatt, S. in: JCI | PubMed | Google Scholar
1Department of Biochemistry and Molecular Biology and
2Sylvester Comprehensive Cancer Center, University of Miami, Miller School of Medicine, Miami, Florida, USA.
3Molecular Pharmacology and Chemistry Program, Sloan Kettering Institute, Memorial Sloan Kettering Cancer Center, New York, New York, USA.
4Department of Internal Medicine, University of Miami, Miller School of Medicine, Miami, Florida, USA.
Address correspondence to: Stephen D. Nimer, 1120 NW 14th Street, CRB 660A (C241), Miami, Florida 33136, USA. Phone: 305.243.1775; E-mail: [email protected].
Find articles by Wang, L. in: JCI | PubMed | Google Scholar
1Department of Biochemistry and Molecular Biology and
2Sylvester Comprehensive Cancer Center, University of Miami, Miller School of Medicine, Miami, Florida, USA.
3Molecular Pharmacology and Chemistry Program, Sloan Kettering Institute, Memorial Sloan Kettering Cancer Center, New York, New York, USA.
4Department of Internal Medicine, University of Miami, Miller School of Medicine, Miami, Florida, USA.
Address correspondence to: Stephen D. Nimer, 1120 NW 14th Street, CRB 660A (C241), Miami, Florida 33136, USA. Phone: 305.243.1775; E-mail: [email protected].
Find articles by Man, N. in: JCI | PubMed | Google Scholar
1Department of Biochemistry and Molecular Biology and
2Sylvester Comprehensive Cancer Center, University of Miami, Miller School of Medicine, Miami, Florida, USA.
3Molecular Pharmacology and Chemistry Program, Sloan Kettering Institute, Memorial Sloan Kettering Cancer Center, New York, New York, USA.
4Department of Internal Medicine, University of Miami, Miller School of Medicine, Miami, Florida, USA.
Address correspondence to: Stephen D. Nimer, 1120 NW 14th Street, CRB 660A (C241), Miami, Florida 33136, USA. Phone: 305.243.1775; E-mail: [email protected].
Find articles by Perna, F. in: JCI | PubMed | Google Scholar
1Department of Biochemistry and Molecular Biology and
2Sylvester Comprehensive Cancer Center, University of Miami, Miller School of Medicine, Miami, Florida, USA.
3Molecular Pharmacology and Chemistry Program, Sloan Kettering Institute, Memorial Sloan Kettering Cancer Center, New York, New York, USA.
4Department of Internal Medicine, University of Miami, Miller School of Medicine, Miami, Florida, USA.
Address correspondence to: Stephen D. Nimer, 1120 NW 14th Street, CRB 660A (C241), Miami, Florida 33136, USA. Phone: 305.243.1775; E-mail: [email protected].
Find articles by Xu, H. in: JCI | PubMed | Google Scholar
1Department of Biochemistry and Molecular Biology and
2Sylvester Comprehensive Cancer Center, University of Miami, Miller School of Medicine, Miami, Florida, USA.
3Molecular Pharmacology and Chemistry Program, Sloan Kettering Institute, Memorial Sloan Kettering Cancer Center, New York, New York, USA.
4Department of Internal Medicine, University of Miami, Miller School of Medicine, Miami, Florida, USA.
Address correspondence to: Stephen D. Nimer, 1120 NW 14th Street, CRB 660A (C241), Miami, Florida 33136, USA. Phone: 305.243.1775; E-mail: [email protected].
Find articles by Tadi, M. in: JCI | PubMed | Google Scholar
1Department of Biochemistry and Molecular Biology and
2Sylvester Comprehensive Cancer Center, University of Miami, Miller School of Medicine, Miami, Florida, USA.
3Molecular Pharmacology and Chemistry Program, Sloan Kettering Institute, Memorial Sloan Kettering Cancer Center, New York, New York, USA.
4Department of Internal Medicine, University of Miami, Miller School of Medicine, Miami, Florida, USA.
Address correspondence to: Stephen D. Nimer, 1120 NW 14th Street, CRB 660A (C241), Miami, Florida 33136, USA. Phone: 305.243.1775; E-mail: [email protected].
Find articles by Luciani, L. in: JCI | PubMed | Google Scholar
1Department of Biochemistry and Molecular Biology and
2Sylvester Comprehensive Cancer Center, University of Miami, Miller School of Medicine, Miami, Florida, USA.
3Molecular Pharmacology and Chemistry Program, Sloan Kettering Institute, Memorial Sloan Kettering Cancer Center, New York, New York, USA.
4Department of Internal Medicine, University of Miami, Miller School of Medicine, Miami, Florida, USA.
Address correspondence to: Stephen D. Nimer, 1120 NW 14th Street, CRB 660A (C241), Miami, Florida 33136, USA. Phone: 305.243.1775; E-mail: [email protected].
Find articles by Nimer, S. in: JCI | PubMed | Google Scholar
Published August 10, 2015 - More info
J Clin Invest. 2015;125(9):3532–3544. https://doi.org/10.1172/JCI81749.
Copyright © 2015, American Society for Clinical Investigation
Received: March 3, 2015; Accepted: June 23, 2015
Related video:
Maintaining adult hematopoiesis
An inability to properly sustain blood cell production results in the development of hematopoietic disorders. Epigenetic regulators have been shown to be involved in the function of hematopoietic stem and progenitor cells; however, dysfunction of these enzymes is associated with development of malignancies. In this episode Stephen Nimer and colleagues identify the type II arginine methyltransferase PRMT5 as an important regulator of adult hematopoiesis. Mice lacking PRMT5 developed fatal pancytopenia due to reduced proliferation of hematopoietic progenitor cells as the results of impaired cytokine signaling and increased p53 signaling. Hematopoietic stem cell populations were not initially reduced in these animals; however, this population was unable to restore blood cell populations in lethally irradiated mice. Together, the results of this study provide important insight into the role of PRMT5-mediated gene expression in maintaining hematopoietic homeostasis.
-
Abstract
Epigenetic regulators play critical roles in normal hematopoiesis, and the activity of these enzymes is frequently altered in hematopoietic cancers. The major type II protein arginine methyltransferase PRMT5 catalyzes the formation of symmetric dimethyl arginine and has been implicated in various cellular processes, including pluripotency and tumorigenesis. Here, we generated Prmt5 conditional KO mice to evaluate the contribution of PRMT5 to adult hematopoiesis. Loss of PRMT5 triggered an initial but transient expansion of hematopoietic stem cells (HSCs); however, Prmt5 deletion resulted in a concurrent loss of hematopoietic progenitor cells (HPCs), leading to fatal BM aplasia. PRMT5-specific effects on hematopoiesis were cell intrinsic and depended on PRMT5 methyltransferase activity. We found that PRMT5-deficient hematopoietic stem and progenitor cells exhibited severely impaired cytokine signaling as well as upregulation of p53 and expression of its downstream targets. Together, our results demonstrate that PRMT5 plays distinct roles in the behavior of HSCs compared with HPCs and is essential for the maintenance of adult hematopoietic cells.
-
Introduction
Homeostasis of blood cell production is sustained by a small pool of hematopoietic stem cells (HSCs), from which all mature blood cells are derived. These pluripotent HSCs must maintain their dual capacity to remain quiescent but also enter the cell cycle to either self-renew or differentiate into multiple lineages. Genes involved in cell cycle regulation, signal transduction, and transcriptional regulation have been shown to play important roles in HSC homeostasis (1), but recently, many epigenetic regulators, including DNA methyltransferases and histone-modifying enzymes (such as EZH1/2, MLL, and p300), have also been shown to regulate HSC function (2–7).
