Advertisement
Research ArticleVirology Free access | 10.1172/JCI45157
Cumulative mechanisms of lymphoid tissue fibrosis and T cell depletion in HIV-1 and SIV infections
Ming Zeng,1 Anthony J. Smith,1 Stephen W. Wietgrefe,1 Peter J. Southern,1 Timothy W. Schacker,2 Cavan S. Reilly,3 Jacob D. Estes,4 Gregory F. Burton,5 Guido Silvestri,6 Jeffrey D. Lifson,4 John V. Carlis,7 and Ashley T. Haase1
1Department of Microbiology and 2Department of Medicine, Medical School, University of Minnesota, Minneapolis, Minnesota, USA. 3Division of Biostatistics, School of Public Health, University of Minnesota, Minneapolis, Minnesota, USA. 4AIDS and Cancer Virus Program, Science Applications International Corporation–Frederick Inc., National Cancer Institute, Frederick, Maryland, USA. 5Department of Chemistry and Biochemistry, Brigham Young University, Provo, Utah, USA. 6Yerkes National Primate Research Center, and Emory University, Atlanta, Georgia, USA. 7Department of Computer Science and Engineering, Institute of Technology, University of Minnesota, Minneapolis, Minnesota, USA.
Address correspondence to: Ashley T. Haase, Department of Microbiology, University of Minnesota, Box 196, 420 Delaware Street SE, Minneapolis, Minnesota 55455, USA. Phone: 612.624.4442; Fax: 612.626.0623; E-mail: [email protected].
Authorship note: Ming Zeng and Anthony J. Smith contributed equally to this work.
Find articles by Zeng, M. in: JCI | PubMed | Google Scholar
1Department of Microbiology and 2Department of Medicine, Medical School, University of Minnesota, Minneapolis, Minnesota, USA. 3Division of Biostatistics, School of Public Health, University of Minnesota, Minneapolis, Minnesota, USA. 4AIDS and Cancer Virus Program, Science Applications International Corporation–Frederick Inc., National Cancer Institute, Frederick, Maryland, USA. 5Department of Chemistry and Biochemistry, Brigham Young University, Provo, Utah, USA. 6Yerkes National Primate Research Center, and Emory University, Atlanta, Georgia, USA. 7Department of Computer Science and Engineering, Institute of Technology, University of Minnesota, Minneapolis, Minnesota, USA.
Address correspondence to: Ashley T. Haase, Department of Microbiology, University of Minnesota, Box 196, 420 Delaware Street SE, Minneapolis, Minnesota 55455, USA. Phone: 612.624.4442; Fax: 612.626.0623; E-mail: [email protected].
Authorship note: Ming Zeng and Anthony J. Smith contributed equally to this work.
Find articles by Smith, A. in: JCI | PubMed | Google Scholar
1Department of Microbiology and 2Department of Medicine, Medical School, University of Minnesota, Minneapolis, Minnesota, USA. 3Division of Biostatistics, School of Public Health, University of Minnesota, Minneapolis, Minnesota, USA. 4AIDS and Cancer Virus Program, Science Applications International Corporation–Frederick Inc., National Cancer Institute, Frederick, Maryland, USA. 5Department of Chemistry and Biochemistry, Brigham Young University, Provo, Utah, USA. 6Yerkes National Primate Research Center, and Emory University, Atlanta, Georgia, USA. 7Department of Computer Science and Engineering, Institute of Technology, University of Minnesota, Minneapolis, Minnesota, USA.
Address correspondence to: Ashley T. Haase, Department of Microbiology, University of Minnesota, Box 196, 420 Delaware Street SE, Minneapolis, Minnesota 55455, USA. Phone: 612.624.4442; Fax: 612.626.0623; E-mail: [email protected].
Authorship note: Ming Zeng and Anthony J. Smith contributed equally to this work.
Find articles by Wietgrefe, S. in: JCI | PubMed | Google Scholar
1Department of Microbiology and 2Department of Medicine, Medical School, University of Minnesota, Minneapolis, Minnesota, USA. 3Division of Biostatistics, School of Public Health, University of Minnesota, Minneapolis, Minnesota, USA. 4AIDS and Cancer Virus Program, Science Applications International Corporation–Frederick Inc., National Cancer Institute, Frederick, Maryland, USA. 5Department of Chemistry and Biochemistry, Brigham Young University, Provo, Utah, USA. 6Yerkes National Primate Research Center, and Emory University, Atlanta, Georgia, USA. 7Department of Computer Science and Engineering, Institute of Technology, University of Minnesota, Minneapolis, Minnesota, USA.
Address correspondence to: Ashley T. Haase, Department of Microbiology, University of Minnesota, Box 196, 420 Delaware Street SE, Minneapolis, Minnesota 55455, USA. Phone: 612.624.4442; Fax: 612.626.0623; E-mail: [email protected].
Authorship note: Ming Zeng and Anthony J. Smith contributed equally to this work.
Find articles by Southern, P. in: JCI | PubMed | Google Scholar
1Department of Microbiology and 2Department of Medicine, Medical School, University of Minnesota, Minneapolis, Minnesota, USA. 3Division of Biostatistics, School of Public Health, University of Minnesota, Minneapolis, Minnesota, USA. 4AIDS and Cancer Virus Program, Science Applications International Corporation–Frederick Inc., National Cancer Institute, Frederick, Maryland, USA. 5Department of Chemistry and Biochemistry, Brigham Young University, Provo, Utah, USA. 6Yerkes National Primate Research Center, and Emory University, Atlanta, Georgia, USA. 7Department of Computer Science and Engineering, Institute of Technology, University of Minnesota, Minneapolis, Minnesota, USA.
Address correspondence to: Ashley T. Haase, Department of Microbiology, University of Minnesota, Box 196, 420 Delaware Street SE, Minneapolis, Minnesota 55455, USA. Phone: 612.624.4442; Fax: 612.626.0623; E-mail: [email protected].
Authorship note: Ming Zeng and Anthony J. Smith contributed equally to this work.
Find articles by Schacker, T. in: JCI | PubMed | Google Scholar
1Department of Microbiology and 2Department of Medicine, Medical School, University of Minnesota, Minneapolis, Minnesota, USA. 3Division of Biostatistics, School of Public Health, University of Minnesota, Minneapolis, Minnesota, USA. 4AIDS and Cancer Virus Program, Science Applications International Corporation–Frederick Inc., National Cancer Institute, Frederick, Maryland, USA. 5Department of Chemistry and Biochemistry, Brigham Young University, Provo, Utah, USA. 6Yerkes National Primate Research Center, and Emory University, Atlanta, Georgia, USA. 7Department of Computer Science and Engineering, Institute of Technology, University of Minnesota, Minneapolis, Minnesota, USA.
Address correspondence to: Ashley T. Haase, Department of Microbiology, University of Minnesota, Box 196, 420 Delaware Street SE, Minneapolis, Minnesota 55455, USA. Phone: 612.624.4442; Fax: 612.626.0623; E-mail: [email protected].
Authorship note: Ming Zeng and Anthony J. Smith contributed equally to this work.
Find articles by Reilly, C. in: JCI | PubMed | Google Scholar
1Department of Microbiology and 2Department of Medicine, Medical School, University of Minnesota, Minneapolis, Minnesota, USA. 3Division of Biostatistics, School of Public Health, University of Minnesota, Minneapolis, Minnesota, USA. 4AIDS and Cancer Virus Program, Science Applications International Corporation–Frederick Inc., National Cancer Institute, Frederick, Maryland, USA. 5Department of Chemistry and Biochemistry, Brigham Young University, Provo, Utah, USA. 6Yerkes National Primate Research Center, and Emory University, Atlanta, Georgia, USA. 7Department of Computer Science and Engineering, Institute of Technology, University of Minnesota, Minneapolis, Minnesota, USA.
Address correspondence to: Ashley T. Haase, Department of Microbiology, University of Minnesota, Box 196, 420 Delaware Street SE, Minneapolis, Minnesota 55455, USA. Phone: 612.624.4442; Fax: 612.626.0623; E-mail: [email protected].
Authorship note: Ming Zeng and Anthony J. Smith contributed equally to this work.
