Advertisement
Research ArticleVirology Free access | 10.1172/JCI63120
Viperin restricts chikungunya virus replication and pathology
Terk-Shin Teng,1 Suan-Sin Foo,1 Diane Simamarta,1 Fok-Moon Lum,1,2 Teck-Hui Teo,1,3 Aleksei Lulla,4 Nicholas K.W. Yeo,1 Esther G.L. Koh,1 Angela Chow,5 Yee-Sin Leo,5 Andres Merits,4 Keh-Chuang Chin,1,6 and Lisa F.P. Ng1,2
1Singapore Immunology Network, Agency for Science, Technology and Research (A*STAR), Biopolis, Singapore. 2Department of Biochemistry, Yong Loo Lin School of Medicine, and 3NUS Graduate School for Integrative Sciences and Engineering, National University of Singapore, Singapore. 4Institute of Technology, University of Tartu, Tartu, Estonia. 5Communicable Disease Centre, Tan Tock Seng Hospital, Singapore. 6Department of Physiology, Yong Loo Lin School of Medicine, National University of Singapore, Singapore.
Address correspondence to: Lisa F.P. Ng, Laboratory of Chikungunya Virus Immunity, Singapore Immunology Network, A*STAR, 8A Biomedical Grove, #04-06 Immunos, Biopolis 138648, Singapore. Phone: 65.64070028; Fax: 65.64642057; E-mail: [email protected]. Or to: Keh-Chuang Chin, Laboratory of Gene Regulation in Inflammation, Singapore Immunology Network, A*STAR, 8A Biomedical Grove, #04-06 Immunos, Biopolis 138648, Singapore. Phone: 65.64070006; Fax: 65.64642057; E-mail: [email protected].
Authorship note: Terk-Shin Teng and Suan-Sin Foo contributed equally to this work.
Find articles by Teng, T. in: JCI | PubMed | Google Scholar
1Singapore Immunology Network, Agency for Science, Technology and Research (A*STAR), Biopolis, Singapore. 2Department of Biochemistry, Yong Loo Lin School of Medicine, and 3NUS Graduate School for Integrative Sciences and Engineering, National University of Singapore, Singapore. 4Institute of Technology, University of Tartu, Tartu, Estonia. 5Communicable Disease Centre, Tan Tock Seng Hospital, Singapore. 6Department of Physiology, Yong Loo Lin School of Medicine, National University of Singapore, Singapore.
Address correspondence to: Lisa F.P. Ng, Laboratory of Chikungunya Virus Immunity, Singapore Immunology Network, A*STAR, 8A Biomedical Grove, #04-06 Immunos, Biopolis 138648, Singapore. Phone: 65.64070028; Fax: 65.64642057; E-mail: [email protected]. Or to: Keh-Chuang Chin, Laboratory of Gene Regulation in Inflammation, Singapore Immunology Network, A*STAR, 8A Biomedical Grove, #04-06 Immunos, Biopolis 138648, Singapore. Phone: 65.64070006; Fax: 65.64642057; E-mail: [email protected].
Authorship note: Terk-Shin Teng and Suan-Sin Foo contributed equally to this work.
Find articles by Foo, S. in: JCI | PubMed | Google Scholar
1Singapore Immunology Network, Agency for Science, Technology and Research (A*STAR), Biopolis, Singapore. 2Department of Biochemistry, Yong Loo Lin School of Medicine, and 3NUS Graduate School for Integrative Sciences and Engineering, National University of Singapore, Singapore. 4Institute of Technology, University of Tartu, Tartu, Estonia. 5Communicable Disease Centre, Tan Tock Seng Hospital, Singapore. 6Department of Physiology, Yong Loo Lin School of Medicine, National University of Singapore, Singapore.
Address correspondence to: Lisa F.P. Ng, Laboratory of Chikungunya Virus Immunity, Singapore Immunology Network, A*STAR, 8A Biomedical Grove, #04-06 Immunos, Biopolis 138648, Singapore. Phone: 65.64070028; Fax: 65.64642057; E-mail: [email protected]. Or to: Keh-Chuang Chin, Laboratory of Gene Regulation in Inflammation, Singapore Immunology Network, A*STAR, 8A Biomedical Grove, #04-06 Immunos, Biopolis 138648, Singapore. Phone: 65.64070006; Fax: 65.64642057; E-mail: [email protected].
Authorship note: Terk-Shin Teng and Suan-Sin Foo contributed equally to this work.
Find articles by Simamarta, D. in: JCI | PubMed | Google Scholar
1Singapore Immunology Network, Agency for Science, Technology and Research (A*STAR), Biopolis, Singapore. 2Department of Biochemistry, Yong Loo Lin School of Medicine, and 3NUS Graduate School for Integrative Sciences and Engineering, National University of Singapore, Singapore. 4Institute of Technology, University of Tartu, Tartu, Estonia. 5Communicable Disease Centre, Tan Tock Seng Hospital, Singapore. 6Department of Physiology, Yong Loo Lin School of Medicine, National University of Singapore, Singapore.
Address correspondence to: Lisa F.P. Ng, Laboratory of Chikungunya Virus Immunity, Singapore Immunology Network, A*STAR, 8A Biomedical Grove, #04-06 Immunos, Biopolis 138648, Singapore. Phone: 65.64070028; Fax: 65.64642057; E-mail: [email protected]. Or to: Keh-Chuang Chin, Laboratory of Gene Regulation in Inflammation, Singapore Immunology Network, A*STAR, 8A Biomedical Grove, #04-06 Immunos, Biopolis 138648, Singapore. Phone: 65.64070006; Fax: 65.64642057; E-mail: [email protected].
Authorship note: Terk-Shin Teng and Suan-Sin Foo contributed equally to this work.
Find articles by Lum, F. in: JCI | PubMed | Google Scholar
1Singapore Immunology Network, Agency for Science, Technology and Research (A*STAR), Biopolis, Singapore. 2Department of Biochemistry, Yong Loo Lin School of Medicine, and 3NUS Graduate School for Integrative Sciences and Engineering, National University of Singapore, Singapore. 4Institute of Technology, University of Tartu, Tartu, Estonia. 5Communicable Disease Centre, Tan Tock Seng Hospital, Singapore. 6Department of Physiology, Yong Loo Lin School of Medicine, National University of Singapore, Singapore.
Address correspondence to: Lisa F.P. Ng, Laboratory of Chikungunya Virus Immunity, Singapore Immunology Network, A*STAR, 8A Biomedical Grove, #04-06 Immunos, Biopolis 138648, Singapore. Phone: 65.64070028; Fax: 65.64642057; E-mail: [email protected]. Or to: Keh-Chuang Chin, Laboratory of Gene Regulation in Inflammation, Singapore Immunology Network, A*STAR, 8A Biomedical Grove, #04-06 Immunos, Biopolis 138648, Singapore. Phone: 65.64070006; Fax: 65.64642057; E-mail: [email protected].
Authorship note: Terk-Shin Teng and Suan-Sin Foo contributed equally to this work.
Find articles by Teo, T. in: JCI | PubMed | Google Scholar
1Singapore Immunology Network, Agency for Science, Technology and Research (A*STAR), Biopolis, Singapore. 2Department of Biochemistry, Yong Loo Lin School of Medicine, and 3NUS Graduate School for Integrative Sciences and Engineering, National University of Singapore, Singapore. 4Institute of Technology, University of Tartu, Tartu, Estonia. 5Communicable Disease Centre, Tan Tock Seng Hospital, Singapore. 6Department of Physiology, Yong Loo Lin School of Medicine, National University of Singapore, Singapore.
Address correspondence to: Lisa F.P. Ng, Laboratory of Chikungunya Virus Immunity, Singapore Immunology Network, A*STAR, 8A Biomedical Grove, #04-06 Immunos, Biopolis 138648, Singapore. Phone: 65.64070028; Fax: 65.64642057; E-mail: [email protected]. Or to: Keh-Chuang Chin, Laboratory of Gene Regulation in Inflammation, Singapore Immunology Network, A*STAR, 8A Biomedical Grove, #04-06 Immunos, Biopolis 138648, Singapore. Phone: 65.64070006; Fax: 65.64642057; E-mail: [email protected].
Authorship note: Terk-Shin Teng and Suan-Sin Foo contributed equally to this work.
Find articles by Lulla, A. in: JCI | PubMed | Google Scholar
1Singapore Immunology Network, Agency for Science, Technology and Research (A*STAR), Biopolis, Singapore. 2Department of Biochemistry, Yong Loo Lin School of Medicine, and 3NUS Graduate School for Integrative Sciences and Engineering, National University of Singapore, Singapore. 4Institute of Technology, University of Tartu, Tartu, Estonia. 5Communicable Disease Centre, Tan Tock Seng Hospital, Singapore. 6Department of Physiology, Yong Loo Lin School of Medicine, National University of Singapore, Singapore.
Address correspondence to: Lisa F.P. Ng, Laboratory of Chikungunya Virus Immunity, Singapore Immunology Network, A*STAR, 8A Biomedical Grove, #04-06 Immunos, Biopolis 138648, Singapore. Phone: 65.64070028; Fax: 65.64642057; E-mail: [email protected]. Or to: Keh-Chuang Chin, Laboratory of Gene Regulation in Inflammation, Singapore Immunology Network, A*STAR, 8A Biomedical Grove, #04-06 Immunos, Biopolis 138648, Singapore. Phone: 65.64070006; Fax: 65.64642057; E-mail: [email protected].
Authorship note: Terk-Shin Teng and Suan-Sin Foo contributed equally to this work.
Find articles by Yeo, N. in: JCI | PubMed | Google Scholar
1Singapore Immunology Network, Agency for Science, Technology and Research (A*STAR), Biopolis, Singapore. 2Department of Biochemistry, Yong Loo Lin School of Medicine, and 3NUS Graduate School for Integrative Sciences and Engineering, National University of Singapore, Singapore. 4Institute of Technology, University of Tartu, Tartu, Estonia. 5Communicable Disease Centre, Tan Tock Seng Hospital, Singapore. 6Department of Physiology, Yong Loo Lin School of Medicine, National University of Singapore, Singapore.
