Advertisement
Research Article Free access | 10.1172/JCI67144
The tumorigenic FGFR3-TACC3 gene fusion escapes miR-99a regulation in glioblastoma
Brittany C. Parker,1,2 Matti J. Annala,1,3 David E. Cogdell,1 Kirsi J. Granberg,1,3 Yan Sun,1,4 Ping Ji,1 Xia Li,1 Joy Gumin,5 Hong Zheng,6 Limei Hu,1 Olli Yli-Harja,3 Hannu Haapasalo,7 Tapio Visakorpi,8 Xiuping Liu,9 Chang-gong Liu,9 Raymond Sawaya,5 Gregory N. Fuller,1 Kexin Chen,6 Frederick F. Lang,2,5 Matti Nykter,3 and Wei Zhang1,2
1Department of Pathology, University of Texas MD Anderson Cancer Center, Houston, Texas, USA. 2The University of Texas Graduate School of Biomedical Sciences, Houston, Texas, USA. 3Institute of Signal Processing, Tampere University of Technology, Tampere, Finland. 4Department of Pathology, Tianjin Medical University Cancer Institute and Hospital, Tianjin, People’s Republic of China. 5Department of Neurosurgery, University of Texas MD Anderson Cancer Center, Houston, Texas, USA. 6Department of Biostatistics and Epidemiology, Tianjin Medical University Cancer Institute and Hospital, Tianjin, People’s Republic of China. 7Department of Neuropathology, Tampere University Hospital, Tampere, Finland. 8Institute of Biomedical Technology, Tampere, Finland. 9Department of Experimental Therapeutics, University of Texas MD Anderson Cancer Center, Houston, Texas, USA.
Address correspondence to: Wei Zhang, Department of Pathology, Unit 85, University of Texas MD Anderson Cancer Center, 1515 Holcombe Blvd., Houston, Texas 77030, USA. Phone: 713.745.1103; Fax: 713.792.5549; E-mail: [email protected]. Or to: Matti Nykter, Institute of Signal Processing, Tampere University of Technology, P.O. Box 553, FI-33101 Tampere, Finland. Phone: 358.40.526.7884; Fax: 358.03.364.1352; E-mail: [email protected]. Or to: Kexin Chen, Department of Epidemiology and Biostatistics, Tianjin Medical University Cancer Institute and Hospital, Ti-Yuan-Bei, Huan-Hu-Xi Road, He Xi District, Tianjin 300060, People’s Republic of China. Phone: 0086.22.23372231; Fax: 0086.22.23372231; E-mail: [email protected].
Authorship note: Brittany C. Parker and Matti J. Annala contributed equally to this work.
Find articles by Parker, B. in: JCI | PubMed | Google Scholar
1Department of Pathology, University of Texas MD Anderson Cancer Center, Houston, Texas, USA. 2The University of Texas Graduate School of Biomedical Sciences, Houston, Texas, USA. 3Institute of Signal Processing, Tampere University of Technology, Tampere, Finland. 4Department of Pathology, Tianjin Medical University Cancer Institute and Hospital, Tianjin, People’s Republic of China. 5Department of Neurosurgery, University of Texas MD Anderson Cancer Center, Houston, Texas, USA. 6Department of Biostatistics and Epidemiology, Tianjin Medical University Cancer Institute and Hospital, Tianjin, People’s Republic of China. 7Department of Neuropathology, Tampere University Hospital, Tampere, Finland. 8Institute of Biomedical Technology, Tampere, Finland. 9Department of Experimental Therapeutics, University of Texas MD Anderson Cancer Center, Houston, Texas, USA.
Address correspondence to: Wei Zhang, Department of Pathology, Unit 85, University of Texas MD Anderson Cancer Center, 1515 Holcombe Blvd., Houston, Texas 77030, USA. Phone: 713.745.1103; Fax: 713.792.5549; E-mail: [email protected]. Or to: Matti Nykter, Institute of Signal Processing, Tampere University of Technology, P.O. Box 553, FI-33101 Tampere, Finland. Phone: 358.40.526.7884; Fax: 358.03.364.1352; E-mail: [email protected]. Or to: Kexin Chen, Department of Epidemiology and Biostatistics, Tianjin Medical University Cancer Institute and Hospital, Ti-Yuan-Bei, Huan-Hu-Xi Road, He Xi District, Tianjin 300060, People’s Republic of China. Phone: 0086.22.23372231; Fax: 0086.22.23372231; E-mail: [email protected].
Authorship note: Brittany C. Parker and Matti J. Annala contributed equally to this work.
Find articles by Annala, M. in: JCI | PubMed | Google Scholar
1Department of Pathology, University of Texas MD Anderson Cancer Center, Houston, Texas, USA. 2The University of Texas Graduate School of Biomedical Sciences, Houston, Texas, USA. 3Institute of Signal Processing, Tampere University of Technology, Tampere, Finland. 4Department of Pathology, Tianjin Medical University Cancer Institute and Hospital, Tianjin, People’s Republic of China. 5Department of Neurosurgery, University of Texas MD Anderson Cancer Center, Houston, Texas, USA. 6Department of Biostatistics and Epidemiology, Tianjin Medical University Cancer Institute and Hospital, Tianjin, People’s Republic of China. 7Department of Neuropathology, Tampere University Hospital, Tampere, Finland. 8Institute of Biomedical Technology, Tampere, Finland. 9Department of Experimental Therapeutics, University of Texas MD Anderson Cancer Center, Houston, Texas, USA.
Address correspondence to: Wei Zhang, Department of Pathology, Unit 85, University of Texas MD Anderson Cancer Center, 1515 Holcombe Blvd., Houston, Texas 77030, USA. Phone: 713.745.1103; Fax: 713.792.5549; E-mail: [email protected]. Or to: Matti Nykter, Institute of Signal Processing, Tampere University of Technology, P.O. Box 553, FI-33101 Tampere, Finland. Phone: 358.40.526.7884; Fax: 358.03.364.1352; E-mail: [email protected]. Or to: Kexin Chen, Department of Epidemiology and Biostatistics, Tianjin Medical University Cancer Institute and Hospital, Ti-Yuan-Bei, Huan-Hu-Xi Road, He Xi District, Tianjin 300060, People’s Republic of China. Phone: 0086.22.23372231; Fax: 0086.22.23372231; E-mail: [email protected].
Authorship note: Brittany C. Parker and Matti J. Annala contributed equally to this work.
Find articles by Cogdell, D. in: JCI | PubMed | Google Scholar
1Department of Pathology, University of Texas MD Anderson Cancer Center, Houston, Texas, USA. 2The University of Texas Graduate School of Biomedical Sciences, Houston, Texas, USA. 3Institute of Signal Processing, Tampere University of Technology, Tampere, Finland. 4Department of Pathology, Tianjin Medical University Cancer Institute and Hospital, Tianjin, People’s Republic of China. 5Department of Neurosurgery, University of Texas MD Anderson Cancer Center, Houston, Texas, USA. 6Department of Biostatistics and Epidemiology, Tianjin Medical University Cancer Institute and Hospital, Tianjin, People’s Republic of China. 7Department of Neuropathology, Tampere University Hospital, Tampere, Finland. 8Institute of Biomedical Technology, Tampere, Finland. 9Department of Experimental Therapeutics, University of Texas MD Anderson Cancer Center, Houston, Texas, USA.
Address correspondence to: Wei Zhang, Department of Pathology, Unit 85, University of Texas MD Anderson Cancer Center, 1515 Holcombe Blvd., Houston, Texas 77030, USA. Phone: 713.745.1103; Fax: 713.792.5549; E-mail: [email protected]. Or to: Matti Nykter, Institute of Signal Processing, Tampere University of Technology, P.O. Box 553, FI-33101 Tampere, Finland. Phone: 358.40.526.7884; Fax: 358.03.364.1352; E-mail: [email protected]. Or to: Kexin Chen, Department of Epidemiology and Biostatistics, Tianjin Medical University Cancer Institute and Hospital, Ti-Yuan-Bei, Huan-Hu-Xi Road, He Xi District, Tianjin 300060, People’s Republic of China. Phone: 0086.22.23372231; Fax: 0086.22.23372231; E-mail: [email protected].
Authorship note: Brittany C. Parker and Matti J. Annala contributed equally to this work.
Find articles by Granberg, K. in: JCI | PubMed | Google Scholar
1Department of Pathology, University of Texas MD Anderson Cancer Center, Houston, Texas, USA. 2The University of Texas Graduate School of Biomedical Sciences, Houston, Texas, USA. 3Institute of Signal Processing, Tampere University of Technology, Tampere, Finland. 4Department of Pathology, Tianjin Medical University Cancer Institute and Hospital, Tianjin, People’s Republic of China. 5Department of Neurosurgery, University of Texas MD Anderson Cancer Center, Houston, Texas, USA. 6Department of Biostatistics and Epidemiology, Tianjin Medical University Cancer Institute and Hospital, Tianjin, People’s Republic of China. 7Department of Neuropathology, Tampere University Hospital, Tampere, Finland. 8Institute of Biomedical Technology, Tampere, Finland. 9Department of Experimental Therapeutics, University of Texas MD Anderson Cancer Center, Houston, Texas, USA.
Address correspondence to: Wei Zhang, Department of Pathology, Unit 85, University of Texas MD Anderson Cancer Center, 1515 Holcombe Blvd., Houston, Texas 77030, USA. Phone: 713.745.1103; Fax: 713.792.5549; E-mail: [email protected]. Or to: Matti Nykter, Institute of Signal Processing, Tampere University of Technology, P.O. Box 553, FI-33101 Tampere, Finland. Phone: 358.40.526.7884; Fax: 358.03.364.1352; E-mail: [email protected]. Or to: Kexin Chen, Department of Epidemiology and Biostatistics, Tianjin Medical University Cancer Institute and Hospital, Ti-Yuan-Bei, Huan-Hu-Xi Road, He Xi District, Tianjin 300060, People’s Republic of China. Phone: 0086.22.23372231; Fax: 0086.22.23372231; E-mail: [email protected].
Authorship note: Brittany C. Parker and Matti J. Annala contributed equally to this work.
Find articles by Sun, Y. in: JCI | PubMed | Google Scholar
1Department of Pathology, University of Texas MD Anderson Cancer Center, Houston, Texas, USA. 2The University of Texas Graduate School of Biomedical Sciences, Houston, Texas, USA. 3Institute of Signal Processing, Tampere University of Technology, Tampere, Finland. 4Department of Pathology, Tianjin Medical University Cancer Institute and Hospital, Tianjin, People’s Republic of China. 5Department of Neurosurgery, University of Texas MD Anderson Cancer Center, Houston, Texas, USA. 6Department of Biostatistics and Epidemiology, Tianjin Medical University Cancer Institute and Hospital, Tianjin, People’s Republic of China. 7Department of Neuropathology, Tampere University Hospital, Tampere, Finland. 8Institute of Biomedical Technology, Tampere, Finland. 9Department of Experimental Therapeutics, University of Texas MD Anderson Cancer Center, Houston, Texas, USA.
Address correspondence to: Wei Zhang, Department of Pathology, Unit 85, University of Texas MD Anderson Cancer Center, 1515 Holcombe Blvd., Houston, Texas 77030, USA. Phone: 713.745.1103; Fax: 713.792.5549; E-mail: [email protected]. Or to: Matti Nykter, Institute of Signal Processing, Tampere University of Technology, P.O. Box 553, FI-33101 Tampere, Finland. Phone: 358.40.526.7884; Fax: 358.03.364.1352; E-mail: [email protected]. Or to: Kexin Chen, Department of Epidemiology and Biostatistics, Tianjin Medical University Cancer Institute and Hospital, Ti-Yuan-Bei, Huan-Hu-Xi Road, He Xi District, Tianjin 300060, People’s Republic of China. Phone: 0086.22.23372231; Fax: 0086.22.23372231; E-mail: [email protected].
Authorship note: Brittany C. Parker and Matti J. Annala contributed equally to this work.
Find articles by Ji, P. in: JCI | PubMed | Google Scholar
1Department of Pathology, University of Texas MD Anderson Cancer Center, Houston, Texas, USA. 2The University of Texas Graduate School of Biomedical Sciences, Houston, Texas, USA. 3Institute of Signal Processing, Tampere University of Technology, Tampere, Finland. 4Department of Pathology, Tianjin Medical University Cancer Institute and Hospital, Tianjin, People’s Republic of China. 5Department of Neurosurgery, University of Texas MD Anderson Cancer Center, Houston, Texas, USA. 6Department of Biostatistics and Epidemiology, Tianjin Medical University Cancer Institute and Hospital, Tianjin, People’s Republic of China. 7Department of Neuropathology, Tampere University Hospital, Tampere, Finland. 8Institute of Biomedical Technology, Tampere, Finland. 9Department of Experimental Therapeutics, University of Texas MD Anderson Cancer Center, Houston, Texas, USA.