Find articles by Estes, J. in: JCI | PubMed | Google Scholar
1Department of Microbiology and 2Department of Medicine, Medical School, University of Minnesota, Minneapolis, Minnesota, USA. 3Division of Biostatistics, School of Public Health, University of Minnesota, Minneapolis, Minnesota, USA. 4AIDS and Cancer Virus Program, Science Applications International Corporation–Frederick Inc., National Cancer Institute, Frederick, Maryland, USA. 5Department of Chemistry and Biochemistry, Brigham Young University, Provo, Utah, USA. 6Yerkes National Primate Research Center, and Emory University, Atlanta, Georgia, USA. 7Department of Computer Science and Engineering, Institute of Technology, University of Minnesota, Minneapolis, Minnesota, USA.
Address correspondence to: Ashley T. Haase, Department of Microbiology, University of Minnesota, Box 196, 420 Delaware Street SE, Minneapolis, Minnesota 55455, USA. Phone: 612.624.4442; Fax: 612.626.0623; E-mail: [email protected].
Authorship note: Ming Zeng and Anthony J. Smith contributed equally to this work.
Find articles by Burton, G. in: JCI | PubMed | Google Scholar
1Department of Microbiology and 2Department of Medicine, Medical School, University of Minnesota, Minneapolis, Minnesota, USA. 3Division of Biostatistics, School of Public Health, University of Minnesota, Minneapolis, Minnesota, USA. 4AIDS and Cancer Virus Program, Science Applications International Corporation–Frederick Inc., National Cancer Institute, Frederick, Maryland, USA. 5Department of Chemistry and Biochemistry, Brigham Young University, Provo, Utah, USA. 6Yerkes National Primate Research Center, and Emory University, Atlanta, Georgia, USA. 7Department of Computer Science and Engineering, Institute of Technology, University of Minnesota, Minneapolis, Minnesota, USA.
Address correspondence to: Ashley T. Haase, Department of Microbiology, University of Minnesota, Box 196, 420 Delaware Street SE, Minneapolis, Minnesota 55455, USA. Phone: 612.624.4442; Fax: 612.626.0623; E-mail: [email protected].
Authorship note: Ming Zeng and Anthony J. Smith contributed equally to this work.
Find articles by Silvestri, G. in: JCI | PubMed | Google Scholar
1Department of Microbiology and 2Department of Medicine, Medical School, University of Minnesota, Minneapolis, Minnesota, USA. 3Division of Biostatistics, School of Public Health, University of Minnesota, Minneapolis, Minnesota, USA. 4AIDS and Cancer Virus Program, Science Applications International Corporation–Frederick Inc., National Cancer Institute, Frederick, Maryland, USA. 5Department of Chemistry and Biochemistry, Brigham Young University, Provo, Utah, USA. 6Yerkes National Primate Research Center, and Emory University, Atlanta, Georgia, USA. 7Department of Computer Science and Engineering, Institute of Technology, University of Minnesota, Minneapolis, Minnesota, USA.
Address correspondence to: Ashley T. Haase, Department of Microbiology, University of Minnesota, Box 196, 420 Delaware Street SE, Minneapolis, Minnesota 55455, USA. Phone: 612.624.4442; Fax: 612.626.0623; E-mail: [email protected].
Authorship note: Ming Zeng and Anthony J. Smith contributed equally to this work.
Find articles by Lifson, J. in: JCI | PubMed | Google Scholar
1Department of Microbiology and 2Department of Medicine, Medical School, University of Minnesota, Minneapolis, Minnesota, USA. 3Division of Biostatistics, School of Public Health, University of Minnesota, Minneapolis, Minnesota, USA. 4AIDS and Cancer Virus Program, Science Applications International Corporation–Frederick Inc., National Cancer Institute, Frederick, Maryland, USA. 5Department of Chemistry and Biochemistry, Brigham Young University, Provo, Utah, USA. 6Yerkes National Primate Research Center, and Emory University, Atlanta, Georgia, USA. 7Department of Computer Science and Engineering, Institute of Technology, University of Minnesota, Minneapolis, Minnesota, USA.
Address correspondence to: Ashley T. Haase, Department of Microbiology, University of Minnesota, Box 196, 420 Delaware Street SE, Minneapolis, Minnesota 55455, USA. Phone: 612.624.4442; Fax: 612.626.0623; E-mail: [email protected].
Authorship note: Ming Zeng and Anthony J. Smith contributed equally to this work.
Find articles by Carlis, J. in: JCI | PubMed | Google Scholar
1Department of Microbiology and 2Department of Medicine, Medical School, University of Minnesota, Minneapolis, Minnesota, USA. 3Division of Biostatistics, School of Public Health, University of Minnesota, Minneapolis, Minnesota, USA. 4AIDS and Cancer Virus Program, Science Applications International Corporation–Frederick Inc., National Cancer Institute, Frederick, Maryland, USA. 5Department of Chemistry and Biochemistry, Brigham Young University, Provo, Utah, USA. 6Yerkes National Primate Research Center, and Emory University, Atlanta, Georgia, USA. 7Department of Computer Science and Engineering, Institute of Technology, University of Minnesota, Minneapolis, Minnesota, USA.
Address correspondence to: Ashley T. Haase, Department of Microbiology, University of Minnesota, Box 196, 420 Delaware Street SE, Minneapolis, Minnesota 55455, USA. Phone: 612.624.4442; Fax: 612.626.0623; E-mail: [email protected].
Authorship note: Ming Zeng and Anthony J. Smith contributed equally to this work.
Find articles by Haase, A. in: JCI | PubMed | Google Scholar
Authorship note: Ming Zeng and Anthony J. Smith contributed equally to this work.
Published February 14, 2011 - More info
J Clin Invest. 2011;121(3):998–1008. https://doi.org/10.1172/JCI45157.
© 2011 The American Society for Clinical Investigation
Received: September 17, 2010; Accepted: December 20, 2010
-
Abstract
The hallmark of HIV-1 and SIV infections is CD4+ T cell depletion. Both direct cell killing and indirect mechanisms related to immune activation have been suggested to cause the depletion of T cells. We have now identified a mechanism by which immune activation-induced fibrosis of lymphoid tissues leads to depletion of naive T cells in HIV-1 infected patients and SIV-infected rhesus macaques. The T regulatory cell response to immune activation increased procollagen production and subsequent deposition as fibrils via the TGF-β1 signaling pathway and chitinase 3-like-1 activity in fibroblasts in lymphoid tissues from patients infected with HIV-1. Collagen deposition restricted T cell access to the survival factor IL-7 on the fibroblastic reticular cell (FRC) network, resulting in apoptosis and depletion of T cells, which, in turn, removed a major source of lymphotoxin-β, a survival factor for FRCs during SIV infection in rhesus macaques. The resulting loss of FRCs and the loss of IL-7 produced by FRCs may thus perpetuate a vicious cycle of depletion of T cells and the FRC network. Because this process is cumulative, early treatment and antifibrotic therapies may offer approaches to moderate T cell depletion and improve immune reconstitution during HIV-1 infection.
-
Introduction
Depletion of CD4+ T cells, the defining characteristic for which the immunodeficiency viruses HIV-1 and SIV were named, has been attributed to direct mechanisms of infection and cell killing and indirect mechanisms such as increased apoptosis accompanying the chronic immune activation associated with HIV-1 and SIV infections. More recently, there has also been increasing evidence that fibrosis induced by immune activation damages lymphoid tissue (LT) niches, thereby contributing to T cell depletion and impaired immune reconstitution upon institution of antiretroviral drug treatment (1, 2). In HIV-1 infection, fibrosis, measured as collagen deposition in LTs, strongly correlates with depletion of naive CD4+ T cells and inversely correlates with the extent of immune reconstitution after suppression of viral replication by antiretroviral therapy (2–6). In pathogenic SIV infection as well, collagen deposition in the early stage of SIV infection of rhesus macaques (Macaca mulatta; RMs) is associated with initial decreases in CD4+ T cells (7).
The mechanisms by which LT fibrosis depletes CD4+ T cells and impairs immune reconstitution in these immunodeficiency virus infections have not been well defined, but one previously advanced hypothesis (2), based on studies in mice (8–11), is that collagen deposition disrupts the architecture of the LT niche, so that T cells have less access to self-antigen/major histocompatibility complex signals and IL-7 on the fibroblastic reticular cell (FRC) network on which they migrate. Since these factors are critical for T cell survival, particularly naive T cells, decreased access to these homeostatic signals would result in increased apoptosis as a mechanism for T cell depletion associated with fibrosis in HIV-1 and SIV infections.