Protein arginine methylation is a common posttranslational modification that affects the function of many proteins, including histones (8). Protein arginine methyltransferases (PRMTs) are classified as type 1 and type 2 enzymes on the basis of their ability to catalyze the formation of asymmetric and symmetric dimethyl arginine, respectively. PRMT5 is the major type 2 enzyme identified thus far, which, together with its cofactor MEP50, mediates the methylation of histones H2A and H4 at R3 and histone H3 at R8 (9). The symmetric dimethylation of these histone residues generally generates repressive marks for gene transcription, and PRMT5 has been found in several transcriptional repressor complexes, including those containing SIN3A/HDAC, MBD2/NURD, and N-CoR/SMRT (9). Moreover, PRMT5 represses globin gene expression through recruitment of DNMT3A, indicating a potential crosstalk between histone arginine methylation and DNA methylation (10, 11). In addition to histones, PRMT5 regulates a number of nonhistone proteins with identifiable functions in RNA processing, translation, signal transduction, apoptosis, and cell cycle regulation. PRMT5 has been shown to maintain pluripotency in both embryonic and adult stem cells and to reprogram mouse embryonic fibroblasts into induced pluripotent stem cells (iPSCs) when introduced with Klf4 and Oct3/4 (12–14).
Growing evidence has indicated a critical role of PRMT5 in tumorigenesis. Although recurrent mutations of PRMT5 have not been observed in cancer cells, PRMT5 expression is upregulated in human leukemia, lymphoma, and in many solid tumors, including gastric, colorectal, and lung cancer tumors (15). PRMT5 promotes the proliferation of lung and ovarian cancer cells, rendering it an attractive therapeutic target in these diseases (16, 17).
The function of PRMT5 in hematopoietic stem and progenitor cells (HSPCs) has not been investigated. In this study, we identify a critical role for PRMT5 in adult hematopoiesis using a Prmt5 conditional KO mouse model. Loss of PRMT5 has a rapid and profound effect on blood cell production with distinct, temporal effects on HSCs and their progenitor cell progeny. The absence of PRMT5 leads to a fatal, very severe aplastic anemia–like (VSAA-like) phenotype. This inability to generate mature blood elements is cell intrinsic and does not result from normal homeostatic mechanisms.
-
Results
Generation of Prmt5 conditional KO mice. To define the role of PRMT5 in normal hematopoiesis, we first determined the levels of Prmt5 mRNA and protein in different populations of mouse BM HSPCs. HSCs and their differentiated progeny were purified according to cell surface marker expression using FACS sorting, and the expression of Prmt5 was determined by quantitative real-time PCR (qPCR) (Figure 1A) and Western blot analysis (Figure 1B). Prmt5 mRNA and protein levels were readily detected in HSPCs, with little change in mRNA levels in the various stem and progenitor cell populations. However, when cells underwent myeloid, erythroid, or lymphoid differentiation, PRMT5 protein levels decreased to 5% to 24% of the levels seen in HSPCs. Although Prmt5 mRNA was maintained in differentiated B cells, its protein levels decreased dramatically, suggesting important posttranscriptional regulation of PRMT5 expression in these cells.
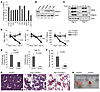 Figure 1
Figure 1Deletion of PRMT5 in adult BM results in severe pancytopenia. (A) Prmt5 mRNA levels decreased when mouse BM cells underwent terminal myeloid and erythroid differentiation. WT BM HSCs and their differentiated progeny were flow sorted on the basis of their cell surface marker expression, and Prmt5 mRNA levels were determined by qPCR (normalized to Hprt1 expression). A representative PCR result from 3 independent experiments (cells in each experiment were pulled together from 3 mice) is shown. MPPs, multipotent progenitors; CMPs, common myeloid progenitors; GMPs, granulocyte-macrophage progenitors. (B) PRMT5 protein levels were determined by Western blot analysis using sorted populations of WT BM cells. Numbers indicate the densitometry of the PRMT5 bands normalized to β-actin. (C) The cellular level of symmetrically dimethylated arginine was detected using an antibody against the Symmetric Di-Methyl Arginine Motif (catalog 13222; Cell Signaling Technology). This antibody recognizes 2 major bands of approximately 25 kDa and 15 kDa. (D) Loss of PRMT5 led to pancytopenia within 15 days. Complete blood count (CBC) analysis of peripheral blood wbc, rbc, and platelet (PLT) counts at 0, 7, and 15 days after injection (d.p.i.) are shown (n = 5). (E) BM cellularity was determined 7 and 15 d.p.i. in Prmt5fl/fl and Prmt5Δ/Δ mice (n = 5). (F) The cellularity of the thymus obtained from Prmt5fl/fl and Prmt5Δ/Δ mice was determined 15 d.p.i. (n = 5). (G) Representative images show H&E-stained cross sections of femurs isolated from the control and Prmt5Δ/Δ mice. Original magnification, ×200. (H) Representative image shows reduced size of the thymus from a Prmt5Δ/Δ mouse compared with that from a Prmt5fl/fl mouse on day 15. All P values were determined by a 2-tailed Student’s t test.
Since straight Prmt5-KO mice die before birth (12), we generated Prmt5–conditional KO mice by first crossing Prmt5 FLIP-OUT mice (obtained from the European Mutant Mouse Archive [EMMA]) with Flp recombinase–expressing transgenic (Tg) mice to generate Prmt5-floxed mice, whereby exon 7 of the Prmt5 gene was flanked by 2 LoxP sites (Supplemental Figure 1A; supplemental material available online with this article; doi:10.1172/JCI81749DS1). We then crossed the Prmt5-floxed mice with Mx1Cre Tg mice to generate Mx1Cre+ Prmt5fl/+ Mx1Cre+ Prmt5fl/fl mice and Mx1Cre-negative littermate controls. To delete Prmt5 in hematopoietic cells, 2 i.p. injections of poly(I:C) (10 mg/kg on days 0 and 1) were given to both the Mx1Cre– and Mx1Cre+ Prmt5-floxed mice. Prmt5 loss was confirmed by PCR analysis of genomic DNA (Supplemental Figure 1B), by quantitative qPCR to detect Prmt5 mRNA (Supplemental Figure 1C), and by Western blot analysis to detect PRMT5 protein (Supplemental Figure 1D). This injection strategy was used in all subsequent experiments. Interestingly, loss of PRMT5 triggered the loss of MEP50 protein (Supplemental Figure 1D), a cofactor that is required for the enzymatic activity of PRMT5 on histones, without changing Mep50 mRNA levels (data not shown). This suggests a probable posttranscriptional effect of PRMT5 on MEP50 protein levels. We also determined the overall level of symmetrically dimethylated arginine in PRMT5-deleted BM cells and found a complete loss of this modification 7 days after Cre induction (Figure 1C), suggesting that PRMT5 is indeed the major type 2 PRMT in these cells.
Deletion of PRMT5 during adult hematopoiesis leads to severe cytopenias. The deletion of PRMT5 in 2-month-old mice had a profound effect on hematopoiesis. Two weeks after the first poly(I:C) injection, the Mx1Cre+ Prmt5fl/fl mice developed severe pallor due to anemia, and most of these PRMT5-deleted mice were moribund 1 to 2 days later, requiring euthanasia. Analysis of the peripheral blood of these mice revealed severe pancytopenia, with a more than 10-fold decrease in wbc, a 5-fold decrease in rbc, and a 100-fold decrease in platelet counts 15 days after Cre induction (Figure 1D). BM cellularity of the PRMT5-deleted mice was reduced by more than 50% on day 7 and by more than 95% on day 15 (Figure 1E), consistent with the development of aplastic anemia (Figure 1G). While PRMT5 loss had little effect on spleen weight (or cellularity) on day 15 (Supplemental Figure 2, A and B), the size and cellularity of the thymus were significantly reduced on day 15 (Figure 1, F and H). Of note, the Prmt5 heterozygous KO mice had normal peripheral blood counts and normal BM, spleen, and thymus cellularity (Figure 1D and data not shown).