Address correspondence to: Lisa F.P. Ng, Laboratory of Chikungunya Virus Immunity, Singapore Immunology Network, A*STAR, 8A Biomedical Grove, #04-06 Immunos, Biopolis 138648, Singapore. Phone: 65.64070028; Fax: 65.64642057; E-mail: [email protected]. Or to: Keh-Chuang Chin, Laboratory of Gene Regulation in Inflammation, Singapore Immunology Network, A*STAR, 8A Biomedical Grove, #04-06 Immunos, Biopolis 138648, Singapore. Phone: 65.64070006; Fax: 65.64642057; E-mail: [email protected].
Authorship note: Terk-Shin Teng and Suan-Sin Foo contributed equally to this work.
Find articles by Koh, E. in: JCI | PubMed | Google Scholar
1Singapore Immunology Network, Agency for Science, Technology and Research (A*STAR), Biopolis, Singapore. 2Department of Biochemistry, Yong Loo Lin School of Medicine, and 3NUS Graduate School for Integrative Sciences and Engineering, National University of Singapore, Singapore. 4Institute of Technology, University of Tartu, Tartu, Estonia. 5Communicable Disease Centre, Tan Tock Seng Hospital, Singapore. 6Department of Physiology, Yong Loo Lin School of Medicine, National University of Singapore, Singapore.
Address correspondence to: Lisa F.P. Ng, Laboratory of Chikungunya Virus Immunity, Singapore Immunology Network, A*STAR, 8A Biomedical Grove, #04-06 Immunos, Biopolis 138648, Singapore. Phone: 65.64070028; Fax: 65.64642057; E-mail: [email protected]. Or to: Keh-Chuang Chin, Laboratory of Gene Regulation in Inflammation, Singapore Immunology Network, A*STAR, 8A Biomedical Grove, #04-06 Immunos, Biopolis 138648, Singapore. Phone: 65.64070006; Fax: 65.64642057; E-mail: [email protected].
Authorship note: Terk-Shin Teng and Suan-Sin Foo contributed equally to this work.
Find articles by Chow, A. in: JCI | PubMed | Google Scholar
1Singapore Immunology Network, Agency for Science, Technology and Research (A*STAR), Biopolis, Singapore. 2Department of Biochemistry, Yong Loo Lin School of Medicine, and 3NUS Graduate School for Integrative Sciences and Engineering, National University of Singapore, Singapore. 4Institute of Technology, University of Tartu, Tartu, Estonia. 5Communicable Disease Centre, Tan Tock Seng Hospital, Singapore. 6Department of Physiology, Yong Loo Lin School of Medicine, National University of Singapore, Singapore.
Address correspondence to: Lisa F.P. Ng, Laboratory of Chikungunya Virus Immunity, Singapore Immunology Network, A*STAR, 8A Biomedical Grove, #04-06 Immunos, Biopolis 138648, Singapore. Phone: 65.64070028; Fax: 65.64642057; E-mail: [email protected]. Or to: Keh-Chuang Chin, Laboratory of Gene Regulation in Inflammation, Singapore Immunology Network, A*STAR, 8A Biomedical Grove, #04-06 Immunos, Biopolis 138648, Singapore. Phone: 65.64070006; Fax: 65.64642057; E-mail: [email protected].
Authorship note: Terk-Shin Teng and Suan-Sin Foo contributed equally to this work.
Find articles by Leo, Y. in: JCI | PubMed | Google Scholar
1Singapore Immunology Network, Agency for Science, Technology and Research (A*STAR), Biopolis, Singapore. 2Department of Biochemistry, Yong Loo Lin School of Medicine, and 3NUS Graduate School for Integrative Sciences and Engineering, National University of Singapore, Singapore. 4Institute of Technology, University of Tartu, Tartu, Estonia. 5Communicable Disease Centre, Tan Tock Seng Hospital, Singapore. 6Department of Physiology, Yong Loo Lin School of Medicine, National University of Singapore, Singapore.
Address correspondence to: Lisa F.P. Ng, Laboratory of Chikungunya Virus Immunity, Singapore Immunology Network, A*STAR, 8A Biomedical Grove, #04-06 Immunos, Biopolis 138648, Singapore. Phone: 65.64070028; Fax: 65.64642057; E-mail: [email protected]. Or to: Keh-Chuang Chin, Laboratory of Gene Regulation in Inflammation, Singapore Immunology Network, A*STAR, 8A Biomedical Grove, #04-06 Immunos, Biopolis 138648, Singapore. Phone: 65.64070006; Fax: 65.64642057; E-mail: [email protected].
Authorship note: Terk-Shin Teng and Suan-Sin Foo contributed equally to this work.
Find articles by Merits, A. in: JCI | PubMed | Google Scholar
1Singapore Immunology Network, Agency for Science, Technology and Research (A*STAR), Biopolis, Singapore. 2Department of Biochemistry, Yong Loo Lin School of Medicine, and 3NUS Graduate School for Integrative Sciences and Engineering, National University of Singapore, Singapore. 4Institute of Technology, University of Tartu, Tartu, Estonia. 5Communicable Disease Centre, Tan Tock Seng Hospital, Singapore. 6Department of Physiology, Yong Loo Lin School of Medicine, National University of Singapore, Singapore.
Address correspondence to: Lisa F.P. Ng, Laboratory of Chikungunya Virus Immunity, Singapore Immunology Network, A*STAR, 8A Biomedical Grove, #04-06 Immunos, Biopolis 138648, Singapore. Phone: 65.64070028; Fax: 65.64642057; E-mail: [email protected]. Or to: Keh-Chuang Chin, Laboratory of Gene Regulation in Inflammation, Singapore Immunology Network, A*STAR, 8A Biomedical Grove, #04-06 Immunos, Biopolis 138648, Singapore. Phone: 65.64070006; Fax: 65.64642057; E-mail: [email protected].
Authorship note: Terk-Shin Teng and Suan-Sin Foo contributed equally to this work.
Find articles by Chin, K. in: JCI | PubMed | Google Scholar
1Singapore Immunology Network, Agency for Science, Technology and Research (A*STAR), Biopolis, Singapore. 2Department of Biochemistry, Yong Loo Lin School of Medicine, and 3NUS Graduate School for Integrative Sciences and Engineering, National University of Singapore, Singapore. 4Institute of Technology, University of Tartu, Tartu, Estonia. 5Communicable Disease Centre, Tan Tock Seng Hospital, Singapore. 6Department of Physiology, Yong Loo Lin School of Medicine, National University of Singapore, Singapore.
Address correspondence to: Lisa F.P. Ng, Laboratory of Chikungunya Virus Immunity, Singapore Immunology Network, A*STAR, 8A Biomedical Grove, #04-06 Immunos, Biopolis 138648, Singapore. Phone: 65.64070028; Fax: 65.64642057; E-mail: [email protected]. Or to: Keh-Chuang Chin, Laboratory of Gene Regulation in Inflammation, Singapore Immunology Network, A*STAR, 8A Biomedical Grove, #04-06 Immunos, Biopolis 138648, Singapore. Phone: 65.64070006; Fax: 65.64642057; E-mail: [email protected].
Authorship note: Terk-Shin Teng and Suan-Sin Foo contributed equally to this work.
Find articles by Ng, L. in: JCI | PubMed | Google Scholar
Authorship note: Terk-Shin Teng and Suan-Sin Foo contributed equally to this work.
Published November 19, 2012 - More info
J Clin Invest. 2012;122(12):4447–4460. https://doi.org/10.1172/JCI63120.
© 2012 The American Society for Clinical Investigation
Received: June 27, 2012; Accepted: September 20, 2012
-
Abstract
Chikungunya virus (CHIKV) is a mosquito-borne arthralgia arbovirus that is reemergent in sub-Saharan Africa and Southeast Asia. CHIKV infection has been shown to be self-limiting, but the molecular mechanisms of the innate immune response that control CHIKV replication remain undefined. Here, longitudinal transcriptional analyses of PBMCs from a cohort of CHIKV-infected patients revealed that type I IFNs controlled CHIKV infection via RSAD2 (which encodes viperin), an enigmatic multifunctional IFN-stimulated gene (ISG). Viperin was highly induced in monocytes, the major target cell of CHIKV in blood. Anti-CHIKV functions of viperin were dependent on its localization in the ER, and the N-terminal amphipathic α-helical domain was crucial for its antiviral activity in controlling CHIKV replication. Furthermore, mice lacking Rsad2 had higher viremia and severe joint inflammation compared with wild-type mice. Our data demonstrate that viperin is a critical antiviral host protein that controls CHIKV infection and provide a preclinical basis for the design of effective control strategies against CHIKV and other reemerging arthrogenic alphaviruses.
-
Introduction
Pattern recognition receptors (PRRs) essential for the detection of specific motifs in pathogens are called pathogen-associated molecular patterns (PAMPs) (1). Upon induction after pathogen invasion, PRRs elicit a cascade of signaling pathways that ultimately converge to produce type I IFNs, namely IFN-α and IFN-β, that coordinate defenses against viruses and other pathogens during infection (1, 2). Following the activation of an antiviral state, type I IFNs bind to IFN-α/IFN-β receptors to activate another essential signaling component, the JAK/STAT pathway (3, 4), for the regulation of a positive feedback loop that amplifies type I IFN production (5). In parallel, activation of type I IFN–mediated signaling pathways result in the induction of numerous IFN-stimulated genes (ISGs) that inhibit viral replication and mediate clearance of viruses from the host (6).
Although type I IFNs are fundamental to the innate immune response, how specific ISGs are regulated by type I IFNs following activation of PRRs to combat different pathogens remains to be fully elucidated, especially for several clinically important reemerging viral diseases, such as chikungunya fever (CHIKF). This disease is caused by chikungunya virus (CHIKV), an alphavirus that has reemerged after a quiescent period of almost 50 years (7, 8) since the first reported outbreak in Tanzania in 1952 (9, 10). Unexpectedly, major outbreaks have been reported globally since late 2005, with millions of CHIKF cases (11–13).