Address correspondence to: Wei Zhang, Department of Pathology, Unit 85, University of Texas MD Anderson Cancer Center, 1515 Holcombe Blvd., Houston, Texas 77030, USA. Phone: 713.745.1103; Fax: 713.792.5549; E-mail: [email protected]. Or to: Matti Nykter, Institute of Signal Processing, Tampere University of Technology, P.O. Box 553, FI-33101 Tampere, Finland. Phone: 358.40.526.7884; Fax: 358.03.364.1352; E-mail: [email protected]. Or to: Kexin Chen, Department of Epidemiology and Biostatistics, Tianjin Medical University Cancer Institute and Hospital, Ti-Yuan-Bei, Huan-Hu-Xi Road, He Xi District, Tianjin 300060, People’s Republic of China. Phone: 0086.22.23372231; Fax: 0086.22.23372231; E-mail: [email protected].
Authorship note: Brittany C. Parker and Matti J. Annala contributed equally to this work.
Find articles by Li, X. in: JCI | PubMed | Google Scholar
1Department of Pathology, University of Texas MD Anderson Cancer Center, Houston, Texas, USA. 2The University of Texas Graduate School of Biomedical Sciences, Houston, Texas, USA. 3Institute of Signal Processing, Tampere University of Technology, Tampere, Finland. 4Department of Pathology, Tianjin Medical University Cancer Institute and Hospital, Tianjin, People’s Republic of China. 5Department of Neurosurgery, University of Texas MD Anderson Cancer Center, Houston, Texas, USA. 6Department of Biostatistics and Epidemiology, Tianjin Medical University Cancer Institute and Hospital, Tianjin, People’s Republic of China. 7Department of Neuropathology, Tampere University Hospital, Tampere, Finland. 8Institute of Biomedical Technology, Tampere, Finland. 9Department of Experimental Therapeutics, University of Texas MD Anderson Cancer Center, Houston, Texas, USA.
Address correspondence to: Wei Zhang, Department of Pathology, Unit 85, University of Texas MD Anderson Cancer Center, 1515 Holcombe Blvd., Houston, Texas 77030, USA. Phone: 713.745.1103; Fax: 713.792.5549; E-mail: [email protected]. Or to: Matti Nykter, Institute of Signal Processing, Tampere University of Technology, P.O. Box 553, FI-33101 Tampere, Finland. Phone: 358.40.526.7884; Fax: 358.03.364.1352; E-mail: [email protected]. Or to: Kexin Chen, Department of Epidemiology and Biostatistics, Tianjin Medical University Cancer Institute and Hospital, Ti-Yuan-Bei, Huan-Hu-Xi Road, He Xi District, Tianjin 300060, People’s Republic of China. Phone: 0086.22.23372231; Fax: 0086.22.23372231; E-mail: [email protected].
Authorship note: Brittany C. Parker and Matti J. Annala contributed equally to this work.
Find articles by Gumin, J. in: JCI | PubMed | Google Scholar
1Department of Pathology, University of Texas MD Anderson Cancer Center, Houston, Texas, USA. 2The University of Texas Graduate School of Biomedical Sciences, Houston, Texas, USA. 3Institute of Signal Processing, Tampere University of Technology, Tampere, Finland. 4Department of Pathology, Tianjin Medical University Cancer Institute and Hospital, Tianjin, People’s Republic of China. 5Department of Neurosurgery, University of Texas MD Anderson Cancer Center, Houston, Texas, USA. 6Department of Biostatistics and Epidemiology, Tianjin Medical University Cancer Institute and Hospital, Tianjin, People’s Republic of China. 7Department of Neuropathology, Tampere University Hospital, Tampere, Finland. 8Institute of Biomedical Technology, Tampere, Finland. 9Department of Experimental Therapeutics, University of Texas MD Anderson Cancer Center, Houston, Texas, USA.
Address correspondence to: Wei Zhang, Department of Pathology, Unit 85, University of Texas MD Anderson Cancer Center, 1515 Holcombe Blvd., Houston, Texas 77030, USA. Phone: 713.745.1103; Fax: 713.792.5549; E-mail: [email protected]. Or to: Matti Nykter, Institute of Signal Processing, Tampere University of Technology, P.O. Box 553, FI-33101 Tampere, Finland. Phone: 358.40.526.7884; Fax: 358.03.364.1352; E-mail: [email protected]. Or to: Kexin Chen, Department of Epidemiology and Biostatistics, Tianjin Medical University Cancer Institute and Hospital, Ti-Yuan-Bei, Huan-Hu-Xi Road, He Xi District, Tianjin 300060, People’s Republic of China. Phone: 0086.22.23372231; Fax: 0086.22.23372231; E-mail: [email protected].
Authorship note: Brittany C. Parker and Matti J. Annala contributed equally to this work.
Find articles by Zheng, H. in: JCI | PubMed | Google Scholar
1Department of Pathology, University of Texas MD Anderson Cancer Center, Houston, Texas, USA. 2The University of Texas Graduate School of Biomedical Sciences, Houston, Texas, USA. 3Institute of Signal Processing, Tampere University of Technology, Tampere, Finland. 4Department of Pathology, Tianjin Medical University Cancer Institute and Hospital, Tianjin, People’s Republic of China. 5Department of Neurosurgery, University of Texas MD Anderson Cancer Center, Houston, Texas, USA. 6Department of Biostatistics and Epidemiology, Tianjin Medical University Cancer Institute and Hospital, Tianjin, People’s Republic of China. 7Department of Neuropathology, Tampere University Hospital, Tampere, Finland. 8Institute of Biomedical Technology, Tampere, Finland. 9Department of Experimental Therapeutics, University of Texas MD Anderson Cancer Center, Houston, Texas, USA.
Address correspondence to: Wei Zhang, Department of Pathology, Unit 85, University of Texas MD Anderson Cancer Center, 1515 Holcombe Blvd., Houston, Texas 77030, USA. Phone: 713.745.1103; Fax: 713.792.5549; E-mail: [email protected]. Or to: Matti Nykter, Institute of Signal Processing, Tampere University of Technology, P.O. Box 553, FI-33101 Tampere, Finland. Phone: 358.40.526.7884; Fax: 358.03.364.1352; E-mail: [email protected]. Or to: Kexin Chen, Department of Epidemiology and Biostatistics, Tianjin Medical University Cancer Institute and Hospital, Ti-Yuan-Bei, Huan-Hu-Xi Road, He Xi District, Tianjin 300060, People’s Republic of China. Phone: 0086.22.23372231; Fax: 0086.22.23372231; E-mail: [email protected].
Authorship note: Brittany C. Parker and Matti J. Annala contributed equally to this work.
Find articles by Hu, L. in: JCI | PubMed | Google Scholar
1Department of Pathology, University of Texas MD Anderson Cancer Center, Houston, Texas, USA. 2The University of Texas Graduate School of Biomedical Sciences, Houston, Texas, USA. 3Institute of Signal Processing, Tampere University of Technology, Tampere, Finland. 4Department of Pathology, Tianjin Medical University Cancer Institute and Hospital, Tianjin, People’s Republic of China. 5Department of Neurosurgery, University of Texas MD Anderson Cancer Center, Houston, Texas, USA. 6Department of Biostatistics and Epidemiology, Tianjin Medical University Cancer Institute and Hospital, Tianjin, People’s Republic of China. 7Department of Neuropathology, Tampere University Hospital, Tampere, Finland. 8Institute of Biomedical Technology, Tampere, Finland. 9Department of Experimental Therapeutics, University of Texas MD Anderson Cancer Center, Houston, Texas, USA.
Address correspondence to: Wei Zhang, Department of Pathology, Unit 85, University of Texas MD Anderson Cancer Center, 1515 Holcombe Blvd., Houston, Texas 77030, USA. Phone: 713.745.1103; Fax: 713.792.5549; E-mail: [email protected]. Or to: Matti Nykter, Institute of Signal Processing, Tampere University of Technology, P.O. Box 553, FI-33101 Tampere, Finland. Phone: 358.40.526.7884; Fax: 358.03.364.1352; E-mail: [email protected]. Or to: Kexin Chen, Department of Epidemiology and Biostatistics, Tianjin Medical University Cancer Institute and Hospital, Ti-Yuan-Bei, Huan-Hu-Xi Road, He Xi District, Tianjin 300060, People’s Republic of China. Phone: 0086.22.23372231; Fax: 0086.22.23372231; E-mail: [email protected].
Authorship note: Brittany C. Parker and Matti J. Annala contributed equally to this work.
Find articles by Yli-Harja, O. in: JCI | PubMed | Google Scholar
1Department of Pathology, University of Texas MD Anderson Cancer Center, Houston, Texas, USA. 2The University of Texas Graduate School of Biomedical Sciences, Houston, Texas, USA. 3Institute of Signal Processing, Tampere University of Technology, Tampere, Finland. 4Department of Pathology, Tianjin Medical University Cancer Institute and Hospital, Tianjin, People’s Republic of China. 5Department of Neurosurgery, University of Texas MD Anderson Cancer Center, Houston, Texas, USA. 6Department of Biostatistics and Epidemiology, Tianjin Medical University Cancer Institute and Hospital, Tianjin, People’s Republic of China. 7Department of Neuropathology, Tampere University Hospital, Tampere, Finland. 8Institute of Biomedical Technology, Tampere, Finland. 9Department of Experimental Therapeutics, University of Texas MD Anderson Cancer Center, Houston, Texas, USA.
Address correspondence to: Wei Zhang, Department of Pathology, Unit 85, University of Texas MD Anderson Cancer Center, 1515 Holcombe Blvd., Houston, Texas 77030, USA. Phone: 713.745.1103; Fax: 713.792.5549; E-mail: [email protected]. Or to: Matti Nykter, Institute of Signal Processing, Tampere University of Technology, P.O. Box 553, FI-33101 Tampere, Finland. Phone: 358.40.526.7884; Fax: 358.03.364.1352; E-mail: [email protected]. Or to: Kexin Chen, Department of Epidemiology and Biostatistics, Tianjin Medical University Cancer Institute and Hospital, Ti-Yuan-Bei, Huan-Hu-Xi Road, He Xi District, Tianjin 300060, People’s Republic of China. Phone: 0086.22.23372231; Fax: 0086.22.23372231; E-mail: [email protected].
Authorship note: Brittany C. Parker and Matti J. Annala contributed equally to this work.
Find articles by Haapasalo, H. in: JCI | PubMed | Google Scholar
1Department of Pathology, University of Texas MD Anderson Cancer Center, Houston, Texas, USA. 2The University of Texas Graduate School of Biomedical Sciences, Houston, Texas, USA. 3Institute of Signal Processing, Tampere University of Technology, Tampere, Finland. 4Department of Pathology, Tianjin Medical University Cancer Institute and Hospital, Tianjin, People’s Republic of China. 5Department of Neurosurgery, University of Texas MD Anderson Cancer Center, Houston, Texas, USA. 6Department of Biostatistics and Epidemiology, Tianjin Medical University Cancer Institute and Hospital, Tianjin, People’s Republic of China. 7Department of Neuropathology, Tampere University Hospital, Tampere, Finland. 8Institute of Biomedical Technology, Tampere, Finland. 9Department of Experimental Therapeutics, University of Texas MD Anderson Cancer Center, Houston, Texas, USA.
Address correspondence to: Wei Zhang, Department of Pathology, Unit 85, University of Texas MD Anderson Cancer Center, 1515 Holcombe Blvd., Houston, Texas 77030, USA. Phone: 713.745.1103; Fax: 713.792.5549; E-mail: [email protected]. Or to: Matti Nykter, Institute of Signal Processing, Tampere University of Technology, P.O. Box 553, FI-33101 Tampere, Finland. Phone: 358.40.526.7884; Fax: 358.03.364.1352; E-mail: [email protected]. Or to: Kexin Chen, Department of Epidemiology and Biostatistics, Tianjin Medical University Cancer Institute and Hospital, Ti-Yuan-Bei, Huan-Hu-Xi Road, He Xi District, Tianjin 300060, People’s Republic of China. Phone: 0086.22.23372231; Fax: 0086.22.23372231; E-mail: [email protected].
Authorship note: Brittany C. Parker and Matti J. Annala contributed equally to this work.
