Advertisement
Research ArticleGastroenterology Free access | 10.1172/JCI76344
Cytosolic HMGB1 controls the cellular autophagy/apoptosis checkpoint during inflammation
Xiaorong Zhu,1 Jeannette S. Messer,1 Yunwei Wang,1 Fanfei Lin,1 Candace M. Cham,1 Jonathan Chang,2 Timothy R. Billiar,3 Michael T. Lotze,3 David L. Boone,1 and Eugene B. Chang1
1Department of Medicine, University of Chicago, Chicago, Illinois, USA.
2Department of Cancer Immunology and AIDS, Dana-Farber Cancer Institute, Boston, Massachusetts, USA.
3Department of Surgery, University of Pittsburgh, Pittsburgh, Pennsylvania, USA.
Address correspondence to: Eugene B. Chang, Knapp Center for Biomedical Discovery, 900 E. 57th Street, 9th Floor, Room 9130, University of Chicago, Chicago, Illinois, USA. Phone: 773.702.6458; E-mail: [email protected].
Authorship note: Xiaorong Zhu and Jeannette S. Messer contributed equally to this work.
Find articles by Zhu, X. in: JCI | PubMed | Google Scholar
1Department of Medicine, University of Chicago, Chicago, Illinois, USA.
2Department of Cancer Immunology and AIDS, Dana-Farber Cancer Institute, Boston, Massachusetts, USA.
3Department of Surgery, University of Pittsburgh, Pittsburgh, Pennsylvania, USA.
Address correspondence to: Eugene B. Chang, Knapp Center for Biomedical Discovery, 900 E. 57th Street, 9th Floor, Room 9130, University of Chicago, Chicago, Illinois, USA. Phone: 773.702.6458; E-mail: [email protected].
Authorship note: Xiaorong Zhu and Jeannette S. Messer contributed equally to this work.
Find articles by Messer, J. in: JCI | PubMed | Google Scholar
1Department of Medicine, University of Chicago, Chicago, Illinois, USA.
2Department of Cancer Immunology and AIDS, Dana-Farber Cancer Institute, Boston, Massachusetts, USA.
3Department of Surgery, University of Pittsburgh, Pittsburgh, Pennsylvania, USA.
Address correspondence to: Eugene B. Chang, Knapp Center for Biomedical Discovery, 900 E. 57th Street, 9th Floor, Room 9130, University of Chicago, Chicago, Illinois, USA. Phone: 773.702.6458; E-mail: [email protected].
Authorship note: Xiaorong Zhu and Jeannette S. Messer contributed equally to this work.
Find articles by Wang, Y. in: JCI | PubMed | Google Scholar
1Department of Medicine, University of Chicago, Chicago, Illinois, USA.
2Department of Cancer Immunology and AIDS, Dana-Farber Cancer Institute, Boston, Massachusetts, USA.
3Department of Surgery, University of Pittsburgh, Pittsburgh, Pennsylvania, USA.
Address correspondence to: Eugene B. Chang, Knapp Center for Biomedical Discovery, 900 E. 57th Street, 9th Floor, Room 9130, University of Chicago, Chicago, Illinois, USA. Phone: 773.702.6458; E-mail: [email protected].
Authorship note: Xiaorong Zhu and Jeannette S. Messer contributed equally to this work.
Find articles by Lin, F. in: JCI | PubMed | Google Scholar
1Department of Medicine, University of Chicago, Chicago, Illinois, USA.
2Department of Cancer Immunology and AIDS, Dana-Farber Cancer Institute, Boston, Massachusetts, USA.
3Department of Surgery, University of Pittsburgh, Pittsburgh, Pennsylvania, USA.
Address correspondence to: Eugene B. Chang, Knapp Center for Biomedical Discovery, 900 E. 57th Street, 9th Floor, Room 9130, University of Chicago, Chicago, Illinois, USA. Phone: 773.702.6458; E-mail: [email protected].
Authorship note: Xiaorong Zhu and Jeannette S. Messer contributed equally to this work.
Find articles by Cham, C. in: JCI | PubMed | Google Scholar
1Department of Medicine, University of Chicago, Chicago, Illinois, USA.
2Department of Cancer Immunology and AIDS, Dana-Farber Cancer Institute, Boston, Massachusetts, USA.
3Department of Surgery, University of Pittsburgh, Pittsburgh, Pennsylvania, USA.
Address correspondence to: Eugene B. Chang, Knapp Center for Biomedical Discovery, 900 E. 57th Street, 9th Floor, Room 9130, University of Chicago, Chicago, Illinois, USA. Phone: 773.702.6458; E-mail: [email protected].
Authorship note: Xiaorong Zhu and Jeannette S. Messer contributed equally to this work.
Find articles by Chang, J. in: JCI | PubMed | Google Scholar
1Department of Medicine, University of Chicago, Chicago, Illinois, USA.
2Department of Cancer Immunology and AIDS, Dana-Farber Cancer Institute, Boston, Massachusetts, USA.
3Department of Surgery, University of Pittsburgh, Pittsburgh, Pennsylvania, USA.
Address correspondence to: Eugene B. Chang, Knapp Center for Biomedical Discovery, 900 E. 57th Street, 9th Floor, Room 9130, University of Chicago, Chicago, Illinois, USA. Phone: 773.702.6458; E-mail: [email protected].
Authorship note: Xiaorong Zhu and Jeannette S. Messer contributed equally to this work.
Find articles by Billiar, T. in: JCI | PubMed | Google Scholar
1Department of Medicine, University of Chicago, Chicago, Illinois, USA.
2Department of Cancer Immunology and AIDS, Dana-Farber Cancer Institute, Boston, Massachusetts, USA.
3Department of Surgery, University of Pittsburgh, Pittsburgh, Pennsylvania, USA.
Address correspondence to: Eugene B. Chang, Knapp Center for Biomedical Discovery, 900 E. 57th Street, 9th Floor, Room 9130, University of Chicago, Chicago, Illinois, USA. Phone: 773.702.6458; E-mail: [email protected].
Authorship note: Xiaorong Zhu and Jeannette S. Messer contributed equally to this work.
Find articles by Lotze, M. in: JCI | PubMed | Google Scholar
1Department of Medicine, University of Chicago, Chicago, Illinois, USA.
2Department of Cancer Immunology and AIDS, Dana-Farber Cancer Institute, Boston, Massachusetts, USA.
3Department of Surgery, University of Pittsburgh, Pittsburgh, Pennsylvania, USA.
Address correspondence to: Eugene B. Chang, Knapp Center for Biomedical Discovery, 900 E. 57th Street, 9th Floor, Room 9130, University of Chicago, Chicago, Illinois, USA. Phone: 773.702.6458; E-mail: [email protected].
Authorship note: Xiaorong Zhu and Jeannette S. Messer contributed equally to this work.
Find articles by Boone, D. in: JCI | PubMed | Google Scholar
1Department of Medicine, University of Chicago, Chicago, Illinois, USA.
2Department of Cancer Immunology and AIDS, Dana-Farber Cancer Institute, Boston, Massachusetts, USA.
3Department of Surgery, University of Pittsburgh, Pittsburgh, Pennsylvania, USA.
Address correspondence to: Eugene B. Chang, Knapp Center for Biomedical Discovery, 900 E. 57th Street, 9th Floor, Room 9130, University of Chicago, Chicago, Illinois, USA. Phone: 773.702.6458; E-mail: [email protected].
Authorship note: Xiaorong Zhu and Jeannette S. Messer contributed equally to this work.
Find articles by Chang, E. in: JCI | PubMed | Google Scholar
Authorship note: Xiaorong Zhu and Jeannette S. Messer contributed equally to this work.
Published February 2, 2015 - More info
J Clin Invest. 2015;125(3):1098–1110. https://doi.org/10.1172/JCI76344.
Copyright © 2015, American Society for Clinical Investigation
Received: March 27, 2014; Accepted: December 26, 2014
-
Abstract
The intracellular protein HMGB1 is released from cells and acts as a damage-associated molecular pattern molecule during many diseases, including inflammatory bowel disease (IBD); however, the intracellular function of HMGB1 during inflammation is poorly understood. Here, we demonstrated that cytosolic HMGB1 regulates apoptosis by protecting the autophagy proteins beclin 1 and ATG5 from calpain-mediated cleavage during inflammation. Colitis in mice with an intestinal epithelial cell–specific Hmgb1 deletion and patients with IBD were both characterized by increased calpain activation, beclin 1 and ATG5 cleavage, and intestinal epithelial cell (IEC) death compared with controls. In vitro cleavage assays and studies of enteroids verified that HMGB1 protects beclin 1 and ATG5 from calpain-mediated cleavage events that generate proapoptotic protein fragments. Together, our results indicate that HMGB1 is essential for mitigating the extent and severity of inflammation-associated cellular injury by controlling the switch between the proautophagic and proapoptotic functions of beclin 1 and ATG5 during inflammation. Moreover, these studies demonstrate that HMGB1 is pivotal for reducing tissue injury in IBD and other complex inflammatory disorders.
-
Introduction
Inflammatory bowel diseases (IBDs) are chronic, debilitating disorders caused by gastrointestinal mucosal damage and inflammation. Two separate but related disease phenotypes make up IBD: Crohn’s disease (CD) and ulcerative colitis (UC). The pathophysiology of IBD is complex, and there are likely numerous mechanisms that are unique but also shared between the 2 conditions. However, both are thought to result from gastrointestinal barrier compromise, leading to inflammation and infiltration with innate and adaptive immune cells (1). The gastrointestinal barrier is primarily composed of intestinal epithelial cells (IECs) and their soluble products (2). IECs also participate in inflammatory responses directly through processes such as autophagy and indirectly through the production of cytokines that recruit innate and adaptive immune cells to sites of mucosal damage (3). The proteins found in the cytosol and released from IECs during inflammation include high-mobility group box 1 (HMGB1).