Individuals infected with CHIKV normally exhibit a sudden onset of high fever together with symptoms including myalgia, rashes, headaches, and incapacitating arthralgia during the acute phase (14, 15). Symptoms are generally resolved within 7–10 days, indicative of innate immune response involvement (7, 12). However, some patients are plagued with chronic arthralgia that could persist for months or years (8, 16). Currently, there are no effective anti-CHIKV therapies or commercial CHIKV vaccines available.
CHIKV was reported to be a potent inducer of type I IFNs during infection as early as the 1960s (17). However, despite the induction of an active type I IFN response in patients during the acute phase of the disease (18, 19), the mechanisms involved remain unknown. Although recent studies have shed light on the interplay between type I IFNs and CHIKV during infection (20–24), how CHIKV replication is controlled by ISGs induced by type I IFNs remains to be elucidated (25, 26). RSAD2 (encoding virus-inhibitory protein, endoplasmic reticulum–associated, interferon-inducible [viperin]; also known as cig5) was first reported as an antiviral gene induced by human cytomegalovirus and IFNs (27). More recently, viperin has been shown to be an important player in the innate immune response against clinically important viruses, including influenza virus, West Nile virus, dengue virus, and HCV, as part of the innate immune response (28–31). Recent studies have shown that viperin is induced during CHIKV infection (32) and that its expression inhibits CHIKV infection in vitro (26). However, the mechanisms involved are unknown.
Here, we demonstrated the antiviral role of viperin in CHIKV infection by studying the longitudinal transcriptional profiles of the innate immune response in PBMCs from a cohort of 24 CHIKV-infected patients (18). An active type I IFN response was characterized by induction of IFNA and IFNB and correlated to induction of IRF3 and IRF7, both crucial regulators of type I IFNs (33). Positive correlations were also observed between viperin and type I IFNs during the acute phase of infection, when levels of viremia were shown to directly influence the expression of these genes. Viperin was highly induced in monocytes, the major PBMC subset targeted by CHIKV in the blood. Further characterization studies revealed that overexpression of viperin significantly suppressed CHIKV infection and replication. Intriguingly, the N-terminal ER targeting domain of viperin was shown to be critical for suppressing CHIKV replication. Furthermore, the absence of viperin expression in Rsad2–/– mice resulted in higher viremia and more pronounced joint inflammation, providing the first direct evidence of viperin inhibition in vivo on CHIKV replication and pathology.
-
Results
Transcriptional profiles of CHIKV-induced innate response in humans. The role of type I IFNs in the innate response against CHIKV infection has been demonstrated in vitro in various cell types and animal models (20, 21, 24, 34, 35). To further understand their functions in patients, we analyzed the transcriptional profiles of type I IFNs and their associated genes in PBMCs collected longitudinally from 24 CHIKV-infected patients. Expression of IFNA, IFNB, and IRF7, a key transcriptional regulator of type I IFN production, were significantly induced in all patients compared with healthy controls (Figure 1 and Supplemental Figure 1; supplemental material available online with this article; doi: 10.1172/JCI63120DS1). The primers used to detect IFNA expression were against a conserved region of all IFNA subtypes (36). Conversely, expression of IRF3, another key regulator of type I IFN expression (33), remained relatively constant (Figure 1B and Supplemental Figure 1). More importantly, we found — for the first time to our knowledge in patients — that after the induction of type I IFN response, numerous genes with known antiviral activities were induced during the acute phase of infection, at median 4 days post illness onset (Figure 1 and Supplemental Figure 1). RIG-I and MDA5, which encode for proteins that act as PRRs and detect viruses to modulate signaling pathways in innate immune response, as well as IPS-1, an adaptor molecule involved in RIG-I– and MDA5-mediated signaling (2), were all significantly induced. Furthermore, CHIKV infection also induced ISGs known to inhibit viral protein translation and replication or to induce apoptosis (6, 35, 37, 38), such as ISG15, ISG54, ISG56, and dsRNA-dependent protein kinase (PKR) (Figure 1 and Supplemental Figure 1).
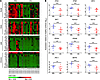 Figure 1
Figure 1Expression profiles of type I IFNs and related ISGs in CHIKV-infected patients. (A) Expression profiles of type I IFNs and related ISGs in PBMCs of CHIKV-infected patients (n = 24) at various time points of the disease were analyzed by qRT-PCR. Data were normalized to GAPDH and presented as expression relative to the mean of healthy controls (n = 10). Data are presented by 2-way hierarchical clustering after dividing the patient cohort according to viral load levels into HVL (n = 11) and LVL (n = 13) groups. Each colored well in the 4 heatmaps represents the relative levels of expression of a particular gene (green, low expression; red, high expression). The different phases of the disease are defined as acute phase (median 4 days post illness onset), early convalescent phase (median 10 days post illness onset), late convalescent phase (4–6 weeks post illness onset) and chronic phase (2–3 months post illness onset). (B) Viral load influenced induction of the innate immune response. The relative expression of the indicated genes was compared between HVL and LVL groups during the acute phase of disease at median 4 days post illness onset. Data are mean ± SEM. **P < 0.01, ***P < 0.001, Mann-Whitney U test.
Interestingly, RSAD2, an ISG with a broad spectrum of antiviral functions (28–31, 39–41), was also significantly induced during CHIKV infection (Figure 1A and Supplemental Figure 1). Expression of IFNA, IFNB, IRF7, RSAD2, ISG15, ISG54, ISG56, RIG-I, MDA5, and PKR generally peaked during the acute phase of the disease and was sustained to the early convalescent phase (median 10 days after illness onset), before declining in the late convalescent (4–6 weeks) and chronic (2–3 months) phases (Figure 1A and Supplemental Figure 1). More importantly, positive correlations were observed between type I IFNs and IRF7, IRF3, RSAD2, ISG15, ISG54, ISG56, PKR, and RIG-I during the acute phase (Supplemental Figure 3), which suggests that these factors are induced as part of the innate immune response against CHIKV infection.
By segregating the cohort into high– and low–viral load (HVL and LVL, respectively) groups during the acute phase (18), we observed a distinct profile in the expression levels in individuals of the HVL group (Figure 1A). Notably, the induction of type I IFN responses and expression of their associated ISGs were significantly influenced by the amount of CHIKV present (Figure 1B). Taken together, these data indicate that viperin is induced as part of the type I IFN–mediated response during CHIKV infection and that its expression is driven by viral load levels.
Viperin is highly induced in monocytes during CHIKV infection. To characterize the type I IFN–mediated antiviral response in various PBMC subsets, we next performed in vitro infections using human whole blood at MOI 10. Heat-inactivated (HI) CHIKV was used as a control. Total PBMCs were isolated; CD14+ monocytes, CD19+ B cells, CD3+ T cells, and CD56+ NK cells were sorted by flow cytometry (Supplemental Figure 3, A and B); and their transcriptional profiles were analyzed. Consistent with our earlier findings (22), CD14+ monocytes were the major subsets targeted by CHIKV, followed by CD19+ B cells (Supplemental Figure 3C and Figure 2A). Control HI CHIKV did not infect and replicate in any subsets (Supplemental Figure 3C). Next, viral load and gene expression analyses were performed using total RNA extracted from both nonsorted and sorted PBMC subsets. Similar to the flow cytometry results, the level of negative-strand nsP1 RNA (an indicator of active viral replication) was detected mainly in monocytes and B cells, but not in T cells or NK cells (Figure 2B). These results clearly indicated that CHIKV infects and replicates actively in monocytes and B cells. Interestingly, expression profiles of the various sorted PBMC subsets revealed that expression of IFNA was induced mainly in monocytes, whereas expression of IFNB was induced in monocytes and B cells (Figure 2C). Interestingly, RSAD2 was induced mainly in monocytes, whereas expression of IRF7, IRF3, RIG-I, MDA5, IPS-1, and PKR was similar among the various subsets. Comparable Ct values were observed for the various genes tested in the different PBMC subset–sorted samples infected with either CHIKV or HI CHIKV, which indicates that induction of viperin in monocytes is specific to CHIKV infection and not caused by a difference in basal expression levels.
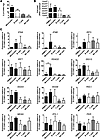 Figure 2
Figure 2Viperin is induced mainly in monocytes during human whole blood infection. (A) Whole blood was collected from healthy donors, and the total leukocyte count was determined before performing infection with HI CHIKV (control) or CHIKV (MOI 10) for 12 hours. Total PBMCs were purified, and a fraction was used for flow cytometry analysis to determine percent CHIKV Ag+ cells in the indicated PBMC subsets (see Supplemental Figure 3). Data are mean ± SD for each subset (n = 3). Horizontal dotted line represents the mean in HI CHIKV–infected controls. (B) Detection of CHIKV viral load in nonsorted and sorted PBMCs. Total RNA was extracted from each indicated cell population, and the amount of CHIKV viral load was determined by qRT-PCR using specific primers against the negative-strand nsP1 RNA. Data are mean ± SD (n = 3). Horizontal dotted line represents the mean in HI CHIKV–infected controls. (C) qRT-PCR was used to determine the expression profiles of type I IFNs and related ISGs in the sorted PBMC subsets as well as the nonsorted PBMC fractions. Data obtained were normalized to GAPDH and presented as expression relative to each cell population from controls infected with HI CHIKV. Data are mean ± SD (n = 3). *P < 0.05, 1-way ANOVA with Tukey’s post-test.