Find articles by Visakorpi, T. in: JCI | PubMed | Google Scholar
1Department of Pathology, University of Texas MD Anderson Cancer Center, Houston, Texas, USA. 2The University of Texas Graduate School of Biomedical Sciences, Houston, Texas, USA. 3Institute of Signal Processing, Tampere University of Technology, Tampere, Finland. 4Department of Pathology, Tianjin Medical University Cancer Institute and Hospital, Tianjin, People’s Republic of China. 5Department of Neurosurgery, University of Texas MD Anderson Cancer Center, Houston, Texas, USA. 6Department of Biostatistics and Epidemiology, Tianjin Medical University Cancer Institute and Hospital, Tianjin, People’s Republic of China. 7Department of Neuropathology, Tampere University Hospital, Tampere, Finland. 8Institute of Biomedical Technology, Tampere, Finland. 9Department of Experimental Therapeutics, University of Texas MD Anderson Cancer Center, Houston, Texas, USA.
Address correspondence to: Wei Zhang, Department of Pathology, Unit 85, University of Texas MD Anderson Cancer Center, 1515 Holcombe Blvd., Houston, Texas 77030, USA. Phone: 713.745.1103; Fax: 713.792.5549; E-mail: [email protected]. Or to: Matti Nykter, Institute of Signal Processing, Tampere University of Technology, P.O. Box 553, FI-33101 Tampere, Finland. Phone: 358.40.526.7884; Fax: 358.03.364.1352; E-mail: [email protected]. Or to: Kexin Chen, Department of Epidemiology and Biostatistics, Tianjin Medical University Cancer Institute and Hospital, Ti-Yuan-Bei, Huan-Hu-Xi Road, He Xi District, Tianjin 300060, People’s Republic of China. Phone: 0086.22.23372231; Fax: 0086.22.23372231; E-mail: [email protected].
Authorship note: Brittany C. Parker and Matti J. Annala contributed equally to this work.
Find articles by Liu, X. in: JCI | PubMed | Google Scholar
1Department of Pathology, University of Texas MD Anderson Cancer Center, Houston, Texas, USA. 2The University of Texas Graduate School of Biomedical Sciences, Houston, Texas, USA. 3Institute of Signal Processing, Tampere University of Technology, Tampere, Finland. 4Department of Pathology, Tianjin Medical University Cancer Institute and Hospital, Tianjin, People’s Republic of China. 5Department of Neurosurgery, University of Texas MD Anderson Cancer Center, Houston, Texas, USA. 6Department of Biostatistics and Epidemiology, Tianjin Medical University Cancer Institute and Hospital, Tianjin, People’s Republic of China. 7Department of Neuropathology, Tampere University Hospital, Tampere, Finland. 8Institute of Biomedical Technology, Tampere, Finland. 9Department of Experimental Therapeutics, University of Texas MD Anderson Cancer Center, Houston, Texas, USA.
Address correspondence to: Wei Zhang, Department of Pathology, Unit 85, University of Texas MD Anderson Cancer Center, 1515 Holcombe Blvd., Houston, Texas 77030, USA. Phone: 713.745.1103; Fax: 713.792.5549; E-mail: [email protected]. Or to: Matti Nykter, Institute of Signal Processing, Tampere University of Technology, P.O. Box 553, FI-33101 Tampere, Finland. Phone: 358.40.526.7884; Fax: 358.03.364.1352; E-mail: [email protected]. Or to: Kexin Chen, Department of Epidemiology and Biostatistics, Tianjin Medical University Cancer Institute and Hospital, Ti-Yuan-Bei, Huan-Hu-Xi Road, He Xi District, Tianjin 300060, People’s Republic of China. Phone: 0086.22.23372231; Fax: 0086.22.23372231; E-mail: [email protected].
Authorship note: Brittany C. Parker and Matti J. Annala contributed equally to this work.
Find articles by Liu, C. in: JCI | PubMed | Google Scholar
1Department of Pathology, University of Texas MD Anderson Cancer Center, Houston, Texas, USA. 2The University of Texas Graduate School of Biomedical Sciences, Houston, Texas, USA. 3Institute of Signal Processing, Tampere University of Technology, Tampere, Finland. 4Department of Pathology, Tianjin Medical University Cancer Institute and Hospital, Tianjin, People’s Republic of China. 5Department of Neurosurgery, University of Texas MD Anderson Cancer Center, Houston, Texas, USA. 6Department of Biostatistics and Epidemiology, Tianjin Medical University Cancer Institute and Hospital, Tianjin, People’s Republic of China. 7Department of Neuropathology, Tampere University Hospital, Tampere, Finland. 8Institute of Biomedical Technology, Tampere, Finland. 9Department of Experimental Therapeutics, University of Texas MD Anderson Cancer Center, Houston, Texas, USA.
Address correspondence to: Wei Zhang, Department of Pathology, Unit 85, University of Texas MD Anderson Cancer Center, 1515 Holcombe Blvd., Houston, Texas 77030, USA. Phone: 713.745.1103; Fax: 713.792.5549; E-mail: [email protected]. Or to: Matti Nykter, Institute of Signal Processing, Tampere University of Technology, P.O. Box 553, FI-33101 Tampere, Finland. Phone: 358.40.526.7884; Fax: 358.03.364.1352; E-mail: [email protected]. Or to: Kexin Chen, Department of Epidemiology and Biostatistics, Tianjin Medical University Cancer Institute and Hospital, Ti-Yuan-Bei, Huan-Hu-Xi Road, He Xi District, Tianjin 300060, People’s Republic of China. Phone: 0086.22.23372231; Fax: 0086.22.23372231; E-mail: [email protected].
Authorship note: Brittany C. Parker and Matti J. Annala contributed equally to this work.
Find articles by Sawaya, R. in: JCI | PubMed | Google Scholar
1Department of Pathology, University of Texas MD Anderson Cancer Center, Houston, Texas, USA. 2The University of Texas Graduate School of Biomedical Sciences, Houston, Texas, USA. 3Institute of Signal Processing, Tampere University of Technology, Tampere, Finland. 4Department of Pathology, Tianjin Medical University Cancer Institute and Hospital, Tianjin, People’s Republic of China. 5Department of Neurosurgery, University of Texas MD Anderson Cancer Center, Houston, Texas, USA. 6Department of Biostatistics and Epidemiology, Tianjin Medical University Cancer Institute and Hospital, Tianjin, People’s Republic of China. 7Department of Neuropathology, Tampere University Hospital, Tampere, Finland. 8Institute of Biomedical Technology, Tampere, Finland. 9Department of Experimental Therapeutics, University of Texas MD Anderson Cancer Center, Houston, Texas, USA.
Address correspondence to: Wei Zhang, Department of Pathology, Unit 85, University of Texas MD Anderson Cancer Center, 1515 Holcombe Blvd., Houston, Texas 77030, USA. Phone: 713.745.1103; Fax: 713.792.5549; E-mail: [email protected]. Or to: Matti Nykter, Institute of Signal Processing, Tampere University of Technology, P.O. Box 553, FI-33101 Tampere, Finland. Phone: 358.40.526.7884; Fax: 358.03.364.1352; E-mail: [email protected]. Or to: Kexin Chen, Department of Epidemiology and Biostatistics, Tianjin Medical University Cancer Institute and Hospital, Ti-Yuan-Bei, Huan-Hu-Xi Road, He Xi District, Tianjin 300060, People’s Republic of China. Phone: 0086.22.23372231; Fax: 0086.22.23372231; E-mail: [email protected].
Authorship note: Brittany C. Parker and Matti J. Annala contributed equally to this work.
Find articles by Fuller, G. in: JCI | PubMed | Google Scholar
1Department of Pathology, University of Texas MD Anderson Cancer Center, Houston, Texas, USA. 2The University of Texas Graduate School of Biomedical Sciences, Houston, Texas, USA. 3Institute of Signal Processing, Tampere University of Technology, Tampere, Finland. 4Department of Pathology, Tianjin Medical University Cancer Institute and Hospital, Tianjin, People’s Republic of China. 5Department of Neurosurgery, University of Texas MD Anderson Cancer Center, Houston, Texas, USA. 6Department of Biostatistics and Epidemiology, Tianjin Medical University Cancer Institute and Hospital, Tianjin, People’s Republic of China. 7Department of Neuropathology, Tampere University Hospital, Tampere, Finland. 8Institute of Biomedical Technology, Tampere, Finland. 9Department of Experimental Therapeutics, University of Texas MD Anderson Cancer Center, Houston, Texas, USA.
Address correspondence to: Wei Zhang, Department of Pathology, Unit 85, University of Texas MD Anderson Cancer Center, 1515 Holcombe Blvd., Houston, Texas 77030, USA. Phone: 713.745.1103; Fax: 713.792.5549; E-mail: [email protected]. Or to: Matti Nykter, Institute of Signal Processing, Tampere University of Technology, P.O. Box 553, FI-33101 Tampere, Finland. Phone: 358.40.526.7884; Fax: 358.03.364.1352; E-mail: [email protected]. Or to: Kexin Chen, Department of Epidemiology and Biostatistics, Tianjin Medical University Cancer Institute and Hospital, Ti-Yuan-Bei, Huan-Hu-Xi Road, He Xi District, Tianjin 300060, People’s Republic of China. Phone: 0086.22.23372231; Fax: 0086.22.23372231; E-mail: [email protected].
Authorship note: Brittany C. Parker and Matti J. Annala contributed equally to this work.
Find articles by Chen, K. in: JCI | PubMed | Google Scholar
1Department of Pathology, University of Texas MD Anderson Cancer Center, Houston, Texas, USA. 2The University of Texas Graduate School of Biomedical Sciences, Houston, Texas, USA. 3Institute of Signal Processing, Tampere University of Technology, Tampere, Finland. 4Department of Pathology, Tianjin Medical University Cancer Institute and Hospital, Tianjin, People’s Republic of China. 5Department of Neurosurgery, University of Texas MD Anderson Cancer Center, Houston, Texas, USA. 6Department of Biostatistics and Epidemiology, Tianjin Medical University Cancer Institute and Hospital, Tianjin, People’s Republic of China. 7Department of Neuropathology, Tampere University Hospital, Tampere, Finland. 8Institute of Biomedical Technology, Tampere, Finland. 9Department of Experimental Therapeutics, University of Texas MD Anderson Cancer Center, Houston, Texas, USA.
Address correspondence to: Wei Zhang, Department of Pathology, Unit 85, University of Texas MD Anderson Cancer Center, 1515 Holcombe Blvd., Houston, Texas 77030, USA. Phone: 713.745.1103; Fax: 713.792.5549; E-mail: [email protected]. Or to: Matti Nykter, Institute of Signal Processing, Tampere University of Technology, P.O. Box 553, FI-33101 Tampere, Finland. Phone: 358.40.526.7884; Fax: 358.03.364.1352; E-mail: [email protected]. Or to: Kexin Chen, Department of Epidemiology and Biostatistics, Tianjin Medical University Cancer Institute and Hospital, Ti-Yuan-Bei, Huan-Hu-Xi Road, He Xi District, Tianjin 300060, People’s Republic of China. Phone: 0086.22.23372231; Fax: 0086.22.23372231; E-mail: [email protected].
Authorship note: Brittany C. Parker and Matti J. Annala contributed equally to this work.
Find articles by Lang, F. in: JCI | PubMed | Google Scholar
1Department of Pathology, University of Texas MD Anderson Cancer Center, Houston, Texas, USA. 2The University of Texas Graduate School of Biomedical Sciences, Houston, Texas, USA. 3Institute of Signal Processing, Tampere University of Technology, Tampere, Finland. 4Department of Pathology, Tianjin Medical University Cancer Institute and Hospital, Tianjin, People’s Republic of China. 5Department of Neurosurgery, University of Texas MD Anderson Cancer Center, Houston, Texas, USA. 6Department of Biostatistics and Epidemiology, Tianjin Medical University Cancer Institute and Hospital, Tianjin, People’s Republic of China. 7Department of Neuropathology, Tampere University Hospital, Tampere, Finland. 8Institute of Biomedical Technology, Tampere, Finland. 9Department of Experimental Therapeutics, University of Texas MD Anderson Cancer Center, Houston, Texas, USA.
Address correspondence to: Wei Zhang, Department of Pathology, Unit 85, University of Texas MD Anderson Cancer Center, 1515 Holcombe Blvd., Houston, Texas 77030, USA. Phone: 713.745.1103; Fax: 713.792.5549; E-mail: [email protected]. Or to: Matti Nykter, Institute of Signal Processing, Tampere University of Technology, P.O. Box 553, FI-33101 Tampere, Finland. Phone: 358.40.526.7884; Fax: 358.03.364.1352; E-mail: [email protected]. Or to: Kexin Chen, Department of Epidemiology and Biostatistics, Tianjin Medical University Cancer Institute and Hospital, Ti-Yuan-Bei, Huan-Hu-Xi Road, He Xi District, Tianjin 300060, People’s Republic of China. Phone: 0086.22.23372231; Fax: 0086.22.23372231; E-mail: [email protected].
