Advertisement
Research Article Free access | 10.1172/JCI61852
Lentiviral vector integration in the human genome induces alternative splicing and generates aberrant transcripts
Arianna Moiani,1 Ylenia Paleari,2 Daniela Sartori,1 Riccardo Mezzadra,2 Annarita Miccio,3 Claudia Cattoglio,1 Fabienne Cocchiarella,3 Maria Rosa Lidonnici,2,4 Giuliana Ferrari,2,4 and Fulvio Mavilio1,3
1Division of Genetics and Cell Biology and 2San Raffaele Telethon Institute for Gene Therapy (HSR-TIGET), Istituto Scientifico H. San Raffaele, Milan, Italy. 3Department of Biomedical Sciences, University of Modena and Reggio Emilia, Modena, Italy. 4Vita-Salute San Raffaele University, Milan, Italy.
Address correspondence to: Fulvio Mavilio, Department of Biomedical Sciences, University of Modena and Reggio Emilia, Via Campi 273, 41125 Modena, Italy. Phone: 39.059.2058076; Fax: 39.059.2058015; E-mail: [email protected]. Or to: Giuliana Ferrari, HSR-TIGET, Istituto Scientifico H. San Raffaele, Via Olgettina 58, 20132 Milan, Italy. Phone: 39.02.26434705; Fax: 39.02.26434668; E-mail: [email protected].
Find articles by Moiani, A. in: JCI | PubMed | Google Scholar
1Division of Genetics and Cell Biology and 2San Raffaele Telethon Institute for Gene Therapy (HSR-TIGET), Istituto Scientifico H. San Raffaele, Milan, Italy. 3Department of Biomedical Sciences, University of Modena and Reggio Emilia, Modena, Italy. 4Vita-Salute San Raffaele University, Milan, Italy.
Address correspondence to: Fulvio Mavilio, Department of Biomedical Sciences, University of Modena and Reggio Emilia, Via Campi 273, 41125 Modena, Italy. Phone: 39.059.2058076; Fax: 39.059.2058015; E-mail: [email protected]. Or to: Giuliana Ferrari, HSR-TIGET, Istituto Scientifico H. San Raffaele, Via Olgettina 58, 20132 Milan, Italy. Phone: 39.02.26434705; Fax: 39.02.26434668; E-mail: [email protected].
Find articles by Paleari, Y. in: JCI | PubMed | Google Scholar
1Division of Genetics and Cell Biology and 2San Raffaele Telethon Institute for Gene Therapy (HSR-TIGET), Istituto Scientifico H. San Raffaele, Milan, Italy. 3Department of Biomedical Sciences, University of Modena and Reggio Emilia, Modena, Italy. 4Vita-Salute San Raffaele University, Milan, Italy.
Address correspondence to: Fulvio Mavilio, Department of Biomedical Sciences, University of Modena and Reggio Emilia, Via Campi 273, 41125 Modena, Italy. Phone: 39.059.2058076; Fax: 39.059.2058015; E-mail: [email protected]. Or to: Giuliana Ferrari, HSR-TIGET, Istituto Scientifico H. San Raffaele, Via Olgettina 58, 20132 Milan, Italy. Phone: 39.02.26434705; Fax: 39.02.26434668; E-mail: [email protected].
Find articles by Sartori, D. in: JCI | PubMed | Google Scholar
1Division of Genetics and Cell Biology and 2San Raffaele Telethon Institute for Gene Therapy (HSR-TIGET), Istituto Scientifico H. San Raffaele, Milan, Italy. 3Department of Biomedical Sciences, University of Modena and Reggio Emilia, Modena, Italy. 4Vita-Salute San Raffaele University, Milan, Italy.
Address correspondence to: Fulvio Mavilio, Department of Biomedical Sciences, University of Modena and Reggio Emilia, Via Campi 273, 41125 Modena, Italy. Phone: 39.059.2058076; Fax: 39.059.2058015; E-mail: [email protected]. Or to: Giuliana Ferrari, HSR-TIGET, Istituto Scientifico H. San Raffaele, Via Olgettina 58, 20132 Milan, Italy. Phone: 39.02.26434705; Fax: 39.02.26434668; E-mail: [email protected].
Find articles by Mezzadra, R. in: JCI | PubMed | Google Scholar
1Division of Genetics and Cell Biology and 2San Raffaele Telethon Institute for Gene Therapy (HSR-TIGET), Istituto Scientifico H. San Raffaele, Milan, Italy. 3Department of Biomedical Sciences, University of Modena and Reggio Emilia, Modena, Italy. 4Vita-Salute San Raffaele University, Milan, Italy.
Address correspondence to: Fulvio Mavilio, Department of Biomedical Sciences, University of Modena and Reggio Emilia, Via Campi 273, 41125 Modena, Italy. Phone: 39.059.2058076; Fax: 39.059.2058015; E-mail: [email protected]. Or to: Giuliana Ferrari, HSR-TIGET, Istituto Scientifico H. San Raffaele, Via Olgettina 58, 20132 Milan, Italy. Phone: 39.02.26434705; Fax: 39.02.26434668; E-mail: [email protected].
Find articles by Miccio, A. in: JCI | PubMed | Google Scholar
1Division of Genetics and Cell Biology and 2San Raffaele Telethon Institute for Gene Therapy (HSR-TIGET), Istituto Scientifico H. San Raffaele, Milan, Italy. 3Department of Biomedical Sciences, University of Modena and Reggio Emilia, Modena, Italy. 4Vita-Salute San Raffaele University, Milan, Italy.
Address correspondence to: Fulvio Mavilio, Department of Biomedical Sciences, University of Modena and Reggio Emilia, Via Campi 273, 41125 Modena, Italy. Phone: 39.059.2058076; Fax: 39.059.2058015; E-mail: [email protected]. Or to: Giuliana Ferrari, HSR-TIGET, Istituto Scientifico H. San Raffaele, Via Olgettina 58, 20132 Milan, Italy. Phone: 39.02.26434705; Fax: 39.02.26434668; E-mail: [email protected].
Find articles by Cattoglio, C. in: JCI | PubMed | Google Scholar
1Division of Genetics and Cell Biology and 2San Raffaele Telethon Institute for Gene Therapy (HSR-TIGET), Istituto Scientifico H. San Raffaele, Milan, Italy. 3Department of Biomedical Sciences, University of Modena and Reggio Emilia, Modena, Italy. 4Vita-Salute San Raffaele University, Milan, Italy.
Address correspondence to: Fulvio Mavilio, Department of Biomedical Sciences, University of Modena and Reggio Emilia, Via Campi 273, 41125 Modena, Italy. Phone: 39.059.2058076; Fax: 39.059.2058015; E-mail: [email protected]. Or to: Giuliana Ferrari, HSR-TIGET, Istituto Scientifico H. San Raffaele, Via Olgettina 58, 20132 Milan, Italy. Phone: 39.02.26434705; Fax: 39.02.26434668; E-mail: [email protected].
Find articles by Cocchiarella, F. in: JCI | PubMed | Google Scholar
1Division of Genetics and Cell Biology and 2San Raffaele Telethon Institute for Gene Therapy (HSR-TIGET), Istituto Scientifico H. San Raffaele, Milan, Italy. 3Department of Biomedical Sciences, University of Modena and Reggio Emilia, Modena, Italy. 4Vita-Salute San Raffaele University, Milan, Italy.
Address correspondence to: Fulvio Mavilio, Department of Biomedical Sciences, University of Modena and Reggio Emilia, Via Campi 273, 41125 Modena, Italy. Phone: 39.059.2058076; Fax: 39.059.2058015; E-mail: [email protected]. Or to: Giuliana Ferrari, HSR-TIGET, Istituto Scientifico H. San Raffaele, Via Olgettina 58, 20132 Milan, Italy. Phone: 39.02.26434705; Fax: 39.02.26434668; E-mail: [email protected].