Viperin inhibits CHIKV infection in vitro. To determine whether viperin plays a role in controlling CHIKV infection, we use HEK 293T cells, a cell line highly susceptible to CHIKV, as a model for characterization. Using a MOI of 10, the kinetics of CHIKV infection in HEK 293T cells peaked at 12 hours post infection (hpi), with about 80% of cells detected as positive for CHIKV antigens (CHIKV Ag+), and reached a plateau at 24 hpi (Supplemental Figure 4A). Using 12 hpi as a reference for the peak of infection, we also demonstrated a MOI-dependent effect of CHIKV infectivity in HEK 293T cells (Supplemental Figure 4B). Next, HEK 293T cells were transiently transfected with a plasmid expressing viperin for 24 hours before being infected with CHIKV at MOI of 1, 2.5, and 5. We observed that overexpression of viperin in HEK 293T cells caused significant inhibition of CHIKV infection at MOI 1 and 2.5 compared with cells transfected with control vector (Supplemental Figure 4C). Furthermore, viral load analysis by negative-strand nsP1 RNA quantification clearly showed that viperin expression significantly suppressed CHIKV replication compared with cells transfected with control vector (Supplemental Figure 4D). Overexpression of viperin was also confirmed by Western blot (Supplemental Figure 4E).
Viperin is composed of 3 distinct domains with different functions (42). It contains an amphipathic α-helix domain at its N-terminus, followed by a conserved central domain with 3 cysteine residues arranged in a CxxxCxxC motif and a conserved C-terminal domain between species (Figure 3A). The amphipathic α-helix domain mediates targeting of viperin to the cytosolic side of ER and lipid droplets (43, 44) The central domain is homologous to the MoaA motif present in the family of radical S-adenosylmethionine (SAM) enzymes, which uses SAM as a cofactor to bind to proteins containing iron-sulfur clusters via the CxxxCxxC motif (45, 46). Herein, we refer to the central domain as the radical SAM catalytic domain, and it has been demonstrated to inhibit virus replication (29, 47). Even though the function of the C-terminal domain is poorly defined, it has been reported to play a role in inhibiting HCV replication (28).
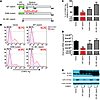 Figure 3
Figure 3Overexpression of viperin controls CHIKV replication. (A) Schematic representation of the domain organization of viperin and the various mutant constructs used. Viperin contains an ER targeting N-terminal amphipathic α-helical domain (aa 1–42) and a radical SAM catalytic domain (aa 77–209). WT viperin, SAM domain mutant of viperin (C83A/C87A/C90A), and a truncated viperin mutant (aa 43–361) are shown. (B) HEK 293T cells were transfected with the various plasmids for 24 hours before infection with HI CHIKV or CHIKV (MOI 2.5). Cells were harvested at 12 hpi and analyzed for CHIKV infectivity by flow cytometry. Histogram plots of percent CHIKV Ag+ cells in various cell populations are representative of 3 independent experiments. (C) Graphical presentation of histogram plots in B. Data are expressed as mean ± SD of percent CHIKV Ag+ cells relative to vector-transfected cells infected with CHIKV (n = 3). *P < 0.05, 1-way ANOVA with Tukey’s post-test. Horizontal dotted line represents the mean percent CHIKV Ag+ cells detected in control. (D) Viral load was determined by qRT-PCR using specific primers against the negative-strand nsP1 RNA. Data are mean ± SD (n = 3). Horizontal dotted line represents the mean amount of RNA detected in control cells. **P < 0.01. (E) Viperin expression was detected with anti–c-Myc antibody. CHIKV nsP2 expression in the infected cells was detected with anti-nsP2 antibody. Detection of α-actin expression served as a loading control. Immunoblots are representative of 3 independent experiments.
To identify the functional domain of viperin required for inhibiting CHIKV replication, we next transfected a series of viperin mutant constructs (Figure 3A) into HEK 293T cells and analyzed for virus infectivity. Flow cytometry analysis showed that antiviral properties of viperin against CHIKV was significantly abolished upon deletion of the N-terminal amphipathic α-helix domain (i.e., aa 43–361 of viperin), whereas mutations to the radical SAM domain (C83A/C87A/C90A) did not significantly affect virus infectivity compared with cells expressing WT viperin (Figure 3, B and C). Consistent with these findings, viral load analysis revealed that expression of either the N-terminal amphipathic α-helix domain deletion mutant or the SAM mutant did not inhibit CHIKV replication compared with cells expressing WT viperin (Figure 3D). These observations were further confirmed at the protein level by Western blot, in which expression of WT viperin was associated with reduced expression of CHIKV nsP2 (Figure 3E), an RNA helicase/protease involved in replication.
The N-terminal amphipathic α-helical domain of viperin controls CHIKV replication. The N-terminal amphipathic α-helical domain is required for targeting viperin to the ER and lipid droplets (43, 44). Therefore, we next sought to determine whether the N-terminal amphipathic α-helix (aa 1–42) domain of viperin is sufficient to suppress CHIKV replication. To verify this, we overexpressed the GFP-tagged N-terminal α-helical domain of viperin as well as another mutant with both the N-terminal amphipathic α-helical and radical SAM domains (Figure 4A) in HEK 293T cells to assess their ability to suppress CHIKV infection. Intriguingly, expression of the GFP–N-terminal amphipathic α-helical domain was able to suppress CHIKV infection and replication significantly — to a level comparable to GFP–WT viperin — versus cells expressing GFP alone (Figure 4, B–D). Moreover, expression of the mutant with both the N-terminal amphipathic α-helical and radical SAM domains (aa 1–220) also inhibited CHIKV replication, similar to cells expressing the GFP–N-terminal amphipathic α-helical domain (Figure 4, B–D). This indicates that the antiviral function of viperin against CHIKV is not dependent on the enzymatic radical SAM domain, as expression of the GFP–N-terminal α-helical domain reduced CHIKV nsP2 expression to an extent similar to that of cells overexpressing GFP–WT viperin (Figure 4, E and F). Furthermore, consistent with these observations, time-lapse movies revealed a difference in the kinetics of CHIKV infection in control cells expressing GFP alone (Supplemental Video 1) compared with cells expressing either GFP–WT viperin (Supplemental Video 2) or the GFP–N-terminal α-helical domain of viperin (Supplemental Video 3). GFP-expressing cells started to exhibit fluorescence of mCherry-tagged CHIKV at about 6 hpi, whereas cells expressing either GFP–WT viperin or GFP–N-terminal α-helical domain of viperin exhibited a delayed fluorescence signal at about 10 hpi. Together, these data defined the importance of the N-terminal α-helical domain of viperin in controlling CHIKV infection and replication, possibly mediated by targeting viperin to the ER.
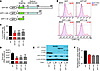 Figure 4
Figure 4The N-terminal amphipathic α-helical domain of viperin controls CHIKV replication. (A) Schematic representation of the domain organization of GFP–WT viperin (GFP-WT), the GFP–N-terminal α-helical domain of viperin (GFP-1-42), and another GFP-tagged viperin mutant with both the N-terminal amphipathic α-helical and radical SAM domains (GFP-1-220). (B) HEK 293T cells were transfected with the various plasmids for 24 hours before infection with either HI CHIKV (control) or CHIKV at MOI 2.5. Cells were analyzed for CHIKV infectivity as described in Figure 3B. Histogram plots of percent CHIKV Ag+ cells in the indicated cell populations are representative of 3 independent experiments. (C) Graphical presentation of histogram plots in B. Data are mean ± SD of percent CHIKV Ag+ cells relative to vector-transfected cells infected with CHIKV (n = 3). ***P < 0.001, 1-way ANOVA with Tukey’s post-test. Horizontal dotted line represents the mean percent CHIKV Ag+ cells in controls. (D) Viral load was determined by qRT-PCR as described in Figure 3D. Data are mean ± SD (n = 3). **P < 0.01, 1-way ANOVA with Tukey’s post-test. Horizontal dotted line represents the mean amount of RNA detected in controls. (E) Viperin expression was detected with anti-GFP antibody. CHIKV nsP2 expression in the infected cells was detected with anti-nsP2 antibody. Detection of α-actin expression served as a loading control. Immunoblots are representative of 2 independent experiments. (F) Densitometric analysis of the nsP2 band in E was performed with NIH ImageJ software and normalized against actin band before being expressed relative to GFP-transfected cells.
CHIKV colocalizes with viperin during infection. To provide insight into how viperin inhibits CHIKV replication, we next investigated the localization of viperin and CHIKV in infected HEK 293T cells. Overexpression of viperin did not affect the cellular distribution of CHIKV during infection, and viperin colocalized with CHIKV around the perinuclear region in infected cells (Figure 5). Moreover, CHIKV nsP2 was observed to localize in the ER region during CHIKV infection (Supplemental Figure 5), further suggesting the importance of viperin localization in the ER for controlling CHIKV replication.
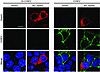 Figure 5
Figure 5Subcellular localization of CHIKV and viperin during CHIKV infection. HEK 293T cells were transfected with either vector or WT viperin for 24 hours before infection with HI CHIKV or CHIKV (MOI 2.5). Cells were fixed at 12 hpi and stained for CHIKV Ag (green), viperin (red), and DAPI. Images are representative of 3 independent experiments. Scale bar: 10 μm.
We showed above that the N-terminal amphipathic α-helical domain, which targets viperin to the ER, was essential for its antiviral function (Figure 4). Moreover, viperin has previously been reported to redistribute from the ER to Golgi and mitochondria during infection by human cytomegalovirus (27, 47). Therefore, to assess whether CHIKV infection affects the localization of viperin in the ER, we next transfected HEK 293T cells with either GFP–WT viperin or GFP–N-terminal amphipathic α-helical domain of viperin and examined their colocalization with ER, using anti-calreticulin antibody as an ER marker, during CHIKV infection (Figure 6). Surprisingly, compared with cells infected with HI CHIKV, CHIKV infection did not disrupt the colocalization of viperin with ER, as both GFP–WT viperin and GFP–N-terminal amphipathic α-helical domain of viperin colocalized distinctly with CHIKV in the ER of infected cells (Figure 6).
 Figure 6
Figure 6CHIKV and viperin colocalize in the ER during CHIKV infection. HEK 293T cells were transfected with GFP, GFP–WT viperin, or GFP–N-terminal α-helical domain of viperin for 24 hours before infection with HI CHIKV or CHIKV (MOI 2.5). Cells were fixed at 12 hpi and stained for GFP (green), calreticulin (red), and CHIKV Ag (blue). Images are representative of 2 independent experiments. Scale bar: 10 μm. Colocalization is indicated by arrowheads.
