Advertisement
Science in Medicine Free access | 10.1172/JCI41246
Chronic myeloid leukemia: mechanisms of blastic transformation
Danilo Perrotti,1 Catriona Jamieson,2 John Goldman,3 and Tomasz Skorski4
1Department of Molecular Virology, Immunology and Medical Genetics and Comprehensive Cancer Center, The Ohio State University, Columbus, Ohio, USA. 2Division of Hematology-Oncology, Department of Internal Medicine, University of California at San Diego, La Jolla, California, USA. 3Department of Haematology, Imperial College at Hammersmith Hospital, London, United Kingdom. 4Department of Microbiology and Immunology, Temple University, Philadelphia, Pennsylvania, USA.
Address correspondence to: Danilo Perrotti, Department of Molecular Virology, Immunology and Medical Genetics and Comprehensive Cancer Center, The Ohio State University, 892 Biomedical Research Tower, 460 West 12th Avenue, Columbus, Ohio 41230, USA. Phone: 614.292.3255; Fax: 614.688.4181; E-mail: [email protected]. Or to: Tomasz Skorski, Department of Microbiology and Immunology, School of Medicine, Temple University, 3400 N. Broad Street, MRB 548, Philadelphia, Pennsylvania 19140, USA. Phone: 215.707.9157; Fax: 215.707.9160; E-mail: [email protected].
Find articles by Perrotti, D. in: JCI | PubMed | Google Scholar
1Department of Molecular Virology, Immunology and Medical Genetics and Comprehensive Cancer Center, The Ohio State University, Columbus, Ohio, USA. 2Division of Hematology-Oncology, Department of Internal Medicine, University of California at San Diego, La Jolla, California, USA. 3Department of Haematology, Imperial College at Hammersmith Hospital, London, United Kingdom. 4Department of Microbiology and Immunology, Temple University, Philadelphia, Pennsylvania, USA.
Address correspondence to: Danilo Perrotti, Department of Molecular Virology, Immunology and Medical Genetics and Comprehensive Cancer Center, The Ohio State University, 892 Biomedical Research Tower, 460 West 12th Avenue, Columbus, Ohio 41230, USA. Phone: 614.292.3255; Fax: 614.688.4181; E-mail: [email protected]. Or to: Tomasz Skorski, Department of Microbiology and Immunology, School of Medicine, Temple University, 3400 N. Broad Street, MRB 548, Philadelphia, Pennsylvania 19140, USA. Phone: 215.707.9157; Fax: 215.707.9160; E-mail: [email protected].
Find articles by Jamieson, C. in: JCI | PubMed | Google Scholar
1Department of Molecular Virology, Immunology and Medical Genetics and Comprehensive Cancer Center, The Ohio State University, Columbus, Ohio, USA. 2Division of Hematology-Oncology, Department of Internal Medicine, University of California at San Diego, La Jolla, California, USA. 3Department of Haematology, Imperial College at Hammersmith Hospital, London, United Kingdom. 4Department of Microbiology and Immunology, Temple University, Philadelphia, Pennsylvania, USA.
Address correspondence to: Danilo Perrotti, Department of Molecular Virology, Immunology and Medical Genetics and Comprehensive Cancer Center, The Ohio State University, 892 Biomedical Research Tower, 460 West 12th Avenue, Columbus, Ohio 41230, USA. Phone: 614.292.3255; Fax: 614.688.4181; E-mail: [email protected]. Or to: Tomasz Skorski, Department of Microbiology and Immunology, School of Medicine, Temple University, 3400 N. Broad Street, MRB 548, Philadelphia, Pennsylvania 19140, USA. Phone: 215.707.9157; Fax: 215.707.9160; E-mail: [email protected].
Find articles by Goldman, J. in: JCI | PubMed | Google Scholar
1Department of Molecular Virology, Immunology and Medical Genetics and Comprehensive Cancer Center, The Ohio State University, Columbus, Ohio, USA. 2Division of Hematology-Oncology, Department of Internal Medicine, University of California at San Diego, La Jolla, California, USA. 3Department of Haematology, Imperial College at Hammersmith Hospital, London, United Kingdom. 4Department of Microbiology and Immunology, Temple University, Philadelphia, Pennsylvania, USA.
Address correspondence to: Danilo Perrotti, Department of Molecular Virology, Immunology and Medical Genetics and Comprehensive Cancer Center, The Ohio State University, 892 Biomedical Research Tower, 460 West 12th Avenue, Columbus, Ohio 41230, USA. Phone: 614.292.3255; Fax: 614.688.4181; E-mail: [email protected]. Or to: Tomasz Skorski, Department of Microbiology and Immunology, School of Medicine, Temple University, 3400 N. Broad Street, MRB 548, Philadelphia, Pennsylvania 19140, USA. Phone: 215.707.9157; Fax: 215.707.9160; E-mail: [email protected].
Find articles by Skorski, T. in: JCI | PubMed | Google Scholar
Published July 1, 2010 - More info
J Clin Invest. 2010;120(7):2254–2264. https://doi.org/10.1172/JCI41246.