HMGB1 is a nuclear, nonhistone DNA–binding protein (4). During cellular stress, it localizes to the cell cytosol and can exit the cell through loss of membrane integrity or active secretion (5). Once it is free from cells, HMGB1 acts as a damage-associated molecular pattern (DAMP) molecule to activate innate immune receptors and drive inflammatory responses (6). Circulating HMGB1 levels are increased in many human inflammatory diseases and their associated experimental models (5). Consistent with this, intestinal HMGB1 expression is elevated in the dextran sodium sulfate (DSS) model of murine colitis (7). Furthermore, HMGB1 antagonism using anti-HMGB1 antibody or ethyl pyruvate ameliorates colitis in the DSS and Il10–/– mouse models, respectively (7, 8). Very little is known about HMGB1 in human IBD, just that children with IBD have increased levels of this protein in their feces (9). These data reflect the fact that the majority of HMGB1 research has focused on its extracellular functions during inflammation, despite it being concurrently found in the cell cytosol under these conditions. The indications that HMGB1 levels were altered in experimental and human colitis and the presence of this protein in IECs, a key cell type in the pathophysiology of IBD, led us to study the intracellular role of this protein in IECs during human and experimental colitis.
-
Results
Loss of HMGB1 exacerbates murine colitis. Mice globally deficient in HMGB1 die within 24 hours of birth, so we generated Hmgb1-floxed mice (Hmgb1fl/fl) to test the role of HMGB1 in physiologic disease models (10). We crossed these mice with mice expressing CRE recombinase under the villin promoter (Vil-Cre) to create mice lacking Hmgb1 solely in IECs (Vil-Cre Hmgb1fl/fl) (Supplemental Figure 1, A–C; supplemental material available online with this article; doi:10.1172/JCI76344DS1). We treated these mice with DSS to induce colitis and found that 100% of Vil-Cre Hmgb1fl/fl mice died by day 11 of the study versus only 25% of Hmgb1fl/fl mice (Figure 1A). DSS administration is commonly used as an acute model of IBD and most closely mimics UC in humans (11). Treatment with a lower dose of DSS resulted in significantly worse colitis in Vil-Cre Hmgb1fl/fl when compared that seen in Hmgb1fl/fl mice (Figure 1, B–E). Vil-Cre Hmgb1fl/fl mice lost more weight and developed worse signs of colitis after DSS administration than did controls (Figure 1, B and C). They also had greater colon shortening and histology consistent with increased intestinal damage in response to DSS administration (Figure 1, D and E).
 Figure 1
Figure 1Loss of HMGB1 exacerbates DSS and Il10–/– colitis. (A) Survival curve for 8-week-old Hmgb1fl/fl (n = 12) and Vil-Cre Hmgb1fl/fl (n = 9) littermates treated with 3% DSS for 5 days. The mice were then followed until day 19. (B) Weight loss (mean ± SEM) of Hmgb1fl/fl (n = 19) and Vil-Cre Hmgb1fl/fl (n = 16) mice expressed as a percentage of their initial body weight during a 5-day treatment with 2.5% DSS and a 14-day recovery period. (C) Disease activity index (DAI) (weight loss, stool consistency, and rectal bleeding; mean ± SEM) generated on day 5 from DSS-treated mice (Hmgb1fl/fl, n = 8; Vil-Cre Hmgb1fl/fl, n = 8). (D) Gross appearance and length (mean ± SEM) of the colon on day 14 after DSS (Hmgb1fl/fl, n = 8; Vil-Cre Hmgb1fl/fl, n = 8). (E) H&E staining of formalin-fixed colons from DSS-treated mice on day 14. Images were obtained 20 mm from the rectum (original magnification, ×100) (Hmgb1fl/fl, n = 3; Vil-Cre Hmgb1fl/fl, n = 3). (F) Vil-Cre Hmgb1fl/fl Il10+/– males were mated with Hmgb1fl/fl Il10+/– females, and the progeny (Hmgb1fl/fl Il10–/–, n = 8; Vil-Cre Hmgb1fl/fl Il10–/–, n = 7) were observed for signs of colitis for 8 to 12 weeks. (G) Mean DAI for Hmgb1fl/fl Il10–/– (n = 8) and Vil-Cre Hmgb1fl/fl Il10–/– (n = 7) mice. (H) Histological images of H&E-stained, formalin-fixed colons from Hmgb1fl/fl Il10–/– (n = 4) and Vil-Cre Hmgb1fl/fl Il10–/– (n = 4) littermate mice (original magnification, ×200). The intestine from the Hmgb1fl/fl Il10–/– mouse was essentially normal, while the Vil-Cre Hmgb1fl/fl Il10–/– mouse showed signs of chronic intestinal inflammation and colitis (loss of goblet cells, crypt abscesses, cellular infiltration, elongation of mucosa, and epithelial erosion). Data were analyzed using 2-tailed Student’s t tests. **P < 0.01; ****P < 0.001.
We also examined the role of IEC HMGB1 in a model of immune-mediated chronic colitis by crossing Vil-Cre Hmgb1fl/fl mice with Il10–/– mice. In the Il10–/– model, chronic colitis develops due to loss of tolerizing DCs in the gastrointestinal tract, and failure to produce functional Tregs (12, 13). One hundred percent of Vil-Cre Hmgb1fl/fl Il10–/– mice developed colitis as opposed to less than 25% of Hmgb1fl/fl Il10–/– mice (Figure 1F). Colitis in Vil-Cre Hmgb1fl/fl Il10–/– mice also developed earlier and was more severe, as measured by clinical signs and histopathology, than that seen in controls (Figure 1, F–H). Therefore, loss of HMGB1 in IECs exacerbated colitis in acute (DSS) and chronic (Il10–/–) mouse models.
The autophagic response to DSS in IECs is blunted in the absence of HMGB1. We next explored the potential mechanisms underlying these effects. In the normal mouse intestine, HMGB1 was found almost exclusively in the nucleus of IECs (Figure 2A). After 3 days of DSS treatment (before the onset of frank inflammation), HMGB1 was primarily localized to the IEC cytosol. HMGB1 translocation was accompanied by an increase in Hmgb1 mRNA and a decrease in HMGB1 protein (Figure 2B and Supplemental Figure 2, A and B). This decrease in HMGB1 protein is transient in the acute colitis model, and the level rises at later time points (7). We also appreciated increased levels of HMGB1 in Hmgb1fl/fl Il10–/– mice after they developed chronic colitis (Figure 2C). The autophagy initiation protein beclin 1 has been reported to interact with HMGB1 in the cell cytosol, and autophagy is diminished in Hmgb1–/– mouse embryonic fibroblasts (14). This led us to evaluate microtubule-associated light chain 3 (LC3) lipidation and levels of the autophagy substrate p62 to characterize autophagy in the absence of HMGB1. Under nonstimulated conditions, the ratio of LC3II to LC3I and p62 levels were similar in Hmgb1fl/fl and Vil-Cre Hmgb1fl/fl mice (Figure 2, D and E, and Supplemental Figure 2, C–E). This indicates that autophagy is not defective in HMGB1-deficient cells under nonstressed conditions. However, following DSS administration, autophagy was decreased in Vil-Cre Hmgb1fl/fl cells. Autophagy is thought to be a cell-survival mechanism, and failure is associated with increased apoptotic death (15). So, we examined cell death by terminal TUNEL staining. Vil-Cre Hmgb1fl/fl mice exhibited more TUNEL-positive cells at day 5 than did Hmgb1fl/fl mice (Figure 2F), suggesting that loss of HMGB1 in IECs leads to decreased autophagy and increased apoptosis during DSS colitis.
 Figure 2
Figure 2The autophagic response to DSS in the intestinal epithelium is blunted in the absence of HMGB1. (A) Confocal microscopic images of colons from untreated (n = 3) or DSS-treated (day 3) (n = 3) Hmgbfl/fl mice stained for the DNA marker Hoechst (blue) and HMGB1 (red) (original magnification, ×400). Colocalization was evaluated using Pearson’s correlation coefficient with the Costes correction (mean ± SEM) (B) qRT-PCR (mean ± SEM) and immunoblot for HMGB1 expression in colonic mucosal scrapings from untreated (n = 4) and DSS-treated (n = 4) Hmgb1fl/fl mice. (C) Immunoblot for HMGB1 in intestinal mucosal scrapings from Hmgb1fl/fl Il10–/– mice with and without signs of colitis (n = 4). (D) Immunoblot for LC3 and p62 in Hmgb1fl/fl (n = 4) and Vil-Cre Hmgb1fl/fl (n = 4) mice on day 3 of DSS treatment. LC3II represents the lipidated form of LC3 and is increased during autophagy. LC3II/LC3I (mean ± SEM) represents the ratio between the lipidated and unlipidated forms of the protein. (E) Confocal microscopic images of endogenous LC3 staining in frozen colonic sections from Hmgb1fl/fl (n = 3) and Vil-Cre Hmgb1fl/fl (n = 3) mice on day 3 of DSS treatment (original magnification, ×630). Autophagosomes have the appearance of LC3-positive punctate structures. (F) TUNEL staining on frozen colonic sections from Hmgb1fl/fl (n = 3) and Vil-Cre Hmgb1fl/fl (n = 3) mice on day 5 of DSS treatment (original magnification, ×400). Data were analyzed using 2-tailed Student’s t tests, except for the data in F, which were analyzed by 2-way Anova with Bonferroni’s multiple comparisons test as well as the 2-tailed Student’s t test for between-genotype comparisons within a treatment group. *P < 0.05; ****P < 0.001.