We tested this hypothesis in SIV-infected RMs, an animal model in which we could analyze LN biopsies obtained in longitudinal and cross-sectional studies. We first show that FRCs are the major source of IL-7, the primary survival factor for naive T cells in RMs, and that LN fibrosis limits lymphocyte access to FRC-derived IL-7, resulting in depletion, particularly of naive CD4+ T cells, through apoptosis. T cell apoptosis, along with other T cell killing mechanisms such as pathological immune activation that can lead to increased apoptosis of T cells, in turn diminishes the availability of T cell–derived lymphotoxin-β (LTβ), on which the FRC network depends. The decreased availability of LTβ-producing cells, along with collagen-restricted access to FRCs, results in loss of both the FRC network and IL-7 production. This further impairs the survival of naive T cells, perpetuating a vicious cycle of continuous and cumulative loss of both T cells and the FRC network (diagrammed in Supplemental Figure 1; supplemental material available online with this article; doi: 10.1172/JCI45157DS1).
These results suggest that therapeutic interventions to avert or moderate this pathological process of fibrosis could improve immune reconstitution after highly active antiretroviral therapy (HAART). To design effective interventions, we investigated the underlying mechanisms of fibrosis in LTs during HIV-1 infection and found that increases in TGF-β1 expression in T regulatory cells activate the TGF-β1 signaling pathway in fibroblasts to trigger the increase of production of procollagen and chitinase 3-like-1 (CHI3L1), an enzyme that could enhance maturation of this procollagen into collagen fibrils in LT fibroblasts. We then show in vitro that targeting the TGF-β1 signaling pathway with the antifibrotic drug pirfenidone dramatically reduces collagen produced by primary human fibroblasts. These data suggest that the TGF-β1 signaling pathway plays a key role in progressive LT fibrosis during HIV-1 infection. Therefore, early initiation of HAART to limit the viral replication-dependent inflammation that drives this process, along with complementary therapeutic interventions directed at the TGF-β1 signaling pathway, could potentially avert or moderate this pathological process and improve immune reconstitution after HAART.
-
Results
The FRC network is the major source of IL-7 for T cells in the T cell zone. We investigated the hypothesis that the mechanism by which LT fibrosis causes depletion of T cells is by decreasing access to the survival factor IL-7 by first showing in uninfected RMs that the FRC network is the major source of IL-7 for T cells. We used antibodies to desmin to label both the FRC network in the T cell zone of LNs and the follicular dendritic cell (FDC) network in B cell follicles, antibodies to IL-7 to label IL-7–positive cells, and antibodies to desmin and in situ hybridization (ISH) to detect and quantify IL7 mRNA. We found that both IL7 mRNA and IL-7 protein predominantly colocalize with the FRC and FDC networks (Figures 1 and 2), analogous to the localization of cytoskeletal and nervous system mRNAs and proteins (12, 13). Quantitatively, we conservatively estimated that approximately 70% of IL7 mRNA colocalizes with the FRC network in the T cell zone and approximately 80% colocalizes with the FDC network in B cell follicles (Supplemental Figure 2A). The colocalization of IL-7 protein and the FRC network is consistent with results reported in the mouse (8, 10), and the small portion of IL-7 we found associated with a subset of dendritic cells and macrophages (Supplemental Figure 2B) is consistent with a study of human LTs (14).
 Figure 1
Figure 1Desmin is a marker of FRCs and FDCs. Confocal images of LN sections from uninfected RMs (representative image for 1 animal out of 9) stained for desmin (green) and markers (red) for FDCs (CD35 and CD21) and FRCs (ER-TR-7). Desmin staining colocalizes with the FDC markers and the FRC marker, respectively. FDCs form a network with higher density than the FRC network in the T cell zone. Scale bar: 50 μm.
 Figure 2
Figure 2FRC and FDC networks are the major producers of IL-7. (A) LN sections (representative image for 1 animal out of 3) stained for desmin (red) and IL-7 (green). Merged confocal images show colocalization of IL-7 and FDC and FRC networks in the B cell follicle and T cell zone, respectively. Scale bar: 20 μm. (B) ISH for IL7 mRNA combined with desmin immunohistochemical staining (brown), showing the colocalization of IL7 mRNA and FRCs. In the image taken in transmitted light, the FRCs and their processes have been stained brown. The red arrow in the left panel corresponds to the position of IL7 mRNA shown with the arrow in the right panel. In developed radioautographs, after hybridization to a S35-labeled IL-7–specific riboprobe, the signals (with epipolarized light) are yellow-appearing silver grains, overlying the brown-stained FRCs and FRC processes (red arrow) shown in the left panel. Original magnification, ×600.
Collagen deposition and loss of the FRC network impede access to IL-7 while also depleting its source. Under physiological conditions, FRCs ensheathe collagen fibers to form a connective tissue and fibrous network on which lymphocytes, antigen-presenting cells, and other cells within LTs migrate (8, 15–17). This architecture allows T cells to efficiently access chemokines and cytokines, including survival factors such as IL-7, “posted” on their path (10, 11, 18). Thus, LT sections from uninfected RMs stained with antibodies to type I collagen, desmin, and T cells reveal that the collagen and desmin colocalize and define the FRC network and 3D space in which the T cells migrate and visibly contact the FRC network (Figure 3A and Supplemental Figure 3A). In marked contrast, in chronically infected RMs (180 days after SIVmac239 infection [dpi]), large deposits of collagen are evident outside the FRC network, while the FRC network itself is greatly decreased (Figure 3B). As a consequence, few T cells visibly contact the FRC network and instead are only in contact with collagen (Figure 3B and Supplemental Figure 3B).
 Figure 3
Figure 3Collagen deposition and loss of the FRC network lead to loss of contact between T cells and FRCs. (A) In the merged confocal image in an uninfected RM (representative image for 1 animal out of 9), collagen (red) colocalizes with the FRC network (green) and the CD3+ T cells (blue) are in contact with the FRC network. (B) In an animal at 180 dpi (representative image for 1 animal out of 4), there is abundant collagen staining that does not colocalize with the FRC network. Note the FRC network (green) is also greatly decreased, so that large numbers of T cells are not in contact with the FRC network but instead mainly contact non-FRC-associated collagen. Scale bar: 10 μm.
Increased naive T cell apoptosis. Since T cells migrate on the FRC network to access survival factors such as IL-7, the physical barrier created by collagen deposition and the loss of the FRC network itself would be expected to restrict IL-7 access and deplete the source of IL-7, thereby increasing apoptosis, particularly in the naive T cell populations (10, 18, 19). We indeed found dramatic increases in apoptotic cells in the T cell zone in chronically infected animals. This was evident by comparison of an uninfected animal (SIV–) with an infected animal at 180 dpi and was even more striking in an infected animal at 462 dpi (Figure 4, A–C). While there were cells clearly undergoing apoptosis even when they were close to the residual FRC network (Figure 4B), consistent with the well-established mechanism of increased apoptosis associated with chronic immune activation, the great majority of activated caspase-3+ apoptotic cells were located in apparent “black holes,” in which the FRC network had been lost and replaced by collagen. As predicted, we found significant increases in naive T cell apoptosis (Figure 4D and Figure 5B), which is inversely associated with the quantity of FRCs and positively associated with the quantity of collagen (P = 0.0004 and P = 0.002, respectively).
 Figure 4
Figure 4Loss of FRC network leads to increased apoptosis of naive T cells. (A–C) Confocal images of LN sections from RMs at different time points after SIV infection immunofluorescently double stained for desmin (green) and activated caspase-3 (red), showing increased apoptosis associated with loss of the FRC network at (B) 180 dpi (representative image for 1 animal out of 5) and (C) 462 dpi (representative image for 1 animal out of 5 animals infected longer than 200 days), compared with (A) uninfected RMs (representative image for 1 animal out of 9). Scale bar: 20 μm. (D) Confocal image of a LN section, showing apoptotic naive T cells (yellow arrows) (TUNEL+ [green]; CD45RA+ [red]; and CD3+ [blue]) at 180 dpi (representative image for 1 animal out of 5). Scale bar: 20 μm.