Authorship note: Brittany C. Parker and Matti J. Annala contributed equally to this work.
Find articles by Nykter, M. in: JCI | PubMed | Google Scholar
1Department of Pathology, University of Texas MD Anderson Cancer Center, Houston, Texas, USA. 2The University of Texas Graduate School of Biomedical Sciences, Houston, Texas, USA. 3Institute of Signal Processing, Tampere University of Technology, Tampere, Finland. 4Department of Pathology, Tianjin Medical University Cancer Institute and Hospital, Tianjin, People’s Republic of China. 5Department of Neurosurgery, University of Texas MD Anderson Cancer Center, Houston, Texas, USA. 6Department of Biostatistics and Epidemiology, Tianjin Medical University Cancer Institute and Hospital, Tianjin, People’s Republic of China. 7Department of Neuropathology, Tampere University Hospital, Tampere, Finland. 8Institute of Biomedical Technology, Tampere, Finland. 9Department of Experimental Therapeutics, University of Texas MD Anderson Cancer Center, Houston, Texas, USA.
Address correspondence to: Wei Zhang, Department of Pathology, Unit 85, University of Texas MD Anderson Cancer Center, 1515 Holcombe Blvd., Houston, Texas 77030, USA. Phone: 713.745.1103; Fax: 713.792.5549; E-mail: [email protected]. Or to: Matti Nykter, Institute of Signal Processing, Tampere University of Technology, P.O. Box 553, FI-33101 Tampere, Finland. Phone: 358.40.526.7884; Fax: 358.03.364.1352; E-mail: [email protected]. Or to: Kexin Chen, Department of Epidemiology and Biostatistics, Tianjin Medical University Cancer Institute and Hospital, Ti-Yuan-Bei, Huan-Hu-Xi Road, He Xi District, Tianjin 300060, People’s Republic of China. Phone: 0086.22.23372231; Fax: 0086.22.23372231; E-mail: [email protected].
Authorship note: Brittany C. Parker and Matti J. Annala contributed equally to this work.
Find articles by Zhang, W. in: JCI | PubMed | Google Scholar
Authorship note: Brittany C. Parker and Matti J. Annala contributed equally to this work.
Published January 9, 2013 - More info
J Clin Invest. 2013;123(2):855–865. https://doi.org/10.1172/JCI67144.
© 2013 The American Society for Clinical Investigation
Received: October 1, 2012; Accepted: November 26, 2012
Related article:
Abstract
RNA sequencing facilitates the discovery of novel gene fusions in cancer. In this issue of the
Authors
Ivan Babic, Paul S. Mischel
-
Abstract
Fusion genes are chromosomal aberrations that are found in many cancers and can be used as prognostic markers and drug targets in clinical practice. Fusions can lead to production of oncogenic fusion proteins or to enhanced expression of oncogenes. Several recent studies have reported that some fusion genes can escape microRNA regulation via 3′–untranslated region (3′-UTR) deletion. We performed whole transcriptome sequencing to identify fusion genes in glioma and discovered FGFR3-TACC3 fusions in 4 of 48 glioblastoma samples from patients both of mixed European and of Asian descent, but not in any of 43 low-grade glioma samples tested. The fusion, caused by tandem duplication on 4p16.3, led to the loss of the 3′-UTR of FGFR3, blocking gene regulation of miR-99a and enhancing expression of the fusion gene. The fusion gene was mutually exclusive with EGFR, PDGFR, or MET amplification. Using cultured glioblastoma cells and a mouse xenograft model, we found that fusion protein expression promoted cell proliferation and tumor progression, while WT FGFR3 protein was not tumorigenic, even under forced overexpression. These results demonstrated that the FGFR3-TACC3 gene fusion is expressed in human cancer and generates an oncogenic protein that promotes tumorigenesis in glioblastoma.
-
Introduction
Fusion genes are hybrid genes that form when a chromosomal rearrangement combines parts of 2 genes. Since their original identification in blood cancers (1), fusion genes have been identified in many solid tumors, such as Ewing sarcoma (2), synovial sarcoma (3), prostate cancer (4, 5), lung cancer (6, 7), breast cancer (8), and ovarian cancer (9). Gene fusions can lead to the production of oncogenic fusion proteins, such as the constitutively active BCR-ABL1 kinase in chronic myelogenous leukemia (10). Alternatively, fusion genes can lead to enhanced expression of oncogenes via promoter switching, as observed in the case of TMPRSS2-ERG fusions in prostate cancer (4). Recent evidence suggests that some fusion genes can bypass microRNA-mediated regulation via 3′–untranslated region (3′-UTR) deletion. For example, protein levels of the transcription factor MYB are typically low in adenoid cystic carcinoma (ACC) due to high expression of the MYB repressors miR-15a and miR-16 (11, 12). However, the 3′-UTR of MYB is lost upon formation of the MYB-NFIB fusion gene, leading to elevated levels of the MYB protein (13).
Molecular therapies targeted at fusion proteins have been successfully used in the treatment of chronic myelogenous leukemia (14) and non–small-cell lung cancer (15), but fusion genes still remain poorly characterized in cancers such as glioma, the most common type of brain tumor. Gliomas, solid tumors that arise from glial cells in the brain or spine, are characterized by poor patient survival and high resistance to current treatment regimens. The most common form of glioma in humans is glioblastoma (GBM), an aggressive grade IV tumor associated with a 14.6-month median survival time for patients treated with surgical resection, radiotherapy, and adjuvant temozolomide (16). GBMs frequently harbor alterations in the receptor tyrosine kinases EGFR, PDGFRA, and MET (17, 18). A recurrent KIAA1549-BRAF fusion has been reported in low-grade astrocytoma (19), and 1 isolated case of LEO1-SLC12A1 fusion has been reported in GBM (20).
In this study, we identified recurrent FGFR3-TACC3 fusions in GBM, a finding that was recently and independently reported by another group (21). We also provided evidence that the FGFR3-TACC3 fusion gene escaped regulation by miR-99a, leading to elevated expression levels.
-
Results
To search for recurrent fusion genes, we performed whole-transcriptome RNA sequencing (RNA-seq) on 8 pools of human glioma tissues and 2 pools of normal brain tissues. We searched for RNA-seq reads spanning exon-exon junctions connecting 2 distinct genes (Supplemental Figure 1; supplemental material available online with this article; doi: 10.1172/JCI67144DS1) and obtained a ranked list of fusion candidates (Supplemental Figure 2). The top fusion candidate, FGFR3-TACC3, was discovered based on 16 reads — 4 of them unique — spanning FGFR3 exon 18 and TACC3 exon 11 in one of the GBM pools (Supplemental Figure 3). We validated the fusion by performing RT-PCR on a total of 91 glioma tissue samples from mixed European descent and Chinese patients. FGFR3-TACC3 was detected in 4 of 48 (8.3%) GBM samples, but not in any of the 43 low-grade glioma samples. Capillary sequencing of fusion junctions in the positive patient samples (GBM-13, GBM-07, GBM-T01, and GBM-T02) revealed 3 fusion variants at the RNA level (Figure 1A). All fusion variants were predicted to produce in-frame fusion proteins. Analysis of RNA sequencing data from The Cancer Genome Atlas (TCGA) GBM project revealed 2 more positive cases (Supplemental Figure 4). Copy number analysis across a cohort of 454 samples suggested mutual exclusivity between FGFR3-TACC3 fusions and high-degree amplification of the receptor tyrosine kinases EGFR, PDGFRA, and MET (P = 0.004, Fisher exact test; Figure 1B).
 Figure 1
Figure 1Detection of FGFR3-TACC3 fusion in GBM at the RNA and protein level. (A) Fusion transcript structures and electropherograms for the 4 fusion-positive samples. GBM-07 and GBM-13 were patients treated at University of Texas MD Anderson Cancer Center (MDACC); GBM-T01 and GBM-T02 were patients treated at Tianjin Medical University Cancer Institute and Hospital (Tianjin). (B) Plot showing mutual exclusivity between FGFR3-TACC3 fusion and high-degree amplification of EGFR, PDGFRA, and MET. **P < 0.01, Fisher exact test. (C) Schematic of protein domains — Ig, transmembrane (TM), and protein kinase (PTKC) — contained within the FGFR3-TACC3 fusion protein. (D) Immunoblot of patient samples and FGFR3-TACC3 fusion– or WT FGFR3–expressing cells. Asterisk denotes stable cell line. (E) FGFR3 immunostaining of patients GBM-T01 and GBM-T02 and a control fusion-negative tumor. Original magnification, ×200.
FGF receptor 3 (FGFR3) encodes a receptor tyrosine kinase that is commonly mutated in bladder and cervical cancers (22). Transforming acidic coiled-coil containing protein 3 (TACC3) encodes a centrosomal protein that is involved in mitosis (23) and is overexpressed in lung and colon carcinomas and in multiple myeloma (24). The predicted fusion protein variants contained the extracellular Ig-like domains, transmembrane domain, and most of the tyrosine kinase domain of FGFR3, fused to the transforming acidic coiled-coil domain of TACC3 (Figure 1C). To determine whether the FGFR3-TACC3 fusion results in chimeric proteins of predicted sizes, we performed immunoblotting on fusion-positive and -negative GBM tissues using an antibody that recognizes the N terminus of FGFR3. As positive controls, we overexpressed FGFR3-TACC3 variant 1 and WT FGFR3 in GBM cell line SNB19. In WT FGFR3 transfected cells, we observed 130- and 120-kDa bands, which corresponded to fully and partially N-glycosylated forms of FGFR3, respectively (25). In FGFR3-TACC3 transfected cells, we observed bands at 145 and 135 kDa, consistent with a predicted 17-kDa difference in molecular weight relative to WT FGFR3. The bands in samples GBM-13 and GBM-T01 matched those in the positive control cell line. In sample GBM-07, we observed bands at 170 and 160 kDa, consistent with a predicted 39-kDa difference in molecular weight between FGFR3-TACC3 variant 2 and WT FGFR3. Endogenous WT FGFR3 was barely detectable in most GBM tissues, whereas the FGFR3-TACC3 fusion protein was abundant in fusion-positive patients (Figure 1D). Immunohistochemical analysis of GBM-T01 and GBM-T02 patient tissues revealed extensive FGFR3 staining that was absent in control tissues (Figure 1E), suggestive of a possible diagnostic measure for fusion-positive patients.
The orientation of the 2 genes on chromosome 4 led us to hypothesize that the fusions were caused by tandem duplication of a 70-kb region on 4p16.3 (Figure 2A). We confirmed this by performing genomic DNA capillary sequencing of tandem duplication boundaries in the 4 samples (Supplemental Figures 5–8). We then hybridized genomic DNA from the fusion-positive samples onto customized comparative genomic hybridization (CGH) microarrays with dense probe coverage for the fusion region. All 4 samples exhibited duplications in the region of interest, and the amplicon breakpoints agreed with DNA sequencing results (Figure 2B). In samples GBM-13 and GBM-T01, amplicons had high copy number and exhibited length variation, with some extending beyond FGFR3 on the centromeric side (Supplemental Figure 9). These longer amplicons give rise to chimeric pre-mRNA containing FGFR3 exon 19, but the last exon is discarded during splicing since it lacks a splicing donor site, thereby producing the observed fusion transcripts. This phenomenon of terminal exon skipping (Supplemental Figure 10) has been shown in previous studies of gene fusions (26).
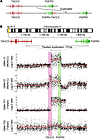 Figure 2
Figure 2FGFR3-TACC3 fusion is formed by a 70-kb tandem duplication on 4p16.3. (A) Schematic illustrating how the FGFR3-TACC3 fusion gene is formed through tandem duplication. (B) Validation of the tandem duplication in fusion-positive patients using customized Agilent CGH arrays with dense coverage for the fusion locus.