Loss of PRMT5 causes aberrant erythroid differentiation and T cell development in thymus. To define the effects of PRMT5 deletion on hematopoiesis, we determined the frequency of erythroid (CD71/Ter119) and myeloid (Mac1/Gr1) cells in the BM and spleens of PRMT5-deleted mice 7 days after poly(I:C) injection. As shown in Figure 2A, PRMT5 deletion dramatically reduced the frequency of CD71/Ter119 double-positive cells in the BM and spleen, at a time when myeloid differentiation was relatively intact (Supplemental Figure 3A). We calculated the absolute numbers of erythroid and myeloid cells on the basis of their frequencies and BM cellularity and observed a 5-fold decrease in Ter119+ erythroid cells (after rbc lysis) and a 2-fold decrease in Mac1+/Gr1+ myeloid cells (Supplemental Figure 3B). We also observed a significant decrease in the frequency of megakaryocyte erythroid progenitors (MEPs) (defined as Lin–, c-Kit+, Sca1–, FcRII/III–, and CD34– cells) in the PRMT5-null mouse BM as early as 7 days after induction (Figure 2B), suggesting that PRMT5 loss initially affects erythroid and megakaryocyte differentiation to a greater degree than it does myeloid differentiation.
 Figure 2
Figure 2PRMT5 loss impairs erythroid differentiation and T cell development. (A) The stage of erythroid differentiation in BM and spleen from Prmt5fl/fl and Prmt5Δ/Δ mice was determined by Ter119/CD71 double staining and FACS analysis at 7 d.p.i. Note that the cells were stained without rbc lysis. Results are representative of 3 experiments, with 2 to 3 mice per group in each experiment. B) Lin–, c-Kit+, and Sca1– progenitor cells isolated from day-7 mouse BM were further stained for CD34 and FcRII/III to distinguish the CMP (CD34+/FcRII/III–), GMP (CD34+/FcRII/III+), and MEP (CD34–/FcRII/III–) cell populations. The percentage of each population is indicated in the plots. Results are representative of 3 experiments, with 2 to 3 mice per group in each experiment. (C) Thymocytes were stained for the CD4, CD8, CD25, and CD44 cell surface markers. The distribution of CD25/CD44 subsets within the CD4/CD8 DN cell population is shown for Prmt5fl/fl , Prmt5+/Δ, and Prmt5Δ/Δ mice. (D) The frequencies of each DN subset (DN1–DN4) on days 9 and 15 are plotted for the Prmt5fl/fl and Prmt5Δ/Δ mice (n = 3). P values were determined by a 2-tailed Student’s t test.
We next determined how PRMT5 loss affects B and T cell development in BM, spleen, and thymus. We observed a slight increase in the frequency and absolute number of B220+ B cells in day-7 PRMT5-null spleens (Supplemental Figure 3C and data not shown), while day-7 PRMT5-null BM showed a significant decrease in pro- and pre-B cells but also a slight increase in mature B cells (despite a 50% decline in BM cellularity; Supplemental Figure 3D). This suggests that the absence of PRMT5 may initially promote B cell maturation. We also determined the frequency and proliferation of common lymphoid progenitor (CLP) cells and found little to no change in the day-7 PRMT5-null BM (Supplemental Figure 3, E and F).
To better understand the decreased cellularity of the PRMT5-null thymus, we examined the cell surface expression of CD4, CD8, CD25, and CD44 on thymocytes. Compared with littermate controls, PRMT5 deletion led to a moderate increase in the frequency of CD4/CD8 double-positive (DP) cells, at the expense of the CD4/CD8 double-negative (DN) and CD8 single-positive (SP) cell populations (Supplemental Figure 3G). Strikingly, we observed a marked accumulation of cells in the DN1 thymic subset (CD44+CD25–), with fewer cells in the DN3 (CD44–CD25+) and DN4 thymic subsets (CD44–CD25–) 9 and 15 days after Cre induction (Figure 2, C and D). Thus, PRMT5 appears to play an essential role in early thymocyte development.
PRMT5 deficiency leads to loss of HSPCs. We next examined how PRMT5 loss affects HSPC biology. FACS analysis of day-7 PRMT5-deficient BM cells revealed a normal frequency of lineage-negative cells (that lack cell surface expression of Ter119, Mac1, Gr1, B220, CD3e, CD4, CD5, CD8, and CD127), a 40%–50% decrease in the frequency of myeloid progenitor cells (Lin–Sca1–c-Kit+ [LK] cells), and a 3- to 4-fold increase in the frequency of stem cell–enriched Lin–Sca1+c-Kit+ (LSK) cells and phenotypic long-term HSCs (LT-HSCs) (CD48–CD150+ LSK cells) (Figure 3A). Even though the total cellularity of the PRMT5-deficient BM was only 50% of that seen in the control mice, there was a transient increase in the absolute number of LSK cells and LT-HSCs on day 7 (Figure 3C). By day 9, the number and frequency of all stem and progenitor cell populations were dramatically reduced (Figure 3C and Supplemental Figure 4) and were even further reduced on day 15 (Figure 3B).
 Figure 3
Figure 3PRMT5 deletion leads to loss of phenotypical HSPCs. (A and B) FACS analysis was performed to determine HSPC frequency. Lineage-negative cells were identified within BM cells isolated from day-7 (A) and day-15 (B) control and PRMT5-deleted mice and further analyzed for cell surface expression of c-Kit, Sca1, and the SLAM markers CD48 and CD150. Representative FACS plots from days 7 and 15 of 5 independent experiments are shown. (C) Absolute numbers of LK cells, LSK cells, and LT-HSCs 7 days (top) and 9 days (bottom) after poly(I:C) injection are shown (n = 5). Data are plotted as the mean ± SD, and P values were determined by a 2-tailed Student’s t test.
We next assessed the function of PRMT5-null HSPCs using several functional assays. PRMT5-deficient BM cells isolated 7 days after poly(I:C) injection failed to form colonies in CFU assays (Figure 4A), and we found no cobblestone area–forming cells (CAFCs) or long-term culture–initiating cells (LTC-ICs) when assessing LT-HSC frequency and function (Figure 4B). Despite the preservation of phenotypic LT-HSCs, PRMT5-null BM cells isolated 5 days after Cre induction failed to rescue lethally irradiated recipient mice in the BM transplantation assays (Figure 4C), indicating that these HSCs are functionally defective. Thus, PRMT5 deletion causes a progressive loss of lineage-committed progenitor cells and a transient expansion of LT-HSCs, which are nonetheless defective and unable to rescue lethally irradiated mice.
 Figure 4
Figure 4PRMT5 deletion leads to loss of functional HSPCs. (A) BM cells isolated from day-7 Prmt5fl/fl and Prmt5Δ/Δ mice were plated in methylcellulose cultures, supplemented with cytokines. Colonies were scored 7 days after plating (n = 3). (B) Left: LTC-IC assays using day-7 BM cells isolated from Prmt5fl/fl and Prmt5Δ/Δ mice. Cells were cocultured with MS5 cells for 4 weeks and then collected and plated in methylcellulose medium. Colonies were counted 1 week after plating. The number of cells plated in MS5 coculture is indicated in the graph. Right: CAFC assays using day-7 BM cells isolated from Prmt5fl/fl and Prmt5Δ/Δ mice. BM cells were cocultured with MS5 stromal cells for 5 weeks, and the number of cobblestone areas at the end of week 5 is plotted (n = 3). (C) Approximately 5 × 106 BM cells isolated from Prmt5fl/fl or Prmt5Δ/Δ mice 5 days after Cre induction were injected into lethally irradiated recipient CD45.1 mice. Survival of the mice was monitored daily (n = 8).