© 2010 The American Society for Clinical Investigation
-
Abstract
The BCR-ABL1 oncoprotein transforms pluripotent HSCs and initiates chronic myeloid leukemia (CML). Patients with early phase (also known as chronic phase [CP]) disease usually respond to treatment with ABL tyrosine kinase inhibitors (TKIs), although some patients who respond initially later become resistant. In most patients, TKIs reduce the leukemia cell load substantially, but the cells from which the leukemia cells are derived during CP (so-called leukemia stem cells [LSCs]) are intrinsically insensitive to TKIs and survive long term. LSCs or their progeny can acquire additional genetic and/or epigenetic changes that cause the leukemia to transform from CP to a more advanced phase, which has been subclassified as either accelerated phase or blastic phase disease. The latter responds poorly to treatment and is usually fatal. Here, we discuss what is known about the molecular mechanisms leading to blastic transformation of CML and propose some novel therapeutic approaches.
-
Introduction
Chronic myeloid leukemia (CML) is a myeloproliferative disorder characterized by excessive accumulation of apparently normal myeloid cells. It occurs with an annual incidence of 1.0–1.5 per 100,000 persons. CML occurs very rarely in children. In the Western world, the median age of onset is 50–60 years, which reflects the average age of the population. Although symptoms at presentation may include lethargy, weight loss, unusual bleeding, sweats, anemia, and splenomegaly, in more developed countries, 50% of patients are asymptomatic and are diagnosed as a consequence of blood tests performed for unrelated reasons. More than 90% of CML patients are diagnosed when their disease is in a relatively early phase known as the chronic phase (CP).
CML-CP is characterized by the presence of the Philadelphia (Ph) chromosome and the oncogene that it encodes in the vast majority of myeloid cells and some lymphocytes. The Ph chromosome results from a (9;22)(q34;q11) reciprocal translocation that juxtaposes the c-abl oncogene 1 (ABL1) gene on chromosome 9 with the breakpoint cluster region (BCR) gene on chromosome 22, generating the BCR-ABL1 fusion oncogene with greatly enhanced ABL1 kinase activity. It is generally accepted that acquisition of the BCR-ABL1 oncogene is the initiating event in the genesis of CML-CP, despite various lines of evidence suggesting that, at least in some cases, hematopoiesis may already be clonal before the acquisition of the Ph chromosome (1). It is believed that acquisition of the BCR-ABL1 gene occurs initially in a single HSC that gains a proliferative advantage and/or aberrant differentiation capacity over its normal counterparts, giving rise to the expanded myeloid compartment (2).
Most CML-CP patients are currently treated with one of three drugs designed to block the enzymatic action of the BCR-ABL1 tyrosine kinase. The first of these to be developed was imatinib. Recent karyotype analyses show that 60%–70% of patients achieve complete disappearance of Ph-positive marrow cells and maintain exclusively Ph-negative bone marrow cells (a state designated as a complete cytogenetic response [CCyR]) 5 years after initiating imatinib treatment. The incidence of progression to a more advanced phase of leukemia in patients responding to imatinib is extremely low beyond the first two years (3). However, a small number of patients fail to respond to imatinib (primary resistance), while others respond initially and then lose their response (secondary resistance) (4). The reasons for imatinib resistance in CML-CP patients are poorly understood. Primary resistance may be related, at least in part, to the intrinsic heterogeneity of the disease (e.g., different BCR-ABL1 levels) in different patients and to the survival of variable numbers of quiescent cells from which the more mature leukemia cells are derived during CP (5). Secondary resistance may have a wide range of causes, of which the best characterized is the acquisition of mutations in the BCR-ABL1 kinase domain (such as the T315I mutation) (6).
In the last few years, two new tyrosine kinase inhibitors (TKIs), dasatinib and nilotinib, have become available, both of which are more potent in vitro inhibitors of the BCR-ABL1 kinase than imatinib. Both of these “second-generation” TKIs are effective at inducing or restoring CCyR in 40%–50% of patients who appear to have failed primary treatment with imatinib. However, approximately 20% of patients presenting with CML-CP fail to respond to both imatinib and a subsequent second-generation TKI; their prognosis is poor because of a higher risk of disease progression.
Before the advent of BCR-ABL1 TKIs, all patients with CML-CP progressed spontaneously to advanced phase CML after a median interval of approximately 5 years. The advanced phase is divided into an initial accelerated phase (AP), during which patients may still respond to treatment for some months or sometimes years, and a subsequent more aggressive blastic phase (BP). Patients with CML-BP have a median survival of approximately 6 months. Some patients progress directly to BP without an intermediate AP. The precise definitions of these three phases have been much debated in recent years (3, 7).
The biological basis of BP is poorly understood. Although the majority of patients have a myeloblastic phenotype, approximately 25% of CML-BP patients show a pre–B lymphoblastic cell phenotype (8). Occasional cases of T lymphoblastic transformation have been identified (9). These findings lend support to the notion that the BCR-ABL1 oncogene arises in a primitive cell, namely a leukemia stem cell (LSC), not yet committed to either myeloid or lymphoid differentiation. Conversely the blastic clone may originate either at the level of the multipotent LSC or at the level of a more committed leukemia progenitor cell (LPC). Here, we discuss genetic and epigenetic mechanisms leading to the transition of CML-CP into CML-BP and propose some novel therapeutic modalities that might prevent malignant progression.