The absence of the 3′–untranslated region (3′-UTR) of FGFR3 in the FGFR3-TACC3 fusion transcript suggested that the high levels of fusion proteins in GBM patient samples could be partly explained by loss of microRNA regulation. TargetScan analysis (27) suggested that FGFR3, but not TACC3, was regulated by miR-99a (Figure 3A), a finding that has been supported experimentally (28). Interestingly, small RNA sequencing results showed that miR-99a was among the 4 most highly expressed microRNAs in both GBM and normal brain (Figure 3B), and in vitro studies showed it was the only highly expressed microRNA to target FGFR3 (Supplemental Figure 11). This finding was consistent with the low FGFR3 protein levels observed in samples of GBM compared with bladder cancer (Supplemental Figure 12), in which FGFR3 is often overexpressed and miR-99a is downregulated (29). Fusion-positive and -negative patient samples did not show a significant difference in miR-99a expression (Supplemental Figure 13). To validate the regulatory function of miR-99a in GBM, we performed a reporter gene assay in SNB19 cells using a plasmid expressing either the WT FGFR3 3′-UTR or a mutant with the majority of the miR-99a binding site deleted. Luciferase activity decreased upon miR-99a mimic transfection only in cells transfected with the WT reporter plasmid, not in those transfected with the mutant (P = 8.3 × 10–5, t test; n = 9; Figure 3C). To verify the ability of miR-99a to decrease FGFR3 mRNA in GBM, we performed quantitative RT-PCR (qRT-PCR) on cells transfected with either miR-99a mimic or anti–miR-99a. After validating successful miR-99a mimic and anti–miR-99a transfection (P = 4.1 × 10–5 and P = 1.7 × 10–4, respectively, Mann-Whitney U test; Figure 3D), we quantified FGFR3 mRNA levels by qRT-PCR, which decreased upon miR-99a mimic transfection and increased upon anti–miR-99a transfection (P = 8.2 × 10–5 and P = 6.2 × 10–4, respectively, Mann-Whitney U test; Figure 3E). FGFR3 protein levels exhibited an even higher increase after anti–miR-99a overexpression (P = 0.0031, t test; Figure 3, F and G). We then added the 3′-UTR back onto WT FGFR3 and FGFR3-TACC3 fusion cDNA constructs. Only constructs containing the 3′-UTR of FGFR3 exhibited decreased expression upon miR-99a transfection (Figure 3, H–K).
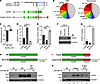 Figure 3
Figure 3Loss of miR-99a binding site in the FGFR3-TACC3 fusion transcript leads to increased levels of FGFR3 protein. (A) Schematic showing the location of the miR-99a binding site in the 3′-UTR of WT FGFR3 and its loss in the FGFR3-TACC3 fusion transcript and construct with the binding site deleted (mutant). (B) Pie chart illustrating high miR-99a expression in both GBM and normal brain. Expression values were calculated based on pooled small RNA sequencing. (C) Luciferase assay of WT FGFR3 3′-UTR versus mutant after miR-99a overexpression. (D) qRT-PCR of miR-99a in parental SNB19 cells after transfection of control, miR-99a mimic, or anti–miR-99a. (E–G) qRT-PCR (E), immunoblotting (F), and densitometry (G) of FGFR3 in parental SNB19 cells after transfection of control, miR-99a mimic, or anti-miR. (H and I) Schematic of WT FGFR3 (H) and FGFR3-TACC3 fusion (I) cDNA, with or without FGFR3 3′-UTR attached. (J) Immunoblot of EV, WT FGFR3, and WT FGFR3 plus FGFR3 3′-UTR after transfection with miR-99a. (K) Immunoblot of EV, FGFR3-TACC3 fusion, or FGFR3-TACC3 fusion plus FGFR3 3′-UTR after miR-99a transfection. Relative densitometry (below) was normalized to β-tubulin. Error bars denote SEM. **P < 0.01, ***P < 0.001, Mann-Whitney U test.
To determine whether the FGFR3-TACC3 fusion protein promotes tumor growth, we first measured cell viability of empty vector (EV) control, fusion, and WT FGFR3–expressing SNB19 cells using a 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay over a 4-day period. Both FGFR3-TACC3 fusion– and WT FGFR3–expressing cells showed greater viability than did control cells (P = 6.7 × 10–10, 2-way ANOVA; Figure 4A). BrdU incorporation assay showed that, compared with control cells, FGFR3-TACC3 fusion– and WT FGFR3–expressing cells proliferated at a faster rate (P = 5.9 × 10–9, 2-way ANOVA; Figure 4B) and formed more numerous colonies in soft agar (P = 1.6 × 10–7, t test; Figure 4C). These results were replicated in another GBM cell line, U251 (Supplemental Figure 14). Taken together, these in vitro findings showed that FGFR3-TACC3 promotes cell proliferation and anchorage-independent cell growth. Microarray experiments followed by pathway analysis revealed significant activation of oncogenic pathways in FGFR3-TACC3 fusion–expressing cells relative to EV control cells (Figure 4D). In contrast, only minor increases in tumorigenic pathways were observed in cells overexpressing WT FGFR3 or WT TACC3 (Supplemental Figure 15). Immunoblotting illustrated that FGFR3-TACC3 fusion overexpression led to increased activation (i.e., phosphorylation) of ERK and STAT3, but not AKT (Supplemental Figure 16), and both FGFR3-TACC3 fusion and WT FGFR3 cells exhibited ligand-dependent firing (Supplemental Figure 17) and were more sensitive to the ERK inhibitor U0126 (Supplemental Figure 18).
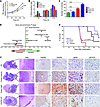 Figure 4
Figure 4FGFR3-TACC3 promotes tumor progression in vivo. (A) MTT assay measuring the viability of FGFR3-TACC3 fusion, WT FGFR3, and EV stable cell lines 48 and 72 hours after cell plating. ***P < 0.001, 2-way ANOVA. (B) BrdU incorporation assay of FGFR3-TACC3 fusion, WT FGFR3, WT TACC3, and EV cell lines 0, 6, 10, and 12 hours after incorporation. (C) Quantification of colony number in FGFR3-TACC3 fusion, WT FGFR3, WT TACC3, and EV cell lines. ***P < 0.001, 2-tailed Student’s t test. (D) Top 10 altered pathways identified by IPA for differential gene expression between SNB19 cell lines transfected with FGFR3-TACC3 fusion and those transfected with EV. Positive regulation z scores indicate increased pathway activation. Only the top 9 pathways are shown because IPA analysis did not allocate a regulation z score for the next gene sets. Statistical analyses were performed using Fisher exact test for gene set enrichment. (E) Kaplan-Meier survival plot showing that higher FGFR3-TACC3 fusion expression led to reduced survival. P values were calculated using the log-rank test. (F) H&E and immunohistochemical staining for the 3 groups of mice, showing increased pSTAT3 and pERK activity in mice injected with FGFR3-TACC3 fusion– or WT FGFR3–expressing cells. In lower-power images, both xenograft tumor tissue (T) and normal mouse brain tissue (N) are indicated. Original magnification, ×40 (H&E); ×100 (FGFR3, left); ×400 (FGFR3, right, pERK, and pSTAT3).
To determine whether the fusion promotes tumor growth and affects host survival in vivo, we orthotopically implanted FGFR3-TACC3 fusion, WT FGFR3, and EV cells into the brains of immunocompromised mice and compared survival patterns. We established stable FGFR3-TACC3 fusion– and WT FGFR3–expressing cell lines and generated stable clones that were evaluated for relative protein expression. We chose FGFR3-TACC3 fusion and WT FGFR3 clones that expressed either high or low levels of protein, as well as the entire mixture, for in vivo studies (Supplemental Figure 19). 1 × 106 cells from each line were injected into the brains of a total of 35 nude mice (n = 5 per group). 5 tumor-free mice died of diarrhea and were censored, while the rest of the mice developed large tumors by the time of their termination. Mice implanted with the high-expressing FGFR3-TACC3 fusion clone died significantly earlier than did EV control mice (within 70–80 days of implantation compared with 110–175 days; P = 0.0072, log-rank test), whereas mice implanted with the FGFR3-TACC3 fusion mixture died in 80–110 days, also faster than the EV controls (P = 0.0071, log-rank test; Supplemental Figure 19). Mice injected with the low-expressing FGFR3-TACC3 fusion clone showed survival similar to that of both EV and WT FGFR3 mice, which suggests that high expression of the FGFR3-TACC3 fusion, as observed in patient samples, is required to influence survival patterns (Supplemental Figure 19). Pooled analysis revealed that mice implanted with the FGFR3-TACC3 fusion mixture or with the high-expressing FGFR3-TACC3 fusion clones died earlier than EV controls (P = 0.0076; Figure 4E). There was no statistical difference in survival observed between EV controls and WT FGFR3 clone–implanted (either high- or low-expressing) mice (Figure 4E and Supplemental Figure 19). Immunohistochemical analysis revealed increased activation of downstream proteins ERK and STAT3 in both FGFR3-TACC3 fusion– and WT FGFR3–expressing tumors (Figure 4F).
-
Discussion
In recent years, the introduction of high-throughput sequencing has led to an explosion in the number of fusion genes identified in solid tumors. In this study, we identified a subset of GBM patients harboring oncogenic FGFR3-TACC3 fusions (Supplemental Figure 20). A recent independent study by Singh et al. also reported the presence of FGFR-TACC fusions in GBM (21). We showed that the FGFR3-TACC3 fusions occurred in patients both of mixed European and of Asian descent and did not present in lower-grade gliomas. Between our present study and that of Singh et al. (21), 12 FGFR-TACC fusions have been identified (11 FGFR3-TACC3 and 1 FGFR1-TACC1; Supplemental Table 1). We also show evidence for mutual exclusivity between FGFR-TACC fusion and amplification of the EGFR, PDGFRA, and MET genes. Amplification of these 3 receptor tyrosine kinases is a common genetic defect in GBM, which suggests that the FGFR3-TACC3 fusion gene may represent an alternative pathway to the same oncogenic endpoint.
By studying the FGFR3-TACC3 fusion gene at the DNA level, we showed that the fusion occurred as a result of a 70-kb tandem duplication on 4p16.3. Intriguingly, the only other known recurrent glioma-associated fusion gene, BRAF-KIAA1549, is also caused by tandem duplication (19). The BRAF-KIAA1549 fusion characterizes 70% of pilocytic astrocytomas. Whether both fusion events being formed via tandem duplication represents a random accident or a characteristic feature of glial cells remains an open question.
In addition to showing that the FGFR3-TACC3 fusion gene was generated by tandem duplication, we showed that the DNA level breakpoints of the tandem duplication varied greatly between tumors. In particular, one case of FGFR3-TACC3 fusion was caused by a tandem duplication that disrupted exons in both genes, yet still resulted in a non-frameshifted fusion protein. This provides evidence of oncogenic selection for a functional chimeric protein. Furthermore, we identified patients whose tandem duplications were of high copy number and exhibited length heterogeneity, particularly on the FGFR3 side. This suggests that some patients harbor copies of the FGFR3-TACC3 fusion in which the DNA level breakpoint is situated downstream of FGFR3. Importantly, such DNA-level fusions still resulted in functional chimeric proteins, as the last exon of FGFR3 is spliced out of the fusion transcript due to its lack of a splicing donor site. This phenomenon has been well characterized in the context of read-through fusion transcripts (26). In combination with the study by Singh et al. (21), our study sheds further light on the diversity of FGFR-TACC fusions present in GBM and may help to identify the protein domains critical for the fusion’s oncogenic function.
Another important discovery in our study was the identification of a mechanism by which the FGFR3-TACC3 fusion escapes regulation by miR-99a. The 3′-UTR of the WT FGFR3 transcript contains a miR-99a binding site that is lost in the fusion, because the last exon of FGFR3 does not participate in the fusion transcript. This allows for stronger expression of the fusion gene, as the FGFR3 promoter’s activity is no longer counteracted by posttranscriptional regulation by miR-99a. Because of high miR-99a levels in both normal brain and GBM, native FGFR3 levels in GBM are low compared with other cancers. Furthermore, it was recently shown that members of the miR-99 family target FGFR3 in several lung cancer cell lines, decreasing cell proliferation and anchorage-independent cell growth (30). We showed that when the 3′-UTR of FGFR3 was added back onto native FGFR3 or the FGFR3-TACC3 fusion construct, protein levels decreased upon miR-99a overexpression.