Find articles by Lidonnici, M. in: JCI | PubMed | Google Scholar
1Division of Genetics and Cell Biology and 2San Raffaele Telethon Institute for Gene Therapy (HSR-TIGET), Istituto Scientifico H. San Raffaele, Milan, Italy. 3Department of Biomedical Sciences, University of Modena and Reggio Emilia, Modena, Italy. 4Vita-Salute San Raffaele University, Milan, Italy.
Address correspondence to: Fulvio Mavilio, Department of Biomedical Sciences, University of Modena and Reggio Emilia, Via Campi 273, 41125 Modena, Italy. Phone: 39.059.2058076; Fax: 39.059.2058015; E-mail: [email protected]. Or to: Giuliana Ferrari, HSR-TIGET, Istituto Scientifico H. San Raffaele, Via Olgettina 58, 20132 Milan, Italy. Phone: 39.02.26434705; Fax: 39.02.26434668; E-mail: [email protected].
Find articles by Ferrari, G. in: JCI | PubMed | Google Scholar
1Division of Genetics and Cell Biology and 2San Raffaele Telethon Institute for Gene Therapy (HSR-TIGET), Istituto Scientifico H. San Raffaele, Milan, Italy. 3Department of Biomedical Sciences, University of Modena and Reggio Emilia, Modena, Italy. 4Vita-Salute San Raffaele University, Milan, Italy.
Address correspondence to: Fulvio Mavilio, Department of Biomedical Sciences, University of Modena and Reggio Emilia, Via Campi 273, 41125 Modena, Italy. Phone: 39.059.2058076; Fax: 39.059.2058015; E-mail: [email protected]. Or to: Giuliana Ferrari, HSR-TIGET, Istituto Scientifico H. San Raffaele, Via Olgettina 58, 20132 Milan, Italy. Phone: 39.02.26434705; Fax: 39.02.26434668; E-mail: [email protected].
Find articles by Mavilio, F. in: JCI | PubMed | Google Scholar
Published April 23, 2012 - More info
J Clin Invest. 2012;122(5):1653–1666. https://doi.org/10.1172/JCI61852.
© 2012 The American Society for Clinical Investigation
Received: November 29, 2011; Accepted: March 7, 2012
Related articles:
Abstract
Gamma-retroviral/lentiviral vectors (γRV/LV) with self-inactivating (SIN) long terminal repeats (LTRs) and internal moderate cellular promoters pose a reduced risk of insertional mutagenesis when compared with vectors with active LTRs. Yet, in a recent LV-based clinical trial for β-thalassemia, vector integration within the HMGA2 gene induced the formation of an aberrantly spliced mRNA form that appeared to cause clonal dominance. Using a method that we developed, cDNA linear amplification-mediated PCR, in combination with high-throughput sequencing, we conducted a whole transcriptome analysis of chimeric LV-cellular fusion transcripts in transduced human lymphoblastoid cells and primary hematopoietic stem/progenitor cells. We observed a surprising abundance of read-through transcription originating outside and inside the provirus and identified the vector sequences contributing to the aberrant splicing process. We found that SIN LV has a sharply reduced propensity to engage in aberrant splicing compared with that of vectors carrying active LTRs. Moreover, by recoding the identified vector splice sites, we reduced residual read-through transcription and demonstrated an effective strategy for improving vectors. Characterization of the mechanisms and genetic features underlying vector-induced aberrant splicing will enable the generation of safer vectors, with low impact on the cellular transcriptome.
Authors
Daniela Cesana, Jacopo Sgualdino, Laura Rudilosso, Stefania Merella, Luigi Naldini, Eugenio Montini
Abstract
The use of integrating vectors for gene therapy — required for stable correction of gene expression — carries the risk of insertional mutagenesis, which can lead to activation of a tumorigenic program. In this issue of the JCI, Moiani et al. and Cesana et al. investigate how viral vectors can induce aberrant splicing, resulting in chimeric cellular-viral transcripts. The finding that this is a general phenomenon is concerning, but some of their results do suggest approaches for the development of safeguards in gene therapy vector design.
Authors
Didier Trono
-
Abstract
Retroviral vectors integrate in genes and regulatory elements and may cause transcriptional deregulation of gene expression in target cells. Integration into transcribed genes also has the potential to deregulate gene expression at the posttranscriptional level by interfering with splicing and polyadenylation of primary transcripts. To examine the impact of retroviral vector integration on transcript splicing, we transduced primary human cells or cultured cells with HIV-derived vectors carrying a reporter gene or a human β-globin gene under the control of a reduced-size locus-control region (LCR). Cells were randomly cloned and integration sites were determined in individual clones. We identified aberrantly spliced, chimeric transcripts in more than half of the targeted genes in all cell types. Chimeric transcripts were generated through the use of constitutive and cryptic splice sites in the HIV 5ι long terminal repeat and gag gene as well as in the β-globin gene and LCR. Compared with constitutively spliced transcripts, most aberrant transcripts accumulated at a low level, at least in part as a consequence of nonsense-mediated mRNA degradation. A limited set of cryptic splice sites caused the majority of aberrant splicing events, providing a strategy for recoding lentiviral vector backbones and transgenes to reduce their potential posttranscriptional genotoxicity.
-
Introduction
Large-scale surveys of retroviral integration in murine and human cells uncovered genomic features systematically associated with retroviral insertions and revealed that each retrovirus type has a unique, characteristic pattern of integration within mammalian genomes (1). Target-site selection depends on both viral and cellular determinants, which are ill-defined for most retroviruses. The Moloney murine leukemia virus (MLV) and its derived vectors integrate preferentially in transcriptionally active promoters and regulatory regions (1–3), while HIV and its derived lentiviral vectors (LVs) target gene-dense regions and the transcribed portion of expressed genes, away from regulatory elements (1, 3, 4). The host cell factor LEDGF/p75 has a major role in tethering HIV preintegration complexes to active genes by directly binding the viral integrase (5), a major viral determinant of target-site selection (6).
Seminal clinical studies have shown the efficacy of retroviral gene transfer for the therapy of genetic diseases (7–11). Some of these studies also showed the genotoxic consequences of retroviral gene transfer technology: insertional activation of proto-oncogenes by MLV-derived vectors caused T cell lymphoproliferative disorders in patients undergoing gene therapy for X-linked severe combined immunodeficiency (12, 13) and Wiskott-Aldrich syndrome (14) and premalignant expansion of myeloid progenitors in patients treated for chronic granulomatous disease (15, 16). The strong transcriptional enhancer present in the MLV long terminal repeat (LTR) played a major role in deregulating gene expression. Preclinical studies showed that enhancer-less (self-inactivating [SIN]), HIV-derived LVs are less likely to cause insertional tumors than MLV-derived vectors (17–20). Transcriptional gene activation, however, is not the only genotoxic event that may result from retroviral vector integration. Preclinical and clinical studies suggested that the insertion of retroviral splicing and polyadenylation signals within transcription units may cause posttranscriptional deregulation of gene expression with a certain frequency (3, 18, 21). This may include aberrant splicing, premature transcript termination, and the generation of chimeric, read-through transcripts originating from vector-borne promoters (21), a classical cause of insertional oncogenesis (22). The propensity of LVs to integrate into the body of transcribed genes increases the probability of such events compared with that of MLV-derived vectors. In addition, the deletion of the U3 region in SIN LVs results in decreased transcriptional termination and increased generation of chimeric transcripts (23). In a clinical context, insertion of a LV caused posttranscriptional activation of a truncated proto-oncogene in one patient treated for β-thalassemia, resulting in benign clonal expansion of hematopoietic progenitors (24). Analyzing the nature and frequency of posttranscriptional genotoxic events in relevant models is therefore crucial to determine the biosafety of clinical gene transfer vectors and to drive intelligent improvement of their design.