-
CML-BP patients: a therapeutic challenge
In the past, CML-BP was often treated with drugs used for acute leukemias, but patients usually relapsed within a few months. The introduction of TKIs has improved prognosis to some degree. The majority of CML-BP patients not previously treated with TKIs do initially respond to treatment with these agents, either alone or in combination with conventional chemotherapeutic drugs, but most still relapse within a few months of achieving a seemingly complete hematologic or even cytogenetic response. Therefore, any CML-BP patient who does respond to modern therapy should proceed, if possible, to allogeneic stem cell transplant prior to relapse. In the 1990s, the results of allografting for CML-BP patients were not impressive — only 5%–10% of patients experienced long-term, leukemia-free survival (10) — but the use of a TKI after transplant may improve these results.
Extrapolating from the good clinical outcomes of treating CML-CP with TKIs and the dismal responses achieved in treating CML-BP, one might reasonably conclude that the best approach to CML-BP would be prevention. Indeed, it appears today that continued use of TKIs to treat CML-CP may prevent BP in a large proportion of patients, but 15%–20% of patients, most of whom will have been classified as nonresponders, may progress to BP (11). Indeed, the GIMEMA Working Party (Italian Group for Adult Hematologic Diseases) reported that the detection of TKI-resistant BCR-ABL1 mutations in CML-CP is associated with a greater likelihood of disease progression (12). These patients may possess genetic/epigenetic abnormalities distinct from the patients with nonmutated BCR-ABL1, the appearance of which could be influenced by the duration of the BCR-ABL1–induced signals. Furthermore, the ability of TKIs to render residual CML cells “inactive” rather than to eradicate them entirely suggests that BP might still occur occasionally even in “responding” patients.
However, as a minority of patients will still progress to CML-BP, the routine use of TKIs may need to be supplemented with other agents, any of which might prevent BP. Possible examples are antioxidants (13), which protect against cancer-causing DNA mutations; farnesyl transferase inhibitors (14), which inhibit RAS signaling; hydroxychloroquine (15), which inhibits autophagy in some situations; sonic hedgehog pathway antagonists (16, 17), which impair self-renewal pathways only when used in combination with TKIs; and activators of protein phosphatase 2A (PP2A) (18), which targets BCR-ABL1 and other downstream oncogenic signaling cascades.
-
Biological complexity of CML-BP
At present, the molecular mechanisms underlying disease progression are still uncertain, but most likely involve activation of oncogenic factors and/or inactivation of tumor suppressors (19). A plausible assumption is that BP is a multistep, time-dependent process initiated by both BCR-ABL1–dependent and –independent DNA damage associated with inefficient and unfaithful DNA repair in CML-CP that, if facilitated by an increased level of BCR-ABL1 activity, leads to selection of one or more CML-BP clones.
The genetic lesions observed in CML-BP patients in the past and now since the introduction of TKIs mostly include the presence of additional chromosomes, gene deletions, gene insertions, and/or point mutations (including BCR-ABL1 mutations) (20–22), but patterns differ in myeloblastic and lymphoblastic transformations (23). At the molecular level, the most common mutations detectable (other than those in the BCR-ABL1 kinase domain) occur at the loci of the tumor suppressor genes P53 (20%–30% of cases) and the runt-related transcription factor gene (RUNX1) (38% of cases) in myeloid BP and at the loci of cyclin-dependent kinase inhibitor 2A/2B (CDKN2A/B) (50% of cases) and Ikaros transcription factor (IKZF1) (55% of cases) in lymphoid BP (22, 24–28). As specific CML-BP–associated genetic alterations are relatively common, no one lesion occurs in the majority of CML-BP patients, and it is unlikely that any one specific secondary genetic aberration can be defined as the “culprit” causing disease progression. More likely, CML-BP results from the accumulation of a critical number or combination of different mutations.
Epigenetic changes are dependent mostly on the pleiotropic effect of constitutive BCR-ABL1 activity (19, 29), the levels of which start to increase in CML-AP (30). In support of this suggestion, expression studies revealed that BCR-ABL1 dramatically perturbs the CML transcriptome (31), resulting in altered expression of genes, some of which (e.g., PRAME, MZF1, EVI-1, WT1, and JUN-B) might play a role in BP (19, 32–34). Nonetheless, the posttranscriptional, translational, and posttranslational effects of high BCR-ABL1 levels result in the constitutive activation of factors with reported mitogenic, antiapoptotic, and antidifferentiation activity (e.g., MAPKERK1/2, MYC, JAK2, YES-1, LYN, hnRNP-E2, MDM2, STAT5, BMI-1, and BCL-2) and inhibition of major key regulators of cellular processes, such as those regulated by the tumor suppressors p53, CCAAT/enhancer binding protein-α (C/EBPα), and PP2A (19, 29, 35). Interestingly, a signature based on six genes (NOB1, DDX47, IGSF2, LTB4R, SCARB1, and SLC25A3) was recently found to accurately discriminate early from late CP, CP from AP, and CP from BP (36); however, the biological role of these genes in disease progression is still unknown.