Viperin directly controls CHIKV replication in vivo and modulates CHIKV-induced inflammation. To investigate viperin’s role in mounting an immune response against CHIKV infection, the viperin-mediated antiviral response was first measured in primary fibroblasts isolated from Rsad2–/– mice. Fibroblasts lacking Rsad2 showed significantly increased susceptibility to CHIKV infection and virus replication compared with WT fibroblasts (Figure 7, A and B). To gain further insights into the mechanism by which viperin induces an antiviral state, the levels of IFN production and ISG expression were analyzed in detail (Figure 7C). Essentially, IFN production was not abolished or reduced in Rsad2–/– fibroblasts, but was significantly higher than in WT fibroblasts (Figure 7C). This observation strongly suggests that the susceptibility to CHIKV infection and replication in Rsad2–/– fibroblasts (Figure 7, A and B) was independent of IFN production. The impaired expression of ISG15, RIG-I, and other IFN-related host factors observed here suggest that the increased in CHIKV infection and replication in Rsad2–/– fibroblasts may be due to a specific defect in the induction of other antiviral genes, as described by other studies (25). To further investigate whether loss of viperin expression affects type I IFN–mediated antiviral responses, we pretreated WT and Rsad2–/– fibroblasts with type I IFNs for 4 hours prior to CHIKV infection. Both IFN-α and IFN-β effectively inhibited CHIKV infection and replication to a comparable level in WT and Rsad2–/– fibroblasts (Figure 7, D and E), which demonstrated the importance of type I IFNs in limiting CHIKV infection.
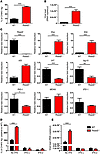 Figure 7
Figure 7Viperin controls CHIKV infection in mice. (A) Primary tail fibroblasts isolated from WT and Rsad2–/– mice were infected with CHIKV (MOI 10) or were mock infected (control) for 12 hours. Percent CHIKV Ag+ cells was measured by flow cytometry. Data (mean ± SD) are representative of 3 independent experiments. ***P < 0.005, unpaired t test. (B) Viral load was determined by qRT-PCR and presented as mean ± SD (n = 3). ***P < 0.005, unpaired t test. (C) qRT-PCR was used to determine expression of the indicated genes in fibroblasts after infection. Data are fold expression ± SD relative to WT. ***P < 0.005, **P < 0.01, *P < 0.05, unpaired t test. (D) WT and Rsad2–/– fibroblasts were pretreated with IFN-α or IFN-β (1,000 IU/ml) for 4 hours before infection with CHIKV (MOI 10) for 12 hours. Mock infection was used as control. Percent CHIKV Ag+ cells was measured as described in A; data are mean ± SD of percent CHIKV Ag+ cells relative to untreated WT (n = 3). ***P < 0.005, unpaired t test. (E) Viral load was quantified as described in B and presented as mean ± SD (n = 3). ***P < 0.005, unpaired t test.
Fundamentally, the importance of viperin in controlling CHIKV replication was confirmed by in vivo CHIKV infection in Rsad2–/– mice. Viral load analysis in the footpad of WT and Rsad2–/– mice revealed that loss of Rsad2 expression resulted in a significant increase of CHIKV at 1 day post infection (dpi; Figure 8A). Comparison of transcriptional profiles in the footpad of WT and Rsad2–/– mice showed that expression levels of Rsad2 and Ifna were markedly induced in WT mice, whereas expression levels of Irf3, Irf7, Isg15, RIG-I, MDA5, and IPS-1 were not affected (Figure 8B). To further investigate the role of viperin in pathology caused by CHIKV infection, infected WT and Rsad2–/– animals were monitored daily for survival, viremia, and swelling at the joint. Although the absence of Rsad2 expression did not affect the mortality of the animals, significantly higher viremia levels were observed during the course of disease in Rsad2–/– versus WT mice (Figure 8C). This was a result of a higher rate of virus replication in the joint prior to virus dissemination. As expected, CHIKV-infected Rsad2–/– mice also exhibited more severe and pronounced swelling in the joint at 2–6 dpi (Figure 8D). Moreover, histological analysis of infected joints during the peak of swelling at 6 dpi revealed that Rsad2–/– mice exhibited more pronounced subcutaneous edema and cellular infiltration (Figure 8E). Immunohistochemistry (IHC) staining of the infected joint with mouse macrophage-specific F4/80 antibody showed an increase infiltration of macrophages into the surrounding connective tissues in Rsad2–/– compared with WT mice (Figure 8E). Collectively, these findings confirmed the critical role of viperin in controlling CHIKV replication and modulating CHIKV-induced inflammation during infection.
 Figure 8
Figure 8Viperin modulates CHIKV replication and disease pathology in mice. (A) WT and Rsad2–/– mice (n = 4–6 per group) were infected with 106 PFU CHIKV in the footpad. Viral RNA was isolated from the footpads a 1 dpi and quantified by qRT-PCR against the negative-strand nsP1 RNA. *P < 0.05, Mann-Whitney U test. (B) qRT-PCR was used to determine the expression of the indicated genes in infected footpads of mice. Data (mean ± SD) were normalized to GAPDH and shown as fold expression relative to WT. *P < 0.05, Mann-Whitney U test. (C) WT and Rsad2–/– mice (n = 6 per group) were infected with 106 PFU CHIKV in the footpad. Blood was collected daily at 0–14 dpi for quantification of viral load as described in A (mean ± SD). *P < 0.05, **P < 0.01, Mann-Whitney U test. (D) Quantification of joint swelling in WT and Rsad2–/– mice. Size of infected joint was measured daily for 14 days and expressed as disease score relative to day 0 (preinfection). Data (mean ± SD) are representative of 2 independent experiments. *P < 0.05, **P < 0.01, Mann-Whitney U test. (E) Histology of CHIKV-induced inflammation in footpads of WT and Rsad2–/– mice was analyzed by H&E staining and IHC staining with anti-F4/80 antibody at 6 dpi. Boxed regions are shown at higher magnification at right. Scale bars: 100 μm.
-
Discussion
The complexity of IFN-mediated signaling, coupled with its regulation of more than 300 ISGs (48, 49), underscored the challenge of revealing how the innate immune response reacts to infection by a pathogen in order to control its pathogenesis. Despite extensive documentation about the induction of type I IFNs during CHIKV infection (22–24, 34), only recently have the mechanisms of type I IFN control of CHIKV replication begun to be unraveled (24, 25, 35, 41, 50). However, the identities of ISGs that directly control and inhibit CHIKV replication still remain elusive. Recently, a comprehensive overexpression screen of more than 380 ISGs conducted against a panel of viruses, including CHIKV, revealed the involvement of several ISGs in controlling virus replication (26, 51).
In our earlier studies, we showed that an innate immune response characterized by IFN-α production is induced in CHIKV-infected patients (18, 19, 22). In the present study, we showed in PBMCs of CHIKV-infected patients that both IFNA and IFNB were highly induced during the acute phase of the disease and that their expression correlated positively with induction of various ISGs, signifying the activation of an anti-CHIKV state. Among the ISGs analyzed, IRF7, RSAD2, ISG15, ISG54, ISG56, RIG-I, and PKR exhibited expression profiles similar to those of both IFNA and IFNB, which indicates that these ISGs are involved in the innate immune response against CHIKV. ISG15 was also recently shown to play a role in controlling CHIKV infection in mice (35). Notably, our data revealed that CHIKV viral load dictated the induction of IFNA, IFNB, IRF7, and RSAD2 during the acute phase. We previously reported that patients in this cohort with higher viral load during the acute phase did not have persistent arthralgia (18). These findings suggest that viperin plays a protective role in limiting CHIKV replication as part of the innate immune response. We also showed previously that CHIKV infection in monocytes triggered an active innate immune response (22). Here, we demonstrated that viperin was highly induced in monocytes, the major PBMC subset targeted by CHIKV during the acute phase of infection.
Viperin has been shown to inhibit replication of different viruses via different mechanisms (42). Previously, it was demonstrated to bind and inhibit farnesyl diphosphate synthase to perturb regulation of the lipid raft on the plasma membrane, thereby inhibiting virus budding and release in influenza A virus (31) and HIV (52) infection. The radical SAM domain of viperin was also shown to be crucial for HCV replication (28). However, even though viperin was recently implicated in controlling CHIKV infection (26), the underlying molecular mechanisms involved remain unclear. We showed here that the N-terminal amphipathic α-helical domain of viperin was critical in suppressing CHIKV replication, whereas mutations that abolished the catalytic activity of the radical SAM domain of viperin (i.e., C83A/C87A/C90A) reduced inhibition of CHIKV replication. Although the SAM mutant contains the N-terminal amphipathic α-helical domain, it did not inhibit CHIKV replication significantly. A recent study demonstrated that the C83A/C87A/C90A mutations at the SAM domain resulted in a loss of conformational stability, inducing partial unfolding and aggregation of viperin that affected antiviral activity (53). This may explain the loss of CHIKV inhibition by the SAM mutant observed in our study. Nevertheless, we demonstrated that expression of the N-terminal amphipathic α-helical domain of viperin, a mere 42–amino acid fragment, was sufficient to inhibit CHIKV infection. This strongly indicates that the N-terminal amphipathic α-helical domain, which mediates targeting and localization of viperin to the ER and lipid droplets (43, 44), is essential for anti-CHIKV action.
Given the function of ER as a crucial cell organelle that regulates the biosynthesis of proteins, lipids, and carbohydrates (54), it is not surprising that ER is targeted as one of the key host organelles by CHIKV to support its replication. This is because CHIKV and other RNA viruses are dependent on the host cell machinery to initiate translation of nonstructural proteins following the release of their coding mRNA after infection (55). Immunofluorescence staining revealed that CHIKV and viperin distinctly colocalized with ER during infection, which further suggests that viperin exerts its control on CHIKV infection in the ER. However, contrary to other infections, such as with human cytomegalovirus (27), viperin was not redistributed after CHIKV infection, indicative of a different mode of antiviral action. Furthermore, replication complexes of alphaviruses have been reported to localize in the proximity of the ER in order to drive synthesis and replication of viral genomes, leading to consequential translation of the viral structural proteins at the ER required for packaging and formation of new virus progeny (56, 57). In addition, we demonstrated that CHIKV nsP2, a RNA helicase/protease mediating replication, colocalized around the periphery of the ER during CHIKV infection.