Since IEC death was the most striking feature of DSS colitis in Vil-Cre Hmgb1fl/fl mice, we investigated whether this death was associated with activation of the adaptive immune system. There were no obvious differences in cellular infiltration of the gut mucosa on day 3 after DSS administration, the time point at which we saw more IEC death in Vil-Cre Hmgb1fl/fl mice by TUNEL staining (Supplemental Figure 3A). Similarly, expression levels of mucosal cytokines were not significantly different at this time point, aside from decreased expression of TNF-α protein in Vil-Cre Hmgb1fl/fl mice (Supplemental Figure 3, B and C). TNF-α is generally considered to be a proinflammatory cytokine, so decreased levels would not be expected to be contributing to tissue damage. Additionally, Tnfa–/– mice develop more severe colitis in response to DSS than do Tnfa+/+ mice, suggesting that this cytokine may also have antiinflammatory or prohealing functions in the intestinal mucosa (16). Taken together, these data suggest that IECs die before mucosal immune cell infiltration into the Vil-Cre Hmgb1fl/fl model, not as a result of immune cell infiltration.
HMGB1 protects beclin 1 and ATG5 from cleavage during inflammation. Identification of an autophagy defect in HMGB1-deficient cells led us to examine beclin 1 in IECs during DSS colitis. Beclin 1 has roles in both cell survival and cell death. Under conditions that favor autophagy, it forms a complex with ambra 1, VPS34, and VPS15 in order to initiate autophagy (17). However, cleavage by cell proteases to remove the N-terminal BCL-2–interacting domain converts it to a proapoptotic protein (18). Beclin 1 expression was increased in Vil-Cre Hmgb1fl/fl mice at both the mRNA and protein levels (Supplemental Figure 4, A and B). This protein was cleaved into fragments of approximately 50, 37, and 35 kDa after DSS treatment in the absence of HMGB1 (Figure 3A). Increased beclin 1 expression and cleavage were also appreciated in Hmgb1fl/fl Il10–/– and Vil-Cre Hmgb1fl/fl Il10–/– mice after development of colitis, but Vil-Cre Hmgb1fl/fl Il10–/– mice had higher levels of the 50-kDa beclin 1 fragment than did Hmgb1fl/fl Il10–/– mice (Figure 3B). The primary 50-kDa fragment seen in Vil-Cre Hmgb1fl/fl mice treated with DSS or lacking IL-10 was consistent with calpain 1 cleavage (19). An in vitro cleavage assay showed that HMGB1 protects beclin 1 from calpain 1–mediated cleavage in a dose-dependent manner (Figure 3, C and D). HMGB1 itself was not cleaved by calpain, therefore it did not diminish cleavage of beclin 1 by acting as a surrogate substrate for the protease (Supplemental Figure 4C). Additionally, the band pattern in vitro and in vivo was consistent with cleavage at aa 63 (EETN[63]SG), a putative cleavage site that would generate a 53-kDa fragment lacking the BCL-2–interacting domain (Figure 3D and ref. 20).
 Figure 3
Figure 3HMGB1 protects beclin 1 and ATG5 from cleavage during murine colitis. (A) Immunoblot for beclin 1 using an antibody that recognizes aa 171–291 of the protein. Blots are representative of Hmgb1fl/fl (n = 4) and Vil-Cre Hmgb1fl/fl (n = 4) mice. (B) Immunoblot for beclin 1 in Hmgb1fl/fl Il10–/– (n = 4) and Vil-Cre Hmgb1fl/fl Il10–/– (n = 4) mice. (C) Beclin 1 immunoblot of the products from an in vitro cleavage assay of beclin 1 by calpain 1 with decreasing amounts of HMGB1. (D) MYC-DDK (FLAG) immunoblot of the products of the beclin 1 in vitro cleavage assay. Recombinant beclin 1 contained a C-terminal MYC-DDK epitope tag. (E) Immunoblot for ATG5 using an antibody that recognizes aa 2–15 of the protein. Blots are representative of Hmgb1fl/fl (n = 4) and Vil-Cre Hmgb1fl/fl (n = 4) mice. (F) Immunoblot for ATG5 in Hmgb1fl/fl Il10–/– (n = 4) and Vil-Cre Hmgb1fl/fl Il10–/– (n = 4) mice. (G) ATG5 immunoblot of the products from an in vitro cleavage assay of ATG5 by calpain 1 with decreasing amounts of HMGB1. (H) Co-IP of HMGB1 and beclin 1 or ATG5 in colonic mucosal lysates from Hmgb1fl/fl (n = 4) and Vil-Cre Hmgb1fl/fl (n = 4) mice treated with DSS for 3 days. (I) Calpain activity assay evaluating cleavage of a fluorogenic calpain 1/2 substrate (Suc-LLVY-AMC) in samples of colonic mucosa from Hmgb1fl/fl (n = 3) and Vil-Cre Hmgb1fl/fl mice (n = 3) on day 3 of DSS treatment (mean ± SEM). (J) Immunoblot for the active p20 fragment of caspase 1 in Hmgb1fl/fl (n = 4) and Vil-Cre Hmgb1fl/fl mice (n = 4) on day 3 of DSS treatment. (K) qRT-PCR for calpastatin in cDNA from Hmgb1fl/fl (n = 6) and Vil-Cre Hmgb1fl/fl mice (n = 6) on day 3 of DSS treatment (mean ± SEM). Data were analyzed by 2-tailed Student’s t tests. *P < 0.05; **P < 0.01; ***P < 0.005; ****P < 0.001. RFU, relative fluorescence units.
After determining that HMGB1 protects beclin 1 from calpain-mediated cleavage, we considered that it could perform a similar function for other proteins. All of the core autophagy proteins, except LC3, have been reported to be cleaved by calpains (20). Two of these proteins, beclin 1 and ATG5, are converted to proapoptotic proteins by this cleavage event. A blast search for homology between beclin 1 and ATG5 revealed that beclin 1 and ATG5 share sequence homology in the region of the calpain cleavage site (Supplemental Figure 4D). However, sequence similarities were not present in the remaining autophagy proteins. This led us to examine ATG5 expression and cleavage in DSS-treated Vil-Cre Hmgb1fl/fl and Vil-Cre Hmgb1fl/fl Il10–/– mice. As with beclin 1, ATG5 expression and cleavage were increased in mice lacking IEC HMGB1 compared with what was observed in controls (Figure 3, E and F, and Supplemental Figure 4E). An in vitro cleavage assay demonstrated that HMGB1 protects ATG5 against calpain 1 cleavage in a dose-dependent manner (Figure 3G). Coimmunoprecipitation (co-IP) experiments performed using mucosal scrapings from DSS-treated Hmgb1fl/fl and Vil-Cre Hmgb1fl/fl mice showed that HMGB1 interacts with beclin 1 and ATG5 during inflammation (Figure 3H). The in vitro calpain cleavage assay showed that HMGB1 can interact with free ATG5, but the physiologically relevant form in vivo is thought to be the ATG5-ATG12 conjugate (21). This conjugate is the form that coimmunoprecipitated with HMGB1 in vivo, showing that HMGB1 interacts with the form of ATG5 important for autophagy. The putative HMGB1-interacting region (aa 197–200) is distant from the site of conjugation with ATG12 (aa 130), suggesting that conjugation would not be expected to interfere with an interaction between HMGB1 and ATG5 (21). Calpains are known to target proteins that are in complexes, so it is also not surprising that the ATG5-ATG12 conjugate would be a target for this protease (22, 23). Immunoblotting for ATG7, a calpain-cleaved protein with no homology with beclin 1 or ATG5, showed that cleavage and expression of this protein were similar between genotypes, suggesting that the homologous sequence in beclin 1 and ATG5 represents a specific HMGB1 interaction motif (Supplemental Figure 4F and ref. 20). To further determine whether HMGB1-mediated protection of beclin 1 and ATG5 from calpain cleavage involved direct, specific interactions, we performed an in vitro calpain cleavage assay incorporating a competitor peptide containing the putative interaction motif or a mock competitor peptide in which this region was mutated to alanine residues (Supplemental Figure 4G). In this assay, the presence of HMGB1 protected beclin 1 from calpain cleavage, and this protection was abrogated by the addition of the competitor peptide, but not the mock peptide, to the reaction mixture. Thus, HMGB1 interacts with beclin 1 and ATG5 and regulates the generation of proapoptotic protein fragments of these proteins during calpain-mediated inflammation.
Calpains are cysteine proteases active in the nucleus, cytosol, and extracellular milieu (24–26). They have been implicated in the pathophysiology of several inflammatory diseases, but calpain activity in IBD or colitis models is not well understood. It has been shown that the administration of a calpain inhibitor ameliorates 2,4,6-trinitrobenzenesulfonic acid–induced (TNBS-induced) colitis and that ethyl pyruvate decreases calpain activity in addition to preventing cytosolic translocation of HMGB1 (27, 28). Measurement of calpain activity in the gastrointestinal mucosa demonstrated calpain activation after DSS administration that was higher in Vil-Cre Hmgb1fl/fl mice than in controls (Figure 3I). In some inflammatory diseases, calpain activation occurs through caspase 1–mediated degradation and downregulation of the endogenous calpain inhibitor calpastatin (29–32). When calpastatin levels decrease sufficiently, calpains are spontaneously activated in the presence of calcium (29–32). We investigated whether this might be the mechanism of calpain activation in HMGB1-deficient IECs by immunoblotting for the p20 fragment of caspase 1. We found that caspase 1 was activated by DSS, and the levels were higher in Vil-Cre Hmgb1fl/fl mice than in controls (Figure 3J). Despite the increase in caspase 1 activity, the levels of IL-1β were comparable in Hmgb1fl/fl and Vil-Cre Hmgb1fl/fl mice (Supplemental Figure 3, B and C). In addition, quantitative reverse transcriptase PCR (RT-PCR) with primers recognizing calpastatin demonstrated decreased expression in Vil-Cre Hmgb1fl/fl mice (Figure 3K). Therefore, calpains are activated during murine colitis secondary to caspase 1–mediated decreases in calpastatin.