Thus, it is highly plausible that unrestrained and increasing BCR-ABL1 activity promotes and/or contributes to clonal evolution, thereby leading to CML-BP (37). This might occur at the level of LSCs, which display innate or acquired TKI resistance, and/or at the level of an LPC population that might have developed resistance and expanded during TKI therapy (38, 39).
Because there is a direct correlation between levels of BCR-ABL1, the frequency of clinically relevant BCR-ABL1 mutations (40, 41), and the differentiation arrest of myeloid progenitors (42), it is likely that disease progression is triggered by the “right” combinations of genetic and epigenetic abnormalities (Figure 1). Thus, we can speculate that prevention or effective treatment of CML-BP will only be achieved if novel therapeutic strategies can be developed that are capable of interfering with the biological processes currently considered critical for the leukemic behavior of CML-BP progenitors.
 Figure 1
Figure 1BCR-ABL1–dependent pathways to blastic transformation. Schematic representation of the potential BCR-ABL1–dependent molecular mechanisms leading to CML disease progression.The relatively high BCR-ABL1 expression/activity in CML-CP CD34+CD38– stem cells and/or CD34+ early progenitors compared with more committed progenitors, which is further markedly increased in CML-BP CD34+ progenitors results in the following: enhancement of proliferation/survival pathways; increased genomic instability; and activation of pathways leading to a block in myeloid differentiation, acquisition of the ability to self renew, and inhibition of tumor suppressors with broad cell regulatory functions. BAD, BCL2 antagonist of cell death; DNA-PKcs, DNA-dependent protein kinase, catalytic subunit; FOXO, forkhead box O; IK6, Ikaros 6; miR-328, microRNA-328; MLH1, mutL homolog 1; PMS2, postmeiotic segregation increased 2; RAD51, RecA homolog in Escherichia coli; RAD52, RAD52 homolog (Saccharomyces cerevisiae); Shh, Sonic Hedgehog; wnt/β-catenin, wingless-int1/beta-catenin.
-
CML-BP LSCs: BCR-ABL1 overexpression, self renewal, and survival
According to the basic concept, LSCs should represent the most primitive cell able to initiate leukemia in animal xenograft limiting dilution experiments, to display self-renewal capacity, and to proliferate and differentiate (43, 44). In CML-CP, LSCs are located in the self-renewing Lin–CD34+CD38– population, but not in the non–self-renewing Lin–CD34+CD38+ population (45), indicating that, in contrast to other oncogenes (e.g., MOZ-TIF2 and MLL-ENL) BCR-ABL1 cannot confer self-renewal properties (46, 47). While most human LSC research has focused on the Lin–CD34+ compartment, a recent study suggests that a Lin–CD34– fraction of CML-CP cells also engrafts immunodeficient mouse strains, underscoring the complexity of the LSC compartment (48).
However, in CML-BP, Lin–CD34+CD38+ granulocyte-macrophage progenitors (GMPs) that overexpress BCR-ABL1 behave like LSCs (49), suggesting that the acquisition of self renewal in GMPs may depend on epigenetic and/or genetic alterations caused by elevated expression of BCR-ABL1. Thus, LSCs in CML-BP patients may reside in at least 3 different subsets: Lin–CD34+CD38– and Lin–CD34+ cells remaining from CML-CP and the disease-driving Lin–CD34+CD38+ GMPs.
Progression to BP is marked by overexpression of BCR-ABL1 in committed progenitors, leading to a multiplicity of genetic and epigenetic events. These cell type– and context-specific molecular events serve to enhance survival and self renewal, leading to impaired differentiation and generation of CML-BP LSCs. To date, the cell type– and context-specific effects of BCR-ABL1 overexpression have not been clearly elucidated in human stem cells, nor has the effect of the microenvironment on LSC maintenance. It seems, however, that increased BCR-ABL1 expression does play a critical role in promoting the genetic instability that drives progression to BP and the molecular evolution of LSCs in CML.
BCR-ABL1 overexpression and aberrant splicing. BCR-ABL1 induces alterations in pre-mRNA splicing in hematopoietic progenitor cells that result in aberrant adhesion, differentiation, survival, and self renewal as well as therapeutic resistance. Ectopic BCR-ABL1 expression in human bone marrow and cord blood CD34+ cells results in induction of factors involved in mRNA processing, export, and translation (50, 51). Interestingly, the metabolism of several mRNAs has been found to be altered in CML-BP (51). Among these, BCR-ABL1 induces alternative splicing of proline-rich tyrosine kinase 2 (PYK2) mRNA, thereby increasing expression of the β1-integrin–responsive PYK2 kinase, which in turn may contribute to aberrant adhesion of CML-BP progenitors (50).
Likewise, BCR-ABL1–induced aberrant splicing might play an important role in those cases of CML-BP without deletion of the IKZF1 gene (25). Indeed, a recent study suggests that BCR-ABL1 may inhibit differentiation and contribute to lymphoid CML-BP by promoting the production of a dominant negative splice isoform (IK6) of IKZF1, a transcription factor gene involved in pre–B cell differentiation (52). When this aberrant, non–DNA-binding splice isoform, IK6, was silenced in Ph-positive pre–B cells using siRNA or its production reduced by imatinib treatment, differentiation along the B cell lineage was partially restored (52). Notably, alternative splicing was also observed for BCR-ABL1. Aberrant BCR-ABL1 mRNA splicing results in the generation of transcripts harboring a 35-kb insertion between ABL1 domain exons 8 and 9, resulting in a frameshift with a truncation that, like IK6 expression, is associated with imatinib resistance (53, 54).