Enhanced apoptosis is not observed in PRMT5-null HSPCs. To determine whether the loss of HSPCs in the PRMT5-deficient mice was due to increased apoptosis, we quantified caspase 3 cleavage in BM cells isolated from control and PRMT5-deleted mice and found no caspase 3 cleavage in bulk BM cells (although caspase 3 cleavage was readily detected in BM cells isolated from LPS-treated mice) (Figure 5A). Furthermore, we performed FACS analysis to examine both lineage-positive cells and Lin–c-Kit+ HSPCs and found no increase in annexin V+ (apoptotic) PRMT5-null cells (Figure 5B and Supplemental Figure 5A), nor did we detect an increase in apoptosis using the TUNEL assay (data not shown). We also used FACS analysis to evaluate whether PRMT5 loss causes global necrosis or necroptosis in BM cells and found no increase in 7AAD+/annexin V– (necrotic) cells (data not shown).
 Figure 5
Figure 5PRMT5 loss has distinct effects on the cell cycle regulation of hematopoietic stem versus progenitor cells. (A) Protein lysates of BM cells isolated on day 7 from control and PRMT5-deleted mice were resolved on SDS-PAGE gels and probed with antibodies recognizing PRMT5, caspase 3, and β-actin. Lysates prepared from BM cells isolated from LPS-treated mice were used as a positive control (Pos). Arrow indicates the cleaved caspase 3. (B) FACS analysis of annexin V in day-7 Lin+, LK, and LSK cells. Percentages of annexin V+ cells are plotted (n = 4). (C) FACS analysis of in vivo BrdU incorporation assays using BM cells isolated from day-7 control, Prmt5 heterozygous, and Prmt5 homozygous KO mice. Cells were first gated on LK or LSK cells; the frequency of BrdU+ cells is indicated on the plots. (D) The percentage of BrdU+ S-phase cells is shown for the Lin+, LK, and LSK cells isolated from day-7 control and Prmt5 homozygous KO mice (n = 6). (E and F) Representative FACS analysis of Ki67 expression in LT-HSCs. BM cells isolated from day-7 control and Prmt5-deleted mice were gated on CD34– and FLT3– LSK (E) or CD48– and CD150+ LSK (F) cells; the frequency of Ki67+ cells is indicated on the plot. (G) The percentage of Ki67+ LT-HSCs in BM cells from Prmt5fl/fl and Prmt5Δ/Δ mice is plotted (n = 5). All P values were determined by 2-tailed Student’s t test.
It remains a possibility that apoptotic PRMT5-null cells are cleared too rapidly for their ready detection, and because PRMT5 deletion has been shown to induce apoptosis via caspase 3 activation in fetal liver cells and neuronal cells (ref. 18 and our unpublished data), we generated Mx1Cre+ Prmt5fl/fl Casp3–/– mice to further address this possibility. We found no effect of caspase 3 deletion on the decreased BM cellularity (Supplemental Figure 5B) or the rapid disappearance of HSPCs that follows PRMT5 deletion (Supplemental Figure 5C). Thus, it appears that increased apoptosis does not account for the loss of HSPCs seen in these mice.
PRMT5 loss has distinct effects on cell cycle regulation of hematopoietic stem versus progenitor cells. To determine whether PRMT5 loss affects the cell cycle of HSPCs, we first performed an in vitro BrdU incorporation assay using BM cells isolated from day-9 control and PRMT5-deleted mice (Supplemental Figure 6A). The PRMT5-null cells showed a profound (5-fold) decrease in the percentage of cells in the S phase compared with the control cells (from 29.6% to 6.0%). In vivo BrdU assays also showed a dramatic decrease in the percentage of Lin– cells in the S phase after PRMT5 deletion (from 30.6% to 1.5%) on day 9 (Supplemental Figure 6A). To make sure that these changes did not simply reflect the loss of Lin–c-Kit+Sca1– progenitor cells, which are the most rapidly cycling cells in the BM, we assessed the cell cycle status on day-7 (Figure 5, C and D) and day-9 (Supplemental Figure 6B) LK and LSK cells using in vivo BrdU assays. We found a 40%–50% decrease in the percentage of PRMT5-null LK cells in the S phase, but also a 2-fold increase in the percentage of PRMT5-null LSK cells in the S phase, suggesting distinct effects of PRMT5 loss on the cell cycle regulation of progenitor cells versus that of stem cells. In addition to its effect on HSPCs, PRMT5 loss also impaired the proliferation of lineage-positive cells (Figure 5D).
To determine whether PRMT5 loss affects LT-HSC quiescence, we measured Ki67 expression in CD34–FLT3– and CD48–CD150+ LSK cells and found a 2-fold increase in Ki67+ PRMT5-deficient LT-HSCs on day 7 (Figure 5, E–G), with nearly all of the PRMT5-null LT-HSCs being Ki67+ by day 9 (Supplemental Figure 6C). Similar results were observed for LT-HSCs in the in vivo BrdU incorporation assay (Supplemental Figure 6D). Thus, PRMT5 loss activates dormant HSCs but impairs the progression of progenitor cells through the cell cycle.
The effects of PRMT5 on hematopoiesis are intrinsic and dependent on its methyltransferase activity. To determine whether the observed hematopoietic phenotype of PRMT5-null mice is cell intrinsic and related or unrelated to homeostatic control, we transplanted approximately 1 × 106 BM cells isolated from Prmt5fl/fl, Mx1Cre+ Prmt5fl/+, or Mx1Cre+ Prmt5fl/fl mice (which are CD45.2), together with the same number of WT CD45.1 competitor cells, into lethally irradiated CD45.1 recipient mice. The transplanted mice were treated with poly(I:C) injections 4 months after transplantation and monitored for 3 weeks. While deletion of 1 allele of Prmt5 had no effect on peripheral blood and BM CD45.2 percentages (Figure 6A and Supplemental Figure 7), 2 weeks after both alleles were deleted, CD45.2 cells were dramatically reduced in the peripheral blood, BM, and thymus of the recipient mice (Figure 6A). In contrast, PRMT5 deletion had no effect on the frequency of CD45.2 cells in the spleen, consistent with what we observed in the primary Prmt5-KO mice (Supplemental Figure 2). The decreased frequency of PRMT5-null CD45.2 LK progenitor cells within the CD45.2 BM cells was again accompanied by a transient increase in LSK cell frequency 7 days after poly(I:C) injection, which returned to normal on day 14 (Figure 6B). We have also found a significant decrease in the absolute number of CD45.2+ LT-HSCs 14 days after induction of PRMT5 deletion (Figure 6C), although the frequencies of these cells were similar in both the control and KO transplanted mice (data not shown).