We further investigated how calpains contribute to colitis in Vil-Cre Hmgb1fl/fl mice by treating them with calpeptin, a calpain inhibitor, during DSS administration. The weight loss experienced by the treated mice was less than that of the untreated mice and did not differ from that seen in Hmgb1fl/fl mice (Figure 4A). Calpeptin treatment also improved the architectural distortion and epithelial cell death seen in Vil-Cre Hmgb1fl/fl mice after DSS administration (Figure 4B). Finally, calpeptin treatment decreased the amount of full-length and cleaved beclin 1 and ATG5 proteins in Vil-Cre Hmgb1fl/fl mice given DSS (Figure 4, C and D). Thus, calpain inhibition decreases beclin 1 and ATG5 cleavage and ameliorates colitis in mice treated with DSS.
 Figure 4
Figure 4Calpain inhibition rescues DSS colitis. Hmgb1fl/fl and Vil-Cre Hmgb1fl/fl mice were administered 2.5% DSS in their drinking water for 5 days and then sacrificed. Vil-Cre Hmgb1fl/fl mice were also treated with vehicle control or the calpain inhibitor calpeptin (5 mg/kg) by daily i.p. injection. (A) Weight loss on day 5 of DSS administration (mean). (B) H&E-stained sections of formalin-fixed, paraffin-embedded intestines from Vil-Cre Hmgb1fl/fl mice treated with vehicle (n = 4) or calpeptin (n = 4). Images were from areas 10 mm proximal to the animal’s rectum (original magnification, ×400). (C) Immunoblot for beclin 1 in intestinal mucosal scrapings from untreated Hmgb1fl/fl (n = 4) and Vil-Cre Hmgb1fl/fl mice treated with vehicle (n = 4) or calpeptin (n = 4). (D) Immunoblot for ATG5 in intestinal mucosal scrapings from untreated Hmgb1fl/fl (n = 4) and Vil-Cre Hmgb1fl/fl mice treated with vehicle (n = 4) or calpeptin (n = 4). Data were analyzed by 1-way ANOVA with Bonferroni’s multiple comparisons test. **P < 0.01.
Vil-Cre Hmgb1fl/fl mice have an IEC-specific deletion of HMGB1, suggesting that loss of a cell-intrinsic function or functions is most likely responsible for the increased IEC death seen with DSS administration. However, in vivo data cannot exclude the possibility that inflammatory mediators produced by other cell types could be acting on IECs to cause their death. To differentiate between cell-intrinsic and cell-extrinsic causes of IEC death, we isolated IEC progenitors from Hmgb1fl/fl and Vil-Cre Hmgb1fl/fl mice and grew them in culture to form intestinal organoids (enteroids) (Supplemental Figure 5A). We then challenged the cells with muramyl dipeptide (MDP), a component of bacterial cell walls. Stimulation of cells with MDP resulted in increased HMGB1 expression in Hmgb1fl/fl enteroids (Figure 5A and Supplemental Figure 5B). Enteroids lacking HMGB1 had increased beclin 1 expression at rest and higher levels of the 50-kDa fragment of beclin 1 after MDP stimulation compared with controls (Figure 5B). The level of active caspase 3, indicative of cell death pathway activation, was also increased in Vil-Cre Hmgb1fl/fl enteroids after MDP stimulation (Figure 5C). Administration of calpeptin in conjunction with MDP prevented caspase 3 activation in both Hmgb1fl/fl and Vil-Cre Hmgb1fl/fl enteroids (Figure 5D). Treatment of enteroids with MDP in the presence of bafilomycin, which allows autophagy activation but inactivates lysosomes so that LC3II accumulates, showed that autophagy was severely compromised in HMGB1-deficient IECs (Figure 5E and Supplemental Figure 5C). Hence, HMGB1 plays a cell-intrinsic role in survival during inflammation by regulating the calpain 1–mediated switch between autophagy and apoptosis.
 Figure 5
Figure 5Loss of cell-intrinsic HMGB1 functions leads to IEC death. IEC progenitors were isolated from the small intestines of Hmgb1fl/fl (n = 4) and Vil-Cre Hmgb1fl/fl (n = 4) mice and grown in culture until intestinal enteroids formed. The cells were then treated with 10 μg/ml L-18 MDP and lysed in 1% Triton lysis buffer. (A) Immunoblot for HMGB1 in lysates from cells treated with MDP for 4 hours. (B) Immunoblot for beclin 1 in lysates from cells treated with MDP for 4 hours. (C) Immunoblot for active caspase 3 in lysates from cells treated with MDP for 4 hours. (D) Immunoblot for active caspase 3 in lysates from cells treated with MDP for 4 hours in the presence of DMSO (vehicle control) or 1 μg/ml calpeptin. (E) Immunoblot for LC3B in lysates treated with MDP for 4 hours in the presence or absence of 100 nM bafilomycin A1. (F) Immunoblot for beclin 1 in lysates from cells treated with MDP at the indicated time points. (G) Immunoblot for ATG5 in lysates from cells treated with MDP at the indicated time points. (H) Immunoblot for cleaved caspase 3 in lysates from cells treated with MDP at the indicated time points. (I) Calpain activity in enteroid lysates at the indicated times after MDP treatment. Data were analyzed by 2-way ANOVA with Bonferroni’s multiple comparisons test as well as the 2-tailed Student’s t test for between-genotype comparisons within a treatment group **P < 0.01; ****P < 0.001.
In the murine colitis and enteroid models, the loss of HMGB1 resulted in higher levels of calpain activation and cleavage of beclin 1 and ATG5. The mechanism whereby HMGB1 protects these proteins from cleavage could either be direct, through protein-protein interactions that sterically inhibit calpain binding or activity, or indirect, through control of calpain activation. The results from the in vitro cleavage assays and in vivo co-IP experiments suggested that HMGB1 acts directly to prevent beclin 1 and ATG5 cleavage, but this did not rule out the possibility that the primary function of HMGB1 in vivo is to indirectly inhibit calpain activity. In order to differentiate between these possibilities, we stimulated enteroids with MDP and followed beclin 1 and ATG5 cleavage, calpain activity, and caspase 3 activation over time. Both beclin 1 and ATG5 cleavage occurred in Vil-Cre Hmgb1fl/fl cells at 60 minutes, while calpain activity was low and not significantly different between the genotypes (Figure 5, F–I). In Hmgb1fl/fl cells, cleavage of both proteins was first appreciated at 240 minutes, as calpain activity began to rise (Figure 5, F–I, and Supplemental Figure 5D). At 120 minutes, active caspase 3 activity was higher in Vil-Cre Hmgb1fl/fl cells than in controls (Figure 5H). Calpain activity was not significantly higher in Vil-Cre Hmgb1fl/fl cells compared with that in controls until 240 minutes, after beclin 1 and ATG5 were cleaved and caspase 3 activated (Figure 5H). These results show that death was triggered prior to massive increases in calpain activity in cells lacking HMGB1, likely due to the proapoptotic effects of beclin 1 and ATG5. They also demonstrate that the differential cleavage of beclin 1 and ATG5 in Hmgb1fl/fl versus Vil-Cre Hmgb1fl/fl cells is not a product of higher calpain activity in the Vil-Cre Hmgb1fl/fl cells.
Our data show that loss of HMGB1 leads to IEC death in vivo and in vitro during cell stress. The death of these cells could be critical to the development of colitis in 2 ways by inducing (a) the loss of the gastrointestinal barrier with exposure of the underlying immune system to the microbial contents of the gut lumen and (b) the release of proinflammatory molecules from dying IECs. To test whether the death of HMGB1-deficient and -replete IECs differs in the ability of these cells to stimulate professional immune cells, we examined cultured mesenteric lymph node (MLN) cells stimulated by supernatants derived from MDP-stimulated Hmgb1fl/fl and Vil-Cre Hmgb1fl/fl enteroids. Production of IFN-γ and IL-17 was similar between groups, whereas production of IL-6 and TNF-α was lower in MLN cultures stimulated with supernatants derived from Vil-Cre Hmgb1fl/fl enteroid cultures (Supplemental Figure 6). This suggests that loss of the gastrointestinal barrier function of IECs, rather than direct immune activation by their death, may be a more important contributor to the increased severity of the colitis seen in Vil-Cre Hmgb1fl/fl mice.
Human IBD is also characterized by altered HMGB1 expression, beclin 1 and ATG5 cleavage, and increased IEC death. Finally, we investigated whether HMGB1 regulates apoptosis during human IBD. Human IBD was associated with higher HMGB1 mRNA levels and a shift in HMGB1 intracellular localization from the nucleus to the cytosol (Figure 6, A and B). Conversely, HMGB1 protein expression was decreased in patients with active colitis compared with that in controls (Figure 6A and Supplemental Figure 7A). When we examined beclin 1 expression in active colitis, we found that it was increased at both the mRNA and protein levels (Figure 6C and Supplemental Figure 7, B–E). Most important, we found that cleavage of beclin 1, cleavage of ATG5, and levels of the active p19 fragment of caspase 3 were increased in patients with active colitis (Figure 6, C–E, and Supplemental Figure 7, C–F). These data show that patients with active colitis have beclin 1 and ATG5 cleavage as well as activation of cell death pathways in cells with decreased levels of intracellular HMGB1. We also observed significantly increased calpain activity, increased active caspase 1, and decreased calpastatin mRNA in patients with active colitis (Figure 6, F–H). Thus, humans with IBD exhibit the same pattern of protease activation, beclin 1 and ATG5 cleavage, and cell death as that seen in DSS-treated mice lacking IEC HMGB1. These data suggest that decreased intracellular HMGB1 contributes to the pathophysiology of IBD.