In this study, we systematically searched for aberrant transcripts in T cells, erythroid cells, and keratinocytes transduced with LVs carrying a “splice trap” or transgene (GFP and β-globin) expression cassettes. Aberrantly spliced transcripts, caused by the usage of constitutive and cryptic splice sites located in the vector or the transgene, were identified in more than 50% of the intragenic integrations in individual cell clones in the absence of selection. Abnormal transcripts were accumulated at a low level compared with constitutively spliced ones. In some cases, we show that nonsense-mediated mRNA degradation is most likely responsible for the low levels of aberrant transcripts in the cytoplasm. We propose systematic identification of cryptic splice sites as a strategy for guiding the recoding of LV backbones and transgenes to reduce their potential genotoxicity.
-
Results
Genes containing an integrated HIV-derived LV produce chimeric transcripts. To test the consequences of LV integration on the expression of targeted genes, Jurkat and SupT1 human T cell lines were transduced at high MOI with a splice trap, HIV-derived SIN LV lacking an internal promoter and carrying the EGFP reporter gene downstream of an internal ribosomal entry site (referred to as the IRES-GFP vector) (Figure 1). Upon vector integration in a transcribed gene in the same orientation, a splicing event trapping the IRES-GFP cassette into a mature transcript may result in the expression of the reporter gene. GFP expression was detected by cytofluorimetry in up to 81.5% of Jurkat and SupT1 cells 3 days after transduction. GFP+ cells were sorted and cloned by limiting dilution. The average vector copy number (VCN) per cell in 57 Jurkat and 24 SupT1 clones ranged between 5 and 7, as assayed by Southern blotting (data not shown). Proviral integration sites were mapped in all clones by linker-mediated PCR (LM-PCR) and sequenced. A majority (55.6%) of the 230 mapped integrations occurred within introns or exons of annotated genes. We focused our analysis on 59 integrations occurring in direct transcriptional orientation in a total of 30 Jurkat and 8 SupT1 clones (Supplemental Table 1; supplemental material available online with this article; doi: 10.1172/JCI61852DS1).
 Figure 1
Figure 1Schematic maps of the SIN LVs in their proviral forms. The promoter-less IRES-GFP vector contains a GFP gene inserted downstream an internal ribosomal entry site (IRES). In the CMV.GFP and K14.GFP vectors, the GFP gene is under the control of an internal, immediate-early CMV promoter or a human K14 enhancer-promoter element (K14). The GLOBE vector contains the human β-globin gene under the control of the β-globin promoter (β-p) and the HS2 and HS3 element of the β-globin LCR, in reverse transcriptional orientation. LTRs are represented by black boxes, where Δ indicates the U3 –18 deletion. RRE, rev-responsive element; SD1, HIV gag major SD site; SA7, HIV gag major SA site; (A)n, polyadenylation signal; wPRE, woodchuck hepatitis posttranscriptional regulatory element. I, II, and III exons are indicated.
Northern blot analysis of poly(A)+ RNA from 15 cell clones (12 Jurkat and 3 SupT1) showed the presence of 1 to 6 transcripts hybridizing to an EGFP probe in each clone (Supplemental Figure 1). The presence of fusion mRNAs was evaluated by reprobing the Northern blots with probes specific for the genes targeted in each clone and annealing to one or more exons upstream of the integration site. We detected transcripts co-hybridizing with EGFP and gene-specific probes in 6 out of 15 clones (Supplemental Figure 1C and data not shown), indicating that polyadenylated, chimeric transcripts generated by read-through transcription and splicing of proviral sequences may be accumulated at levels detectable by Northern blotting.
Chimeric transcripts are generated by alternative splicing through the usage of HIV constitutive and cryptic splice sites. To characterize the chimeric transcripts generated by the IRES-GFP vector, nested 5′ rapid amplification of cDNA ends (RACE) PCR or RT-PCR were performed on poly(A)+ RNA obtained from the 38 selected Jurkat and SupT1 clones, using forward primers annealing to the exons upstream of the LV integration sites (E-for, Figure 2A) and a reverse primer (Lenti-rev, Figure 2A) annealing to the provirus downstream the HIV gag major splice acceptor (SA) site SA7 (25). Fusion transcripts were detected for more than 60% of the mapped integration sites (Figure 2B). In many cases, we detected amplicons of different molecular weight with the same primer pairs, indicating the existence of multiple gene-vector fusion transcripts from the same gene (Figure 2B). Sequencing of the PCR products allowed the identification of the SA sites used in combination with splice donor (SD) sites in the upstream exon to generate the fusion transcripts. We detected transcripts spliced to the LV SA7 site (aberrant transcripts type 1, Figure 3A) or to cryptic SA sites located at the 5′ end of the provirus (aberrant transcripts type 2, Figure 3A). In particular, type-2 transcripts were generated by the use of 8 different cryptic SA sites located in the U5 region of the 5′ LTR (sites B, C, and D, Figure 3B), in the primer binding site (sites E and F, Figure 3B), in the sequence immediately downstream (site G, Figure 3B), and in the packaging signal (site H and I, Figure 3B). In a few cases, splicing occurred through the use of cryptic SA sites located in the intron upstream the proviral integration site (aberrant transcripts type 3, Figure 3A, and site A, Figure 3B). Overall, type-2 transcripts occurred most frequently (30 out of 37 sequenced transcripts, Table 1). The most frequently used SA sites were C and H, detected in 17 and 9 out of 37 splice junctions, respectively, whereas the LV SA7 site was used at a lower frequency (6 out of 37 junctions). Other sites were rarely used (1 or 2 junctions) (Table 2).
 Figure 2
Figure 25′RACE and RT-PCR analysis of aberrantly spliced transcripts in clones of Jurkat and SupT1 T cells, primary T cells, HaCaT keratinocytes, and myeloid HEL cells. (A) Schematic structure of the integrated provirus. The “expression cassette” represents the IRES-GFP splice trap in Jurkat and SupT1 cells, the CMV-GFP cassette in transduced T cells, the K14-GFP cassette in HaCaT cells, and the β-globin gene in reverse orientation in HEL cells (see Figure 1). E(n) indicates the exon upstream of the lentiviral integration, and E(n+1) indicates the downstream exon. SD1, gag major SD site; SA7, gag major SA site. Arrows indicate the vector-specific primer used to reverse transcribe the mRNAs into cDNAs (Lenti-RT), the upstream exon-specific forward primer (E-for), and the vector-specific reverse primer (Lenti-rev) used in the RT-PCR reaction. (B) 5′ RACE and RT-PCR products in representative SupT1/Jurkat, HaCaT, T lymphocytes, and HEL clones stained by ethidium bromide on 1% agarose gels. Transcripts were amplified using the Lenti-rev primer and a primer specific for the upstream exon of the gene identified on top of each lane. Molecular weight markers (sizes in bp) are indicated on the left of each gel.
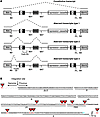 Figure 3
Figure 3Analysis of splicing variant types and mapping of cryptic HIV SA sites. (A) Schematic view of the families of chimeric transcripts generated by alternative splicing to HIV constitutive (type 1) or cryptic (type 2) SA sites or to cryptic SA sites located in the upstream intron (type 3), as identified from sequencing of the PCR products shown in Figure 2B. Exons are indicated by continuous lines, spliced sequences are indicated by dotted lines. (B) Mapping of the cryptic HIV SA sites (red triangles) identified (A) in the intron upstream of the vector integration site and (B–D) in the HIV U5 region of the LTR, (E and F) primer binding site, (G) packaging signal (Ψ), and (H and I) gag gene (SA7). The SA7 site is the gag constitutive, REV-sensitive acceptor site. The dinucleotides at the end of a spliced sequence are indicated in red. The GT dinucleotide at the beginning of the constitutive gag intron is indicated in blue. The U3, R, and U5 region of the 5′LTR are underlined. The frequency of SA site usage in all the sequenced transcripts is reported in Table 2.