 Figure 6
Figure 6PRMT5 functions in HSCs are cell intrinsic and require the methyltransferase activity of PRMT5. (A) CD45.2 BM cells isolated from either Prmt5fl/fl, Mx1Cre+ Prmt5fl/+, or Mx1Cre+ Prmt5fl/fl mice were transplanted into recipient mice, together with WT CD45.1 BM cells. The ratio of CD45.2 to CD45.1 in peripheral blood mononuclear cells and the percentages of CD45.2 cells in BM, spleen, and thymus were determined by FACS analysis 7, 14, and 21 d.p.i. (n ≥4). (B) The frequency of CD45.2+ LK and LSK cells in BM from transplanted mice was determined by FACS analysis 7 and 14 d.p.i. (n = 5). (C) The absolute number of CD45.2 LT-HSCs (CD48–CD150+ LSK cells) in the transplanted mice 14 d.p.i. was calculated and plotted as the mean ± SD (n = 4). (D) The percentage of Ki67+ LT-HSCs in CD45.2 and CD45.1 BM cells was determined by FACS analysis. Results are representative of 2 independent transplantation experiments, with 3 to 4 mice per group in each experiment. (E) CFU assays were performed using day-7 BM cells transduced with either empty lentivirus (pBGJR) or viruses expressing PRMT5wt or PRMT5EQ proteins. GFP+ cells were flow sorted and plated in methylcellulose cultures in duplicate. Colonies were scored 7 days after plating. Results are representative of 3 independent experiments using cells from 2 mice of the same genotype pulled together for viral transduction. (F) CAFC assays were performed using day-5 BM cells isolated from Prmt5fl/fl or ERCre+ Prmt5fl/fl mice. Cells were transduced with pBGJR or PRMT5wt-, or PRMT5EQ-expressing lentiviruses and were cocultured with MS5 cells in the presence of 0.1 μM 4-OHT. The number of cobblestone areas at the end of the coculture is plotted as the mean ± SD (n = 3). All P values were determined by a 2-tailed Student’s t test.
We determined that the loss of HSC quiescence is an intrinsic property of the PRMT5-null HSCs; as shown in Figure 6D, deletion of PRMT5 led to a 2-fold increase in Ki67+ CD45.2 LT-HSCs (CD48–CD150+ LSK cells), even though the peripheral blood counts and the total number of BM cells were relatively constant (data not shown). The WT CD45.1 cells showed a proliferative response to the loss of CD45.2 cells, with a slight increase in the frequency of Ki67+ CD45.1 LT-HSCs (28.6% vs. 20.5%).
To determine whether the methyltransferase activity of PRMT5 is required for its essential role in hematopoiesis, we transduced Prmt5fl/fl and Prmt5Δ/Δ BM cells with lentiviruses expressing either WT PRMT5 (PRMT5wt) or an enzymatically dead version of PRMT5 (PRMT5EQ). While expression of PRMT5wt completely rescued the colony-forming ability of Prmt5Δ/Δ BM cells in CFU assays (with some overshoot), expression of PRMT5EQ had no effect (Figure 6E). We also performed CAFC assays using an ERCre-mediated KO system, so PRMT5 deletion could be induced in vitro. Once again, reexpression of PRMT5wt rescued the ability of PRMT5-null HSCs to form cobblestone areas, while reexpressing PRMT5EQ did not (Figure 6F). Thus, the enzymatic activity of PRMT5 is required for its function in normal HSPCs.
PRMT5 is required for the maintenance of cytokine signaling in BM cells. To gain mechanistic insight into how PRMT5 loss negatively impacts HSPC function, we performed paired-end RNA-sequencing (RNA-seq) using HSPCs isolated from day-3 Prmt5 control and KO mouse BM, after confirming PRMT5 loss and downregulation of H4R3 symmetric dimethylation at this time point (Figure 7A). Functional classification analysis using DAVID (Database for Annotation, Visualization and Integrated Discovery) identified 332 genes as being differentially expressed by at least 2-fold between the control and PRMT5-null HSPCs. We found 216 genes upregulated and 116 genes downregulated, with signal transduction pathways being among the primary gene sets affected by PRMT5 deletion (Figure 7B). We did not observe a molecular signature consistent with apoptosis in PRMT5-null HSPCs, confirming that apoptosis is not the cause of the disappearance of these cells. In addition, we identified 1,550 intron retention events in the PRMT5-deficient cells, suggesting that PRMT5 loss significantly impairs the constitutive splicing machinery. The genes affected by aberrant splicing were also primarily involved in signal transduction (Figure 7C).
 Figure 7
Figure 7Impaired cytokine signaling pathways in PRMT5-deficient BM cells. (A) HSPCs were isolated from day-3 control and Prmt5 KO mice, and the levels of PRMT5, H4R3 symmetric dimethylation, and histone H3 were determined by Western blot analysis. (B) DAVID functional pathway analysis of the differentially expressed gene set between day-3 control and Prmt5-KO HSPCs. (C) A total of 1,500 genes with intron retention in day-3 PRMT5-null HSPCs were analyzed using the DAVID functional annotation tool. (D) Total BM cells isolated from day-7 Prmt5fl/fl and Prmt5Δ/Δ mice were stimulated with GM-CSF (50 ng/ml) for 10 minutes at 37°C. Blots were probed for ERK1/2, p-ERK1/2, STAT5, p-STAT5, AKT, and p-AKT. Data are representative of 3 independent experiments. (E) Purified HSPCs from control and Prmt5 homozygous KO mice were stimulated with cytokine mixture, including murine SCF (100 ng/ml), IL-3 (20 ng/ml), IL-6 (20 ng/ml), and FLT3 ligand (100 ng/ml) for 10 minutes at 37°C. Phosphorylation of ERK1/2, STAT5, and AKT was determined as in D. Three independent experiments were performed using cells pulled together from 2 mice per genotype in each experiment. (F) Paraffin-embedded femurs and spleens isolated from day-7 control and Prmt5Δ/Δ mice were immunohistochemically stained with antibodies against p-ERK1/2. Original magnification, ×200.
PRMT5 has been shown to regulate the MAPK and NF-κB signal transduction pathways in various cancer cell lines (19, 20). To determine whether PRMT5 loss affects normal hematopoietic cytokine signaling, we examined the level of phosphorylated ERK1/2 (p-ERK1/2), p-STAT5, and p-AKT in bulk BM cells (Figure 7D) and purified HSPCs (Figure 7E). To activate signaling, the bulk BM cells were stimulated with granulocyte-macrophage CSF (GM-CSF), while the HSPCs were stimulated with SCF, IL-3, IL-6, and FLT3 ligand. As shown in Figure 7, D and E, ERK1/2 phosphorylation was lost in the PRMT5-null BM mononuclear cells (BMMCs) and HSPCs, without a change in ERK1/2 protein levels. Similarly, p-STAT5 and p-AKT were markedly decreased in the PRMT5-null bulk BM cells and purified HSPCs. We also determined by IHC that p-ERK1/2 levels were significantly reduced in day-7 PRMT5-null BM and splenic cells (Figure 7F). This suggests that PRMT5 is required for cytokine-mediated signal transduction in both HSPCs and more mature hematopoietic cells.
PRMT5 loss decreases the cell surface expression of important cytokine receptors. To further explore the mechanisms underlying the effect of PRMT5 loss on cytokine signaling, we used flow cytometry to examine the cell surface expression of several cytokine receptors that are important for HSPC function. We found a significant reduction in the expression of FLT3, IL-6Rα, and IL-3Rα in day-7 PRMT5-null LK and LSK cells (Figure 8A). Although the frequency of c-Kit+ HSPCs was comparable between the control and PRMT5-null mice at this time point, the level of c-Kit expression, as measured by mean fluorescence intensity (MFI), was significantly reduced in PRMT5-null HSPCs (Figure 8, B and C). We next determined the levels of mRNA that encode these receptors using qPCR (Figure 9, A–D) and found significant decreases in Il6ra and Il3ra expression in LK cells, where these receptors are generally highly expressed. In contrast, Flt3 and c-KIT mRNA levels were not affected by PRMT5 deletion. We next determined whether the decreased expression of FLT3 and c-Kit on PRMT5-null HSPCs is due to impaired RNA splicing. For the Flt3 gene, we found that 6 of over 20 introns had higher retention levels than normal in the PRMT5-null BM cells (other introns were either undetectable or comparable between control and KO) (Figure 9E). In contrast, increased intron retention was not observed in c-KIT or Il6ra mRNA (data not shown). These results suggest that PRMT5 controls the cell surface expression of these important receptors through multiple, distinct mechanisms.