 Figure 5
Figure 5Loss of FRCs is associated with loss of naive T cells within LTs. (A) Confocal images of LN sections from RMs at different time points after SIV infection immunofluorescently double stained for CD45RA (green) and CD3 (red), showing gradual loss of CD45RA+CD3+ naive T cells within LTs during SIV infection (total n = 38; n = 9 for uninfected RMs, n = 5 for 7-dpi RMs, n = 5 for 14-dpi RMs, n = 5 for 30-dpi RMs, n = 4 for 90-dpi RMs, n = 5 for 180-dpi RMs, and n = 5 for 300-dpi RMs). Scale bar: 30 μm. (B) Quantification of the number of apoptotic naive T cells (TUNEL+CD45RA+CD3+) and the number of naive T cells (CD45RA+CD3+), showing significant inverse association (total n = 38, P = 0.0001). The time point “300 dpi” represents the mean measurement of 5 animals, which were infected for 222 days, 280 days, 294 days, 462 days, and 560 days, respectively. (C) QIA of the percentage of area staining positive for desmin and the percentage of area staining positive for IL-7, showing significant association between the quantity of IL-7 and the quantity of the FRC network (total n = 38, P < 0.0001). (D) QIA of the percentage of area staining positive for IL-7 and the number of CD45RA+CD3+ naive T cells within LTs, showing the kinetics of IL-7 and the kinetics of naive T cell number. Error bars represent SD. (E) QIA of the percentage of area staining positive for IL-7 and the number of CD45RA+CD3+ naive T cells for each animal, showing that loss of naive T cells is associated with loss of IL-7 production (total n = 38, P = 0.0004).
Depletion of naive CD4+ and CD8+ T cell populations by increased apoptosis. The increased apoptosis in naive T cell populations was significantly associated with their depletion (Figure 5, A and B; P = 0.0001). Significant increases were detectable after the acute stage of SIV infection and accelerated markedly in the later chronic stages of infection. In the acute stage of infection, the FRC network and IL-7 were slightly increased (Figure 5C and Supplemental Figure 4, A and B), coincident with the LT hyperplasia in this stage of infection (Supplemental Figure 4B), potentially compensating for collagen deposition occurring during the acute phase, so that there was only a small decrease in naive T cell numbers. However, after 90 dpi, there was a rapid and parallel decline in the FRC network, IL-7 (Figure 5C and Supplemental Figure 4, A and B; P < 0.0001), and naive T cells (Figure 5, D and E; P = 0.0004). Most of the surviving naive T cells resided at the border of B cell and T cell zones (Supplemental Figure 4C), where the majority of the residual FRC network and IL-7 was still detectable (Supplemental Figure 4, A and B, arrow). Lastly, as our hypothesis predicts, the destruction of LT architecture not only decreased the naive CD4+ T cell number in LNs but also had a similar impact on naive CD8+ T cells, with similar and parallel losses of both naive CD4+ and CD8+ cells (Supplemental Figure 4, D–F).
Survival interdependencies — loss of FRCs caused by loss of LTβ+ T cells. Collagen deposition and loss of the FRC network decreased both access to IL-7 and the major source of IL-7 in the T cell zone, resulting in increased apoptosis and loss of T cells. However, what explains the loss of the FRC network itself? We hypothesized that the loss of the FRC network was reciprocally related to depletion of T cells as a major source of LTβ (20), based on ex vivo studies in the mouse, in which maintenance of the FRC network has been shown to be dependent on LTβ signaling from T cells (21). We confirmed this dependency in vivo by documenting loss of the FRC network in mice by separately blocking either the LTβ pathway or by depleting T cells (Supplemental Figure 5) and then showed that T cells are a major source of LTβ (~80% of the LTβ+ cells are T cells) in the T cell zone of RMs (Figure 6A). Furthermore, the loss of LTβ+ T cells followed a parallel time course, consistent with the loss of the FRC network in SIV infection (Figure 6, B–D), further supporting the conclusion that the loss of FRCs is a consequence of the loss of LTβ-producing T cells.
 Figure 6
Figure 6LTβ+ T cells are depleted during SIV infection in RMs. (A) Confocal images (representative image for 1 animal out of 5) showing CD3+ T cells (red) are the major producers of LTβ (green) in the T cell zone in an uninfected RM LN. Scale bar: 30 μm. (B and C) Progressive depletion of LTβ+ T cells (green) in RM LTs at (C) 180 dpi (representative image for 1 animal out of 5) compared with those at (B) 30 dpi and (A) those from an uninfected animal. Scale bar: 30 μm. (D) QIA of the number of LTβ+ T cells and the percentage of area staining positive for desmin for each animal, showing the correlation between LTβ+ T cells and FRCs (P < 0.0001; n = 9 for uninfected RMs, n = 5 for 30-dpi RMs, n = 5 for 180-dpi RMs, n = 5 for 300-dpi RMs).
Fibroblasts are the major producers of collagen. These findings establish a cooperative and cumulative mechanism of collagen deposition and naive T cell depletion but leave unanswered the questions of the cellular sources and mechanisms of collagen production and deposition. For answers to these questions that could provide insights to design interventions in HIV-1 infection to minimize depletion and improve immune reconstitution, we turned to HIV-1 infection in studies of LT and in vitro models.
To determine the cellular source of collagen within LT in HIV-1 infection, we used antibodies to CD3 (T cells), CD68 (macrophages), CD20 (B cells), CD11c (dendritic cells), and vimentin (fibroblasts) to identify which cell types were producing procollagen, the precursor of mature collagen fibers (22–24). As shown in a representative image in Supplemental Figure 6A, procollagen mainly colocalized with vimentin+ fibroblasts. To corroborate this finding, we used ISH combined with immunohistochemistry to show that type I collagen (COL1A2) mRNA primarily colocalized with vimentin+ fibroblasts (Supplemental Figure 6B). Lastly, we isolated leukocyte and fibroblast subpopulations from human tonsils and compared their ability to produce collagen in an ex vivo culture system. As shown in Supplemental Figure 6C, cultured primary fibroblasts were able to synthesize type I collagen, while leukocytes produced very little type I collagen (data not shown). Taken together, these data confirm that fibroblasts are the major producers of collagen within LTs.
Enhanced TGF-β1 signaling leads to increased production of collagen. The TGF-β1 signaling pathway is a known, key mediator of fibrosis (25–27), while TGF-β1+ regulatory T cells have been shown to be an important factor in increased fibrosis of LT during acute SIV infection (7). We therefore examined the expression of various components of the TGF-β1 signaling pathway in the LTs from a cohort of HIV-1–infected patients (Supplemental Table 1), including TGF-β1, TGF-β1 receptor II (TGF-β1 RII), and downstream signaling molecules, such as phosphorylated smad2/3 (p-smad2/3). We did detect an increase in TGF-β1+ T cells during HIV-1 infection (Figure 7, A and B, and Supplemental Figure 7A), identified mainly as CD3+/CD4+/Foxp3+ regulatory cells (Supplemental Figure 7A and Supplemental Figure 8). This increase in TGF-β1+ regulatory T cells was accompanied by a parallel increase in TGF-β1 RII expression in vimentin+ fibroblasts and increases in the downstream signaling molecules p-smad2/3 (Figure 7A and Supplemental Figure 7, B and C).
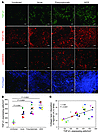 Figure 7
Figure 7The TGF-β1 signaling pathway is activated in LTs during HIV-1 infection. (A) Immunofluorescent images of TGF-β1 (green staining; first row), TGF-β1 RII (red staining; second row), p-SMAD2/3 (red staining; third row), and procollagen (blue staining; fourth row) in LTs from HIV-1–infected individuals, showing a parallel increase in TGF-β1 and its cognate receptor, leading to activation of the TGF-β1 signaling pathway (p-SMAD2/3) and resulting synthesis of procollagen (representative image for 1 out of 4 subjects at each stage). Scale bars: 50 μm. (B) TGF-β1+ cells were quantified in each LN biopsy and are reported as the number of TGF-β1–expressing cells per mm2 of tissue. Mean values for each group are indicated by horizontal black bars. (C) TGF-β1–expressing cells were significantly correlated with collagen type I deposition in the inguinal LN. Each individual colored symbol represents an individual subject.