Our results showed that the fusion protein was highly oncogenic both in vitro and in vivo. FGFR3 has been shown to be mutated and amplified in various cancers, such as multiple myeloma (31), bladder cancer (32), and lung cancer (30). As expected, both WT FGFR3– and FGFR3-TACC3 fusion–overexpressing cells showed increased cell proliferation and anchorage-independent cell growth in vitro, as well as increased STAT3 and ERK activation compared with controls. However, FGFR3-TACC3 fusion–overexpressing cells showed the greatest tumorigenicity in our in vivo mouse model. The differences observed between FGFR3-TACC3 fusion– and WT FGFR3–overexpressing cells could be due to the TACC domain present on the fusion gene, or the observed FGFR3 truncation within the fusion. Recent work performed by Singh et al. showed that the fusion is localized to mitotic spindles and contributes to abnormal chromosomal separation, a function associated with TACC3 (21). TACC3 overexpression has been observed in breast cancer (33) and non–small-cell lung cancer (34). However, we showed that WT TACC3 overexpression did not confer increased cell proliferation or anchorage-independent cell growth in vitro, which suggests that the fusion of TACC3 and FGFR3 results in a gain of function. Taken together, our in vitro and in vivo results suggest that both FGFR3 and TACC components must be fused together to confer the greatest oncogenic phenotype. Future work should involve teasing apart the FGFR3-TACC3 fusion to better understand how the genes work together to promote gliomagenesis.
-
Methods
Tumor samples. 40 human glioma samples (20 GBM; 5 anaplastic astrocytoma; 6 anaplastic oligodendroglioma; 9 oligodendroglioma) were acquired from University of Texas MD Anderson Cancer Center’s Brain Tumor Center tissue bank. 51 human glioma samples (28 GBM; 23 low-grade glioma) were acquired from the Tumor Tissue Bank of the Tianjin Medical University Cancer Institute and Hospital. Tissues were obtained from surgery and snap frozen.
RNA extraction for transcriptome sequencing. While frozen, each tissue (weighing up to 50 mg) was transferred to a liquid nitrogen–cooled mortar and pestle, crushed into powder, and dissolved in 1 ml TRIzol reagent (Invitrogen). Chloroform (200 μl) was added to the sample, which was then vortexed at high speed for 15 seconds and centrifuged at 12,000 g for 15 minutes at 4°C. The aqueous phase was transferred to a fresh 1.5-ml Eppendorf tube, and an equal volume of 70% ethanol was added and the contents mixed by tube inversion. A Qiagen RNeasy mini column (Qiagen) was used to further purify the sample. The column was washed twice with 500 μl RPE buffer (Qiagen), and RNA was eluted with 50 μl nuclease-free water. The RNA was quantified using a spectrophotometer. RNA integrity was verified using an Agilent 2100 BioAnalyzer. Poly-A selection was not performed on any of the sample pools.
Sample pooling. Samples were pooled for SOLiD sequencing according to tumor type. GBM samples were divided into 4 pools of 5 tumor samples. Anaplastic astrocytoma and anaplastic oligodendroglioma samples were pooled into 2 separate pools, and the oligodendroglioma samples were further split into 2 pools. 2 pools of commercial normal brain RNA (Ambion) were also acquired: 1 pool contained RNA from adult brain, the other from fetal brain.
Library preparation for whole transcriptome sequencing. Libraries for both whole transcriptome and small RNA sequencing were prepared using the small RNA expression kit from Applied Biosystems Inc. (PN 4397682; Life Technologies Corp.), based on the SOLiD System whole transcriptome and small RNA sequencing protocols provided by Applied Biosystems. rRNA was depleted from total RNA using the Invitrogen Ribominus Eukaryotic Kit (PN A1083708; Life Technologies Corp.), and 0.5–1.0 μg rRNA-depleted total RNA was fragmented using RNase III. The fragmented rRNA-depleted total RNA was hybridized and ligated with truncated adaptor mix A from the SOLiD small RNA expression kit. Next, reverse transcription was performed to generate cDNA templates. cDNA was size selected from Novex 6% TBE-urea gel (Invitrogen). The excised gel piece containing DNA 100–200 bp in length was split vertically into 4 pieces using a razor blade. The size-selected cDNA was further amplified using the supplied primer set containing a 6-base-long sequence-specific barcode in approximately 12–15 cycles of PCR. The purified PCR products with barcodes served as a library. Libraries ranged in size from 150 to approximately 250 bp and contained 50- to 150-bp cDNA inserts, quantitated and qualitated by Agilent Bioanalyzer 2100.
Library preparation for small RNA sequencing. The sample containing small RNA was hybridized with truncated adaptor mix A provided in the small RNA expression kit. The adaptor mixes were sets of RNA/DNA oligonucleotides with a single-stranded degenerate sequence at one end and a defined sequence required for SOLiD sequencing at the other. Hybridizing and ligating the sample with adaptor mix A sequentially yielded the template for SOLiD sequencing from the 5′ end of the small RNA. The small RNA population of 18–40 nt ligated with adaptor mix A was reverse transcribed to generate cDNAs. To meet the sample quantity requirement for SOLiD sequencing and to append the required terminal sequences to each molecule, the cDNA libraries were amplified using one of the supplied primer sets containing a 6-base-long sequence-specific barcode in approximately 12–15 cycles of PCR. The individual library PCR products, containing small RNA of 18–40 nt with barcode(s), were purified and size-selected for 108–130 bp by electrophoresis on 6% polyacrylamide gel.
Template bead preparation. The individual prepared library was quantitated and titrated, before multiplexing, as pooled templates for emulsion PCR; the template molecules were attached to 1 μm beads as described in the Applied Biosystems SOLiD emulsion PCR protocol. After emulsion PCR, the template beads with amplified monoclonal templates were enriched by adaptor P2-affinity binding of polystyrene beads in 60% glycerol gradient by centrifugation. The P2-enriched template beads were further modified by terminal transferase with oligo linker for immobilization to the slide and prepared for deposit on substrate-coated glass slides, as described in the Applied Biosystems protocol.
Sequencing. Sequencing runs were performed on SOLiD version 3.5 for both whole transcriptome RNA sequencing and small RNA sequencing. The library template beads were titrated by workflow analysis to determine the percentage of P2-positive beads in the total template before they were deposited onto slides for sequencing. The number of P2-positive template beads deposited in full chambers of slides with multiplexed 5 barcoded whole transcriptome libraries and performed in 50 nt of whole transcriptome sequencing, and of P2-positive beads of 10 pooled barcoded libraries on full slide for small RNA sequencing in 35 nt small RNA sequencing, was determined. The average depth of colorspace reads per sample pool was 6.7 × 107.
Fusion gene discovery. To identify fusion gene candidates, RNA-seq reads from each sample pool were aligned against transcripts from NCBI RefSeq 38. Reads that aligned to known transcripts were assumed to result from normal transcription and were discarded from further analysis. The 5′ and 3′ ends of unaligned reads were split into 2 anchors, each 18 colors in length. The paired anchors were aligned against human exon sequences from NCBI RefSeq 38. Anchors with more than 3 alignments against the exome were discarded as noninformative. Since anchors were short, no mismatches against the reference exon sequences were allowed in anchor alignments. For each read with alignments for both anchors, a list of anchor-based exon-exon junctions was generated by taking a Cartesian product of the 2 sets of exons to which the anchors aligned. If an anchor pair aligned to an exon-exon junction between 2 exons from the same gene, all junctions for that anchor pair were discarded, because the read was assumed to originate from some form of unannotated transcription or splicing. We were then left with fusion candidates represented by exon-exon junctions between distinct genes. Once we had the lists of anchor-based junctions for every sample, we combined them and verified the junctions by aligning the full transcriptome-unaligned reads against them.
To reduce the number of false positives, each fusion candidate was required to fulfill the following requirements (Supplemental Figure 1). (a) The fusion candidate must not involve rRNA genes or other highly abundant genes, and must not involve genes located at hypervariable genomic sites. The list of blacklisted gene name patterns included RN18S1, RN28S1, RPPH1, SNORD*, SNORA*, RNY*, RN7SL, RN7SK, RNU*, and HLA-*. (b) Full 50-base reads aligning to the fusion candidate must together cover at least 15 bases on both sides of the fusion junction. (c) The fusion candidate must not be found in Ambion commercial normal brain samples. (d) The fusion candidate’s supporting reads must not contain more than an average of 0.7 nt mismatches per read against the reference transcriptome. (e) The 3′ side of the fusion junction must have no perfect alignments against any sequence within 50 kb of the genomic alignment for the 5′ side of the fusion junction. The 5′ side of the fusion junction must have no perfect alignments against any sequence within 50 kb of the genomic alignment for the 3′ side of the fusion junction.
For each fusion fulfilling these requirements, we counted the number of reads in each of the 8 tumor sample pools and calculated a P value using the Pearson χ2 test to compare goodness-of-fit against a uniform reference distribution. Fusions were then ranked in ascending order by P value, so that fusions whose read distributions deviated most significantly from a uniform distribution were ranked at the top. In total, we identified 17,564 putative fusion junctions supported by 1 or more RNA-seq reads; filtering yielded a ranked list of 52 fusion candidates.
Identification of FGFR3-TACC3 fusions in TCGA samples. We downloaded TCGA GBM transcriptome sequencing data for 169 patients using the CGHub system. To look for evidence of FGFR3-TACC3–positive cases, we executed our fusion gene discovery algorithm on the samples and identified 2 patients (TCGA-27-1835 and TCGA-76-4925) with hundreds of reads overlapping an FGFR3-TACC3 fusion junction. Patient samples with 1–4 reads overlapping an FGFR3-TACC3 fusion junction were ignored because all such samples exclusively originated from the same sequencing batch as TCGA-27-1835 and exhibited the same fusion variant as TCGA-27-1835. We concluded that these weakly expressing fusion genes resulted from nucleic acid contamination between samples. RPKM gene expression values were calculated for FGFR3 and TACC3 to show their overexpression in fusion-positive samples relative to fusion-negative controls.
Assessment of mutual exclusivity between FGFR3-TACC3 and RTK amplifications. Custom Agilent CGH microarrays on 4 fusion-positive and 3 fusion-negative GBM samples from the University of Texas MD Anderson Cancer Center and the Tianjin Medical University Cancer Institute and Hospital were analyzed for EGFR, PDGFRA, and MET amplification by calculating the median log ratio of all probes within each gene relative to commercial pooled reference DNA.
Agilent HG CGH 244A microarray data for 444 GBM samples was downloaded from TCGA GBM project. These data included fusion-positive samples TCGA-27-1835 and TCGA-06-6390, reported by Singh et al. (21). EGFR, PDGFRA, and MET amplification was assessed by calculating the median log ratio of all probes within each gene relative to commercial pooled reference DNA.
TCGA GBM exome sequencing data was downloaded for the fusion-positive patients TCGA-06-6390, TCGA-19-5958, and TCGA-76-4925 reported by Singh et al. (21). Paired tumor DNA and normal blood DNA was available for all 3 cases. EGFR, PDGFRA, and MET amplification was assessed by calculating the number of reads overlapping each gene in tumor versus normal blood samples.
A sample was considered to harbor high-level EGFR, PDGFRA, or MET amplification if data indicated 5 or more extra copies of the gene. Mutual exclusivity of FGFR3-TACC3 fusion and EGFR, PDGFRA, and MET amplification was assessed using Fisher exact test.
Small RNA expression analysis based on sRNA-seq. Small RNA sequencing reads were trimmed to a length of 18 colors and aligned against mature microRNA sequences from miRBase version 14 using Bowtie (35). Mature microRNA expression levels were calculated by counting reads that aligned completely within an annotated mature microRNA site in the pre-microRNA sequence. Read distributions of selected microRNAs are shown in Supplemental Figure 3. Trimming of reads to 18 colors prior to alignment was necessary because the sRNA-sequence protocol resulted in reads that included a 3′ adapter sequence. The trimming step also allowed us to effectively calculate the expression level of each microRNA as the sum of its iso-miR expressions. MicroRNA expression levels were normalized by the total number of mappable reads per sequencing experiment (36).
Array CGH analysis. Customized CGH microarrays with high probe density at the fusion region were designed using the Agilent eArray tool. The microarray design was based on the Agilent 105A backbone profile with 105,000 probes and 22-kbp median spacing. Extra probes (22,500 at 200-bp intervals) were designed for cytoband 4p16.3 (containing FGFR3-TACC3). Customized microarrays were ordered from Agilent. To analyze copy numbers, all microarray probes were remapped against the hg19 genome assembly, and log ratios were calculated between Cy3 and Cy5 channels. The log ratios were then segmented using circular binary segmentation (37). Microarray files have been deposited in GEO (accession no. GSE42400).
RT-PCR validation of fusion transcripts. To create a mammalian expression vector, cDNA was synthesized using SuperScript III (catalog no. 18080-051; Invitrogen) and random hexamers followed by PCR amplification using Advantage HD polymerase mix (639241; Clontech) with FGFR3 primer TCGCCAGTCTCCCGAGC and TACC3 primer GACAGCGGCTCCGTGGAGG. The fusion PCR products were TOPO cloned (45-0640; Invitrogen). Finally, the MSC enzymes BamHI and XbaI were used to move the fusion genes into pcDNA3.1+. All final constructs were sequence verified.