 Figure 8
Figure 8PRMT5 loss impairs the cell surface expression of cytokine receptors. (A) BM cells were isolated from day-7 control and Prmt5-KO mice and stained for HSPC markers and several cytokine receptors. Percentages of FLT3-, IL-6Rα–, and IL-3Rα–positive cells in LK and LSK cell populations were plotted. (B and C) Day-7 control and Prmt5-KO BM cells were stained for HSPC cell surface marker expression and subjected to FACS analysis. Representative histogram shows decreased c-Kit expression in day-7 PRMT5-null HSPCs (B). The MFI of c-Kit staining in control and PRMT5-null LK and LSK cells was plotted as the mean ± SD (n = 5) (C). P values were determined by a 2-tailed Student’s t test.
 Figure 9
Figure 9PRMT5 loss impairs the expression of cytokine receptors via both transcriptional and posttranscriptional mechanisms. (A–D) Flt3, c-KIT, Il6ra, and Il3ra mRNA levels were determined in day-7 LK and LSK cells using qPCR and normalized to Hprt1 expression. Cont, Prmt5fl/fl; KO, Prmt5Δ/Δ. Representative PCR results from 3 independent experiments are shown. (E) Top: Diagram of primer design to detect unspliced Flt3 mRNA. PCR #1, intron PCR; PCR #2, total mRNA PCR; PCR #3, mature mRNA PCR used in Figure 7C. Bottom: qPCR was used to determine the levels of unspliced introns in Flt3 mRNA in day-7 BM cells. The intron levels were first normalized to the total mRNA level in each sample, then the intron levels in PRMT5-null cells were normalized to levels in control cells (n = 3). Introns with a higher level of retention in PRMT5-deleted cells are shown in red in the diagram.
Activation of p53 in PRMT5-null BM cells is a later event and is independent of DNA damage. PRMT5 has been shown to regulate p53 activity (18, 21–23). However, we were unable to detect a p53 gene signature in our day-3 RNA-seq data (Figure 7B); instead, we found a significant increase in p53 protein levels in the day-7 PRMT5-null HSPCs (and moderately increased p53 protein levels in PRMT5-null total BM cells) (Supplemental Figure 8, A and B), together with increased expression of several p53 target genes (Cdkn1a [p21], Noxa1, Bbc3 [PUMA], and Mdm2) (Supplemental Figure 8, C and D). We did not detect an increase in γ-H2AX in the PRMT5-null HSPCs (Supplemental Figure 8A), suggesting that increased DNA damage is not present in these cells and thus could not be responsible for the observed increased p53 levels.
-
Discussion
We have demonstrated an essential role of PRMT5 in steady-state adult hematopoiesis, in which loss of 1 Prmt5 allele had no effect on hematopoiesis, while the complete loss of Prmt5 led to BM aplasia and lethal pancytopenia. It appears that the primary effect is in the stem/progenitor cell compartment, as we observed an early and dramatic decrease in hematopoietic progenitor cells (HPCs) and a transient expansion of HSCs, followed by a complete loss of both HPCs and HSCs. The loss of these cells is likely due to the combined effects of impaired cytokine signaling and increased p53 signaling, which result from PRMT5 deletion.
Adult HSCs are maintained in a quiescent, or dormant, state, which is important for their longevity and function. Therefore, genetic lesions that lead to loss of this quiescence can result in HSC exhaustion (1, 24). The loss of HSCs in Prmt5-KO mice could relate, at least in part, to their impaired quiescence, as PRMT5-null HSCs incorporate 2 times more BrdU and stain more positively for Ki67 than normal. Our chimeric BM transplant experiments demonstrate that the loss of HSC quiescence is cell intrinsic and is independent of the loss of HPCs or of the decrease in peripheral blood cells that occurs in the primary conditional KO mice. Intriguingly, PRMT5-deficient HSCs show increased proliferation in the presence of increased p53 activity, which has been shown to promote HSC quiescence (25); this suggests that other factors may play a more dominant role in coaxing PRMT5-deficient HSCs into the cell cycle. Indeed, we observed increased levels of ROS, specifically in PRMT5-null HSCs (data not shown), which may account for the increased cycling seen in these cells. Although the number of PRMT5-null phenotypic LT-HSCs was increased on day 7, these cells failed to form CAFCs or rescue lethally irradiated recipient mice, indicating that they are severely functionally defective.
In contrast to its initial effect on HSCs, PRMT5 loss in transient amplifying cells (the Lin–c-Kit+Sca1– HPCs) impairs their proliferation. PRMT5 is known to repress the expression of a number of cell cycle regulatory genes, including cyclin E1, RB, p27, CDKN2A, and SMAD7 (26–29). It can also control cell cycle progression by methylating nonhistone proteins, such as p53, E2F1, and FEN1 (21, 30, 31), and we found a significant increase in p53 protein in HSPCs, with increased expression of several p53 target genes, including Cdkn1a (which increased 40-fold in the progenitor cells and 5-fold in the LSK cells), Bbc3, and Mdm2. This dramatic increase in p21 expression could impair the cycle progression of these cells.
We have identified another important mechanism by which PRMT5 loss impairs the proliferation of progenitor cells and lineage-positive cells, and that is by impairing cytokine signaling. Our day-3 RNA-seq data in HSPCs already showed alterations in signal transduction pathways in HSPCs, suggesting that signal transduction is likely the primary biological process affected by PRMT5 deletion. Although PRMT5 has been reported to negatively regulate MAPK signaling in various epithelial cancer cell lines (19), we found that PRMT5 loss impaired ERK1/2 phosphorylation in normal primary hematopoietic cells, suggesting cell type–specific effects. In fact, PRMT5 deletion also impaired cytokine-driven STAT5 and AKT signaling in hematopoietic cells. Although the homozygous deletion of Erk1 and Erk2 results in the rapid loss of HSPCs as well as severe anemia and early lethality in mice (32), similar to what we observed in Prmt5-KO mice, the impairment in JAK/STAT and AKT/PI3K signaling pathways probably also contributes to the profound phenotype that we observed.
The remarkable decrease in cell surface expression of important cytokine receptors on HSPCs, such as c-Kit, FLT3, IL-6Rα, and IL-3Rα, likely accounts for the globally impaired cytokine signaling that occurs following PRMT5 deletion. PRMT5 loss appears to affect these cytokine receptors via multiple mechanisms, including intron retention. PRMT5 has numerous nonhistone targets, including several spliceosome components, and loss of PRMT5 has been shown to impair both constitutive splicing and alternative splicing (18, 33). We found intron retention events in more than 1,500 genes, many of which are involved in signal transduction. For instance, we detected several unspliced introns in Flt3 mRNA, but not in c-KIT or Il6ra mRNA, suggesting that PRMT5 loss specifically impairs RNA splicing of some genes in HSPCs.