Finally, BCR-ABL1 overexpression is associated with mis-splicing of glycogen synthase kinase 3β (GSK3β) (55), a key component of the β-catenin destruction complex, leading to enhanced self renewal of GMPs that behave like LSCs (49). Lentiviral overexpression of wild-type GSK3β in CML-BP progenitors inhibits their capacity to engraft leukemia in immunocompromised mice (55).
Stem cell self renewal. Self renewal refers to division without differentiation and is a property normally ascribed to long-term HSCs. In mouse models, loss of junB/AP-1 enhances HSC proliferation and myeloid progenitor expansion, setting the stage for BP (56). In CML-BP, committed progenitors subvert this stem cell property of self renewal, lack the capacity to regulate it, and are able to propagate leukemia more readily. Various self-renewal pathways — including Wnt/β-catenin, sonic hedgehog, and Notch signaling — have been implicated in the generation and maintenance of CML-BP LSCs. Indeed, BCR-ABL1–independent (57) and –dependent (49, 58) mechanisms both seem to contribute to the acquisition of self renewal by CD34+CD38+CD45RA+CD123+Lin– CML-BP GMPs. In fact, CML-BP is associated with accumulation of β-catenin, a key stem cell self-renewal mediator, in the nucleus of GMPs, thereby endowing them with self-renewal potential (49, 59). BCR-ABL1 stabilizes β-catenin through phosphorylation of tyrosines 86 and 654, which inhibits binding to axin/GSK3β, thereby enabling binding to T cell factor 4 (TCF4) and activation of transcription.
By inhibiting BCR-ABL1, imatinib prevents tyrosine phosphorylation of β-catenin and thus prevents nuclear translocation and transcriptional activation (58). Loss of β-catenin in a CML mouse model impairs self renewal of both normal HSCs and CML-BP LSCs, although the effects of decreased nuclear β-catenin on human normal HSC and CML-BP LSC maintenance remains to be established in xenograft models (60).
Decreased expression of functional GSK3β enhances CML progenitor self renewal by activating β-catenin and by elevating levels of sonic hedgehog pathway–mediators such as GLI family zinc finger 1 (GLI1) and GLI2 (32, 55). Recently, two independent studies demonstrated that overexpression of smoothened homolog (Drosophila) (Smo), an essential activator of sonic hedgehog signaling, enhanced LSC maintenance in mouse models of CML (16, 17). Conversely, Smo inhibition reduced LSC, but not normal HSC engraftment (16, 17), suggesting that the sonic hedgehog pathway is preferentially utilized by LSCs for self renewal.
Another recent study confirmed that sonic hedgehog signaling is dispensable for normal adult mouse HSC function, suggesting the possibility of targeting leukemic GMP without damaging residual HSCs (61). These findings provide the impetus for preclinical testing of a combination of Smo and BCR-ABL1 inhibitors to determine whether LSCs can be eradicated both in vitro and in xenogeneic transplantation models.
LSC survival. Resistance to apoptosis, an intrinsic property of normal HSCs, is also a hallmark of LSCs. In vivo inactivation of Dok-1 or Dok-2 decreases apoptosis, resulting in a myeloproliferative disorder (62). Moreover, the promyelocytic leukemia (PML) gene, a tumor suppressor that was first shown to be deregulated in acute promyelocytic leukemia, was recently found to play a pivotal role in LSC maintenance in a CML mouse model (63). Other investigators demonstrated that enhanced progenitor cell survival driven by B cell leukemia/lymphoma 2 (BCL2) and BCR-ABL1 overexpression promoted CML-BP development in a transgenic mouse model (64), underscoring the importance of resistance to apoptosis in BP evolution.
Although extensive SNP marker analyses demonstrated that a SNP (rs1801018) in the BCL2 gene was associated with susceptibility to CML (65), the role of BCL2 in CML-BP progenitor survival remains to be elucidated. In CML-BP cell lines, expression levels of BCL2-interacting mediator of cell death (BIM), a proapoptotic BCL2 family member, are low and can be induced by BCR-ABL1 inhibition (66). In recent studies, induction of apoptosis correlated with the magnitude and duration of BCR-ABL1 kinase inhibition (67). Transient, potent BCR-ABL1 inhibition was associated with BIM activation and induction of apoptosis, underscoring the importance of BCR-ABL1 gene dosage in regulating apoptotic responses (67). In addition, JAK2-mediated activation of LYN kinase through the suppressor of variegation, enhancer of zeste, and Trithorax (SET)/PP2A/SHP1 pathway (68) may be important in promoting CML-BP LSC survival during imatinib therapy and disease progression. Pharmacologic inhibition of JAK2 induced apoptosis in imatinib-resistant CML-BP cells to a greater degree than in normal progenitors (68). Recently, targeted inhibition of arachidonate 5-lipoxygenase (ALOX5) with a 5-lipoxygenase inhibitor was shown to impair LSC survival in a CML mouse model, a finding that warrants further investigation into the role of ALOX5 in CML-BP pathogenesis (69).