 Table 1
Table 1
Aberrant transcripts generated by the usage of cryptic and constitutive SA sites in the LV backbone
LV integration causes aberrantly spliced transcripts at high frequency in different cell types. We then investigated the frequency by which chimeric transcripts are generated by the integration of conventional SIN LVs, harboring an internal gene expression cassette, in the absence of selection. To this aim, human peripheral blood primary T lymphocytes and a keratinocyte cell line (HaCaT) were transduced with LVs in which EGFP expression is driven by a constitutive CMV promoter or a tissue-specific keratin 14 (K14) promoter, respectively (CMV-GFP and K14-GFP; Figure 1). In addition, a human erythroleukemia cell line (HEL) was transduced with a LV carrying a human β-globin minigene driven by a β-globin promoter and a minimal locus-control region (LCR) in reverse transcriptional orientation (GLOBE) (Figure 1). Cells were transduced, cloned by limiting dilution, and scored as positive by FACS analysis (GFP vectors) or vector-specific PCR (GLOBE). Southern Blot analysis revealed an average VCN of 5 to 7 in HaCaT and T cell clones and 2 in HEL clones. A total of 591 integration sites (259 in HaCaT clones, 73 in HEL clones, and 259 in T lymphocytes) were mapped by LM-PCR in all clones, 382 of which (65%) (178 in HaCaT clones, 41 in HEL clones, and 163 in T lymphocytes) landed in an intron or exon of a gene. Sixty-three intragenic, forward-oriented proviruses from 49 cell clones were selected for further analysis (Supplemental Table 1). RT-PCR analysis using exon and vector-specific primers (Figure 2A) detected fusion transcripts for a total of 45 out of 63 proviruses. Aberrant transcripts were detected at high frequency in all cell types (Figure 2B). In particular, chimeric transcripts were generated in 50% of the analyzed integration events (8 out of 16) in HEL clones, 68% of the analyzed integration events (21 out of 31) in HaCaT clones, and 100% of the analyzed integration events (16 out of 16) in primary T lymphocyte clones, a significant difference with respect to cell lines (P = 5 × 10–4, Fisher’s exact test). The PCR products were cloned and sequenced to map the SA sites in the proviruses used to generate the fusion transcripts. The 3 families of transcripts generated by the IRES-GFP vector in Jurkat and SupT1 cells (types 1, 2, and 3, Figure 3A) were represented in all cell types. Again, type-2 transcripts were recovered most frequently, generated by 19 out of 28 sequenced transcripts in primary T lymphocytes, 17 out of 24 in HaCaT cells, and 13 out of 19 in HEL cells, while type-1 and type-3 transcripts were rarer (Table 1). As observed in T cell lines, the cryptic SA sites C (LTR U5 region) and H (packaging signal) were the most frequently used in primary T lymphocyte and HaCaT clones (28 out of 52 transcripts), while the SA site D (LTR U5 region) and the HIV SA7 site were the most frequently used in HEL clones (9 out of 19 transcripts) (Table 2). Two additional SA sites (E and I) were mapped in HEL cells, and one site (B) was mapped in primary T lymphocytes (Figure 3B).
Chimeric, alternatively spliced transcripts are less abundant than wild-type transcripts. To estimate the relevance of the aberrant splicing events, in terms of potential perturbation of gene expression, we estimated the relative abundance of aberrantly spliced transcripts compared with that of wild-type constitutively spliced transcripts by semiquantitative RT-PCR. RNA was reverse transcribed from all cell clones using random hexamer primers. Wild-type transcripts were amplified using E-for and E-rev primers annealing to the exons immediately upstream and downstream of the integration site, while the chimeric transcripts were amplified with the E-for and the vector-specific Lenti-rev primer (Figure 4). Aberrantly spliced transcripts were divided in 4 arbitrary classes of abundance, i.e., high, intermediate, low, and rare, depending on their relative expression level compared with that of the wild-type transcripts after 24, 28, or 33 PCR cycles (Figure 4 and Supplemental Figure 2). A transcript was classified as high when it was detected at the same PCR cycle compared with the respective wild-type transcript, intermediate when detected 4 cycles later, low when detected 8 cycles later, and rare when it was undetectable by PCR of RNA reverse transcribed with random hexamer even though it was sequenced from RNA reverse transcribed with the Lenti-RT primer (Figure 2A). We found that the majority of integrations produced rare chimeric transcripts in all cell lines, while in primary T cells the majority of integrations resulted in low and intermediate levels of aberrant splicing. Abundant transcripts were generated in less than 10% of the integrations in cell lines and in a statistically nonsignificant higher proportion (<20%; P > 0.05, Fisher’s exact test) in primary T cells (Figure 5A). We observed no correlation between abundance of alternatively spliced transcripts and the use of specific SA sites within the provirus.
 Figure 4
Figure 4Semiquantitative PCR analysis of wild-type and aberrantly spliced transcripts from the HCFC2, PLEKHA7, and DOCK5 genes in HaCaT clones transduced with the K14-GFP vector. (A) cDNAs were prepared using random hexamer primers from Poly(A)+ RNA. Wild-type transcripts were amplified using the E-for and E-rev primers (arrows) annealing to the exons upstream and downstream of the provirus. Fusion transcripts were amplified using the E-for and Lenti-rev primers. (B) PCR reactions were arrested at 24, 28, and 33 cycles and run on 1% agarose gels in the following order (from left): wild-type transcript amplified from the HaCaT clone; wild-type transcript amplified from a HaCaT bulk culture; fusion transcript(s) in the HaCaT clone; and GAPDH transcript in the HaCaT clone, used for signal normalization. The GAPDH transcript in the HaCaT bulk culture was run on each gel but shown only once at the right of all panels. Transcripts were ranked in 4 arbitrary classes of relative abundance, i.e., low, when fusion transcripts were detected 8 PCR cycles later than wild-type transcripts; intermediate, when fusion transcripts were detected 4 PCR cycles later than wild-type transcripts; and high, when chimeric and wild-type transcripts were detected after the same number of PCR cycles. A fusion transcript was classified as rare (data not shown) when it was undetectable after 33 PCR cycles starting from RNA reverse transcribed with random hexamers, although it was detected and sequenced using RNA reverse transcribed with the vector-specific Lenti-RT primer (Figure 2).
 Figure 5
Figure 5Summary of the relative frequency (percentage) of the 4 classes of abundance of aberrantly spliced transcripts recovered in all analyzed cell clones. (A) Aberrant transcripts generated from LV proviruses integrated in forward orientation in Jurkat/SupT1, primary T cell, HaCaT, and HEL clones, ranked in the high (red), intermediate (yellow), low (blue), rare (gray), and absent (black) abundance classes, as defined by the semiquantitative PCR assay shown in Figure 4. (B) Aberrant transcripts generated from the GLOBE proviruses integrated in reverse orientation in HEL clones. The total number of analyzed proviruses (n) is indicated above each bar.
To validate the semiquantitative RT-PCR test, we estimated the relative abundance of chimeric transcripts compared with that of constitutive transcripts by qPCR in 15 cell clones (Supplemental Table 5). Transcripts classified as high, intermediate, or low by semiquantitative PCR were estimated as having a relative abundance of 11.6% ± 3.7%, 4.8% ± 1.7%, and 1.5% ± 0.8%, respectively, with respect to wild-type transcripts by qPCR, although with a somewhat overlapping distribution of values, confirming the results obtained by the semiquantitative assay. Examples of direct comparisons between the 2 assays are shown in Supplemental Figure 3.