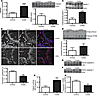 Figure 6
Figure 6IBD in humans is associated with decreased HMGB1, beclin 1, and ATG5 cleavage and increased cell death. (A) HMGB1 expression by qRT-PCR in controls (n = 6) and in patients with active UC (n = 5) or indeterminate colitis (IC) (n = 1) (mean ± SEM). Immunoblot for HMGB1 protein in lysates of snap-frozen endoscopic biopsies. Lanes 1–4, controlled CD patients; lanes 5–8, active CD patients (mean ± SEM). (B) Confocal microscopic images of endoscopic biopsies stained for Hoechst (blue) and HMGB1 (red) in controls (n = 3) and in patients with active UC (n = 4) (original magnification, ×400). Colocalization evaluated using Pearson’s correlation coefficient with the Costes correction (mean ± SEM). (C) Beclin 1 immunoblot. Lane 1, control/normal; lane 2, control/normal; lane 3, control/quiescent CD; lane 4, moderate UC; lane 5, moderate UC; and lane 6, severe CD (mean ± SEM). (D) Immunoblot for ATG5 in lysates of snap-frozen endoscopic biopsies (mean ± SEM). Samples were loaded as in A. (E) Immunoblot with antibody recognizing the active p19/p17 fragments of cleaved caspase 3. Samples were loaded as in Figure 4D. (F) Immunoblot for the active p20 fragment of cleaved caspase 1. Samples were loaded as in Figure 4D. (G) Calpain activity as evaluated by cleavage of the fluorescent substrate in samples from controls (n = 3) and from patients with active UC (n = 3) (mean ± SEM). (H) Calpastatin levels analyzed by qRT-PCR in controls (n = 6) and in patients with active UC (n = 5) or IC (n = 1) (mean ± SEM). Data were analyzed by 2-tailed Student’s t tests, except for data in G, which were determined by the 1-tailed Student’s t test. *P < 0.05; **P < 0.01; ****P < 0.001.
-
Discussion
HMGB1 controls the autophagy/apoptosis checkpoint in IECs by protecting beclin 1 and ATG5 from calpain 1 cleavage (Figure 7). The calpain activity seen in DSS-treated mice and IBD patients appears to result from caspase 1–mediated degradation and downregulation of calpastatin, the endogenous calpain inhibitor. Active caspase 1 is generated by inflammasome activation in response to microorganisms or cell stress (33). This protease cleaves pro–IL-1β, pro–IL-18, or pro–IL-33 to generate the active forms of these inflammatory cytokines, and inflammasome signaling is terminated by autophagic degradation of assembled inflammasomes (34). Caspase 1 is also essential for tissue healing in the intestine and contributes to autophagy through upregulation of beclin 1 expression during oxidative stress (35, 36). Active caspase 1 was increased in Vil-Cre Hmgb1fl/fl mice after DSS and in humans with active colitis, consistent with either increased inflammasome activation or decreased clearance. Our observation that autophagy was compromised suggests that decreased clearance of activated inflammasomes is the more likely explanation. Despite increased levels of active caspase 1, levels of IL-1β were similar in DSS-treated Hmgb1fl/fl and Vil-Cre Hmgb1fl/fl mice. This suggests that while caspase 1 activation was increased, caspase 1–mediated activation of inflammatory cytokines was not responsible for the cell death seen in this colitis model. Instead, caspase 1–mediated activation of calpains in the absence of HMGB1 led to the inflammation and apoptosis seen in this model.
 Figure 7
Figure 7HMGB1 protects beclin 1 and ATG5 from calpain-mediated cleavage and conversion from proautophagic to proapoptotic functions. (A) Calpain activation occurs secondarily to inflammasome-mediated caspase 1 activation with subsequent degradation and downregulation of the calpain inhibitor calpastatin. Low levels of calpastatin in the presence of calcium lead to autoactivation of calpains. Inflammasome-mediated caspase 1 activation is terminated by autophagic degradation of inflammasomes. (B) HMGB1 interacts with beclin 1 and ATG5 to prevent calpain-mediated cleavage of these proteins, allowing autophagy to proceed. (C) In the absence of HMGB1, beclin 1 and ATG5 are cleaved by calpain, generating protein fragments that localize to the mitochondria and trigger cell death.
Calpain activation does not degrade target proteins but rather cleaves them into stable fragments with functions that are different from the parent protein (37). The calpain proteases are ubiquitously expressed and play roles in cell division, cell movement, signal transduction, and apoptotic pathways (26). Calpain activity is also involved in the activation or termination of autophagy. Calpain-deficient cells are defective in autophagy in response to starvation, rapamycin, etoposide, and ceramide, and there is a dramatic increase in apoptosis in these cells after autophagy induction (38). Furthermore, calpains can block autophagy through cleavage of beclin 1 and ATG5 (19, 39–41). In Vil-Cre Hmgb1fl/fl IECs, autophagy proceeded normally under conditions of low stress, while beclin 1 and ATG5 were cleaved, and autophagosome formation was decreased during high stress compared with that seen in Hmgb1fl/fl IECs. Our use of mice conditionally deficient in HMGB1 for in vivo autophagy studies and of primary IECs deficient in HMGB1 for in vitro autophagy studies allowed us to examine both basal and induced autophagy in nontransformed IECs. Not surprisingly, there were some subtle differences from previous studies using mouse embryonic fibroblasts or transformed cells knocked down for HMGB1. Those studies primarily focused on inducible autophagy and showed that HMGB1 was required for beclin 1 dissociation from BCL-2 and autophagy initiation (14). This finding is in agreement with our findings in stress-induced autophagy, but the fact that autophagy proceeded under basal conditions in HMGB1-deficient cells suggests that this may not be an absolute requirement in nontransformed cells. Our findings are also in agreement with those recently reported by other groups using mouse models conditionally deficient in HMGB1. Under basal conditions, HMGB1-deficient hepatocytes showed no autophagy or mitophagy defects (42). However, in studies using models of infectious or inflammatory disease, conditional HMGB1 deficiency resulted in increased cell death (43–45). This suggests that either basal autophagy does not require HMGB1 or that cells are able to compensate for the loss of this protein under nonstressed conditions. However, under high-stress, calpain-activated conditions, beclin 1 and ATG5 cleavage not only compromises autophagosome formation at 2 different steps of the process, but also generates de novo proapoptotic proteins. Therefore, under these conditions, cell death is not a passive process due to autophagy failure but rather is an active process of organized cell death in response to generation of these cleavage fragments. This conclusion is supported by studies showing that the truncated products of these cleavage events cause mitochondria-mediated cell death in several different cell types (18, 39).
IBD patients with active colitis have features similar to those of DSS-treated Vil-Cre Hmgb1fl/fl mice. HMGB1 is one of the most evolutionarily conserved proteins in the eukaryotic kingdom and has never been implicated in a genome-wide association scan for IBD or any other disease (46). The small number of polymorphisms identified in this protein have been studied in the context of sepsis, a disease of intense, calpain-mediated inflammation (47, 48). Experimental models of sepsis have been associated with high levels of circulating HMGB1, and anti-HMGB1 antibody has been proposed as a treatment for this disease (49–53). While HMGB1 is increased in sepsis patients compared with that in noninfected controls, the human data suggest a more nuanced view of the role of HMGB1 in this disease (52). Lower HMGB1 levels were associated with decreased survival in a prospective study of 2 cohorts of sepsis patients, and an HMGB1 polymorphism associated with decreased serum HMGB1 was also associated with increased mortality in sepsis patients (50, 54).
Our data from Vil-Cre Hmgb1fl/fl mice suggest that these nuances may be due to the differences between the intracellular and extracellular functions of HMGB1. Levels of intracellular HMGB1 are increased during inflammation to counter calpain activity, and failure to counter this activity leads to cell death and HMGB1 release. Cells could compensate for low levels of intracellular HMGB1 under conditions of low calpain activity by upregulating target proteins. However, high levels of calpain activation would be expected to overwhelm this compensation, leading to beclin 1 and ATG5 cleavage and production of the proapoptotic versions of the proteins.
In conclusion, we have identified a novel function for HMGB1 in cell survival. Extracellular HMGB1 is proinflammatory, whereas intracellular HMGB1 is effectively antiinflammatory. It acts to minimize tissue inflammation by mitigating cell death, which prevents release of intracellular inflammatory mediators and maintains tissue barrier integrity. In the gastrointestinal tract, tissue barrier integrity is particularly important, since loss of the mucosal barrier leads to increased exposure of underlying tissues to inflammatory ligands in the gut lumen (1). We have also described the mechanism whereby HMGB1 promotes cell survival during inflammation and shown that this mechanism applies to both human and experimental colitis. Calpain activation is a feature of many human diseases, including cardiomyopathy, type 2 diabetes, ischemia-reperfusion, microbial infections, and cancers (55–58). Calpains are also important for normal cell division and tissue repair and remodeling programs (59). Our data suggest that HMGB1 may be a crucial factor in all of these processes, since it determines whether cells undergo apoptosis or autophagy during calpain activation. As such, it may represent a useful therapeutic target in a diverse range of human diseases.