Interestingly, a recent study has shown that imatinib induces autophagy in CML-BP primitive progenitors through a mechanism that is independent of imatinib-induced, caspase-dependent apoptosis but is associated with ER stress and is suppressed by intracellular Ca2+ depletion (15). Suppression of autophagy genes enhanced imatinib-induced death of Ph-positive cells (15). Critically, the combination of TKIs with autophagy inhibitors resulted in killing of CML LSCs (15). Thus, autophagy inhibitors may enhance the therapeutic effects of TKIs in the treatment of CML-BP.
-
Global interference with BCR-ABL1–dependent and –independent signals
Since the late 1980s, when BCR-ABL1 was identified as a constitutively active tyrosine kinase, an impressive series of in vitro and in vivo studies have indicated a direct causal role of BCR-ABL1 activity in the acquisition of the molecular changes that characterize the phenotype of CML-BP progenitors (19).
In vivo resistance and in vitro sensitivity of CML-BP progenitors to TKI treatment: a biological paradox. Compelling research shows that CML-CP LSCs are resistant to imatinib as a result of various collaborating factors. These factors include quiescence, high BCR-ABL1 levels, lack of “oncogene addiction,” increased activity of the drug efflux pumps ATP-binding cassette sub-family B member 1 (ABCB1) and transporter G2 (ABCG2), and decreasing organic cation transporter 1 (OCT1) expression (5, 70, 71).
In CML-BP, increased BCR-ABL1 expression (49, 72) accounts for activation of pathways transducing mitogenic, antiapoptotic signals and for differentiation arrest of the Ph-positive progenitors (42, 49, 73, 74). However, BCR-ABL1–independent mechanisms (e.g., LYN kinase–dependent mechanisms) also contribute to disease progression and imatinib resistance in some CML-BP cases with no BCR-ABL1 amplification/overexpression (75–78). In this regard, the paradoxical in vitro and in vivo response of CML-BP progenitors to TKIs needs to be taken into consideration. While most CML-BP patients do not show long-term responses to TKIs and relapse within 12–24 months, CML-BP progenitors from these patients are still sensitive to the proapoptotic effects of imatinib when administered ex vivo. Thus, it is possible that the bone marrow environment elicits BCR-ABL1–independent signals conferring TKI resistance and sustaining in vivo survival of CML-BP blasts. In this scenario, BCR-ABL1–dependent and –independent signals likely synergize in inducing and maintaining the CML-BP phenotype. Furthermore, from this consideration, the concept emerges clearly that TKI treatment, especially at high dosage, might exert a selective pressure allowing clonal expansion of genetically unstable CML-BP progenitor cell clones that are more prone to acquire secondary chromosomal abnormalities and/or clinically relevant mutations in the BCR-ABL1 oncogene itself and, likely, in other kinases targeted by TKIs. As CML-BP is also characterized by the loss of function of tumor suppressors, a rational and alternative therapeutic approach might envision the use of drugs capable of reactivating a tumor suppressor or suppressors.
Pharmacologic reactivation of the PP2A tumor suppressor gene. The notion that the serine-threonine phosphatase PP2A is inhibited in several types of cancer, through mechanisms that either involve the loss of expression/activity of one or more subunits or the enhanced expression of the endogenous PP2A inhibitors SET (79, 80) and cancerous inhibitor of PP2A (CIP2A) (81), led to the recognition of PP2A as a true tumor suppressor. In fact, loss of PP2A activity plays a central role in the pathophysiology of BCR-ABL1–driven leukemias. PP2A activity is slightly reduced in CML-CP CD34+ progenitors but becomes markedly inhibited in CML-BP through the BCR-ABL1 dose- and kinase-dependent induction of SET (74, 82) (Figure 2). Remarkably, several targets that are shared by BCR-ABL1 and PP2A are either essential for BCR-ABL1 leukemogenesis or are altered in CML-BP (19).
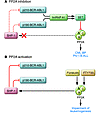 Figure 2
Figure 2BCR-ABL1 and PP2A interplay. (A) In CML-BP and Ph-positive ALL CD34+ progenitors, p210 and p190 BCR-ABL1 oncoproteins inhibit PP2A activity by inducing hnRNP-A1, which, in turn, enhances expression of SET. In BCR-ABL–positive myeloid progenitor cells, suppression of PP2A phosphatase activity is required for sustained activation of mitogenic and survival signals. (B) Restored PP2A activity, achieved by treatment with PP2A activators (e.g., Forskolin or FTY720), impairs in vitro and in vivo wild-type and T315I BCR-ABL1 leukemogenesis by antagonizing the effects of BCR-ABL1 on its downstream signal transducers (not shown) and promoting SHP-1–mediated BCR-ABL1 inactivation and proteasome-dependent degradation.