The increased frequency and spatial proximity of TGF-β1+ regulatory T cells and TGF-β1 R+ fibroblasts (Supplemental Figure 7D) suggested a potential mechanism for increased collagen deposition in which interactions between T cells and fibroblasts activate the TGF-β1 signaling pathway. In support of this hypothesis, we found a significant association between TGF-β1+ cells and fibrosis, as measured by collagen type I deposition (r = 0.68, P = 0.0001) (Figure 7C). Moreover, addition of TGF-β1 to primary fibroblasts cultured ex vivo resulted in increased production of type I collagen over basal levels (Supplemental Figure 9).
Blocking TGF-β1 signaling decreases production of collagen. These results, linking LT fibrosis to activation of the TGF-β1 pathway during viral replication-related inflammation, with consequent disruption of the FRC network and depletion of naive T cells, suggested that interventions that disrupt these processes, even without directly affecting viral replication, might confer therapeutic benefits. To begin to evaluate approaches for inhibition of LT fibrosis as a possible adjunctive therapy in HIV-1 infection, we investigated the effects of the antifibrotic drug pirfenidone on the TGF-β1 signaling pathway. Addition of pirfenidone to TGF-β1–stimulated fibroblasts effectively blocked this signaling pathway and concomitantly decreased collagen production in an ex vivo culture system (Supplemental Figure 9).
Role of CHI3L1 in collagen deposition and relationship to TGF-β1 signaling. In a previous microarray analysis of HIV-1–infected LT (28), we identified CHI3L1 as one of the genes with the greatest increase in expression during HIV-1 infection. Since CHI3L1 has been shown to play an important role in collagen formation, both by increasing the rate of fibrillogenesis during the collagen maturation process (29, 30) and decreasing the rate of collagen degradation by inhibiting MMP-1 (30), we explored the role that CHI3L1 might play in increasing collagen deposition during HIV-1 infection. We found significant increases in the CHI3L1 expression in LTs during HIV-1 infection, consistent with the previously documented increases in mRNA (P = 0.034) (Supplemental Figure 10, A and B), and identified vimentin+ fibroblasts as the cells that express CHI3L1 (Supplemental Figure 10C). As a number of cytokines induce CHI3L1 expression (31, 32), we asked whether increased TGF-β1 expression was associated with increased CHI3L1 expression during HIV-1 infection and found that there was a significant, positive association between TGF-β1+ cells and CHI3L1 expression in LT (r = 0.49, P = 0.0092) (Figure 8A). In addition, TGF-β1 treatment of primary LT fibroblasts resulted in enhancement of CHI3L1 expression compared with that of untreated cultures (Figure 8, B and C).
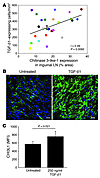 Figure 8
Figure 8TGF-β1 stimulates the production of CHI3L1 by LT fibroblasts. (A) TGF-β1–expressing cells were significantly correlated with CHI3L1 expression in the inguinal LN. Each individual colored symbol represents an individual subject. (B) Immunofluorescent images of CHI3L1 (green staining) in primary human fibroblasts, treated with or without TGF-β1 (250 ng/ml) for 48 hours (representative image of 10). Cell nuclei appear blue (DAPI staining). Scale bars: 50 μm. (C) The extracellular CHI3L1 networks of primary human fibroblasts were quantified for each condition and are reported as CHI3L1 mean fluorescence intensity. Data are expressed as mean ± SD, where 3 independent experiments were performed in quadruplicate. The results are shown with significance where applicable (P < 0.05).
As we had now shown that CHI3L1 and procollagen are both produced in LT fibroblasts and both increased during HIV-1 infection, we hypothesized that CHI3L1 directly contributed to collagen deposition by facilitating the formation of collagen fibrils (29, 30). Consistent with this hypothesis, we found (a) CHI3L1 levels and collagen type I in LT are significantly correlated (r = 0.85, P < 0.0001) (Figure 9A); (b) addition of exogenous CHI3L1 to cultures of primary fibroblasts enhanced the formation of mature collagen type I (Figure 9B); and (c) CHI3L1-specific blocking antibodies reduced this effect. We also found that pirfenidone treatment blocked the effects of CHI3L1 on collagen formation (Figure 9B), suggesting that the antifibrotic effects of the drug are cooperative: blocking the TGF-β1 signaling pathway inhibits both production of procollagen and CHI3L1, thereby blocking fibril formation.
 Figure 9
Figure 9CHI3L1 facilitates type I collagen formation. (A) CHI3L1 expression was significantly correlated with collagen type I deposition in the inguinal LN. Each individual colored symbol represents an individual subject. (B) The extracellular collagen type I networks of primary human fibroblasts were quantified for fibroblasts, treated with or without CHI3L1 (500 ng/ml) for 48 hours. Addition of the antifibrotic drug pirfenidone (0.5 mg/ml) or a CHI3L1-blocking antibody decreased collagen type I production. The extracellular collagen type I networks were quantified for each condition and are reported as collagen type I mean fluorescence intensity. Data are expressed as mean ± SD, where 3 independent experiments were performed in quadruplicate. The results are shown with significance where applicable (P < 0.05).
-
Discussion
The hallmark of untreated immunodeficiency virus infections is the cumulative depletion of CD4+ T cells that is most marked in the later stages of infection by the predominant depletion in blood and LT of naive CD4+ T cells (3–5). This has been attributed to impaired output from the thymus (33–35) combined with loss in the periphery, in which T cells die by direct and indirect mechanisms related to chronic immune activation: (a) activated naive T cells become memory T cells that are susceptible to infection in which they succumb or are killed by virus-specific CD8+ T cells and (b) activated cells undergo activation-induced cell death (1, 36, 37).
To these established mechanisms we now add another indirect mechanism of T cell depletion in which chronic immune activation elicits a T regulatory counter response with the adverse consequence of increased collagen production and deposition. This, in turn, sets in motion cooperative and cumulative mechanisms that contribute to the depletion of T cells, particularly naive T cells in SIV-infected RMs (Supplemental Figure 1A): collagen disrupts T cell access to IL-7 provided on the FRC network, leading to increased apoptosis of T cells, particularly naive T cells. The depletion of LTβ+ T cells, in turn, deprives the FRC network of the major source of LTβ+ (20), on which it depends (21). This, along with the collagen-restricted access of LTβ+ T cells to the FRC network, results in loss of the FRC network and further decreases in IL-7 availability for T cells. Thus, a positive loop mechanism (Supplemental Figure 1) is established that progressively and cumulatively causes T cell depletion, particularly in the naive T cell populations.
This mechanism provides a broadened perspective on the preferential depletion of naive CD4+ T cells in late HIV-1 infection in both peripheral blood and LTs (3–5) and helps explain why both naive CD4+ and CD8+ T cells are depleted in HIV-1 infection (38, 39), thereby impairing the ability to respond to new infections and malignancies. Because of resistance to viral infection by naive CD8+ T cells and relative resistance to viral infection and killing of naive CD4+ T cells (40, 41) compared with that of memory CD4+ T cells (42, 43), depletion of these populations has heretofore been attributed to indirect mechanisms of apoptosis associated with chronic immune activation (1, 36, 37) and to decreased thymic output (33–35), rather than direct mechanisms. However, these indirect mechanisms do not fully account for the continued apoptosis and slow and incomplete restoration of the naive CD4+ T cell populations after HAART has largely suppressed direct losses attributable to viral infection and after immune activation has largely subsided with commensurate decreases in activation-induced cell death (ref. 44 and Supplemental Figure 1A). On the other hand, collagen deposition, which impairs access to IL-7 on the FRC network and causes loss of the FRC network, itself an important source of IL-7, provides mechanisms for (a) depletion of both naive CD4+ and CD8+ T cells (38, 39); (b) the continued high level of apoptosis in LTs while on HAART; and (c) the correlation between the extent of fibrosis of LTs before the initiation of HAART and the extent of restoration of naive CD4 T cells after long-term HAART (ref. 6 and Supplemental Figure 1B).