The opposite effects of PRMT5 on the cell cycle of HSCs versus progenitor cells likely reflects this specificity due to its involvement in different multiprotein complexes and possibly to differences in its subcellular localization. PRMT5 appears to be primarily localized in the cytoplasm of proliferating (tumor) cells, while it shuttles between the cytoplasm and nucleus in pluripotent embryonic stem cells and germ cells (12, 23, 34). It has been reported that cytoplasmic PRMT5 is essential for prostate cancer cell proliferation, while nuclear PRMT5 can inhibit prostate cancer cell growth (35). Thus, it is possible that PRMT5 primarily resides in the nucleus in HSCs, where it functions to maintain pluripotency and quiescence in these cells. Its translocation into the cytoplasm could enable the differentiation of these cells into progenitor cells.
The enzymatically dead version of PRMT5 (PRMT5EQ) is unable to rescue PRMT5-deficient HSPCs in in vitro functional assays, demonstrating that the methyltransferase activity of PRMT5, and not just its presence in the cell, is required for its function in hematopoiesis. While some functions of nonmammalian PRMT5 are independent of its enzymatic activity (e.g., the methyltransferase activity of HSL7, the homolog of mammalian PRMT5, is dispensable to stabilize SWE1 and regulate cell cycle progression in Xenopus) (36), it appears that PRMT5 enzymatic activity is required for HSPC maintenance.
PRMT5 has pleiotropic effects in the cell, thus PRMT5 inhibitors may also have cell type–specific effects on gene expression and cell growth. While PRMT5 overexpression is found in several malignancies, treating cancer patients with PRMT5 inhibitors could result in important myelosuppressive side effects. Nonetheless, the clinical activity and side effects of chemical inhibitors of PRMT5 will need to be defined in vivo.
-
Methods
Further information can be found in the Supplemental Methods.
Mice. B6.SJL CD45.1 recipient mice and Mx1Cre and ERCre Tg mice were purchased from The Jackson laboratory. Prmt5 homozygous floxed mice (Prmt5fl/fl) were generated by crossing Prmt5 FLIP-OUT mice (obtained from EMMA) with Flp1 recombinase Tg mice (The Jackson Laboratory). Prmt5 conditional KO mice were generated by crossing Prmt5-floxed mice with Mx1Cre or ERCre Tg mice. Prmt5 gene excision in Mx1Cre+ Prmt5fl/fl mice was induced by poly(I:C) HMW (InvivoGen), given by i.p. injection at a dose of 10 mg/kg on 2 consecutive days. Mice used for all experiments except for the chimeric transplantation studies were 8 to 12 weeks of age. In vitro deletion of Prmt5 in ERCre+ Prmt5fl/fl BMMCs was induced by treating cultured cells with 0.1 μM 4-hydroxytamoxifen (4-OHT; Sigma-Aldrich) for 3 days. Prmt5/Casp3 double-KO mice were generated by crossing Mx1Cre+ Prmt5fl/fl mice with Casp3–/– mice (provided by Richard A. Flavell, Yale School of Medicine, New Haven, Connecticut, USA).
Flow cytometry. For all FACS analyses (except the experiment shown in Figure 2A), isolated BM cells were treated with rbc lysis buffer before staining. Cell surface marker staining for HSPCs was performed as previously described (25). BrdU incorporation was detected using the FITC BrdU Flow Kit (BD Biosciences), and apoptosis was determined using the PE Apoptosis Detection Kit (BD Biosciences), according to the manufacturers’ instructions. All flow cytometric analyses were performed using either a BD LSRII or LSR Fortessa Cell Analyzer (BD Biosciences). Data were analyzed using FlowJo software. Flow antibodies, including APC–c-Kit, APC-B220, PEcy7-sca1, PerCPcy5.5-Gr1, PE-Mac1, PE-IgM, and PacBlue-Ki67, were purchased from eBioscience. Other antibodies, including streptavidin–APC-cy7, PerCPcy5.5-CD3, FITC-CD45.1, APC-CD45.2, and PEcy7-CD4, were purchased from BioLegend.
Mouse BM cell culture and lentiviral infection. BM cells were isolated from tibiae and femurs by flushing the cells out of the bone using syringes. Cells were cultured in 20% FBS DMEM, supplemented with murine cytokines (50 ng/ml SCF, 10 ng/ml IL-3, and 10 ng/ml IL-6). Control lentivirus or viruses expressing either PRMT5wt or PRMT5EQ were produced as described (37). After 24 hours of growth, BM cells were infected with high-titer concentrated lentiviral suspensions in the presence of 4 μg/ml polybrene (Sigma-Aldrich). GFP+ cells were FACS sorted 2 days after infection.
CFU, CAFC, and LTC-IC assays. For CFU assays, 1 × 104 BM cells or 1 × 103 transduced GFP+ BM cells were plated in duplicate in methylcellulose media (MethoCult GF M3434; STEMCELL Technologies). Colonies were scored 7 days after seeding.
For CAFC assays, approximately 1.5 × 105 untransduced or sorted GFP+ cells were seeded onto MS5 stromal cells cultured in αMEM containing 12.5% FBS, 12.5% horse serum, 1 μM hydrocortisone, and 1 mM glutamine. The media were semireplenished weekly, and the cobblestone colonies were scored at week 5.
For the LTC-IC assay, 1.2 × 105, 2.4 × 104, or 1.2 × 104 BM cells were cultured on MS5 stromal cells. After 4 weeks of weekly semireplenishment of the media, cells were harvested and plated in methylcellulose media to perform CFU assays. Colonies were scored 1 week after plating.
BM transplantation assays. Approximately 5 × 106 BM cells isolated from control and Prmt5Δ/Δ mice 5 days after poly(I:C) injection were injected into lethally irradiated (9.5 Gy) B6.SJL recipient mice. Survival of these mice was monitored daily.
For chimeric BMT experiment, 1 × 106 BM cells were isolated from Prmt5fl/fl, Mx1Cre+ Prmt5fl/+, or Mx1Cre+ Prmt5fl/fl mice (CD45.2) and injected into lethally irradiated recipient mice (6- to 8-week-old female CD45.1 mice), together with 1 × 106 WT BM cells (CD45.1). PRMT5 deletion was induced by 2 poly(I:C) injections (10 mg/kg) given on consecutive days 4 months after transplantation.
RNA-seq and data analysis. HSPCs were isolated from control and KO mice 3 days after poly(I:C) injection, and total RNA was purified using an RNeasy Micro Kit (QIAGEN). To minimize the differences between individual mice and to ensure adequate high-quality RNA, RNA samples were pooled from 2 mice with the same genotype; duplicate samples were used for each genotype. The library preparation and 50-bp paired-end RNA-seq were performed on the Illumina HiSeq 2000 at the Epigenetics Core Facility of Weill Cornell Medical College (New York, New York, USA). The functional annotation of differentially expressed gene sets and aberrantly spliced gene sets was performed using the DAVID functional annotation tool. The RNA-seq data are deposited in the NCBI’s Gene Expression Omnibus (GEO) database (GEO GSE69937).
Statistics. Experimental results are displayed as the mean ± SD. A 2-tailed Student’s t test was used to determine significant differences between control and experimental groups. P values of less than 0.05 were considered statistically significant.
Study approval. All animal studies were conducted in compliance with NIH guidelines for the care and use of laboratory animals and were approved by the IACUC of the University of Miami.
-
Acknowledgments
We thank the members of the Nimer laboratory for their assistance and thoughtful input and the Flow Cytometry Core of the Sylvester Comprehensive Cancer Center. This work was supported by National Cancer Institute (NCI) grant RO1 CA166835-01 (to S.D. Nimer).
Address correspondence to: Stephen D. Nimer, 1120 NW 14th Street, CRB 660A (C241), Miami, Florida 33136, USA. Phone: 305.243.1775; E-mail: [email protected].