Restoration of PP2A activity, either by chemical PP2A activators (e.g., forskolin and FTY720) (Figure 2) or by interfering with SET/PP2A interplay, promotes Src homology region 2–domain phosphatase 1 (SHP-1) tyrosine phosphatase–dependent BCR-ABL1 dephosphorylation (inactivation) which, in turn, triggers its degradation (74, 82, 83). Notably, SHP-1 expression is diminished in most leukemias and lymphomas (84, 85). Restoring normal PP2A activity induces marked apoptosis of CD34+ CML (CP and BP) progenitors and suppresses in vivo leukemogenesis regardless of sensitivity to imatinib/dasatinib (74, 82) (Figure 2).
Loss of PP2A activity is also a feature of imatinib/dasatinib-insensitive CD34+CD38–BCR-ABL1+ HSCs from CML (CP and BP) patients (57). Clonogenic, colony-forming cell (CFC)/replating, long-term culture–initiating cell (LTC-IC), and CFSE-mediated cell division–tracking assays revealed that FTY720 suppresses survival and self renewal and triggers apoptosis of BCR-ABL1+ stem cells in a BCR-ABL1 kinase–independent and β-catenin–mediated manner (57). Notably, normal quiescent stem cells are not sensitive to FTY720 (57).
Because of the central role of PP2A in the regulation of survival, proliferation, self renewal, and differentiation of CML stem/progenitor cells, it is highly plausible that its loss of function contributes to BP. In this scenario, PP2A may have the role of a “gatekeeper,” as its activation may control and restrain BCR-ABL1 expression/activity, whereas its inhibition allows increased oncogene activity and induces a cascade of events that promotes disease development and progression. Thus, incorporating PP2A-activating drugs into current therapeutic protocols for CML-BP and imatinib/dasatinib-resistant (including T315I) patients has not only the potential to treat CML-BP but also to eradicate CML at the stem cell level.
-
Impaired myeloid differentiation in CML-BP: a true BCR-ABL1 dosage effect
Blastic transformation is phenotypically associated with the clonal expansion of the GMP pool (49), members of which have lost the ability to differentiate in response to cytokine stimuli. If we exclude the 20%–30% of CML-BP cases with P53 mutations (28), the 11% of CML-BP cases with GATA-binding protein 2 (GATA2) mutations (86), and the 1%–2% of CML-BP cases with the t(3;21)(q26;q22) and t(7;11)(p15;p15) translocations associated with expression of AME (AML-1 [acute myeloid leukemia 1], MDS/EVI1 [Myelodysplastic syndrome–associated gene 1]) (87) and NUP98-HOXA9 (88) chimeric proteins, we can safely state that impaired myeloid maturation of Ph-positive GMPs is the consequence of increased BCR-ABL1 dosage. Indeed, low BCR-ABL1 levels allow G-CSF–induced granulocytic maturation, while high oncogene expression impedes differentiation of Lin– progenitors (89).
BCR-ABL1 levels and C/EBPα inhibition. Different genetic and epigenetic mechanisms may act alone or in cooperation to enhance BCR-ABL1 expression and activity. Among them, BCR-ABL1 gene amplification (90, 91), increased BCR promoter activity (92), decreased miR-203 expression (93), impaired PP2A activity (74), and genetic/epigenetic inhibition of SHP-1 phosphatase (74, 94) may all account for increased BCR-ABL1 expression/activity observed during disease progression (72). Interestingly, restoration of PP2A activity in myeloid precursors expressing high BCR-ABL1 levels restores G-CSF–driven differentiation (74), suggesting that PP2A loss of function might play a central role in impairing maturation of Ph-positive GMPs.
The inhibitory effect of high BCR-ABL1 levels on differentiation depends on marked downregulation of C/EBPα (42), a transcription factor essential for granulocytic differentiation. The importance of the loss of C/EBPα activity as a central mechanism leading to differentiation arrest of myeloid CML blasts is supported by evidence that ectopic C/EBPα expression induces maturation of differentiation-arrested BCR-ABL1+ myeloid precursors and CD34+ CML-BP progenitors (42, 95, 96) and that a CML-BP–like process emerges in mice transplanted with BCR-ABL1–transduced Cebpa-null fetal liver cells (97). In CD34+ CML-BP GMPs, loss of C/EBPα does not depend on CEBPA gene mutations (98), but results from the BCR-ABL1 dose–dependent induction of the RNA-binding protein heterogenous nuclear ribonucleoprotein E2 (hnRNP-E2) that, upon interaction with the CEBPA upstream open reading frame (uORF)/spacer element, inhibits CEBPA translation (42). hnRNP-E2 expression is high in CD34+ CML-BP progenitors, where it suppresses C/EBPα and inhibits differentiation (42). Highlighting the importance of loss of C/EBPα expression in CML-BP, coexpression of BCR-ABL1 and AME also suppresses CEBPA translation and induces accumulation of blasts through activation of the CEBPA uORF-binding protein calreticulin (99, 100). Notably, C/EBPβ is also repressed in CML-BP (101), suggesting that loss of C/EBP activity contributes to differentiation arrest and aggressive behavior of CML-BP cells. In this regard, suppression of C/EBP proteins in CML-BP may also depend on BCR-ABL1–induced preferentially expressed antigen in melanoma (PRAME) expression, which impairs myeloid differentiation when ectopically expressed in CD34+ progenitors (34).