Our findings suggest the potential benefit of IL-7 treatment during HIV-1 infection. Indeed, studies have shown that complementing HAART with IL-7 during both SIV and HIV infection significantly increases the circulating naive CD4+ T cell number (45–47). Furthermore, ex vivo and in vivo studies with T cells from HIV-1–infected patients showed that IL-7 treatment could significantly upregulate Bcl-2 levels to normalize the extent of apoptosis in CD4+ and CD8+ T cells from HIV-1–infected individuals (48, 49). These data consistently suggest that insufficient IL-7 is a key contributor in the impaired T cell homeostasis in SIV/HIV infection.
However, the immediate decline of the absolute numbers of both naive CD4+ and CD8+ T cells after termination of IL-7 therapy (45–47) suggests that complementing IL-7 only provides transient survival benefit for naive CD4 and CD8 T cells and strongly argues for the development of therapeutic interventions to provide long-term survival benefit for naive T cells. Our findings here clearly suggest that collagen deposition and the loss of FRCs as the major source of IL-7 play critical roles in compromising homeostasis of naive T cells, and thus restoration of the LT niche could potentially provide long-term survival benefit for naive T cells.
Our findings on mechanisms of fibrosis in LTs during HIV-1 infection also support a cooperative and cumulative mechanism of collagen deposition driven by immune activation (Supplemental Figure 1A). The increased expressions of both TGF-β1 in regulatory T cells (responding to immune activation) and its cognate receptor in spatially proximate fibroblasts chronically activate the TGF-β1 signaling pathway. This results in increased expression of procollagen, the immature, unprocessed form of collagen (Supplemental Figure 11). TGF-β1 also enhances the expression of CHI3L1, so that in this model, TGF-β1 plays a dual role; it is a cytokine that increases both an upstream substrate (procollagen) and its downstream effector (CHI3L1) within the same cell type (fibroblast), thereby establishing a cooperative process of collagen deposition and cumulative fibrosis in LT during HIV-1 infection.
The link established here among fibrosis, damage to the LT niche through loss of access and a source of IL-7 on the FRC network, and naive T cell depletion clearly argues for therapeutic approaches targeting fibrosis and maintaining and restoring a functional FRC network. Because the underlying mechanisms of both fibrosis and damage to the FRC network operate cumulatively, the most straightforward way to do this would be through earlier initiation of HAART to limit the inflammation associated with viral replication that drives the process. In addition, development of adjunctive antifibrosis treatment might additionally avert or lessen the consequences of damage to the LT niche and improve immune reconstitution. As a proof of principle, we targeted the TGF-β1 signaling pathway with the antifibrotic drug pirfenidone, currently in a phase II clinical trial in the treatment of pulmonary fibrosis (50), and showed that we could dramatically reduce collagen production by primary human fibroblasts. Thus, therapeutic interventions directed at the TGF-β1 signaling pathway or other pathways involved in collagen formation, deposition, and degradation could potentially avert or moderate LT fibrosis and improve immune reconstitution after HAART.
-
Methods
Ethics statement. The human study was conducted according to the principles expressed in the Declaration of Helsinki. The study was approved by the Institutional Review Board of the University of Minnesota. All patients provided written informed consent for the collection of samples and subsequent analysis.
LN biopsy specimens. Inguinal LN biopsies from 4 HIV-negative individuals and 23 untreated HIV-1–infected individuals at different clinical stages (Supplemental Table 1) were obtained for this University of Minnesota Institutional Review Board–approved study. Viral load measurements were obtained on the same day as biopsies. Each LN biopsy was immediately placed in fixative (4% neutral buffered paraformaldehyde or Streck’s tissue fixative) and paraffin embedded.
Animals, SIV infection, and LN biopsy specimens. Adult RMs used in these studies were housed in accordance with the regulations of the American Association of Accreditation of Laboratory Animal Care and the standards of the Association for Assessment and Accreditation of Laboratory Animal Care International. These studies were approved by the Institutional Animal Care and Use Committees of Emory University (Atlanta, Georgia, USA) and the University of Pennsylvania (Philadelphia, Pennsylvania, USA). LTs were obtained in longitudinal studies from 5 RMs who were inoculated intravenously with 10,000 TCID50 of SIVmac239 (gift from R. Desrosiers, New England Primate Research Center, Harvard Medical School, Southborough, Massachusetts, USA), and additional LTs from 11 RMs were obtained in previously described cross-sectional studies (51). Each LN biopsy was immediately placed in fixative (4% neutral buffered paraformaldehyde or Streck’s tissue fixative) and paraffin embedded.
Immunofluorescence, immunohistochemistry, ISH, and quantitative image analysis. All staining procedures were performed as previously described (51, 52), using 5- to 30-μm tissue sections mounted on glass slides. Tissues were deparaffinized and rehydrated in deionized water. Heat-induced epitope retrieval was performed using a high-pressure cooker (125°C) in either DIVA Decloaker (Biocare Medical) or EDTA Decloaker (Biocare Medical), followed by cooling to room temperature. Tissues for collagen type I staining required pretreatment with 20 μg/ml proteinase K (Roche Diagnostics) in proteinase K buffer (0.2 M Tris, pH 7.4, 20 mM CaCl2) for 15–20 minutes at room temperature. Tissue sections were then blocked with SNIPER Blocking Reagent (Biocare Medical) for 30 minutes at room temperature. Primary antibodies were diluted in TNB (0.1 M Tris-HCl, pH 7.5; 0.15 M NaCl; 0.05% Tween 20 with Dupont blocking buffer) and incubated overnight at 4°C. After the primary antibody incubation, sections were washed with PBS and then incubated with fluorochrome-conjugated secondary antibodies in TNB for 2 hours at room temperature. For sections using 2 mouse primary antibodies, one of the antibodies was first biotinylated using the One-step Antibody Biotinylation Kit (Miltenyi Biotec) according to the manufacturer’s instructions; an anti-biotin fluorophore-conjugated antibody was then used as the secondary reagent. Finally, sections were washed with PBS, nuclei were counterstained blue with TOTO-3 or DAPI, and sections were mounted using Aqua Poly/Mount (Polysciences Inc.). Immunofluorescent micrographs were taken using an Olympus BX61 Fluoview confocal microscope with the following objectives: ×20 (0.75 NA), ×40 (0.75 NA), and ×60 (1.42 NA); images were acquired and mean fluorescence intensity was analyzed by using Olympus Fluoview software (version 1.7a).
For combined immunohistochemistry/ISH, collagen type I (COL1A2) cDNA (IMAGE clone 740269 from ATCC, vector PT7T3D-Pac) was cut with EcoR I and transcribed with T3 polymerase (Promega) in the presence of S35 UTP to make the anti-sense probe. The cDNA was cut with Not I and transcribed with T7 polymerase to make the control sense probe, according to the manufacturer’s instructions. IL-7 cDNA (IMAGE clone 5748841; Invitrogen) was cut with Sal I and transcribed with T7 polymerase (Promega) in the presence of S35 UTP. Tissues were deparaffinized in an incubator at 60°C for 2 hours and rehydrated through graded ethanols. Heat-induced epitope retrieval was performed using a water bath (95°C for 25 minutes), followed by treatment with acetic anhydride. Sections were hybridized with S35-labeled anti-sense or control sense RNA probes overnight at 45°C, washed in 2X standard saline citrate (SSC) at 37°C, treated with RNase A at 37°C for 60 minutes, and washed in 50% formamide/2X SSC at 50°C for 5 minutes, 1X SSC at 50°C for 10 minutes, and 0.5X SSC at 50°C for 15 minutes. Tissues were then blocked with SNIPER Blocking Reagent (Biocare Medical) and incubated overnight at 4°C with the primary antibody. Tissues were then washed. Endogenous peroxidase was inactivated with 3% (vol/vol) H2O2 in methanol and signal detected with Mach-3 (Biocare Medical) and DAB Kits (Vector Laboratories). Slides were dehydrated in ethanols containing 0.1 M ammonium acetate and air dried. Slides were dipped in nuclear track emulsion, dried, exposed at 4°C, developed, fixed, counterstained with Harris Hematoxylin (Surgipath), and mounted with Permount (Fisher Scientific). Stained sections were examined by light microscopy at ambient temperatures. Light micrographs were taken using an Olympus BX60 upright microscope with the following objectives: ×10 (0.3 NA), ×20 (0.5 NA), and ×40 (0.75 NA); images were acquired using a SPOT Color Mosaic camera (model 11.2; Diagnostic Instruments) and SPOT acquisition software (version 4.5.9; Diagnostic Instruments). Isotype-matched IgG/IgM-negative control antibodies in all instances yielded negative staining results (see Supplemental Table 2, which lists the primary antibodies and antigen retrieval methodologies).