-
Methods
Generation of Hmgb1fl/fl mice. Hmgb1fl/fl mice were generated on a C57B/6 genetic background with the help of Ozgene (Ozgene Pty Ltd.). Gene targeting resulted in exons 2 and 3 of the Hmgb1 gene being flanked by LoxP sites. Exon 2 contains 14 bp of the 5′-UTR, the start codon, and the first 49 aa of the protein. Exon 3 contains the coding sequence for aa 50–98 of the protein. Deletion of these 2 exons eliminated the first 99 aa of the protein and caused an ORF shift, resulting in early termination of translation. Mice were genotyped using the primers 5′-GAGGCCTCCGTGAGTATGAP-3′ and 5′-TTGCAACATCACCAATGGAT-3′. Hmgb1fl/fl mice were bred with Vil-Cre mice provided by Sylvie Robine (INSERM, Institut Curie, Paris, France) to generate Vil-Cre Hmgb1fl/fl mice. Each figure pertaining to experimental colitis represents at least 3 independent experiments, wherein the mice were sex and age matched (littermates were used whenever possible). Approximately 8-week-old mice (unless otherwise noted) of both sexes were used in all experiments.
DSS colitis model. Colitis was induced in mice using 2.5% DSS (MP Biomedicals), except for the initial survival studies, which used 3% DSS. DSS was added to the drinking water for 5 days, and then mice were given fresh water and allowed to recover. Mice were monitored for weight changes, diarrhea, bloody stools, and overall health. They were removed from the study when their weight loss exceeded 25% of their original body weight or when they developed rectal prolapse of more than 0.5 cm.
Il10–/– colitis model. Il10–/– mice on a C57B/6 genetic background were obtained from The Jackson Laboratory and bred with Vil-Cre Hmgb1fl/fl mice. Monitoring and removal criteria were the same as those for the mice used in the DSS colitis model.
Measurement of mucosal cytokine levels. Mucosal scrapings from the colons of Hmgb1fl/fl and Vil-Cre Hmgb1fl/fl mice were homogenized in cell lysis buffer (Cell Signaling Technology), centrifuged at top speed for 15 minutes, and then supernatants were collected and assayed for protein concentrations using the bichronic acid method. Cytokine levels were then determined using the MILLIPLEX MAP Mouse Cytokine Assay (EMD Millipore) according to the manufacturer’s instructions.
IEC isolation. Colonic crypts were isolated either by mucosal scraping or by methods modified from Dekaney et al. (60) and Wilson et al. (61). The latter involves dissociating the colonic crypts from the colon by EDTA perfusion. Mice were anesthetized, and the cardiovascular system was perfused with 2 ml warm PBS containing 30 mM EDTA and 25 mM NaHCO3 and 1.5 mM DTT. The colon was removed, everted, rinsed briefly in PBS, and placed over a Pasteur pipette. The colon was then spun with a homogenizer motor into ice-cold PBS containing 30 mM EDTA and 1.5 mM DTT. The isolated material was examined using phase-contrast microscopy, which showed that it consisted mainly of intact crypts and sheets of epithelial cells.
Immunoblot analysis of mouse and human samples. Samples for immunoblotting were obtained from mucosal scrapings, EDTA dissociation, or snap-frozen endoscopic biopsies. These were lysed in Cell Lysis Buffer (Cell Signaling Technology) containing Complete Protease Inhibitor (Roche) and PMSF and subjected to separation on reducing SDS-PAGE gels. The following antibodies were used: ATG5 (2630), beclin 1 (3738s), cleaved caspase 1 (human, 4199), cleaved caspase (9664), LC3B (2775s), and ATG7 (D12B11) (all from Cell Signaling Technology); cleaved beclin 1 (612113; BD Biosciences); HMGB1 (ab18256) and MBL (p62 M162-3) (both from Abcam); cleaved ATG5 (A2859; Sigma-Aldrich ); and caspase 1 (casper 1 [mouse]; Adipogen).
qRT-PCR. RNA isolation was performed on mucosal scrapings (mouse), EDTA-isolated IECs (mouse), or intestinal biopsies (human) using TRIzol (Life Technologies) and standard procedures. qRT-PCR was performed using SYBR Green Master Mix (Bio-Rad) and a Roche LightCycler 48 II. Mouse primer sequences were as follows: Hmgb1, 5′-GGCGAGCATCCTGGCTTATC-3′ and 5′-GGCTGCTTGTCATCTGCTG-3′; Gapdh, 5′-GGCAAATTCAACGGCACAGT-3′ and 5′-AGATGGTGATGGGCTTCCC-3′; villin, 5′-TCAAAGGCTCTCTCAACATCA-3′ and 5′-AGCAGTCACCATCGAAGAAGC-3′; beclin 1, 5′-ATGGAGGGGTCTAAGGCGTC-3′ and 5′-TCCTCTCCTGAGTTAGCCTCT-3′; calpastatin, 5′-CAATCAAGTGAGCAACCTGTGG-3′ and 5′-CGGAAGATTTTTGGGCTCTGA-3′. For human samples, the following primers were used: GAPDH, 5′-ACAACTTTGGTATCGTGGAAGG-3′ and 5′-GCCATCACGCCACAGTTTC-3′; HMGB1, 5′-TATGGCAAAAGCGGACAAGG-3′ and 5′-CTTCGCAACATCACCAATGGA-3′; beclin 1, CCATGCAGGTGAGCTTCGT-3′ and 5′-GAATCTGCGAGAGACACCATC-3′; and calpastatin, 5′-TACAGGAAGTAACGATGCTCACA-3′ and 5′-CACCCGGCTTGCCAGATATT-3′.
The following primer pairs were used to determine mouse mucosal cytokine expression: Il1b, 5′-ATCTTTTGGGGTCCGTCAACT and 5′-GCAACTGTTCCTGAACTCAACT; Il12p40, 5′-TGGTTTTCCATCGTTTTGCTG and 5′-ACAGTTGAGGTTCACTGTTTCT; Tnfa, 5′-CCCTCACACTCAGATCATCTTCT and 5′-GCTACGACGTGGTCTACAG; Il6, 5′-TAGTCCTTCCTACCCCAATTTCC and 5′-TTGGTCCTTAGCCACTCCTTC; Ifng, 5′-ATGAACGCTACACACTGCATC and 5′-CCATCCGTTTGCCAGTTCCTC; Il17, 5′-TTTAACTCCCTTGGCGCAAAA and 5′-CTTTCCCTCCGCATTTGACAC; Il10, 5′-TGCTATGCTGCCTGCTCTTA and 5′-GTTCCTTTGGTTGCTGGAAG; Il23p19, 5′-ATGCTGGATTGCAGAGCAGTA and 5′-ACGTGGCACATTATTTTTAGTCG; and Mcp1, 5′-TTAAAAACCTGGATCGGAACCAA and 5′-GCATTAGCTTCAGATTTACGGGT.
Immunofluorescence staining and microscopy. Confocal microscopic images were captured using a Leica SP5 II STED-CW Super-Resolution Laser-Scanning Confocal instrument and analyzed using ImageJ software (NIH).
GFP-LC3 reporter mice. GFP-LC3 reporter mice were purchased from RIKEN. Procedures for visualization of LC3-GFP in the mouse intestine were adapted from Mizushima (62). Briefly, intestines were cut longitudinally, washed in PBS, and fixed in 4% paraformaldehyde for 20 minutes. They were then incubated in 15% sucrose for 4 hours, followed by 30% sucrose overnight before embedding in OCT. Samples were cryosectioned at 5-μm thickness and allowed to dry at room temperature for 30 minutes. Sections were stained with Hoechst (Life Technologies) for 15 minutes then mounted with Prolong Gold (Life Technologies).
Cell death evaluation. The In Situ Cell Death Detection Kit (Roche) was used for TUNEL staining. Images were captured with a Leica DM2500 epifluorescence microscope.
In vitro calpain cleavage assay. Recombinant active calpain 1 (Sigma-Aldrich), beclin 1 (OriGene), ATG5 (Abcam), and HMGB1 (R&D Systems) were used for the in vitro calpain 1 cleavage assay. The assay conditions were adopted from Norman et al. (20) and Bano et al. (63). The buffer system contained 50 mM NaCl, 10 mM EGTA, 0.1% Triton, and 100 mM HEPES (pH 7.5). CaCl2 was added to a final concentration of 20 mM immediately before reaction. In a reaction volume of 20 μl, 0.2 units of active calpain 1 and 0.1 μg beclin 1 or ATG5 were added with various amounts of HMGB1 protein. The reaction mixture was incubated at 30°C for 30 minutes, and then stopped by adding Laemmli buffer and incubating at 90°C for 5 minutes. The samples were resolved using a 15% SDS-PAGE system and blotted with antibodies recognizing beclin 1 (612113; BD Transduction Laboratories), FLAG (2368; Cell Signaling Technology), ATG5 (A2859; Sigma-Aldrich), or HMGB1 (ab18256; Abcam).
For the competitive in vitro calpain cleavage assay, the previously described reaction buffer was used with the addition of 0.05 units of active calpain, 0.1 μg beclin 1, 0.25 μg HMGB1 (beclin 1/HMGB1 ratio of 1:6), and a 100 μM concentration of either the competitor or the mock competitor peptide. The reaction mixture was incubated at 30°C for 15 minutes, and then the reaction was stopped and the products analyzed as previously described.