A β-globin transgene provides alternative splicing signals when integrated into active genes. A popular way to express an intron-containing transgene in a LV is to insert it in reverse transcriptional orientation with respect to the vector backbone. The paradigm antisense LVs were those expressing the human β-globin gene, such as GLOBE (Figure 1). To analyze the consequences of integrating a transgene containing natural intron-exon junctions on target gene expression, we analyzed HEL clones transduced with GLOBE and harboring reverse-oriented proviruses, in which the β-globin transgene is in the same transcriptional orientation as the target gene. RT-PCR analysis was performed by using a forward primer annealing to the exon upstream of the β-globin transgene (E-for, Figure 6A) and a reverse primer specific for the β-globin third exon (Globin-rev, Figure 6A). We were able to detect chimeric transcripts in 55% (12 out of 22) of the analyzed proviruses in 13 HEL clones. Cloning and sequencing of the PCR products identified 4 species of transcripts: type-4 transcripts, splicing the upstream exon SD site to the constitutive SA site of the second intron of the β-globin gene; type-5 transcripts, splicing the upstream exon SD to a cryptic SA site located in the first exon of the β-globin; type-6 transcripts, splicing the upstream exon SD to cryptic SA sites located in the LCR HS3 element and cryptic SD sites in HS3 to the constitutive SA site of the β-globin second intron; and type-7 transcripts, splicing cryptic SD sites in HS3 to the cryptic SA site in the β-globin first exon (Figure 6B). Constitutive splicing of the β-globin second and third exons occurred in all transcript types, while the first exon was retained in type-5 and type-7 transcripts (Figure 6B). In terms of relative frequency, aberrant type-4, -6, and -7 transcripts were all found in approximately 27% of the cases (9, 9, and 8 out of 29 sequenced transcripts, respectively), while type-5 transcripts were detected in only 3 cases (Table 3). Sequencing of the splice junctions identified 6 different cryptic SD sites (sites A–F, Figure 6C) and 3 SA sites (sites J–L, Figure 6C) in the HS3 element and 1 cryptic SA site in the 5′ UTR of the β-globin first exon (site M, Figure 6C). The most frequently used SA sites were J and M, identified in 10 and 13 out of 27 sequenced transcripts, respectively (Table 4). The most frequently used SD sites were B, C, and F, mapped in 10, 6, and 4 out of 24 transcripts, respectively, while the A, D, and E sites were found in only 1 or 2 cases (Table 5).
 Figure 6
Figure 6RT-PCR analysis of aberrantly spliced transcripts in random, unselected clones of HEL cells transduced with the GLOBE vector. (A) Schematic structure of the GLOBE provirus integrated between exons E(n) and E(n+1) in reverse transcriptional orientation. SD, SD site; SA, SA site. E-for, E-rev, and Globin-rev primers are indicated by arrows. The β-globin HS3 and HS2 LCR elements; promoter; I, II, and III exons; and polyadenylation signal are indicated. (B) Schematic view of the families of chimeric transcripts generated by alternative splicing to the β-globin first intron constitutive SA site (type 4) or to the β-globin promoter (type 5) and HS3 (type 6 and 7) cryptic SA sites, as identified from sequencing of the PCR products. Exons are indicated by continuous lines, spliced sequences are indicated by dotted lines. (C) Mapping of the cryptic SA (red triangles) and SD (blue triangles) sites identified in the HS3 element and the 5′ UTR of the β-globin gene. The dinucleotides at the beginning or end of a spliced sequence are indicated in blue and red, respectively. The frequency of SD and SA site usage in all the sequenced transcripts is reported in Table 3.
 Table 3
Table 3
Aberrant transcripts generated by the usage of cryptic and constitutive SA and SD sites in the β-globin expression cassette
To determine the relevance of alternative splicing generated by the use of splice 1 and 2 signals in the β-globin gene, we estimated the relative abundance of alternatively spliced transcripts compared with that of wild-type constitutive spliced transcripts by the semiquantitative RT-PCR assay described above. We used E-for and E-rev primers to amplify constitutive transcripts, and E-for and Globin-rev primers to amplify chimeric transcripts (Figure 6A and Figure 7). Aberrantly spliced transcripts were divided in the 4 arbitrary classes of abundance, as described above. The majority of transcripts were rare, while less than 15% of the transcripts belonged to the high class (Figure 5B).
 Figure 7
Figure 7Semiquantitative PCR analysis of wild-type and aberrantly spliced transcripts from the EFR3A, QARS, and HMG20A genes in HEL clones transduced with the GLOBE vector. cDNAs were prepared using random hexamer primers from Poly(A)+ RNA. Wild-type transcripts were amplified using the E-for and E-rev primers, annealing to the exons immediately upstream and downstream of the provirus. Fusion transcripts were amplified using the E-for and Globin-rev primers. PCR reactions were stopped at 24, 28, and 33 cycles and run on 1% agarose gels in the following order: wild-type transcript amplified from the HEL clone (first lane from left); wild-type transcript amplified from a HEL bulk culture (second lane); fusion transcript in the HEL clone (third lane); GAPDH transcript in the HEL clone, used for signal normalization (fourth lane); and GAPDH transcript in the HEL bulk culture (fifth lane). Transcripts were ranked in 4 arbitrary classes of relative abundance, i.e., rare, low, intermediate, and high, as described in Figure 4.
The strength of the vector splice signals determines the frequency of alternative splicing. Alternative splicing is favored by the presence of weak splice signals, i.e., departing from the optimal consensus sequences of SD, branch point, and SA sites. To test whether LV cryptic and canonical splicing signals are preferentially used in the presence of weak cellular splice sites, we used ESEfinder and Splice Analyzer Tool (SAT) software to predict the strength of SA signals on 100 bp of genomic sequence encompassing the upstream polypyrimidine tract and the canonical AG dinucleotide at the intron-exon boundary of the exons downstream of all 144 analyzed LV integration sites. SA sites of genes associated with high and intermediate levels of aberrant splicing were compared with SA sites of less affected genes (rare or absent by semiquantitative PCR). No significant difference was observed in the strength scores between the 2 groups (ESEfinder, 8.7 vs. 9.3, P > 0.2; SAT, 86.1 vs. 86.7, P > 0.6), both containing SA sites perfectly fitting the human consensus SA sequence (Figure 8). These results show that the strength of the constitutive splice signals in the targeted genes does not predict the extent of vector-induced alternative splicing.
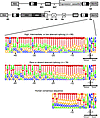 Figure 8
Figure 8Analysis of the strength of the constitutive SA signals in the introns targeted by LV integration. enoLOGOS plots of the consensus sequences (49 bp) encompassing the SA sites of the exons downstream of the integration sites of proviruses generating rare (top panel) or relatively abundant (middle panel) fusion transcripts. The human SA consensus sequence (23 bp) is shown in the bottom panel. Nucleotide positions are conventionally numbered, starting from the intron/exon boundary (intron, –1 to –46; exon, +1 to +3). For each position, the height of the letter represents the frequency of the corresponding base at that position. A schematic structure of the integrated provirus with upstream and downstream exons is shown in the top panel.
On the contrary, the sequences encompassing the forward-strand proviral constitutive or cryptic SA sites showed lower scores (ESEfinder, 7.7 and SAT, 84.1) than the SA sites of the downstream exons and departed substantially from the human consensus SA sequence (Supplemental Table 2). The SA 3′ end contained the canonical AG dinucleotide in the most frequently used C, D, and H cryptic sites located in the LTR and primer-binding region (Figure 3B and Table 2), which collectively accounted for 59% (69 out of 118) of the sequenced junctions. Of these 3 sites, only H featured a pyrimidine-rich tract (Supplemental Table 2). In other sites, the AG dinucleotide was replaced respectively by AC (B, F, G, and I sites) and GC (E site), while a loose polypyrimidine tract was present only in the B and I sites. On the opposite strand, the most used SA sites were J in the globin LCR HS3 element and M in the 5ι UTR of the β-globin first exon (Figure 6C and Table 4), both featuring an AG dinucleotide and a pyrimidine-rich tract (Supplemental Table 3). Interestingly, the 2 most used SD sites in the HS3 element (sites B and C, Figure 6C and Table 5) contained the canonical GU dinucleotide and fit well (6 out of 8 and 5 out of 8 bases, respectively) the mammalian consensus sequence AGGURAGU (conserved GU dinucleotide in bold) (Supplemental Table 4). These data indicate that the relative usage of cryptic SD and SA splice sites in an integrated provirus is proportional to their homology to the human splice consensus sequences. The fact that we sequenced more than one transcript species containing the same noncanonical splice sites and that each species was found in more than one cell clone excluded that junctions containing noncanonical splice sites were the consequence of RT-PCR artifacts.