Quantitative image analysis (QIA) was performed using 10–20 randomly acquired, high-powered images (×200 or ×400 magnification) by either manually counting the cells in each image or by determining the percentage of LT area occupied by a positive chromogen or fluorescence signal using an automated action program in Adobe Photoshop CS, with tools from Reindeer Graphics.
Ex vivo culture system. The experimental protocols used here for human tissue samples had full Institutional Review Board approval (Human Subjects Committee, Research Subjects’ Protection Program, University of Minnesota), and informed written consent was obtained from individual patients, or the legal guardians of minors, for the use of tissue in research applications prior to the initiation of surgery. Fresh human palatine tonsil tissues were obtained from routine tonsillectomies and processed within 1–2 hours of completion of surgery. Viable tonsil lymphocyte suspensions were prepared by forcing cut tissue pieces through a metal sieve and collecting the released single cell suspension in complete RPMI medium (10% heat-inactivated fetal calf serum, 1x l-glutamine, and penicillin and streptomycin; Invitrogen). The cells were washed and immediately cryopreserved. The remaining stromal cells left on the sieve were cultured in complete RPMI medium at 37°C/5% CO2 and adherent, proliferating fibroblasts were visible after 2–5 days in culture; confluent monolayers developed after 10–25 days. These primary stromal populations were readily released with trypsin and further expanded/passaged using routine procedures for adherent cells.
Subconfluent primary stromal cells (2 × 104 cells/well) were grown on Lab-Tek II chamber slides 2 days before experimental manipulation. TGF-β1, CHI3L1, and other agents were added to the culture and incubated for 48 hours before analysis. For vimentin and p-smad2/3 staining, cells were first fixed by Streck’s tissue fixative and then subjected to secondary antibody staining as previously described. For live cell staining, samples were directly incubated with primary antibodies against collagen type I and/or CHI3L1 at 4°C overnight, without antigen retrieval treatment; samples were then subject to secondary antibody staining as previously described. To quantify levels of collagen type I or CHI3L1, 10 randomly stained confocal images from each specimen and condition were captured, and the mean fluorescence intensities were calculated by FV10-ASW software.
LTβ signaling blockade and CD3+ T cell depletion in mice. In order to block LTβ signaling, mice were injected intra-peritoneally each week with 100 μg of LTβR-Ig (gift from Jeffrey Browning, Biogen Idec, Inc., Cambridge, Massachusetts, USA) or control IgG and were sacrificed 1 week after the second injection of LTβR-Ig. LTs were sectioned and stained for ER-TR7 using rat monoclonal anti-ER-TR-7 (clone ER-TR-7, Acris, Germany). For the CD3+ T cell depletion study, mice were injected intra-peritoneally each week with rat anti-CD3 antibody (clone 30H12, 500 μg per injection) or vehicle control. LTs were isolated after the eighth injection and sections were stained for ER-TR7.
Statistics. All associations involving SIV-infected monkeys were tested using linear mixed models. Restricted maximum likelihood was used to obtain parameter estimates, and Wald tests of regression coefficients were used to test for an association between each independent variable and the response variable of interest. Random intercepts were used to model the animal correlation in the response variable. More specifically, 3 different variables were used as response variables (the percentage of area staining positive for IL-7, the number of naive T cells, and the number of naive T cells undergoing apoptosis), and all other variables (namely, the percentage of area staining positive for desmin, the percentage of area staining positive for collagen, and days after infection) were used as explanatory variables. Backward variable selection was used to remove nonsignificant covariates. This process led to models in which (a) days after infection and the percentage of area staining positive for desmin (P < 0.0001) were predictive of the percentage of area staining positive for IL-7; (b) the percentage of area staining positive for collagen (P = 0.002) and the percentage of area staining positive for desmin (P = 0.0004) were predictive of the number of naive T cells undergoing apoptosis; (c) the percentage of area staining positive for IL-7 was predictive of the number of naive T cells (P = 0.0004); and (d) the number of naive T cells undergoing apoptosis was predictive of the number of naive T cells (P = 0.0001). P values of less than 0.05 were considered statistically significant.
Associations between continuous variables involving HIV-infected humans were estimated using Pearson’s correlation coefficient (after transforming to the log scale), and tests were conducted using the usual t test for a correlation. Differences in CHI3L1 protein levels between HIV-negative and -positive subjects were tested using a 2-sample t test (after transforming to the logarithmic scale to stabilize the variance). All calculations were performed with the software R, version 2.10.1.
-
Acknowledgments
We thank the Yerkes National Primate Research Center; D. Masopust and V. Vezys for providing CD3-depleting antibodies; C. O’Neill and T. Leonard for help in preparing the manuscript and figures; Jeffrey Browning for providing soluble LTβR-Ig and control IgG; and Ben Ng from Carl Zeiss for help in generating the 3D images. We also thank all of the donor participants in this study. This work was supported in part by NIH research grants AI028246, AI048484, and AI056997 to A.T. Haase; in part by Yerkes base grant P51-00165 to G. Silvestri; and in part by federal funds from the National Cancer Institute, NIH, under contract no. HHSN261200800001E.
Address correspondence to: Ashley T. Haase, Department of Microbiology, University of Minnesota, Box 196, 420 Delaware Street SE, Minneapolis, Minnesota 55455, USA. Phone: 612.624.4442; Fax: 612.626.0623; E-mail: [email protected].
-
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Reference information: J Clin Invest. 2011;121(3):998–1008. doi:10.1172/JCI45157.
-
References
- Haase AT. Population biology of HIV-1 infection: viral and CD4+ T cell demographics and dynamics in lymphatic tissues. Annu Rev Immunol. 1999;17:625–656.View this article via: PubMed Google Scholar
- Estes JD, Haase AT, Schacker TW. The role of collagen deposition in depleting CD4+ T cells and limiting reconstitution in HIV-1 and SIV infections through damage to the secondary lymphoid organ niche. Semin Immunol. 2008;20(3):181–186.
- Diaz A, et al. Factors associated with collagen deposition in lymphoid tissue in long–term treated HIV–infected patients. AIDS. 2010;24(13):2029–2039.
- Schacker TW, et al. Lymphatic tissue fibrosis is associated with reduced numbers of naive CD4+ T cells in human immunodeficiency virus type 1 infection. Clin Vaccine Immunol. 2006;13(5):556–560.
- Schacker TW, et al. Collagen deposition in HIV-1 infected lymphatic tissues and T cell homeostasis. J Clin Invest. 2002;110(8):1133–1139.
- Schacker TW, et al. Amount of lymphatic tissue fibrosis in HIV infection predicts magnitude of HAART-associated change in peripheral CD4 cell count. AIDS. 2005;19(18):2169–2171.
- Estes JD, et al. Simian immunodeficiency virus-induced lymphatic tissue fibrosis is mediated by transforming growth factor beta 1-positive regulatory T cells and begins in early infection. J Infect Dis. 2007;195(4):551–561.
- Bajenoff M, et al. Stromal cell networks regulate lymphocyte entry, migration, and territoriality in lymph nodes. Immunity. 2006;25(6):989–1001.
- Fletcher AL, et al. Lymph node fibroblastic reticular cells directly present peripheral tissue antigen under steady-state and inflammatory conditions. J Exp Med. 2010;207(4):689–697.
- Link A, et al. Fibroblastic reticular cells in lymph nodes regulate the homeostasis of naive T cells. Nat Immunol. 2007;8(11):1255–1265.
- Mueller SN, Ahmed R. Lymphoid stroma in the initiation and control of immune responses. Immunol Rev. 2008;224:284–294.
- Lawrence JB, Singer RH. Intracellular localization of messenger RNAs for cytoskeletal proteins. Cell. 1986;45(3):407–415.
- St. Johnston D. The intracellular localization of messenger RNAs. Cell. 1995;81(2):161–170.
- Napolitano LA, et al. Increased production of IL-7 accompanies HIV-1-mediated T-cell depletion: implications for T-cell homeostasis. Nat Med. 2001;7(1):73–79.