CHIKV nsP2 interacts with multiple host proteins to induce cellular shutoff and promote viral replication (58). We showed here that overexpression of viperin was strongly associated with reduced CHIKV nsP2 expression levels, but it is still unclear whether the inhibitory effect of viperin on CHIKV nsP2 is direct or indirect. Interestingly, CHIKV nsP2 has previously been demonstrated to counteract the inhibitory effects on type I IFN (21), which further illuminates the continuous tug-of-war evolution of host/pathogen interactions.
Although it is well established that production of viral dsRNA intermediates triggers type I IFN induction during virus replication, it could also lead to an apparent opposing event via activation of PKR to phosphorylate translation initiation factor 2α (EIF2A) and inhibit translation of most host cellular and viral mRNA (1, 59). This intricate interplay between the innate host response and virus infection was also demonstrated to involve GADD34, a subunit of protein phosphatase 1 (PP1) induced in response to CHIKV and ER stress to control virus infection (60, 61). Thus, it is plausible that targeting of the N-terminal amphipathic α-helical domain of viperin to ER could represent a mechanism to control CHIKV, as viperin mutants lacking the ER-targeting N-terminal amphipathic α-helical domain displayed a compromised ability to inhibit CHIKV infection. Furthermore, targeting the N-terminal amphipathic α-helical domain of viperin alone to the ER was demonstrated to be sufficient for inhibition of protein secretion, possibly due to the disrupted ER structure and integrity (43). However, whether the antiviral function of viperin against CHIKV is mediated in a similar manner in order to hamper release of new virions and spread of infection will require further exploration to address the molecular mechanisms involved.
We further demonstrated the direct role of viperin in controlling CHIKV replication in primary fibroblasts isolated from Rsad2–/– mice. Loss of viperin expression resulted in a significant increase in CHIKV infectivity and viral load compared with fibroblasts isolated from WT mice. More importantly, the significant increase of viral load detected during the early phase of infection in the footpad of Rsad2–/– mice further confirmed the critical role for viperin in limiting CHIKV replication in vivo. Loss of viperin expression in Rsad2–/– mice also significantly increased disease susceptibility and pathology, as reflected by increased viremia and CHIKV-induced joint inflammation. Taking these observations together, we propose a model by which viperin may control CHIKV replication following the activation of the innate immune response after infection (Figure 9). This model further defines another role for viperin as a multifunctional protein with antiviral activities against CHIKV. It will be important to assess the association between RSAD2 polymorphisms (and other ISGs) and disease-induced arthritis intensity in genome-wide association studies of patient cohorts. Due to the resurgence of CHIKV and other reemerging viruses, the identification of new antiviral strategies of viperin has regained importance, as each virus has a different antidote. Such baseline data would become integral for future development of novel immune-based control strategies.
 Figure 9
Figure 9Proposed model for viperin as an anti-CHIKV ISG during the acute phase of infection. Upon infection of target cells, the presence of CHIKV RNA in the cytoplasm can be detected by pathogen receptors such as RIG-I and MDA5 to trigger the innate immune response. Consequentially, this leads to the activation of signaling pathways that regulate IRF3 and IRF7 to induce type I IFN expression. Secretion of type I IFNs to their receptors then induces the JAK/STAT pathway for the regulation of a positive feedback loop for type I IFN production via IRF7. In parallel, induction of type I IFN signaling leads to the expression of various ISGs regulated by ISGF3 to control CHIKV infection. Viperin is induced and targeted to the ER via the N-terminal amphipathic α-helical domain. This triggers an ER stress response that activates PKR to interfere with the synthesis of viral proteins essential for CHIKV replication. Synthesis of CHIKV nsP2, a helicase/protease, acts to contravene the type I IFN–mediated JAK/STAT signaling pathway. However, viperin induction could circumvent CHIKV nsP2 and control CHIKV replication, emphasizing its role as a multifunctional protein.
-
Methods
Patients. 24 patients that were admitted with acute CHIKF to the Communicable Disease Centre at Tan Tock Seng Hospital (CDC/TTSH) during the outbreak from August 1 to September 23, 2008 (14, 15), were included in this study. PBMC samples were isolated at 4 different time points: (a) acute phase (median 4 days post illness onset), (b) early convalescent phase (median 10 days post illness onset), (c) late convalescent phase (4–6 weeks post illness onset), and (d) chronic phase (2–3 months post illness onset). Patients were classified into HVL (n = 11) and LVL (n = 13) groups based on their viral load, as described previously (18). In addition, PBMC samples were isolated from 10 healthy volunteers and used as a control. Specimens were stored at –80°C until use.
Viruses and antibodies. CHIKV isolate SGP 011 was isolated from the outbreak in Singapore and propagated in Vero-E6 cells as described previously (22). Virus titer was estimated by standard plaque assays using Vero-E6 cells as described previously (22). A CHIKV variant expressing mCherry fluorescent protein was constructed using a full-length infectious cDNA clone of CHIKV LR2006-OPY1 isolate as described previously (62). The infectious cDNA was subsequently cloned into a modified pMA vector (Geneart) with a SP6 promoter 5′ upstream of CHIKV cDNA. Sequencing was conducted to ensure that no mutations were introduced. A mCherry insert was introduced at 5′ to the structural genes of infectious cDNA clone followed by duplication of a second subgenomic promoter as described previously (63). Viruses were rescued from infectious cDNA clones as described previously (62) before being propagated and titrated using Vero-E6 cells. Rabbit polyclonal antibodies against nsP2 protein of SFV were obtained by immunization of rabbits with recombinant protein corresponding to the C-terminal (protease) domain of nsP2. The antiserum was then subjected to affinity purification. Antibodies against GFP were purchased from Santa Cruz Biotechnology. Antibody against calreticulin was purchased from Abcam.
In vitro CHIKV infection. 50 ml whole blood was obtained from healthy donors and mixed with citrate phosphate dextrose according to the manufacturer’s instructions. Total leukocyte count was determined using a Beckman Coulter Z2 cell and particle counter, and blood was split into 2 equal volumes before performing infection with CHIKV or HI CHIKV (MOI 10) in 150-mm dishes. Samples were incubated at 37°C for 12 hours with intermittent shaking. For transfected HEK 293T cells, CHIKV infection was performed at 24 hours after transfection with either CHIKV or HI CHIKV (MOI 2.5) in serum-free DMEM. Virus overlay was removed after 1.5 hours of incubation at 37°C and replaced with DMEM supplemented with 10% FBS. Cells were harvested at 12 hpi. Primary fibroblasts were isolated from tails of WT and Rsad2–/– mice using a previously described protocol (64), and infections were performed with CHIKV (MOI 10) as described for HEK 293T cells. Mock infections were performed in parallel with controls. Mouse recombinant IFN-α and IFN-β (Merck Millipore) were used for pretreatment of WT and Rsad2–/– fibroblasts prior to CHIKV infection.
Isolation of PBMC subsets. PBMCs isolated from whole blood infection were stained with a panel of antibodies — CD3-FITC, CD14–PerCP-Cy5.5, CD19-PeCy7, and CD56-PE against T cells, monocytes, B cells, and NK cells, respectively — to obtain live cell subsets. CD14 antibodies were purchased from Biolegend, CD3 and CD56 antibodies were purchased from Miltenyi Biotech, and other antibodies were from BD Biosciences. The various PBMC subsets were sorted as described in Supplemental Figure 3 by FACS Aria (BD Biosciences). For all cell populations, a minimum of 250,000 cells was collected and stored in TRIzol at –80°C. Total RNA from each subset was extracted and analyzed by quantitative real-time PCR (qRT-PCR).
Flow cytometry. For detection of CHIKV Ag+ cells in PBMCs by FACS, a 2-step indirect intracellular staining protocol was used as described previously (22). To identify the various PBMC subsets, an additional surfacing staining step was performed with the same panel of antibodies described above for live-cell sorting. Data were acquired using BD LSR II (BD Biosciences) and BD FACSDiva software. HEK 293T cells were stained using the 2-step indirect intracellular staining protocol to detect CHIKV Ag+ cells, and data were acquired using BD FACSCalibur (BD Biosciences) and BD FACS CellQuest Pro software. All analyses were performed with Flowjo software.
Total RNA extraction and qRT-PCR. Total RNA was extracted using RNeasy Mini Kit (QIAGEN) according to the manufacturer’s instructions. Eluted RNA was stored at –80°C. Quantification of total RNA was measured by NanoDrop 1000 Spectrophotometer (Thermo Scientific). Extracted total RNA (10 ng/μl) was subjected to RT-PCR using QuantiFast SYBR Green RT-PCR Kit (QIAGEN) according to the manufacturer’s recommendations in 12.5 μl reaction volume. qRT-PCR was performed in Applied Biosystems (ABI) 7900HT Fast Real-Time PCR System in 384-well plates, with the following conditions: (a) reverse transcription step (50°C for 10 minutes; 1 cycle); (b) PCR initial activation step (95°C for 5 minutes; 1 cycle); (c) 2-step cycling (95°C for 10 seconds, followed by 60°C for 30 seconds; 40 cycles). The fold change relative to healthy donors for each gene was determined using the ΔΔCt method using Microsoft Excel 2010. Briefly, ΔΔCt was calculated as ΔCtpatient – ΔCthealthy donor, with ΔCt determined as Ct[gene of interest] – CtGAPDH (the latter used as housekeeping gene); the fold change for each gene between the patient and healthy donor was calculated as 2–ΔΔCt. See Supplemental Table 1 for forward and reverse primers used.