Coimmunoprecipitation. Mucosal scrapings from Hmgb1fl/fl and Vil-Cre Hmgb1fl/fl mouse colons were homogenized in cell lysis buffer (Cell Signaling Technology), and lysate containing 100 μg of total protein was incubated with 5 μl of the designated antibody overnight at 4°C. For coimmunoprecipitation (Co-IP) of HMGB1 and beclin 1, anti-HMGB1 (Ab18256; Abcam) and anti–beclin 1 (612113; BD Biosciences) were used. For HMGB1-ATG5 Co-IP, anti-HMGB1 (D090-3; MBL) and anti-ATG5 (2630; Cell Signaling Technology) were used. The antibody-antigen complex was precipitated using 50 μl cell lysis buffer balanced protein A agarose (Thermo Scientific), and Laemmli buffer (50 μl) was added to the precipitated complexes prior to heating to 90°C for 5 minutes. The supernatant was resolved on a 15% SDS-PAGE gel and blotted for the respective proteins.
Calpain activity assay. Calpain activity was measured as the cleavage of a fluorogenic substrate (Calbiochem) using a fluorescence plate reader (Biotek Synergy 2). Briefly, 10 mg of tissue was homogenized in lysis buffer and incubated on ice for 30 minutes. The samples were than centrifuged at top speed in a prechilled tabletop centrifuge for 15 minutes. The protein concentration was determined by the bicinchoninic acid method. Fifty microliters of sample (about 100 μg total protein) was used for each assay. The calpain activity in each sample was expressed as units per milligram of protein per minute.
Calpeptin treatment in vivo. Vil-CreHmgb1fl/fl mice receiving 2.5% DSS in their drinking water were treated with either vehicle or 5 mg/kg calpeptin (benzyloxycarbonylleucyl-norleucinal; Calbiochem) daily through i.p. injection. After 5 days of DSS administration, mice were sacrificed, and tissues were collected for immunoblot and histological analyses as previously described.
Intestinal epithelial organoid (enteroids) culture. IEC progenitors were isolated and cultured according to methods described in Sato et al. (64). Briefly, the small intestine was removed from each mouse, cleaned, and then cut into 1-mm pieces. After multiple washes with ice-cold PBS, intestinal pieces were incubated in EDTA-containing PBS, with agitation at 4°C for 30 minutes. Cells were then collected in Advanced DMEM/F12 (ADF; Life Technologies), and a single-cell suspension was ensured by passing the cells through a 70-μm cell strainer. Cells were resuspended in 100 μl complete ADF media (ADF supplemented with GlutaMAX; Life Technologies); HEPES buffer (Life Technologies); penicillin and streptomycin (Life Technologies); N2 supplement (Life Technologies); B-27 Supplement Minus Vitamin A (Life Technologies); murine EGF (50 ng/ml; Life Technologies); Noggin (100 ng/ml; Peprotech); jagged 1 (1 μM; Anaspec); Y27632 (10 nM; Cayman Scientific); and R-spondin 1 (500 ng/ml; Peprotech)). Cells were then combined with 200 μl Matrigel (BD Biosciences) and plated onto 6-well plates with or without collagen-coated coverslips. Matrigel beads containing crypts and cells were allowed to solidify for 1 hour at 5% CO2 and 37°C before adding 2 ml complete ADF media to each well. Fresh growth factors were added every 2 days, and complete ADF media were changed every 4 days.
MDP treatment of enteroids. Enteroid cultures were were plated in Matrigel and ADF media without growth factors. The following day, they were treated with 10 μg/ml L-18 MDP for the indicated times (Figure 5) prior to lysis. Cells were isolated from Matrigel using low-speed centrifugation and were then lysed in 1% Triton lysis buffer as previously described.
IEC stimulation of adaptive immune responses. MLNs were harvested from 8-week-old male and female C57BL/6 WT mice and transferred to 10-mm Petri dishes to remove residual surrounding fat tissues using a razor blade. Single-cell suspensions were obtained by passing cells through a 70-μm nylon strainer (BD Biosciences). The mixture of MLN cells (2 × 106 cells in 800 μl complete medium [RPMI 1640 plus 10% FBS plus 100 U/ml penicillin and streptomycin]) was cultured with 200 μl supernatant derived from MDP-stimulated Hmgb1fl/fl or Vil-Cre Hmgb1fl/fl enteroids or from unstimulated controls for 16 hours. Supernatants were harvested and stored at –80°C prior to assaying for TNFA, IL-6, IL-17, and IFN-γ by ELISA (ELISA Ready-Set-Go! Kits; eBioscience).
Calpeptin treatment of enteroids. Enteroid cultures were plated in Matrigel and ADF media without growth factors. The following day, they were treated with 10 μg/ml L-18 MDP in the presence of DMSO (vehicle control) or 1 μg/ml calpeptin for 4 hours. The enteroids were harvested by centrifugation in ice-cold PBS and homogenized in cell lysis buffer (Cell Signaling Technology). Total protein (30 μg) was then immunoblotted for active caspase 3 as previously described.
Autophagic flux in organoids. Enteroids were plated as described above. The following day, they were treated with 10 μg/ml L-18 MDP in the presence or absence of 100 nm bafilomycin A1 (Tocris) for 4 hours, and the lysates were assayed for LC3B and p62 as previously described.
Statistics. Prism software (GraphPad Software) was used for statistical evaluation. Data are reported as the mean ± SEM, unless otherwise noted. Pairwise comparisons were made using the 1- or 2-tailed unpaired Student’s t test, while 1- or 2-way ANOVA with Bonferroni’s post-hoc testing was used for multiple comparisons. A P value of less than 0.05 was considered statistically significant.
Study approval. NIH guidelines were followed for the care and handling of mice in this study. The IACUC of the University of Chicago approved all animal studies and procedures (Animal Protocol 71629). Protocols for obtaining human tissue samples were approved by the IRB of the University of Chicago (approval 15573A), and all patients provided written informed consent.
-
Acknowledgments
We would like to thank Karl B. Hansen for his assistance with figure preparation.
Address correspondence to: Eugene B. Chang, Knapp Center for Biomedical Discovery, 900 E. 57th Street, 9th Floor, Room 9130, University of Chicago, Chicago, Illinois, USA. Phone: 773.702.6458; E-mail: [email protected].
-
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Reference information: J Clin Invest. 2015;125(3):1098–1110. doi:10.1172/JCI76344.
This work was supported by the National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK) of the NIH through Digestive Disease Research Core Center grants DK-42086 (to E.B. Chang), DK47722 (to E.B. Chang), DK097268 (to E.B. Chang), 1F32DK082104 (to J.S. Messer), and P30 DK42086 (to J.S. Messer). Support was also provided by grants from the National Institute of Allergy and Infectious Diseases (NIAID) (AI083375, to D.L. Boone); the Gastrointestinal Research Foundation; the Crohn’s and Colitis Foundation of America (to J.S. Messer and Y. Wang); the Broad Medical Research Program (to D.L. Boone); the Peter and Carol Goldman Family Research Fund; and the Harry and Leona Helmsley Charitable Trust (SHARE).
-
References
- Jäger S, Stange EF, Wehkamp J. Inflammatory bowel disease: an impaired barrier disease. Langenbecks Arch Surg. 2013;398(1):1–12.
- Antoni L, Nuding S, Wehkamp J, Stange EF. Intestinal barrier in inflammatory bowel disease. World J Gastroenterol. 2014;20(5):1165–1179.
- Asselin C, Gendron FP. Shuttling of information between the mucosal and luminal environment drives intestinal homeostasis. FEBS Lett. 2014;588(22):4148–4157.
- Harris HE, Andersson U, Pisetsky DS. HMGB1: a multifunctional alarmin driving autoimmune and inflammatory disease. Nat Rev Rheumatol. 2012;8(4):195–202.
- Andersson U, Tracey KJ. HMGB1 is a therapeutic target for sterile inflammation and infection. Annu Rev Immunol. 2011;29:139–162.
- Lotze MT, Tracey KJ. High-mobility group box 1 protein (HMGB1): nuclear weapon in the immune arsenal. Nat Rev Immunol. 2005;5(4):331–342.
- Yamasaki H, et al. Roles of high-mobility group box 1 in murine experimental colitis. Mol Med Rep. 2009;2(1):23–27.View this article via: PubMed Google Scholar
- Davé SH, et al. Ethyl pyruvate decreases HMGB1 release and ameliorates murine colitis. J Leukoc Biol. 2009;86(3):633–643.
- Vitali R, et al. Fecal HMGB1 is a novel marker of intestinal mucosal inflammation in pediatric inflammatory bowel disease. Am J Gastroenterol. 2011;106(11):2029–2040.
- Calogero S, et al. The lack of chromosomal protein Hmg1 does not disrupt cell growth but causes lethal hypoglycaemia in newborn mice. Nat Genet. 1999;22(3):276–280.
- Solomon L, et al. The dextran sulphate sodium (DSS) model of colitis: an overview. Comp Clin Path. 2010;19(3):235–239.
- Monteleone I, Platt AM, Jaensson E, Agace WW, Mowat AM. IL-10-dependent partial refractoriness to Toll-like receptor stimulation modulates gut mucosal dendritic cell function. Eur J Immunol. 2008;38(6):1533–1547.
- Asseman C, Mauze S, Leach MW, Coffman RL, Powrie F. An essential role for interleukin 10 in the function of regulatory T cells that inhibit intestinal inflammation. J Exp Med. 1999;190(7):995–1004.
- Tang D, et al. Endogenous HMGB1 regulates autophagy. J Cell Biol. 2010;190(5):881–892.
- Gordy C, He YW. The crosstalk between autophagy and apoptosis: where does this lead? Protein Cell. 2012;3(1):17–27.