-
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Reference information: J Clin Invest. 2015;125(9):3532–3544. doi:10.1172/JCI81749.
-
References
- Rossi L, et al. Less is more: unveiling the functional core of hematopoietic stem cells through knockout mice. Cell Stem Cell. 2012;11(3):302–317.
- Challen GA, et al. Dnmt3a is essential for hematopoietic stem cell differentiation. Nat Genet. 2012;44(1):23–31.View this article via: PubMed Google Scholar
- Tadokoro Y, Ema H, Okano M, Li E, Nakauchi H. De novo DNA methyltransferase is essential for self-renewal, but not for differentiation, in hematopoietic stem cells. J Exp Med. 2007;204(4):715–722.
- Hidalgo I, et al. Ezh1 is required for hematopoietic stem cell maintenance and prevents senescence-like cell cycle arrest. Cell Stem Cell. 2012;11(5):649–662.
- Kamminga LM, et al. The Polycomb group gene Ezh2 prevents hematopoietic stem cell exhaustion. Blood. 2006;107(5):2170–2179.
- McMahon KA, et al. Mll has a critical role in fetal and adult hematopoietic stem cell self-renewal. Cell Stem Cell. 2007;1(3):338–345.
- Sandberg ML, et al. c-Myb and p300 regulate hematopoietic stem cell proliferation and differentiation. Dev Cell. 2005;8(2):153–166.
- Bedford MT, Clarke SG. Protein arginine methylation in mammals: who, what, and why. Mol Cell. 2009;33(1):1–13.
- Karkhanis V, Hu YJ, Baiocchi RA, Imbalzano AN, Sif S. Versatility of PRMT5-induced methylation in growth control and development. Trends Biochem Sci. 2011;36(12):633–641.
- Rank G, Cerruti L, Simpson RJ, Moritz RL, Jane SM, Zhao Q. Identification of a PRMT5-dependent repressor complex linked to silencing of human fetal globin gene expression. Blood. 2010;116(9):1585–1592.
- Zhao Q, et al. PRMT5-mediated methylation of histone H4R3 recruits DNMT3A, coupling histone and DNA methylation in gene silencing. Nat Struct Mol Biol. 2009;16(3):304–311.
- Tee WW, et al. Prmt5 is essential for early mouse development and acts in the cytoplasm to maintain ES cell pluripotency. Genes Dev. 2010;24(24):2772–2777.
- Chittka A, Nitarska J, Grazini U, Richardson WD. Transcription factor positive regulatory domain 4 (PRDM4) recruits protein arginine methyltransferase 5 (PRMT5) to mediate histone arginine methylation and control neural stem cell proliferation and differentiation. J Biol Chem. 2012;287(51):42995–43006.
- Nagamatsu G, et al. A germ cell-specific gene, Prmt5, works in somatic cell reprogramming. J Biol Chem. 2011;286(12):10641–10648.
- Yang Y, Bedford MT. Protein arginine methyltransferases and cancer. Nat Rev Cancer. 2013;13(1):37–50.View this article via: PubMed Google Scholar
- Gu Z, et al. Protein arginine methyltransferase 5 is essential for growth of lung cancer cells. Biochem J. 2012;446(2):235–241.
- Bao X, Zhao S, Liu T, Liu Y, Liu Y, Yang X. Overexpression of PRMT5 promotes tumor cell growth and is associated with poor disease prognosis in epithelial ovarian cancer. J Histochem Cytochem. 2013;61(3):206–217.
- Bezzi M, et al. Regulation of constitutive and alternative splicing by PRMT5 reveals a role for Mdm4 pre-mRNA in sensing defects in the spliceosomal machinery. Genes Dev. 2013;27(17):1903–1916.
- Andreu-Perez P, et al. Protein arginine methyltransferase 5 regulates ERK1/2 signal transduction amplitude and cell fate through CRAF. Sci Signal. 2011;4(190):ra58.View this article via: PubMed Google Scholar
- Wei H, et al. PRMT5 dimethylates R30 of the p65 subunit to activate NF-kappaB. Proc Natl Acad Sci U S A. 2013;110(33):13516–13521.
- Jansson M, et al. Arginine methylation regulates the p53 response. Nat Cell Biol. 2008;10(12):1431–1439.
- Scoumanne A, Zhang J, Chen X. PRMT5 is required for cell-cycle progression and p53 tumor suppressor function. Nucleic Acids Res. 2009;37(15):4965–4976.
- Kim S, et al. PRMT5 protects genomic integrity during global DNA demethylation in primordial germ cells and preimplantation embryos. Mol Cell. 2014;56(4):564–579.
- Pietras EM, Warr MR, Passegue E. Cell cycle regulation in hematopoietic stem cells. J Cell Biol. 2011;195(5):709–720.
- Liu Y, et al. p53 regulates hematopoietic stem cell quiescence. Cell Stem Cell. 2009;4(1):37–48.
- Fabbrizio E, et al. Negative regulation of transcription by the type II arginine methyltransferase PRMT5. EMBO Rep. 2002;3(7):641–645.
- Wang L, Pal S, Sif S. Protein arginine methyltransferase 5 suppresses the transcription of the RB family of tumor suppressors in leukemia and lymphoma cells. Mol Cell Biol. 2008;28(20):6262–6277.
- Nicholas C, et al. PRMT5 is upregulated in malignant and metastatic melanoma and regulates expression of MITF and p27(Kip1). PLoS One. 2013;8(9):e74710.
- Le Guezennec X, et al. MBD2/NuRD and MBD3/NuRD, two distinct complexes with different biochemical and functional properties. Mol Cell Biol. 2006;26(3):843–851.
- Cho EC, et al. Arginine methylation controls growth regulation by E2F-1. EMBO J. 2012;31(7):1785–1797.
- Guo Z, et al. Methylation of FEN1 suppresses nearby phosphorylation and facilitates PCNA binding. Nat Chem Biol. 2010;6(10):766–773.
- Chan G, Gu S, Neel BG. Erk1 and Erk2 are required for maintenance of hematopoietic stem cells and adult hematopoiesis. Blood. 2013;121(18):3594–3598.
- Friesen WJ, et al. The methylosome, a 20S complex containing JBP1 and pICln, produces dimethylarginine-modified Sm proteins. Mol Cell Biol. 2001;21(24):8289–8300.
- Ancelin K, et al. Blimp1 associates with Prmt5 and directs histone arginine methylation in mouse germ cells. Nat Cell Biol. 2006;8(6):623–630.
- Gu Z, Li Y, Lee P, Liu T, Wan C, Wang Z. Protein arginine methyltransferase 5 functions in opposite ways in the cytoplasm and nucleus of prostate cancer cells. PLoS One. 2012;7(8):e44033.
- Yamada A, Duffy B, Perry JA, Kornbluth S. DNA replication checkpoint control of Wee1 stability by vertebrate Hsl7. J Cell Biol. 2004;167(5):841–849.
- Liu F, et al. JAK2V617F-mediated phosphorylation of PRMT5 downregulates its methyltransferase activity and promotes myeloproliferation. Cancer Cell. 2011;19(2):283–294.
-
Version history
- Version 1 (August 10, 2015): No description
- Version 2 (September 1, 2015): No description
Article tools
-
View PDF

- Download citation information
- Send a comment
- Terms of use
- Standard abbreviations
- Need help? Email the journal
Related posts
Metrics



Copyright © 2024 American Society for Clinical Investigation
ISSN: 0021-9738 (print), 1558-8238 (online)