Viral RNA extraction and viral load quantification. Aliquots of 140 μl of cell suspension or blood (diluted in PBS containing citrate) from each experimental sample were collected for viral RNA extraction using QIAamp Viral RNA Kit (QIAGEN). Viral load quantification was performed by qRT-PCR using QuantiTect Probe RT-PCR Kit (QIAGEN) modified from a previously described method to detect negative-strand nsP1 RNA (65). qRT-PCR reaction mixes were done in 12.5 μl reaction volume and performed in Applied Biosystems (ABI) 7900HT Fast Real-Time PCR System in 384-well plates, with the following conditions: (a) reverse transcription step (50°C for 30 minutes; 1 cycle); (b) PCR initial activation step (95°C for 15 minutes; 1 cycle); (c) 2-step cycling (94°C for 15 seconds, follow by 60°C for 1 minute; 45 cycles). Viral load was estimated from a standard curve generated using serial dilution of synthetic CHIKV negative-strand nsP1 RNA transcripts as described previously (66).
Cell culture, plasmid construction, and transfection. HEK 293T cells were cultured in DMEM supplemented with 10% FBS. All mutant constructs were generated by standard PCR amplification using appropriate sets of primers from the full-length human RSAD2 (27). Transient transfection was done with Lipofectamine 2000 (Invitrogen) according to the manufacturer’s instructions.
Western blot. Transfected cells were harvested and washed once with PBS and incubated for 5 minutes with lysis buffer containing 20 mM Tris-HCl (pH 7.4), 1 mM EDTA, 50 mM KCL, 1 mM DTT, 10 mM Na3VO4, 5 mM MgCl2, 5 mM NaF, 10% glycerol, 1% Triton X-100, and protease inhibitors (Roche). Protein concentrations were quantified using DC protein assay (Bio-Rad), and absorbance readings were taken at 750 nm using Magellan microplate reader. Equal amounts of proteins were separated by SDS-PAGE gels and transferred onto nitrocellulose membranes (Bio-Rad). Membranes were blocked using 5% (w/v) skim milk in Tris-buffered saline containing 0.05% Tween 20, prior to incubation with appropriate secondary antibodies conjugated with horseradish peroxidase. Protein bands were visualized by chemiluminescence (Amersham Biosciences) on autoradiography films (Thermo Scientific).
Immunofluorescence staining. Cells were seeded on fibronectin-coated coverslips for staining. Cells were fixed with 4% paraformaldehyde, permeabilized in PBS containing 0.2% Triton X-100, and blocked with 10% FBS in PBS. Cell were incubated with primary antibodies in PBS containing 1% BSA, followed by incubation with the appropriate secondary antibodies conjugated with Alexa Fluor 488, Alexa Fluor 594, or Alexa Fluor 647 (Invitrogen). Cells were washed, mounted, and examined with a confocal laser-scanning microscope (Fluoview FV1000; Olympus) using a ×100 NA 1.40 objective. Images were collected using FV1000-ASW software and processed with Adobe Photoshop software.
Animal studies. 3-week-old female WT or Rsad2–/– C57BL/6 mice were inoculated subcutaneously in the ventral side of the right hind footpad toward the ankle with 106 PFU CHIKV in 50 μl PBS. Viremia analysis was performed by collecting 10 μl blood from the tail for CHIKV RNA extraction and quantified by viral load qRT-PCR. To quantify foot swelling, mice hind footpads were measured using a Vernier caliper daily from 0 to 14 dpi. Measurements were done for the height (thickness) and breath of the foot and quantified as the product of height and breadth. Degree of inflammation was expressed as relative increase compared with preinfection (0 dpi), calculated as (x dpi – 0 dpi)/0 dpi. For foot joint extraction, mice were anesthetized with 150 mg/kg ketamine and 10 mg/kg xylazin and perfused with PBS. Feet were removed and preserved in TRIzol (Invitrogen) at –80°C. For tissue homogenization, tissue samples were transferred to gentleMACS M Tubes (Miltenyi Biotec) and homogenized using a rotor-stator homogenizer (Xiril Dispomix) at approximately 500 g for 15 seconds. Homogenized tissues were transferred to clean eppendorf tubes and were mixed thoroughly with 230 μl chloroform. After 2 minutes of incubation, tissue mixtures were centrifuged at 13,523 g for 15 minutes at 4°C. The aqueous phase was collected and isolated for total RNA and viral RNA. Tissues were fixed with 4% paraformaldehyde and embedded in paraffin wax before being subjected to sectioning and H&E or IHC staining with mouse anti-F4/80 antibody.
Statistics. All statistical analyses were performed using Prism 5.01 (GraphPad Software). Analysis of all gene expression profiles among different phases of CHIKV infection in patients was done using Kruskall-Wallis test followed by Dunn’s post-test. Comparison of gene expression profiles between HVL and LVL patient groups was analyzed by Mann-Whitney U test. Effects of viperin overexpression on CHIKV infectivity and viral load analysis was performed using 1-way ANOVA with Tukey’s post-test. Comparison of CHIKV infectivity, viral load, and gene expression profiles in WT and Rsad2–/– fibroblasts was performed using 2-tailed Student’s t test. Comparison of viral load, gene expression profiles, and CHIKV-induced joint swelling in WT and Rsad2–/– mice was performed using Mann-Whitney U test. P values less than 0.05 were considered statistically significant. Correlation analyses between expression of various genes were determined using Pearson’s correlation analysis, in which R greater than 0.6 was considered a strong correlation.
Study approval. This study was conducted according to Declaration of Helsinki principles. The use of human samples was approved by the National Healthcare Group’s Domain-Specific Ethics Review Board (DSRB reference no. B/08/026), and written informed consent was obtained from all participants. All animal protocols were approved by the Institutional Animal Care and Use Committee (no. 080383) at the Biological Resource Center (Biopolis, Singapore).
-
Acknowledgments
We acknowledge the study participants and healthy volunteers for their participation in the study; the research staff from the Communicable Disease Centre/Tan Tock Seng Hospital for patient enrollment, study coordination, and data entry; and the clinical staff of the Communicable Disease Centre/Tan Tock Seng Hospital for patient enrollment and care. We thank the SIgN Flow Cytometry Facility for assistance with cell sorting experiments and the Advanced Molecular Pathology Laboratory (IMCB, A*STAR) for the histology experiments. We thank L. Renia (SIgN, A*STAR) for critically reading the manuscript and Yi-lin Wang (SIgN, A*STAR) for assistance with video images. The work was supported by the Biomedical Research Council of A*STAR. Part of this work was also supported by European Union FP7 project Integration of Chikungunya RESearch (ICRES; grant no. 261202). F.-M. Lum is supported by a NUS postgraduate scholarship, and T.-H. Teo is supported by an A*STAR postgraduate scholarship. The sponsors were not involved in study design, data gathering or analysis, or drafting of the manuscript.
Address correspondence to: Lisa F.P. Ng, Laboratory of Chikungunya Virus Immunity, Singapore Immunology Network, A*STAR, 8A Biomedical Grove, #04-06 Immunos, Biopolis 138648, Singapore. Phone: 65.64070028; Fax: 65.64642057; E-mail: [email protected]. Or to: Keh-Chuang Chin, Laboratory of Gene Regulation in Inflammation, Singapore Immunology Network, A*STAR, 8A Biomedical Grove, #04-06 Immunos, Biopolis 138648, Singapore. Phone: 65.64070006; Fax: 65.64642057; E-mail: [email protected].
-
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Reference information: J Clin Invest. 2012;122(12):4447–4460. doi:10.1172/JCI63120.
Suan-Sin Foo’s present address is: Emerging Viruses and Inflammation Research Group, Institute for Glycomics, Griffith University, Gold Coast, Queensland, Australia.
-
References
- Stetson DB, Medzhitov R. Type I interferons in host defense. Immunity. 2006;25(3):373–381.
- McCartney SA, Colonna M. Viral sensors: diversity in pathogen recognition. Immunol Rev. 2009;227(1):87–94.
- Decker T, Stockinger S, Karaghiosoff M, Muller M, Kovarik P. IFNs and STATs in innate immunity to microorganisms. J Clin Invest. 2002;109(10):1271–1277.
- Levy DE, Garcia-Sastre A. The virus battles: IFN induction of the antiviral state and mechanisms of viral evasion. Cytokine Growth Factor Rev. 2001;12(2–3):143–156.
- Marie I, Durbin JE, Levy DE. Differential viral induction of distinct interferon-alpha genes by positive feedback through interferon regulatory factor-7. EMBO J. 1998;17(22):6660–6669.
- Sadler AJ, Williams BR. Interferon-inducible antiviral effectors. Nat Rev Immunol. 2008;8(7):559–568.
- Kam YW, Ong EK, Renia L, Tong JC, Ng LF. Immuno-biology of Chikungunya and implications for disease intervention. Microbes Infect. 2009;11 (14-15): :1186–1196.
- Schwartz O, Albert ML. Biology and pathogenesis of chikungunya virus. Nat Rev Microbiol. 2010;8(7):491–500.
- Lumsden WHR. An epidemic of virus disease in Southern Province, Tanganyika territory, in 1952-1953 II. General description and epidemiology. Trans R Soc Trop Med Hyg. 1955;49(1):33–57.
- Robinson MC. An epidemic of virus disease in Southern Province, Tanganyika territory, in 1952-1953. Trans R Soc Trop Med Hyg. 1955;49(1):28–32.
- Her Z, Kam YW, Lin RT, Ng LF. Chikungunya: a bending reality. Microbes Infect. 2009;11(14–15):1165–1176.
- Powers AM, Logue CH. Changing patterns of chikungunya virus: re-emergence of a zoonotic arbovirus. J Gen Virol. 2007;88(9):2363–2377.
- Schuffenecker I, et al. Genome microevolution of chikungunya viruses causing the Indian Ocean outbreak. PLoS Med. 2006;3(7):0001–0013.
- Ng KW, et al. Clinical features and epidemiology of chikungunya infection in Singapore. Singapore Med J. 2009;50(8):785–790.View this article via: PubMed Google Scholar
- Win MK, Chow A, Dimatatac F, Go CJ, Leo YS. Chikungunya fever in Singapore: acute clinical and laboratory features, and factors associated with persistent arthralgia. J Clin Virol. 2010;49(2):111–114.