- Naito Y, et al. Enhanced intestinal inflammation induced by dextran sulfate sodium in tumor necrosis factor-alpha deficient mice. J Gastroenterol Hepatol. 2003;18(5):560–569.
- Marquez RT, Xu L. Bcl-2:Beclin 1 complex: multiple, mechanisms regulating autophagy/apoptosis toggle switch. Am J Cancer Res. 2012;2(2):214–221.View this article via: PubMed Google Scholar
- Wirawan E, et al. Caspase-mediated cleavage of Beclin-1 inactivates Beclin-1-induced autophagy and enhances apoptosis by promoting the release of proapoptotic factors from mitochondria. Cell Death Dis. 2010;1(1):e18.
- Russo R, et al. Calpain-mediated cleavage of Beclin-1 and autophagy deregulation following retinal ischemic injury in vivo. Cell Death Dis. 2011;2:e144.
- Norman JM, Cohen GM, Bampton ET. The in vitro cleavage of the hAtg proteins by cell death proteases. Autophagy. 2010;6(8):1042–1056.
- Mizushima N, Sugita H, Yoshimori T, Ohsumi Y. A new protein conjugation system in human. The counterpart of the yeast Apg12p conjugation system essential for autophagy. J Biol Chem. 1998;273(51):33889–33892.
- Bhatt A, Kaverina I, Otey C, Huttenlocher A. Regulation of focal complex composition and disassembly by the calcium-dependent protease calpain. J Cell Sci. 2002;115(17):3415–3425.View this article via: PubMed Google Scholar
- Franco SJ, et al. Calpain-mediated proteolysis of talin regulates adhesion dynamics. Nat Cell Biol. 2004;6(10):977–983.
- Letavernier E, et al. The role of calpains in myocardial remodelling and heart failure. Cardiovasc Res. 2012;96(1):38–45.
- Tremper-Wells B, Vallano ML. Nuclear calpain regulates Ca2+-dependent signaling via proteolysis of nuclear Ca2+/calmodulin-dependent protein kinase type IV in cultured neurons. J Biol Chem. 2005;280(3):2165–2175.View this article via: PubMed Google Scholar
- Goll DE, Thompson VF, Li H, Wei W, Cong J. The calpain system. Physiol Rev. 2003;83(284):731–801.View this article via: PubMed Google Scholar
- Cuzzocrea S. Calpain inhibitor I reduces colon injury caused by dinitrobenzene sulphonic acid in the rat. Gut. 2001;48(4):478–488.
- Shen H, et al. Ethyl pyruvate protects against hypoxic-ischemic brain injury via anti-cell death and anti-inflammatory mechanisms. Neurobiol Dis. 2010;37(3):711–722.
- Wang KK, et al. Caspase-mediated fragmentation of calpain inhibitor protein calpastatin during apoptosis. Arch Biochem Biophys. 1998;356(2):187–196.
- Pörn-Ares MI, Samali A, Orrenius S. Cleavage of the calpain inhibitor, calpastatin, during apoptosis. Cell Death Differ. 1998;5(12):1028–1033.
- Barnoy S, Kosower NS. Caspase-1-induced calpastatin degradation in myoblast differentiation and fusion: cross-talk between the caspase and calpain systems. FEBS Lett. 2003;546(2–3):213–217.View this article via: PubMed Google Scholar
- Shi Y, Melnikov VY, Schrier RW, Edelstein CL. Downregulation of the calpain inhibitor protein calpastatin by caspases during renal ischemia-reperfusion. Am J Physiol Ren. 2000;279(3):F509–F517.View this article via: PubMed Google Scholar
- Latz E, Xiao TS, Stutz A. Activation and regulation of the inflammasomes. Nat Rev Immunol. 2013;13(6):397–411.
- Shi C-S, et al. Activation of autophagy by inflammatory signals limits IL-1β production by targeting ubiquitinated inflammasomes for destruction. Nat Immunol. 2012;13(3):255–263.
- Becker C, Watson AJ, Neurath MF. Complex roles of caspases in the pathogenesis of inflammatory bowel disease. Gastroenterology. 2013;144(2):283–293.
- Sun Q, et al. Caspase 1 activation is protective against hepatocyte cell death by up-regulating beclin 1 protein and mitochondrial autophagy in the setting of redox stress. J Biol Chem. 2013;288(22):15947–15958.
- Carragher NO. Calpain inhibition: a therapeutic strategy targeting multiple disease states. Curr Pharm Des. 2006;12(5):615–638.
- Demarchi F, et al. Calpain is required for macroautophagy in mammalian cells. J Cell Biol. 2006;175(4):595–605.
- Yousefi S, et al. Calpain-mediated cleavage of Atg5 switches autophagy to apoptosis. Nat Cell Biol. 2006;8(10):1124–1132.
- Lépine S, Allegood JC, Edmonds Y, Milstien S, Spiegel S. Autophagy induced by deficiency of sphingosine-1-phosphate phosphohydrolase 1 is switched to apoptosis by calpain-mediated autophagy-related gene 5 (Atg5) cleavage. J Biol Chem. 2011;286(52):44380–44390.
- Shi M, et al. Calpain, Atg5 and Bak play important roles in the crosstalk between apoptosis and autophagy induced by influx of extracellular calcium. Apoptosis. 2013;18(4):435–451.
- Huebener P, et al. High-mobility group box 1 is dispensable for autophagy, mitochondrial quality control, and organ function in vivo. Cell Metab. 2014;19(3):539–547.
- Huang H, et al. Hepatocyte specific HMGB1 deletion worsens the injury in liver ischemia/reperfusion: A role for intracellular HMGB1 in cellular protection. Hepatology. 2014;59(5):1984–1997.
- Kang R, et al. Intracellular Hmgb1 inhibits inflammatory nucleosome release and limits acute pancreatitis in mice. Gastroenterology. 2014;146(4):1097–1107.
- Yanai H, et al. Conditional ablation of HMGB1 in mice reveals its protective function against endotoxemia and bacterial infection. Proc Natl Acad Sci U S A. 2013;110(51):20699–20704.
- Zhang S, Zhong J, Yang P, Gong F, Wang C-Y. HMGB1, an innate alarmin, in the pathogenesis of type 1 diabetes. Int J Clin Exp Pathol. 2009;3(1):24–38.View this article via: PubMed Google Scholar
- Kornblit B, Munthe-Fog L, Petersen SL, Madsen HO, Vindeløv L, Garred P. The genetic variation of the human HMGB1 gene. Tissue Antigens. 2007;70(2):151–156.
- Callahan LA, Supinski GS. Sepsis-induced myopathy. Crit Care Med. 2009;37(10 suppl):S354–S367.View this article via: PubMed Google Scholar
- Wang H, et al. HMG-1 as a late mediator of endotoxin lethality in mice. Science. 1999;285(5425):248–251.
- Sundén-Cullberg J, et al. Persistent elevation of high mobility group box-1 protein (HMGB1) in patients with severe sepsis and septic shock. Crit Care Med. 2005;33(3):564–573.
- Ueno H, et al. Contributions of high mobility group box protein in experimental and clinical acute lung injury. Am J Respir Crit Care Med. 2004;170(12):1310–1316.
- Gaïni S, Pedersen SS, Koldkjaer OG, Pedersen C, Møller HJ. High mobility group box-1 protein in patients with suspected community-acquired infections and sepsis: a prospective study. Crit Care. 2007;11(2):R32.
- van Zoelen MA, et al. Systemic and local high mobility group box 1 concentrations during severe infection. Crit Care Med. 2007;35(12):2799–2804.
- Kornblit B, et al. Association of HMGB1 polymorphisms with outcome in patients with systemic inflammatory response syndrome. Crit Care. 2008;12(3):R83.
- Zatz M, Starling A. Calpains and disease. N Engl J Med. 2005;352(23):2413–2423.
- Chakraborti S, Alam MN, Paik D, Shaikh S, Chakraborti T. Implications of calpains in health and diseases. Indian J Biochem Biophys. 2012;49(5):316–328.View this article via: PubMed Google Scholar
- Storr SJ, Carragher NO, Frame MC, Parr T, Martin SG. The calpain system and cancer. Nat Rev Cancer. 2011;11(5):364–374.
- Soong G, Chun J, Parker D, Prince A. Staphylococcus aureus activation of caspase 1/calpain signaling mediates invasion through human keratinocytes. J Infect Dis. 2012;205(10):1571–1579.
- Frame MC, Fincham VJ, Carragher NO, Wyke JA. v-Src’s hold over actin and cell adhesions. Nat Rev Mol Cell Biol. 2002;3(4):233–245.
- Dekaney CM, Rodriguez JM, Graul MC, Henning SJ. Isolation and characterization of a putative intestinal stem cell fraction from mouse jejunum. Gastroenterology. 2005;129(5):1567–1580.
- Wilson KT, et al. Production and localization of cGMP and PGE2 in nitroprusside-stimulated rat colonic ion transport. Am J Physiol. 1996;270(3 pt 1):C832–C840.View this article via: PubMed Google Scholar
- Mizushima N. Methods for monitoring autophagy using GFP-LC3 transgenic mice. Methods Enzymol. 2009;452:13–23.View this article via: PubMed Google Scholar
- Bano D, et al. Cleavage of the plasma membrane Na+/Ca2+ exchanger in excitotoxicity. Cell. 2005;120(2):275–285.
- Sato T, et al. Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature. 2009;459(7244):262–265.
-
Version history
- Version 1 (February 2, 2015): No description
- Version 2 (March 2, 2015): No description



Copyright © 2024 American Society for Clinical Investigation
ISSN: 0021-9738 (print), 1558-8238 (online)