Immunoblot validation of fusion proteins. Total protein was isolated from tumor tissue by subjecting it to lysis buffer (1× RIPA containing 0.1% Halt protease) and phosphatase inhibitor cocktail (Fisher Scientific) and brief sonication (Branson Digital Sonifer Model 450) at 10% amplitude, followed by rotation at 40°C for 30 minutes. Total protein was isolated by centrifugation at 1,214 g for 15 minutes, and lysates were stored at –80°C.
Detection of fusion breakpoint at the DNA level. Genomic DNA was isolated from fusion-positive patient samples and used as a template for PCR with Extensor Long Range PCR Polymerase (Thermo Scientific). Primers were designed against exon 18 (5′-CCCTCCCAGAGGCCCACCTT-3′) of FGFR3 and exon 11 (5′-CCTGCTCCTCAGCTCCCGGT-3′) or the intronic region after exon 4 (5′-GCAGACCCACGGCCAAGACC-3′) of TACC3. PCR products were gel purified and cloned with TOPO TA Cloning Kit for Sequencing with chemically One Shot TOP10 Chemically Competent E. coli (Invitrogen). At least 2 fusion-positive clones from each patient were subjected to capillary sequencing with plasmid-specific primers.
Cloning. cDNA (random primed, superscript III) was made from GBM-13, the patient sample with the highest detected fusion level. The complete 3.0-kb fusion transcript was amplified using the forward primer 5′-TCGCCAGTCTCCCGAGC-3′ (upstream of the FGFR3 start codon) and the reverse primer 5′-GACAGCGGCTCCGTGGAGG-3′ (downstream of the TACC3 stop codon) with the Clontech Advantage — LA polymerase kit (catalog no. 639152). The PCR product was cloned into pCR2.1 (catalog no. K4500; Invitrogen). An error-free subclone was created in pcDNA3.1 (by way of a pBluescript II intermediate to pick up required HindIII-Xba cloning sites). WT FGFR3 and TACC3 constructs were purchased (Origene) and subcloned into the pcDNA3.1 expression vector.
Cell line generation and immunoblotting. All tissue cultures were maintained in DMEM/F12 medium supplemented with 10% FBS in a 37°C humidified incubator containing 5% CO2. Control cell lines containing pcDNA3.1 expression vector only were obtained as previously described (38). The FGFR3-TACC3 construct sequenced from GBM-13 was inserted into the pcDNA3.1 expression plasmid (Invitrogen) under control of the cytomegalovirus promoter. 6 × 105 SNB19 and U251 cells were transfected with 10 mg FGFR3-TACC3, WT FGFR3, or WT TACC3 cDNA with Lipofectamine (Invitrogen) per the manufacturer’s instructions. Stably transfected cells were selected for with 0.4 mg/ml G418 (Invitrogen) for 2 weeks, after which SNB19 clones were selected and amplified. Relative expression of either WT FGFR3 or the FGFR3-TACC3 fusion between SNB19 cell clones and the mixed population was measured by immunoblot analysis using a mouse monoclonal antibody probing for FGFR3 amino acids 25–124 (sc-13121; Santa Cruz Biotechnology Inc.). Downstream analysis after FGFR3-TACC3 fusion overexpression was performed, probing for β-tubulin (9F3), phosphorylated STAT3 (Tyr705; 3E2), total STAT3 (79D7), phosphorylated p44/42 MAPK (ERK1/2; Thr202/Tyr204), total p44/42 MAPK (ERK1/2), and total TACC3 (Santa Cruz Biotechnology Inc.)
Cell viability, proliferation, apoptosis, and soft-agar assays. An MTT assay was performed to measure cell viability. Cells were seeded at 650 cells/well in a 96-well plate in quadruplicate and allowed to attach overnight. Cell viability was measured by incubating cells with 0.5 mg/ml MTT reagent in PBS (Sigma-Aldrich) for 2 hours. MTT reagent was then aspirated, and cells were subjected to lysis with 100% DMSO. Plates were read at 590 nm using the Tecan SpectraFluor Microplate Reader and Magellan 6 software (Tecan Group Ltd.) at 48, 72, or 96 hours after cell plating. Cell proliferation was measured via BrdU incorporation assay at 7, 11, and 13 hours in triplicate. Briefly, cells were synchronized at G1 by starvation for 3 days. Fresh medium was then added, and the cells were incubated for 12 hours, followed by pulse labeling with 20 mm BrdU for 1 hour at the indicated time points. The cells were stained with FITC-conjugated anti-BrdU antibody, incubated with 7-aminoactinomycin-D (7-AAD), and quantified via flow cytometry. To determine whether FGFR3-TACC3 promotes colony formation in soft agar, empty vector, FGFR3-TACC3, WT FGFR3, and WT TACC3 SNB19 or U251 stable cells were cultured in DMEM/F12 medium with 10% FBS in the log phase. The soft agar was prepared by mixing 1.5% sterile low melting point agarose in PBS with fresh medium at a 1:2 dilution. Soft agar (1 ml of 0.5%) was added to each well of a 6-well plate and kept at room temperature for 15 minutes. Cells were then harvested and suspended in 0.375% soft agar at 1,000 cells/ml. This cell suspension (1 ml) was added on top of the prepared base agar layer. The plates were incubated for 2 weeks and fed 2 times per week. Colonies were counted under a microscope, and relative colony size was measured using AxioVision 3.1 software.
Gene expression microarray analysis on fusion clones. FGFR3-TACC3 fusion– or WT FGFR3–transfected SNB19 mixtures or clones were hybridized onto dual-channel Agilent Whole Human Genome 4 × 44K v1 microarrays. RNA from parental SNB19 cells was hybridized onto the reference channel. Microarray slides were imaged, and background adjusted probe intensities were calculated using Agilent Feature Extraction Software version 9.1.3.1. Probe sequences were aligned against RefSeq 38 transcript sequences, and the probes were arranged into probesets based on the genes they aligned against. Background-adjusted probe intensities were quantile normalized and summarized using the robust multiarray analysis (RMA) algorithm (39). Differential gene expression was calculated by comparing against the reference channel. Pathway analysis was performed using Ingenuity Pathway Analysis (IPA; version 11904312). Microarray files have been deposited in GEO (accession no. GSE42401).
miR-99a reporter gene assay. The 3′-UTR of FGFR3 in the pMirTarget reporter vector was purchased from Origene Technologies Inc. The miR-99a binding site was mutated using QuikChange Lightning Site-Directed Mutagenesis Kit (Agilent Technologies), creating a deleted mutant from 5′-AAUACGGGUA-3′ to 5′-AA––––––UA-3′. To test the ability of miR-99a to target the 3′-UTR of FGFR3, WT and mutant constructs were transfected into SNB19 cells concurrently with TK Renilla luciferase reporter vector (Promega) and either control (scrambled) microRNA or miR-99a mimic (Thermo Fisher Scientific). At 48 hours after transfection, cells were assayed for relative luciferase activity using the Dual-Luciferase Reporter Assay System (Promega). Transfections were replicated in 9 independent experiments. In each experiment, relative luciferase activity following miR-99a overexpression was normalized to scrambled controls and converted to a log ratio. An unpaired 1-tailed t test was used to determine whether the log ratios were different between WT FGFR3 UTR and deletion mutant cells.
qRT-PCR validation of successful miR-99a transfection in SNB19 cells. To confirm successful transfection, miR-99a mimic (300516-03), anti–miR-99a (IH-300516-05), and scrambled (control) microRNA (Thermo Fisher Scientific) were transfected into SNB19 parental cells with 9 biological replicates (transfections) per group. For each of the biological replicates, miR-99a (assay ID 000435; Applied Biosystems) levels were quantified using TaqMan qRT-PCR (microRNA protocol 4364031 revision B; Applied Biosystems) with 3 technical replicates that were averaged and normalized to endogenous U6 (assay ID 001093; Applied Biosystems). A 2-tailed Mann-Whitney U test was used to assess whether miR-99a/U6 log ratios differed between groups.
qRT-PCR validation of FGFR3 mRNA level regulation by miR-99a, miR-21, and miR-125b. miR-99a mimic (300516-03), anti–miR-99a (IH-300516-05), miR-21 mimic (C-301023-01), miR-125b mimic (C-300595-03), or scrambled (control) microRNA (Thermo Fisher Scientific) were transfected into SNB19 parental cells with 9 biological replicates (transfections) per group. 0.5 μg total RNA from each transfection was reverse transcribed in 20-μl reactions. A ratio of 1 μg total RNA to 0.4 μg random hexamers was maintained, and the mixtures were heated at 70°C for 10 minutes. The tubes were then incubated at room temperature for 10 minutes, and the following components were added: 1× Superscript II RT Buffer (Invitrogen), 10 mM dithiothreitol (Invitrogen), 0.5 mM dNTPs (ISC Bioexpress), 20 U RNase Inhibitor (Ambion), and 200 U Superscript II Reverse Transcriptase (Invitrogen). The reaction was again incubated for 10 minutes at room temperature and then held at 37°C for 1 hour. The reaction was incubated at 42°C for 1.5 hours and then at 50°C for 30 minutes. Real-time PCR was performed on the Applied Biosystems Prism 7900 using an FGFR3 assay (Hs00179829_m1; Applied Biosystems) and human cyclophilin A (4326317e) Vic-labeled Pre-Developed Assay Reagent (Applied Biosystems); the 15-μl final reaction volume contained 1× TaqMan Universal PCR Master Mix (Applied Biosystems) and 1× Assay-on-Demand. cDNA (25 ng/well) was amplified with the following cycling conditions: 10 minutes at 95°C, followed by 50 cycles at 95°C for 15 seconds and 60°C for 1 minute. Each qRT-PCR measurement was performed in 3 technical replicates that were averaged and normalized to cyclophilin A. A 2-tailed Mann-Whitney U test was used to assess whether FGFR3/cyclophilin A log ratios differed between groups.
Measurement of FGFR3 protein level regulation by miR-99a, miR-21, and miR-125b. To determine whether overexpression of miR-99a mimic or anti-miR, miR-21 mimic, and miR-125b mimic affected FGFR3 protein expression, each respective microRNA was transfected into SNB19 parental cells, and relative FGFR3 expression was measured 48 hours later via immunoblot analysis (Santa Cruz Biotechnology Inc.). Band intensity was quantified and normalized to β-tubulin expression using Image J software (NIH). The experiment was replicated 3 times.
Generation of FGFR3 WT and fusion constructs containing the 3′-UTR of FGFR3. To generate WT FGFR3 and FGFR3-TACC3 fusion constructs containing the 3′-UTR of FGFR3, the 3′-UTR of FGFR3 was amplified by PCR from a FGFR3 3′-UTR plasmid (SC215711; Origene Technologies) to introduce the NheI cloning sites at both sides (underlined below), using the forward primer 5′-GCTAGCGGGCTCGCGGACGTGAAG-3′ and the reverse primer 5′-GCTAGCGGTTAGCAACCAGGTGTC-3′. The PCR product was cloned into pCR2.1 and verified by DNA sequencing. FGFR3 3′-UTR was then digested with NheI from pCR2.1–FGFR3 3′-UTR and subcloned into the XbaI sites downstream of pcDNA3.1+ WT FGFR3 and pcDNA3.1+FGFR3-TACC3 vectors. All constructs were verified by DNA sequencing. To determine the ability of miR-99a to regulate expression of FGFR3 3′-UTR–containing constructs, miR-99a or scrambled control was transfected and assayed via immunoblot as described previously.
bFGF stimulation. To determine whether the FGFR3-TACC3 fusion exhibits ligand dependence or independent firing, EV, FGFR3-TACC3 fusion, and WT FGFR3 cells were seeded in 6-well plates and incubated with 50 ng/ml bFGF ligand for 30 minutes at 37°C. Cells were then harvested and subjected to immunoblot, probing for FGFR3, phosphorylated ERK, total ERK, and actin (Santa Cruz Biotechnology Inc.).
Intracranial xenograft implantation and tissue preparation. Male athymic mice (nu/nu) were implanted in the brain with EV, FGFR3-TACC3 fusion, or WT FGFR3 cells. Briefly, mice were anesthetized with 0.25 ml of a cocktail of 10 mg/ml ketamine and 1 mg/ml xylazine, and cells were implanted using cranial guide screws as previously described (40). A Hamilton syringe and microinfusion syringe pump (0.5 ml/min; Harvard Apparatus) were used to implant 1 × 106 cells into the brain of 10 mice simultaneously, as described previously (41). Upon detection of an external tumor or obvious declining health, mice were sacrificed by intracardiac perfusion of PBS and 4% paraformaldehyde. Brains were extracted and fixed in 10% formalin for 24 hours, embedded in paraffin, and sectioned into 5-mm slices.