The abundance of aberrantly spliced transcripts is limited by nonsense-mediated mRNA decay. Fusion transcripts between endogenous, coding exons and proviral or β-globin sequences may be susceptible to recognition and degradation by the nonsense-mediated mRNA decay (NMD) cytoplasmic machinery, which normally degrades mature transcripts containing mutations that result in a premature termination codon. To test whether exon-provirus fusion transcripts are susceptible to regulation by the NMD system, we first computationally translated the aberrant transcript sequences and found that the vast majority of them do contain premature termination codons. Then, we performed semiquantitative PCR of 3 selected transcripts from HEL cell clones and 4 from HaCaT cell clones (n = 2 and n = 4, respectively) treated for 8 hours with 50–100 μM cycloheximide, an inhibitor of NMD degradation. We observed an increase in the relative amount of chimeric transcripts in 3 out of 6 clones after cycloheximide treatment (Figure 9) and, in one case (PLEKHA7), the appearance of a new, unspliced RNA species (Figure 9A). Sequencing of this transcript revealed a less-spliced species containing the entire LTR as well as the intron upstream the integration site (results not shown). These results indicate that mRNAs generated by aberrant splicing caused by proviral insertion are likely to be rapidly degraded by the NMD complex, thus reducing their accumulation compared with that of wild-type mRNAs.
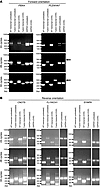 Figure 9
Figure 9RT-PCR analysis of aberrantly spliced transcripts from the PBX4, PLEKHA7, CNOT6, FLYWCH1, and SYMPK genes in cycloheximide-treated cell clones. (A) HaCaT and (B) HEL clones were treated with 100 or 50 μg/ml cycloheximide (CHX), respectively, and cDNAs were prepared from poly(A)+ RNA using random hexamer primers. Wild-type transcripts were amplified using E-for and E-rev primers (Figure 4), fusion transcripts were amplified using the E-for and Lenti-rev primers (Figure 4) in HaCaT clones and E-for and Globin-rev primers (Figure 6A) in HEL clones. PCR reactions were stopped at 24, 28, and 33 cycles and run on 1% agarose gels in the following order: wild-type transcript amplified from the untreated HaCaT/HEL clone (first lane from left); fusion transcript in the untreated HaCaT/HEL clone (second lane); wild-type transcript amplified from the cycloheximide-treated HaCaT/HEL clone (third lane); fusion transcript in the cycloheximide-treated HaCaT/HEL clone (fourth lane); and GAPDH transcript in the untreated (fifth lane) and cycloheximide-treated (sixth lane) HaCaT/HEL clone, used for signal normalization.
-
Discussion
Retroviruses and retroviral vectors integrate in the cell genome by mechanisms that couple integration with target-site selection, according to virus-specific patterns. MLV-derived gamma-retroviral vectors integrate preferentially in active regulatory regions (1, 3), thereby increasing their chance to deregulate host gene expression at the transcriptional level. Insertional activation of proto-oncogenes has in fact been observed in patients treated with MLV-derived vectors (12, 13, 15, 16). On the contrary, HIV-derived LVs integrate away from regulatory elements (1, 3, 4), a characteristic that significantly reduces transcriptional gene activation and consequently their genotoxicity (17, 19, 20). On the other hand, the propensity of LVs to target the transcribed portion of expressed genes increases their chances to deregulate gene expression by interfering with splicing and polyadenylation of primary transcripts. Posttranscriptional genotoxicity cannot be prevented by the use of regulated promoters or enhancers, as it depends on the endogenous activity of the targeted gene. Preclinical and clinical studies showed that MLV and HIV insertion may indeed deregulate gene expression at the posttranscriptional level (3, 18, 21, 24), leading to both clonal loss (26) and clonal expansion (24) of transduced cells in patients. In particular, LV-borne internal promoters were reported to generate read-through transcripts extending into downstream genes, favored by the suboptimal characteristics of the 3ι LTR polyadenylation signals (21).
In this study, we show the considerable potential of HIV-derived LVs to generate abnormally spliced transcripts upon integration in the human genome. The use of a splice trap allowed selection for T cell clones in which LV integration caused aberrant splicing and detection of fusion transcripts between endogenous genes and vector sequences in the majority of the integration events. Strikingly, in unselected HEL erythroid cells, HaCaT keratinocytes, and primary T cells, more than 50% of the proviruses integrated in either orientation within a transcription unit caused some splicing alteration and the generation of fusion transcripts. In primary T cells, such transcripts were detected at a frequency of more than 80%. Aberrant splicing was caused by all analyzed LVs, containing either a conventional cDNA expression cassette in forward orientation or a complete β-globin gene and minimal LCR in reverse orientation. Splicing signals carried by LVs thus cause alternative splicing at very high frequency upon integration in transcribed genes, an event that accounts for more than 70% of LV integrations in most cell types (1, 3, 4, 26).
We sequenced a substantial number of fusion transcripts from all analyzed cell types and mapped the SA and SD sites used by the cell splicing machinery on both vector strands. Surprisingly, cryptic sites located in the LTRs, primer binding site and the gag gene on one strand, and in the β-globin promoter and LCR on the opposite strand generated fusion transcripts at higher frequency compared with the constitutive sites located in the HIV gag and β-globin genes. In fact, the gag intron and the 2 β-globin introns were fully spliced in most fusion transcripts, indicating that the splicing machinery removes canonical introns efficiently by using their native SD and SA sites and that most of the aberrant splicing events are caused by uncoupled, cryptic splice signals. As expected, strong cryptic sites were used more frequently than weak ones, as defined by their degree of homology with the human SA/branch site and SD consensus sequences.
The clonal nature of the analyzed cells allowed for estimating the relative abundance of aberrant transcripts compared with that of wild-type transcripts by a relatively simple, semiquantitative assay based on PCR amplification, starting from reverse transcribed RNA. The assay did not provide an absolute measure of transcript accumulation but only a relative one, which was independent from the expression level of the gene hit by the integration event. qPCR validation indicated that the semiquantitative assay predicts the relative abundance of the aberrantly spliced transcripts well and allows the simultaneous detection of multiple splicing variants generated by the same provirus. Interestingly, most of the fusion transcripts were far less abundant than wild-type ones for all cell types, vectors, and orientations, indicating that alternative splicing generated by the use of proviral sites is less efficient than constitutive splicing of upstream and downstream exons. In some cases, we obtained evidence that nonsense-mediated RNA decay has a remarkable role in reducing the abundance of aberrant transcripts, thus limiting the potentially negative consequences of this type of insertional mutagenesis. Contrary to our expectations, the strength of a downstream exon SA site was not predictive of the frequency of an alternative splicing event: proviruses generating either rare or abundant fusion transcripts were inserted upstream of exons containing SA sites featuring the same degree of homology to the optimal consensus sequence. This indicates that the interaction between upstream SD sites and proviral SA sites is not simply regulated by competition with the downstream constitutive SA site.
The relative inefficiency of provirus-induced alternative splicing has important implications in terms of vector genotoxicity. Based on our observations, the insertion of a LV in a transcription unit is expected to cause only a moderate loss of mature mRNA and not a monoallelic gene knockout, at least in most cases. Integration into “dangerous” genes, such as tumor suppressors, is therefore unlikely to lead to true loss-of-function mutations. We did observe integration in cancer-related genes for more than 20% of the mapped proviruses (Supplemental Table 1), e.g., PTEN, a tumor suppressor gene, in 2 independent T cell clones and KDM5A (encoding an Rb-binding protein) in another clone. These integrations generated fusion transcripts detectable by Northern blotting, though no significant reduction of wild-type mRNA accumulation (Supplemental Figure 1). However, in more than 10% of the mapped proviruses, particularly in primary T cells, fusion transcripts were apparently as abundant as constitutive transcripts by semiquantitative PCR, suggesting that in some cases gene expression could be substantially downregulated by vector integration. We previously reported downregulation of genes hit by LVs and MLV vectors in primary T cells (18) and selective loss of T cell clones carrying forward-oriented, intragenic proviruses in patients treated with transduced T cells (26, 27). These studies suggested that downregulation of gene expression caused by vector insertion does have consequences on cell homeostasis, although they appeared to be reduced cell fitness rather than clonal expansion or transformation (26, 27). Moreover, aberrant splicing caused by cryptic proviral signals may occasionally lead to gain-of-function mutations, as observed for the HMGA2 proto-oncogene in myeloid cells transduced by a β-globin LV (24). Removing the cryptic splice signals from vectors designed for clinical application appears therefore as a desirable safety measure. This study shows that it is relatively easy to map the cryptic splice signals contained on both strands of a given vector by analyzing a small number of random cell clones. The sites can then be removed by careful recoding of the vector sequence. The fact that constitutive introns appear to interfere only marginally with cellular gene splicing suggests that intron-containing genes may still be incorporated in a recoded vector backbone if necessary for a specific therapeutic application, as in the case of thalassemia.