- Gretz JE, Kaldjian EP, Anderson AO, Shaw S. Sophisticated strategies for information encounter in the lymph node: the reticular network as a conduit of soluble information and a highway for cell traffic. J Immunol. 1996;157(2):495–499.View this article via: PubMed Google Scholar
- Hayakawa M, Kobayashi M, Hoshino T. Direct contact between reticular fibers and migratory cells in the paracortex of mouse lymph nodes: a morphological and quantitative study. Arch Histol Cytol. 1988;51(3):233–240.
- Sixt M, et al. The conduit system transports soluble antigens from the afferent lymph to resident dendritic cells in the T cell area of the lymph node. Immunity. 2005;22(1):19–29.
- Schluns KS, Kieper WC, Jameson SC, Lefrançois L. Interleukin-7 mediates the homeostasis of naive and memory CD8 T cells in vivo. Nat Immunol. 2000;1(5):426–432.View this article via: PubMed Google Scholar
- Rathmell JC, Farkash EA, Gao W, Thompson CB. IL-7 enhances the survival and maintains the size of naive T cells. J Immunol. 2001;167(12):6869–6876.View this article via: PubMed Google Scholar
- Ohshima Y, et al. Naive human CD4+ T cells are a major source of lymphotoxin alpha. J Immunol. 1999;162(7):3790–3794.View this article via: PubMed Google Scholar
- Katakai T, Hara T, Sugai M, Gonda H, Shimizu A. Lymph node fibroblastic reticular cells construct the stromal reticulum via contact with lymphocytes. J Exp Med. 2004;200(6):783–795.
- Hulmes DJ. Building collagen molecules, fibrils, and suprafibrillar structures. J Struct Biol. 2002;137(1–2):2–10.View this article via: PubMed Google Scholar
- Lamande SR, Bateman JF. Procollagen folding and assembly: the role of endoplasmic reticulum enzymes and molecular chaperones. Semin Cell Dev Biol. 1999;10(5):455–464.View this article via: PubMed Google Scholar
- Silver FH, Freeman JW, Seehra GP. Collagen self-assembly and the development of tendon mechanical properties. J Biomech. 2003;36(10):1529–1553.
- Border WA, Noble NA. Transforming growth factor beta in tissue fibrosis. N Engl J Med. 1994;331(19):1286–1292.
- Border WA, Ruoslahti E. Transforming growth factor-beta in disease: the dark side of tissue repair. J Clin Invest. 1992;90(1):1–7.
- Gressner AM, Weiskirchen R, Breitkopf K, Dooley S. Roles of TGF-beta in hepatic fibrosis. Front Biosci. 2002;7:d793–d807.
- Li Q, et al. Microarray analysis of lymphatic tissue reveals stage-specific, gene expression signatures in HIV-1 infection. J Immunol. 2009;183(3):1975–1982.
- Bigg HF, Wait R, Rowan AD, Cawston TE. The mammalian chitinase-like lectin, YKL-40, binds specifically to type I collagen and modulates the rate of type I collagen fibril formation. J Biol Chem. 2006;281(30):21082–21095.
- Iwata T, et al. YKL-40 secreted from adipose tissue inhibits degradation of type I collagen. Biochem Biophys Res Commun. 2009;388(3):511–516.
- Mizoguchi E. Chitinase 3-like-1 exacerbates intestinal inflammation by enhancing bacterial adhesion and invasion in colonic epithelial cells. Gastroenterology. 2006;130(2):398–411.
- Recklies AD, Ling H, White C, Bernier SM. Inflammatory cytokines induce production of CHI3L1 by articular chondrocytes. J Biol Chem. 2005;280(50):41213–41221.View this article via: PubMed Google Scholar
- Bonyhadi ML, et al. HIV induces thymus depletion in vivo. Nature. 1993;363(6431):728–732.
- Douek DC, et al. Changes in thymic function with age and during the treatment of HIV infection. Nature. 1998;396(6712):690–695.
- Stanley SK, et al. Human immunodeficiency virus infection of the human thymus and disruption of the thymic microenvironment in the SCID-hu mouse. J Exp Med. 1993;178(4):1151–1163.
- Giorgi JV, et al. Predictive value of immunologic and virologic markers after long or short duration of HIV-1 infection. J Acquir Immune Defic Syndr. 2002;29(4):346–355.View this article via: PubMed Google Scholar
- Hazenberg MD, et al. Persistent immune activation in HIV-1 infection is associated with progression to AIDS. AIDS. 2003;17(13):1881–1888.
- Rabin RL, Roederer M, Maldonado Y, Petru A, Herzenberg LA, Herzenberg LA. Altered representation of naive and memory CD8 T cell subsets in HIV-infected children. J Clin Invest. 1995;95(5):2054–2060.
- Roederer M, Dubs JG, Anderson MT, Raju PA, Herzenberg LA, Herzenberg LA. CD8 naive T cell counts decrease progressively in HIV-infected adults. J Clin Invest. 1995;95(5):2061–2066.
- Roederer M, Raju PA, Mitra DK, Herzenberg LA, Herzenberg LA. HIV does not replicate in naive CD4 T cells stimulated with CD3/CD28. J Clin Invest. 1997;99(7):1555–1564.
- Woods TC, Roberts BD, Butera ST, Folks TM. Loss of inducible virus in CD45RA naive cells after human immunodeficiency virus-1 entry accounts for preferential viral replication in CD45RO memory cells. Blood. 1997;89(5):1635–1641.View this article via: PubMed Google Scholar
- Nie C, et al. Selective infection of CD4+ effector memory T lymphocytes leads to preferential depletion of memory T lymphocytes in R5 HIV-1-infected humanized NOD/SCID/IL-2Rgammanull mice. Virology. 2009;394(1):64–72.
- Schnittman SM, Lane HC, Greenhouse J, Justement JS, Baseler M, Fauci AS. Preferential infection of CD4+ memory T cells by human immunodeficiency virus type 1: evidence for a role in the selective T-cell functional defects observed in infected individuals. Proc Natl Acad Sci U S A. 1990;87(16):6058–6062.
- Zhang ZQ, et al. Kinetics of CD4+ T cell repopulation of lymphoid tissues after treatment of HIV-1 infection. Proc Natl Acad Sci U S A. 1998;95(3):1154–1159.
- Fry TJ, et al. IL-7 therapy dramatically alters peripheral T-cell homeostasis in normal and SIV-infected nonhuman primates. Blood. 2003;101(6):2294–2299.
- Leone A, et al. Increased CD4+ T cell levels during IL-7 administration of antiretroviral therapy-treated simian immunodeficiency virus-positive macaques are not dependent on strong proliferative responses. J Immunol. 2010;185(3):1650–1659.
- Levy Y, et al. Enhanced T cell recovery in HIV-1-infected adults through IL-7 treatment. J Clin Invest. 2009;119(4):997–1007.
- Sportes C, et al. Administration of rhIL-7 in humans increases in vivo TCR repertoire diversity by preferential expansion of naive T cell subsets. J Exp Med. 2008;205(7):1701–1714.
- Vassena L, Proschan M, Fauci AS, Lusso P. Interleukin 7 reduces the levels of spontaneous apoptosis in CD4+ and CD8+ T cells from HIV-1-infected individuals. Proc Natl Acad Sci U S A. 2007;104(7):2355–2360.
- http://clinicaltrials.gov/ct2/show/NCT00080223.
- Estes JD, et al. Early resolution of acute immune activation and induction of PD-1 in SIV-infected sooty mangabeys distinguishes nonpathogenic from pathogenic infection in rhesus macaques. J Immunol. 2008;180(10):6798–6807.View this article via: PubMed Google Scholar
- Li Q, et al. Peak SIV replication in resting memory CD4+ T cells depletes gut lamina propria CD4+ T cells. Nature. 2005;434(7037):1148–1152.View this article via: PubMed Google Scholar
- Haase AT. Population biology of HIV-1 infection: viral and CD4+ T cell demographics and dynamics in lymphatic tissues. Annu Rev Immunol. 1999;17:625–656.
-
Version history
- Version 1 (February 14, 2011): No description
- Version 2 (March 1, 2011): No description



Copyright © 2024 American Society for Clinical Investigation
ISSN: 0021-9738 (print), 1558-8238 (online)