Immunohistochemical staining. For immunohistochemical staining, Dako Envision+System–horseradish peroxidase and diaminobenzidine (DAB) were used (Dako). Briefly, after antigen retrieval for 10 minutes in 0.1 M citrate buffer (pH 6.0) or 1 mM EDTA (pH 8.0) and subsequent incubation in peroxidase block solution for 5 minutes at room temperature, the sections were incubated overnight at 4°C with rabbit anti-human FGFR3 (1:600; Abcam), pSTAT3 (1:200, Tyr705 D3A7; Cell Signaling Technology), and pERK (1:200, T202/Y204; Cell Signaling Technology). The sections were incubated in peroxidase-labeled polymer for 30 minutes at room temperature, and the signals were revealed with DAB+ substrate–chromogen solution. For negative controls, primary antibodies were replaced with PBS.
Drug treatment studies. For studies measuring cell viability after U0126 drug treatment, EV, WT FGFR3, and FGFR3-TACC3 fusion cells were plated at 100,000 cells/well in a 96-well plate. 12 hours after plating time, “0” was read to ensure cells were plated in equal numbers. At this time, drugs were added in increasing concentrations, with DMSO only as control. Plates were assayed 48 hours later, and values were normalized to DMSO controls. Cells were also seeded in 6-well plates and incubated with each inhibitor for 1 hour. These cells were assayed via immunoblot to determine the potency of each drug concentration.
Statistics. Unless otherwise indicated, data are represented as mean ± SEM. Statistical analyses were performed using Fisher exact test, Mann-Whitney U test, 2-way ANOVA, 1- or 2-tailed Student’s t test, and log-rank test, as indicated. Activation z scores were calculated as defined in the Ingenuity white paper “Ingenuity Downstream Effects Analysis in IPA.” Mice that died of causes other than cancer were considered censored. A P value of 0.05 or less was considered significant.
Study approval. Glioma tissue samples from the Brain Tumor Center tissue bank of the University of Texas MD Anderson Cancer Center were collected under an institutional review board–approved protocol. Glioma tissue samples from the Tumor Tissue Bank of the Tianjin Medical University Cancer Institute and Hospital were collected with approval from the institutional review board. All subjects provided informed consent of their participation in the study. Implantation of cells into the brains of male athymic mice were performed according to institution-approved protocols.
-
Acknowledgments
We thank Kathryn Hale (Department of Scientific Publications, University of Texas MD Anderson Cancer Center) for editing this manuscript. This study was partially supported by a grant from the NIH (U24CA143835, to W. Zhang); by a grant from the Paul and Joann Oreffice Fund for Brain Tumor Research (to R. Sawaya, G.N. Fuller, and W. Zhang); by the Academy of Finland (project no. 132877), Sigrid Juselius Foundation, and Finnish Funding Agency for Technology and Innovation Finland Distinguished Professor programme (to O. Yli-Harja and M. Nykter); and by the Program for Changjiang Scholars and Innovative Research Team in University (PCSIRT) in China and National Key Scientific and Technological Project (2011ZX0 9307-001-04, to K. Chen) and Tianjin Science and Technology Committee Foundation (09ZCZDSF04700, to K. Chen). The Tumor Tissue Bank of Tianjin Cancer Hospital is partially supported by a grant from the National Foundation for Cancer Res. The genomic studies were supported in part by the Cancer Genomics Core Laboratory and the Sequencing Facility through University of Texas MD Anderson Cancer Center Support Grant CA016672.
Address correspondence to: Wei Zhang, Department of Pathology, Unit 85, University of Texas MD Anderson Cancer Center, 1515 Holcombe Blvd., Houston, Texas 77030, USA. Phone: 713.745.1103; Fax: 713.792.5549; E-mail: [email protected]. Or to: Matti Nykter, Institute of Signal Processing, Tampere University of Technology, P.O. Box 553, FI-33101 Tampere, Finland. Phone: 358.40.526.7884; Fax: 358.03.364.1352; E-mail: [email protected]. Or to: Kexin Chen, Department of Epidemiology and Biostatistics, Tianjin Medical University Cancer Institute and Hospital, Ti-Yuan-Bei, Huan-Hu-Xi Road, He Xi District, Tianjin 300060, People’s Republic of China. Phone: 0086.22.23372231; Fax: 0086.22.23372231; E-mail: [email protected].
-
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Reference information: J Clin Invest. 2013;123(2):855–865. doi:10.1172/JCI67144.
See the related article at Multiple functions of a glioblastoma fusion oncogene.
-
References
- Croce CM. Chromosome translocations and human cancer. Cancer Res. 1986;46:6019–6023.View this article via: PubMed Google Scholar
- Delattre O, et al. Gene fusion with an ETS DNA-binding domain caused by chromosome translocation in human tumours. Nature. 1992;359(6391):162–165.
- Clark J, et al. Identification of novel genes, SYT and SSX, involved in the t (X; 18)(p11.2; q11.2) translocation found in human synovial sarcoma. Nat Genet. 1994;7(4):502–508.View this article via: PubMed Google Scholar
- Tomlins SA, et al. Recurrent fusion of TMPRSS2 and ETS transcription factor genes in prostate cancer. Science. 2005;310(5748):644–648.
- Palanisamy N, et al. Rearrangements of the RAF kinase pathway in prostate cancer, gastric cancer and melanoma. Nat Med. 2010;16(7):793–798.
- Soda M, et al. Identification of the transforming EML4-ALK fusion gene in non-small-cell lung cancer. Nature. 2007;448(7153):561–566.
- Rikova K, et al. Global survey of phosphotyrosine signaling identifies oncogenic kinases in lung cancer. Cell. 2007;131(6):1190–1203.
- Tognon C, et al. Expression of the ETV6-NTRK3 gene fusion as a primary event in human secretory breast carcinoma. Cancer Cell. 2002;2(5):367–376.
- Salzman J, et al. ESRRA-C11orf20 is a recurrent gene fusion in serous ovarian carcinoma. PLoS Biol. 2011;9(9):e1001156.
- Shtivelman E, Lifshitz B, Gale RP, Canaani E. Fused transcript of abl and bcr genes in chronic myelogenous leukaemia. Nature. 1985;315(6020):550–554.
- Chung EY, et al. c-Myb oncoprotein is an essential target of the dleu2 tumor suppressor microRNA cluster. Cancer Biol Ther. 2008;7(11):1758–1764.View this article via: PubMed Google Scholar
- Zhao H, Kalota A, Jin S, Gewirtz AM. The c-myb proto-oncogene and microRNA-15a comprise an active autoregulatory feedback loop in human hematopoietic cells. Blood. 2009;113(3):505–516.
- Persson M, Andrén Y, Mark J, Horlings HM, Persson F, Stenman G. Recurrent fusion of MYB and NFIB transcription factor genes in carcinomas of the breast and head and neck. Proc Natl Acad Sci U S A. 2009;106(44):18740–18744.
- Druker BJ, et al. Five-year follow-up of patients receiving imatinib for chronic myeloid leukemia. N Engl J Med. 2006;355(23):2408–2417.
- Shaw AT, et al. Effect of crizotinib on overall survival in patients with advanced non-small-cell lung cancer harbouring ALK gene rearrangement: a retrospective analysis. Lancet Oncol. 2011;12(11):1004–1012.
- Stupp R, et al. Effects of radiotherapy with concomitant and adjuvant temozolomide versus radiotherapy alone on survival in glioblastoma in a randomised phase III study: 5-year analysis of the EORTC-NCIC trial. Lancet Oncol. 2009;10(5):459–466.
- The Cancer Genome Atlas Research Network. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455(7216):1061–1068.
- Liu KW, Hu B, Cheng SY. Platelet-derived growth factor receptor alpha in glioma: a bad seed. Chin J Cancer. 2011;
30
(9):590–602.View this article via: PubMed Google Scholar
- Jones D, et al. Tandem duplication producing a novel oncogenic BRAF fusion gene defines the majority of pilocytic astrocytomas. Cancer Res. 2008;68(21):8673–8677.
- Bralten LB, et al. Integrated genomic profiling identifies candidate genes implicated in glioma-genesis and a novel LEO1-SLC12A1 fusion gene. Genes Chromosomes Cancer. 2010;49(6):509–517.View this article via: PubMed Google Scholar
- Singh D, et al. Transforming fusions of FGFR and TACC genes in human glioblastoma. Science. 2012;337(6099):1231–1235.
- Cappellen D, et al. Frequent activating mutations of FGFR3 in human bladder and cervix carcinomas. Nat Genet. 1999;23(1):18–20.View this article via: PubMed Google Scholar
- Gergely F, Kidd D, Jeffers K, Wakefield JG, Raff JW. D-TACC: a novel centrosomal protein required for normal spindle function in the early Drosophila embryo. EMBO J. 2000;19(2):241–252.
- Still IH, Vince P, Cowell JK. The third member of the transforming acidic coiled coil-containing gene family, TACC3, maps in 4p16, close to translocation breakpoints in multiple myeloma, and is upregulated in various cancer cell lines. Genomics. 1999;58(2):165–170.
- Chen F, Hristova K. The physical basis of FGFR3 response to fgf1 and fgf2. Biochemistry. 2011;50(40):8576. -8582.
- Akiva P, et al. Transcription-mediated gene fusion in the human genome. Genome Res. 2006;16(1):30–36.
- Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120(1):15–20.
- Oneyama C, et al. MicroRNA-mediated downregulation of mTOR/FGFR3 controls tumor growth induced by Src-related oncogenic pathways. Oncogene. 2011;30(32):3489–3501.
- Song T, et al. Differential miRNA expression profiles in bladder urothelial carcinomas. Asian Pac J Cancer Prev. 2010;11(4):905–911.View this article via: PubMed Google Scholar
- Kang J, et al. microRNA-99b acts as a tumor suppressor in non-small cell lung cancer by directly targeting fibroblast growth factor receptor 3. Exp Ther Med. 2012;3(1):149–153.View this article via: PubMed Google Scholar
- Chang H, Stewart AK, Qi XY, Li ZH, Yi QL, Trudel S. Immunohistochemistry accurately predicts FGFR3 aberrant expression and t(4;14) in multiple myeloma. Blood. 2005;106(1):353–355.
- Gómez-Román JJ, et al. Fibroblast growth factor receptor 3 is overexpressed in urinary tract carcinomas and modulates the neoplastic cell growth. Clin Cancer Res. 2005;11(2 pt 1):459–465.View this article via: PubMed Google Scholar
- Ma XJ, et al. Gene expression profiles of human breast cancer progression. Proc Natl Acad Sci U S A. 2003;100(10):5974–5979.
- Jung CK, Jung JH, Park GS, Lee A, Kang CS, Lee KY. Expression of transforming acidic coiled-coil containing protein 3 is a novel independent prognostic marker in non-small cell lung cancer. Pathol Int. 2006;56(9):503–509.
- Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10(3):R25.
- Moore L, et al. Transcriptome and small RNA deep sequencing reveals deregulation of miRNA biogenesis in human glioma [published online ahead of print September 24, 2012]. J Pathol. doi: 10.1002/path.4109 .
- Olshen AB, Venkatraman ES, Lucito R, Wigler M. Circular binary segmentation for the analysis of array-based DNA copy number data. Biostatistics. 2004;5(4):557–572.
- Wang H, et al. Insulin-like Growth Factor Binding Protein 2 enhances glioblastoma invasion by activating invasion-enhancing genes. Cancer Res. 2003;63(15):4315–4321.View this article via: PubMed Google Scholar
- Irizarry RA, et al. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4(2):249–264.
- Lal S, Lacroix M, Tofilon P, Fuller GN, Sawaya R, Lang FF. An implantable guide-screw system for brain tumor studies in small animals. J Neurosurg. 2000;92(2):326–333.View this article via: PubMed Google Scholar
- Nakamizo A, et al. Human bone marrow-derived mesenchymal stem cells in the treatment of gliomas. Cancer Res. 2005;65(8):3307–3318.View this article via: PubMed Google Scholar
- Croce CM. Chromosome translocations and human cancer. Cancer Res. 1986;46:6019–6023.
-
Version history
- Version 1 (January 9, 2013): No description
- Version 2 (January 9, 2013): No description
- Version 3 (February 1, 2013): No description



Copyright © 2024 American Society for Clinical Investigation
ISSN: 0021-9738 (print), 1558-8238 (online)