-
Methods
Vectors and cells. To generate the IRES-GFP vector, an EcoRV-SmaI fragment containing the internal ribosomal entry site of the encephalomyocarditis virus was introduced into the EcoRV-SmaI sites of the pRRLppt.PGK.GFP.WPRE-18 SIN-LV plasmid (28). The CMV-GFP, K14-GFP, and GLOBE vectors were previously described (29–31). VSV-G pseudotyped viral stocks were produced by transient transfection in 293-T cells and concentrated and titrated as previously described (2). Jurkat, SupT1, HaCaT, and HEL cells were maintained in DMEM or RPMI 1640 (HEL) supplemented with 10% fetal bovine serum, 100 U/ml penicillin, 0.1 mg/ml streptomycin, and 2 mM l-glutamine and transduced with viral supernatant at an MOI of 10 in the presence of 4 μg/ml polybrene. Transduced cells were enriched by FACS, cloned by limiting dilution (0.3 cells per well), and screened for GFP expression by cytofluorimetry. Primary human lymphocytes were obtained from Ficoll-Hypaque mononuclear cell fractions isolated from cord blood or peripheral blood from healthy donors and stimulated in culture with X-VIVO-15 (BioWhittaker) supplemented with 10% human serum (Cambrex BioScience), 50 U/ml IL-2 (Chiron), and 25 U/ml IL-7 (ImmunoTools) in the presence of CD3/CD28 T cell expander (Dynal) at a ratio of 0.5 bead per cell for 3 days. Lymphocytes were transduced by spinoculation, cloned by limiting dilution in 96-well plates at a concentration of 0.3 to 1 cells per well as previously described (32), and screened for GFP expression by cytofluorimetry. Average VCN was assessed by Southern blotting on genomic DNA using a 32P-labeled GFP probe or by qPCR on genomic DNA, using primers and probe annealing to the proviral rev-responsive element (RRE) region, as previously described (31).
For NMD inhibition studies, HEL and HaCaT cells were seeded at 5 × 105 cells/ml in medium containing 50 or 100 μM cycloheximide, respectively (Sigma-Aldrich) and harvested after 8 hours for RNA extraction.
Analysis of lentiviral integration sites. Integration sites were determined in individual cell clones by LM-PCR, as previously described (2). Briefly, genomic DNA was extracted from cell clones, digested with MseI, and ligated to an MseI double-stranded linker. LM-PCR was performed with nested primers specific for the linker and the 3′ HIV LTR (2). PCR products were shotgun cloned (TOPO TA Cloning Kit, Invitrogen) into libraries of integration junctions, which were then sequenced to saturation. Sequences were mapped on the human genome (UCSC Human Genome Project Working Draft, hg18) by the BLAT genome browser ( http://genome.ucsc.edu/cgi-bin/hgBlat?command=start), requiring a 98% identity over the entire sequence length and selecting the best hit. We annotated an integration as intragenic when occurring inside the transcribed portion of a known gene (UCSC definition). In case of multiple transcripts from the same gene, we arbitrarily chose the longest isoform.
RNA extraction and Northern blot analysis. Total cellular RNA was extracted using TRI Reagent (Sigma-Aldrich), and polyadenylated transcripts were magnetically isolated using Dynabeads Oligo dT (Invitrogen). Poly(A)+ RNA was run on a 1.2% agarose-formaldehyde gel, blotted onto nylon membranes, and hybridized with a 32P-labeled GFP probe. To detect specific chimeric transcripts, the membrane was stripped and rehybridized with 32P-labeled probes specific for the exons upstream from the vector integration site.
5′ RACE-PCR and RT-PCR. Poly(A)+ RNA was reverse transcribed using an internal provirus-specific reverse primer annealing downstream of the HIV gag SA7 (Lenti-RT, Figure 2A) or random hexamers, following the manufacturer’s instructions (ROCHE First-Strand cDNA Synthesis Kit). 5′ RACE-PCR was carried out according to the manufacturer’s instructions (Invitrogen) using RNA reverse transcribed with the Lenti-RT primer. RT-PCR was performed using the Lenti-rev reverse primer (Figure 3A) or a primer annealing to the third exon of the β-globin transgene (Globin-rev, Figure 6A), in combination with a forward primer annealing to an exon upstream of the vector integration site (E-for, Figure 2A) or a forward primer annealing to the exon upstream the β-globin transgene (E-for, Figure 6A). Primer sequences are listed in Supplemental Table 6. PCR products were shotgun cloned (TOPO TA Cloning Kit, Invitrogen) and sequenced.
Semiquantitative and quantitative RT-PCR. Poly(A)+ RNA was reverse transcribed using random hexamer primers. To detect constitutively spliced transcripts, PCR was carried out using primers annealing to the exons immediately upstream and downstream of the vector integration site (E-for/E-rev in Figure 4 and Figure 6A). To detect fusion transcripts, the E-for primer was used in combination with either the Lenti-rev (forward strand) or the Globin-rev (reverse strand) primer (Figure 4 and Figure 6A). PCR reactions were run for 24, 28, and 33 cycles. PCR products were run onto 1% to 1.5% agarose gels and visualized by ethidium bromide staining. Real-time qPCR was carried out using the SYBR Green method on an ABI instrument. In each experiment, duplicates of a standard dilution series of specific PCR fragments for the endogenous and aberrant transcript were amplified in a 25-μl reaction containing 1X SYBR Green Master Mix (Applied Biosystems) and 200 nM of primer pair E-for/E-rev for the endogenous transcript or E-for–GSP7 for the aberrant transcript (Figure 4). The thermal profile consisted of 1 cycle at 95°C for 10 minutes, followed by 40 cycles at 95°C for 30 seconds and at 60°C for 1 minute. The relative expression of each transcript was assessed by considering the Ct and efficiency values and normalized to the GAPDH expression level.
Sequence analysis. To assess the strength of cellular and proviral SA sites, we used the publicly available ESEfinder (33) and SAT (34) software. Fifty-nucleotide-long sequences encompassing the SA site located downstream of each proviral integration were aligned, and the splice scores were obtained. The sequences starting 40 nucleotides upstream the AG dinucleotide were aligned using the enoLOGOS web-based tool (35), which provides a graphic representation of the consensus sequences.
Statistics. Statistical significance of data comparisons was determined using a 2-tailed Student’s t test or a 2-sided Fisher’s exact test. The threshold for statistical significance was set at a P value of less than 0.05.
Study approval. All human studies were approved by the San Raffaele Scientific Institute Ethical Committee. Written informed consent was received from participants prior to inclusion in the study.
-
Acknowledgments
This work was supported by grants from the Italian Ministry of Health, the Italian Ministry of University and Research, the European Commission (FP7-PERSIST), and Telethon (TIGET core grant). A. Moiani is a student of the PhD program in Cellular and Molecular Biology at Vita-Salute San Raffaele University, Milan, Italy.