- Pialoux G, Gauzere BA, Jaureguiberry S, Strobel M. Chikungunya, an epidemic arbovirosis. Lancet Infect Dis. 2007;7(5):319–327.
- Gifford GE, Heller E. Effect of actinomycin D on interferon production by ‘active’ and ‘inactive’ chikungunya virus in chick cells. Nature. 1963;200:50–51.
- Chow A, et al. Persistent arthralgia induced by chikungunya virus infection is associated with interleukin-6 and granulocyte macrophage colony-stimulating factor. J Infect Dis. 2010;203(2):149–157.
- Ng LFP, et al. IL-1beta, IL-6, and RANTES as biomarkers of chikungunya severity. PLoS One. 2009;4(1):e4261.
- Couderc T, et al. A mouse model for chikungunya: Young age and inefficient type-I interferon signaling are risk factors for severe disease. PLoS Pathog. 2008;4(2):e29.
- Fros JJ, et al. Chikungunya virus nonstructural protein 2 inhibits type I/II interferon-stimulated JAK-STAT signaling. J Virol. 2010;84(20):10877–10887.
- Her Z, et al. Active infection of human blood monocytes by Chikungunya virus triggers an innate immune response. J Immunol. 2010;184(10):5903–5913.
- Hoarau JJ, et al. Persistent chronic inflammation and infection by Chikungunya arthritogenic alphavirus in spite of a robust host immune response. J Immunol. 2010;184(10):5914–5927.
- Schilte C, et al. Type I IFN controls chikungunya virus via its action on nonhematopoietic cells. J Exp Med. 2010;207(2):429–442.
- Schilte C, Buckwalter MR, Laird ME, Diamond MS, Schwartz O, Albert ML. Cutting edge: independent roles for IRF-3 and IRF-7 in hematopoietic and nonhematopoietic cells during host response to Chikungunya infection. J Immunol. 2012;188(7):2967–2971.
- Schoggins JW, et al. A diverse range of gene products are effectors of the type I interferon antiviral response. Nature. 2011;472(7344):481–485.
- Chin KC, Cresswell P. Viperin (cig5), an IFN-inducible antiviral protein directly induced by human cytomegalovirus. Proc Natl Acad Sci U S A. 2001;98(26):15125–15130.
- Jiang D, et al. Identification of three interferon-inducible cellular enzymes that inhibit the replication of hepatitis C virus. J Virol. 2008;82(4):1665–1678.
- Jiang D, et al. Identification of five interferon-induced cellular proteins that inhibit west nile virus and dengue virus infections. J Virol. 2010;84(16):8332–8341.
- Szretter KJ, Brien JD, Thackray LB, Virgin HW, Cresswell P, Diamond MS. The interferon-inducible gene viperin restricts west nile virus pathogenesis. J Virol. 2011;85(22):11557–11566.
- Wang X, Hinson ER, Cresswell P. The interferon-inducible protein viperin inhibits influenza virus release by perturbing lipid rafts. Cell Host Microbe. 2007;2(2):96–105.
- Mattijssen S, Pruijn GJ. Viperin, a key player in the antiviral response. Microbes Infect. 2011;14(5):419–426.
- Honda K, Takaoka A, Taniguchi T. Type I Interferon gene induction by the interferon regulatory factor family of transcription tactors. Immunity. 2006;25(3):349–360.
- Labadie K, et al. Chikungunya disease in nonhuman primates involves long-term viral persistence in macrophages. J Clin Invest. 2010;120(3):894–906.
- Werneke SW, et al. ISG15 is critical in the control of chikungunya virus infection independent of UbE1L mediated conjugation. PLoS Pathog. 2011;7(10):e1002322.
- Collado-Hidalgo A, Sung C, Cole S. Adrenergic inhibition of innate anti-viral response: PKA blockade of Type I interferon gene transcription mediates catecholamine support for HIV-1 replication. Brain Behav Immun. 2006;20(6):552–563.
- Stawowczyk M, Van Scoy S, Kumar KP, Reich NC. The interferon stimulated gene 54 promotes apoptosis. J Biol Chem. 2011;286(9):7257–7266.
- Terenzi F, Saikia P, Sen GC. Interferon-inducible protein, P56, inhibits HPV DNA replication by binding to the viral protein E1. EMBO J. 2008;27(24):3311–3321.
- Helbig KJ, et al. The antiviral protein viperin inhibits hepatitis C virus replication via interaction with nonstructural protein 5A. Hepatology. 2011;54(5):1506–1517.
- Wang S, et al. Viperin inhibits hepatitis C virus replication by interfering with the binding of NS5A to host protein hVAP-33. J Gen Virol. 2012;93(pt 1):83–92.
- Zhang Y, Burke CW, Ryman KD, Klimstra WB. Identification and characterization of interferon-induced proteins that inhibit alphavirus replication. J Virol. 2007;81(20):11246–11255.
- Seo JY, Yaneva R, Cresswell P. Viperin: A multifunctional, interferon-inducible protein that regulates virus replication. Cell Host Microbe. 2011;10(6):534–539.
- Hinson ER, Cresswell P. The N-terminal amphipathic alpha-helix of viperin mediates localization to the cytosolic face of the endoplasmic reticulum and inhibits protein secretion. J Biol Chem. 2009;284(7):4705–4712.
- Hinson ER, Cresswell P. The antiviral protein, viperin, localizes to lipid droplets via its N-terminal amphipathic alpha-helix. Proc Natl Acad Sci U S A. 2009;106(48):20452–20457.
- Duschene KS, Broderick JB. The antiviral protein viperin is a radical SAM enzyme. FEBS Lett. 2010;584(6):1263–1267.
- Shaveta G, Shi J, Chow VT, Song J. Structural characterization reveals that viperin is a radical S-adenosyl-L-methionine (SAM) enzyme. Biochem Biophys Res Commun. 2010;391(3):1390–1395.
- Seo JY, Yaneva R, Hinson ER, Cresswell P. Human cytomegalovirus directly induces the antiviral protein viperin to enhance infectivity. Science. 2011;332(6033):1093–1097.
- de Veer MJ, et al. Functional classification of interferon-stimulated genes identified using microarrays. J Leukoc Biol. 2001;69(6):912–920.View this article via: PubMed Google Scholar
- Der SD, Zhou A, Williams BR, Silverman RH. Identification of genes differentially regulated by interferon alpha, beta, or gamma using oligonucleotide arrays. Proc Natl Acad Sci U S A. 1998;95(26):15623–15628.
- White LK, et al. Chikungunya virus induces IPS-1-dependent innate immune activation and protein kinase R-independent translational shutoff. J Virol. 2011;85(1):606–620.
- Schoggins JW, Rice CM. Interferon-stimulated genes and their antiviral effector functions. Curr OpIn Virol. 2011;1(6):519–525.
- Nasr N, et al. HIV-1 infection of human macrophages directly induces viperin which inhibits viral production. Blood. 2012;120(4):778–788.
- Haldar S, Paul S, Joshi N, Dasgupta A, Chattopadhyay K. The presence of the iron-sulfur motif is important for the conformational stability of the antiviral protein, Viperin. PLoS One. 2012;7(2):e31797.
- Roy CR, Salcedo SP, Gorvel JP. Pathogen-endoplasmic-reticulum interactions: in through the out door. Nat Rev Immunol. 2006;6(2):136–147.
- Salonen A, Ahola T, Kaariainen L. Viral RNA replication in association with cellular membranes. Curr Top Microbiol Immunol. 2005;285:139–173.View this article via: PubMed Google Scholar
- Jose J, Snyder JE, Kuhn RJ. A structural and functional perspective of alphavirus replication and assembly. Future Microbiol. 2009;4(7):837–856.
- Strauss JH, Strauss EG. The alphaviruses: gene expression, replication, and evolution. Microbiol Rev. 1994;58(3):491–562.View this article via: PubMed Google Scholar
- Bourai M, et al. Mapping of chikungunya virus interactions with host proteins identified nsP2 as a higly connected viral component. J Virol. 2012;86(6):3121–3134.
- He B. Viruses, endoplasmic reticulum stress, and interferon responses. Cell Death Differ. 2006;13(3):393–403.
- Clavarino G, et al. Induction of GADD34 Is necessary for dsRNA-dependent interferon-β production and participates in the control of chikungunya virus infection. PLoS Pathog. 2012;8(5):e1002708.
- Novoa I, Zeng H, Harding HP, Ron D. Feedback inhibition of the unfolded protein response by GADD34-mediated dephosphorylation of eIF2alpha. J Cell Biol. 2001;153(5):1011–1022.
- Pohjala L, et al. Inhibitors of alphavirus entry and replication identified with a stable chikungunya replicon cell line and virus-based assays. PLoS One. 2011;6(12):e28923.
- Tsetsarkin K, Higgs S, McGee CE, De Lamballerie X, Charrel RN, Vanlandingham DL. Infectious clones of Chikungunya virus (La Reunion isolate) for vector competence studies. Vector Borne Zoonotic Dis. 2006;6(4):325–337.
- Salmon AB, Murakami S, Bartke A, Kopchick J, Yasumura K, Miller RA. Fibroblast cell lines from young adult mice of long-lived mutant strains are resistant to multiple forms of stress. Am J Physiol Endocrinol Metab. 2005;289(1):E23–E29.
- Plaskon NE, Adelman ZN, Myles KM. Accurate strand-specific quantification of viral RNA. PLoS One. 2009;4(10):e7468.
- Lee CY, et al. Chikungunya virus neutralization antigens and direct cell-to-cell transmission are revealed by human antibody-escape mutants. PLoS Pathog. 2011;7(12):e1002390.
-
Version history
- Version 1 (November 19, 2012): No description
- Version 2 (December 3, 2012): No description



Copyright © 2024 American Society for Clinical Investigation
ISSN: 0021-9738 (print), 1558-8238 (online)