Address correspondence to: Fulvio Mavilio, Department of Biomedical Sciences, University of Modena and Reggio Emilia, Via Campi 273, 41125 Modena, Italy. Phone: 39.059.2058076; Fax: 39.059.2058015; E-mail: [email protected]. Or to: Giuliana Ferrari, HSR-TIGET, Istituto Scientifico H. San Raffaele, Via Olgettina 58, 20132 Milan, Italy. Phone: 39.02.26434705; Fax: 39.02.26434668; E-mail: [email protected].
-
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Reference information: J Clin Invest. 2012;122(5):1653–1666. doi:10.1172/JCI61852.
Fulvio Mavilio’s present address is: Genethon, Evry, France.
Claudia Cattoglio’s present address is: Department of Molecular and Cell Biology, University of California at Berkeley, Berkeley, California, USA.
See the related articles at Whole transcriptome characterization of aberrant splicing events induced by lentiviral vector integrations and Gene therapy: too much splice can spoil the dish.
-
References
- Bushman F, et al. Genome-wide analysis of retroviral DNA integration. Nat Rev Microbiol. 2005;3(11):848–858.
- Cattoglio C, et al. Hot spots of retroviral integration in human CD34+ hematopoietic cells. Blood. 2007;110(6):1770–1778.
- Cattoglio C, et al. High-definition mapping of retroviral integration sites identifies active regulatory elements in human multipotent hematopoietic progenitors. Blood. 2010;116(25):5507–5517.
- Wang GP, Ciuffi A, Leipzig J, Berry CC, Bushman FD. HIV integration site selection: analysis by massively parallel pyrosequencing reveals association with epigenetic modifications. Genome Res. 2007;17(8):1186–1194.
- Engelman A, Cherepanov P. The lentiviral integrase binding protein LEDGF/p75 and HIV-1 replication. PLoS Pathog. 2008;4(3):e1000046.
- Lewinski MK, et al. Retroviral DNA integration: viral and cellular determinants of target-site selection. PLoS Pathog. 2006;2(6):e60.
- Aiuti A, et al. Gene therapy for immunodeficiency due to adenosine deaminase deficiency. N Engl J Med. 2009;360(5):447–458.
- Hacein-Bey-Abina S, et al. Sustained correction of X-linked severe combined immunodeficiency by ex vivo gene therapy. N Engl J Med. 2002;346(16):1185–1193.View this article via: PubMed Google Scholar
- Mavilio F, et al. Correction of junctional epidermolysis bullosa by transplantation of genetically modified epidermal stem cells. Nat Med. 2006;12(12):1397–1402.
- Cartier N, et al. Hematopoietic stem cell gene therapy with a lentiviral vector in X-linked adrenoleukodystrophy. Science. 2009;326(5954):818–823.
- Boztug K, et al. Stem-cell gene therapy for the Wiskott-Aldrich syndrome. N Engl J Med. 2010;363(20):1918–1927.
- Hacein-Bey-Abina S, et al. Insertional oncogenesis in 4 patients after retrovirus-mediated gene therapy of SCID-X1. J Clin Invest. 2008;118(9):3132–3142.
- Howe SJ, et al. Insertional mutagenesis combined with acquired somatic mutations causes leukemogenesis following gene therapy of SCID-X1 patients. J Clin Invest. 2008;118(9):3143–3150.
- Krause D. Gene therapy for Wiskott–Aldrich syndrome: benefits and risks. The Hematologist. 2011;8(2):10.
- Ott MG, et al. Correction of X-linked chronic granulomatous disease by gene therapy, augmented by insertional activation of MDS1-EVI1, PRDM16 or SETBP1. Nat Med. 2006;12(4):401–409.
- Stein S, et al. Genomic instability and myelodysplasia with monosomy 7 consequent to EVI1 activation after gene therapy for chronic granulomatous disease. Nat Med. 2010;16(2):198–204.
- Montini E, et al. Hematopoietic stem cell gene transfer in a tumor-prone mouse model uncovers low genotoxicity of lentiviral vector integration. Nat Biotechnol. 2006;24(6):687–696.
- Maruggi G, et al. Transcriptional enhancers induce insertional gene deregulation independently from the vector type and design. Mol Ther. 2009;17(5):851–856.
- Modlich U, et al. Insertional transformation of hematopoietic cells by self-inactivating lentiviral and gammaretroviral vectors. Mol Ther. 2009;17(11):1919–1928.
- Montini E, et al. The genotoxic potential of retroviral vectors is strongly modulated by vector design and integration site selection in a mouse model of HSC gene therapy. J Clin Invest. 2009;119(4):964–975.
- Almarza D, Bussadori G, Navarro M, Mavilio F, Larcher F, Murillas R. Risk assessment in skin gene therapy: viral-cellular fusion transcripts generated by proviral transcriptional read-through in keratinocytes transduced with self-inactivating lentiviral vectors. Gene Ther. 2011;18(7):674–681.
- Nilsen TW, et al. c-erbB activation in ALV-induced erythroblastosis: novel RNA processing and promoter insertion result in expression of an amino-truncated EGF receptor. Cell. 1985;41(3):719–726.
- Yang Q, Lucas A, Son S, Chang LJ. Overlapping enhancer/promoter and transcriptional termination signals in the lentiviral long terminal repeat. Retrovirology. 2007;4:4.
- Cavazzana-Calvo M, et al. Transfusion independence and HMGA2 activation after gene therapy of human beta-thalassaemia. Nature. 2010;467(7313):318–322.
- Purcell DF, Martin MA. Alternative splicing of human immunodeficiency virus type 1 mRNA modulates viral protein expression, replication, and infectivity. J Virol. 1993;67(11):6365–6378.View this article via: PubMed Google Scholar
- Cattoglio C, et al. High-definition mapping of retroviral integration sites defines the fate of allogeneic T cells after donor lymphocyte infusion. PLoS One. 2010;5(12):e15688.
- Recchia A, et al. Retroviral vector integration deregulates gene expression but has no consequence on the biology and function of transplanted T cells. Proc Natl Acad Sci U S A. 2006;103(5):1457–1462.
- De Palma M, Venneri MA, Naldini L. In vivo targeting of tumor endothelial cells by systemic delivery of lentiviral vectors. Hum Gene Ther. 2003;14(12):1193–1206.
- Follenzi A, Sabatino G, Lombardo A, Boccaccio C, Naldini L. Efficient gene delivery and targeted expression to hepatocytes in vivo by improved lentiviral vectors. Hum Gene Ther. 2002;13(2):243–260.View this article via: PubMed Google Scholar
- Di Nunzio F, et al. Correction of laminin-5 deficiency in human epidermal stem cells by transcriptionally targeted lentiviral vectors. Mol Ther. 2008;16(12):1977–1985.
- Miccio A, et al. In vivo selection of genetically modified erythroblastic progenitors leads to long-term correction of beta-thalassemia. Proc Natl Acad Sci U S A. 2008;105(30):10547–10552.View this article via: PubMed Google Scholar
- Marktel S, et al. Immunologic potential of donor lymphocytes expressing a suicide gene for early immune reconstitution after hematopoietic T-cell-depleted stem cell transplantation. Blood. 2003;101(4):1290–1298.
- Cartegni L, Wang J, Zhu Z, Zhang MQ, Krainer AR. ESEfinder: A web resource to identify exonic splicing enhancers. Nucleic Acids Res. 2003;31(13):3568–3571.View this article via: PubMed Google Scholar
- The AST lab of Tel Aviv University. Splice Analyzer Tool. Tel Aviv University Web site. http://ibis.tau.ac.il/ssat/SpliceSiteFrame.htm . Accessed March 8, 2012.
- Workman CT, Yin Y, Corcoran DL, Ideker T, Stormo GD, Benos PV. enoLOGOS: a versatile web tool for energy normalized sequence logos. Nucleic Acids Res. 2005;33(Web Server issue):W389–W392.
-
Version history
- Version 1 (April 23, 2012): No description
- Version 2 (May 1, 2012): No description



Copyright © 2024 American Society for Clinical Investigation
ISSN: 0021-9738 (print), 1558-8238 (online)