The BCR-ABL1/hnRNP-E2/MAPK inhibitory pathway. The ability of hnRNP-E2 to suppress C/EBPα requires the constitutive activation of the MAPKs ERK1 and ERK2 (89, 102), which directly increases hnRNP-E2 stability (89). This is consistent with the observation that enhanced expression of various RNA-binding proteins is among the many imatinib-sensitive changes found in myeloid CML-BP (51). The involvement of ERK1/2 in the regulation of hnRNP-E2 is not surprising, as constitutive MAPK activation is readily detectable in CD34+ CML-BP (102), while CML-CP progenitors show transient MAPK activation in response to mitogenic/survival signals induced by extracellular growth factors (103). Accordingly, levels of activated ERK1/2 in the absence of exogenous cytokines were similar in normal and CD34+ CML-CP progenitors and were not affected by imatinib (103). Graded BCR-ABL1 expression correlates with a progressive increase in ERK1/2 activity (102), and ERK1/2 suppression rescues C/EBPα expression and allows G-CSF–driven maturation of differentiation-arrested progenitors expressing high BCR-ABL1 levels (89). Thus, constitutive ERK1/2 activation in CML-BP is not only essential for transduction of mitogenic/survival signals but also promotes the activation of antidifferentiation signals leading to translational (42) and, perhaps, posttranslational (104) inactivation of C/EBPα. Notably, a decrease in monophosphorylated ERK2 in imatinib-responsive but not -resistant patients suggests that ERK signaling may be important for transformation of TKI-resistant CML (105).
miR-328: a molecular relay in CML disease progression. A few miRNAs are aberrantly regulated in CML (93, 106, 107), but their involvement in disease progression is unclear. Interestingly, the correct functioning of the BCR-ABL1/MAPK/hnRNP-E2 inhibitory axis requires the inhibition of miR-328, which, otherwise, would bind hnRNP-E2 and prevent its interaction with CEBPA mRNA, thus restoring CEBPA mRNA translation. Loss of miR-328 occurs in CD34+ CML-BP but not CML-CP myeloid progenitors, and forced miR-328 expression at levels resembling those observed in CML-CP rescues C/EBPα expression and reverses the CML-BP–like leukemia to a disease that resembles a myeloproliferative disorder in mice transplanted with BCR-ABL1–expressing myeloid precursors (108).
-
Genomic instability facilitates blastic transformation
Genomic instability usually results from an aberrant cellular response to enhanced DNA damage. In CML cells, these mechanisms can be modulated by BCR-ABL1 kinase (Figure 3) or may be kinase-independent.
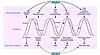 Figure 3
Figure 3BCR-ABL1 regulates DNA damage and DNA repair, the 2 major components of genomic instability. BCR-ABL1–positive leukemia cells accumulate more DNA lesions, such as 7,8-dihydro-8-oxo-2′-deoxyguanosine (8-oxoG), and DNA DSBs induced by ROS, AID, and genotoxic agents (e.g., γ-radiation, cisplatin, mitomycin C, hydroxyurea, and UV light) in comparison with normal cells. In addition, BCR-ABL1 inhibits MMR and stimulates mutagenic NER to generate point mutations including those causing TKI resistance. Moreover, BCR-ABL1 activates unfaithful DSB repair mechanisms, HRR, NHEJ, and SSA, which contribute to chromosomal aberrations. The effect of BCR-ABL1 on base excision repair (BER) and O(6)-methylguanine–DNA methyltransferase (MGMT) is not known. Altogether, elevated levels of DNA damage combined with inefficient/unfaithful DNA repair cause genomic instability in CML-CP and facilitate CML-BP.
Enhanced DNA damage. Much endogenous DNA damage arises from ROS such as superoxide radical anion (•O2–), which may lead to the production of hydrogen peroxide (H2O2) and hydroxyl radical (•OH). BCR-ABL1–transformed cell lines and CD34+ CML cells contain, on average, 2–6 times more ROS than their normal counterparts (CML-BP > CML-CP > normal) (37, 109, 110); the mitochondrial respiratory chain, enhanced glucose uptake, and NADPH oxidase may play a role in this phenomenon (111). ROS can cause damage to all nucleobases and deoxyribose residues in DNA and free nucleotides, generating oxidized bases and DNA double-strand breaks (DSBs) (112). The number of oxidative “hits” to DNA per normal human cell per day is about 104, and normal cells contain approximately 50 DSBs per cell per cell cycle. CD34+ CML cells display 3–8 times more oxidized nucleobases and 4–8 times more DSBs (37, 109, 110).
DNA damage could also be directly induced by ionizing radiation and genotoxic drugs, which are used as part of a conditioning regimen in hematopoietic transplantation for CML patients. BCR-ABL1–positive cells, in comparison with normal cells, accumulate more irradiation- and drug-induced DNA lesions, thus generating more chromosomal aberrations (113, 114).
Unfaithful and inefficient DNA repair. Unfaithful and/or inefficient repair of ROS-induced oxidized DNA bases and DSBs may lead to a variety of point mutations and chromosomal aberrations (115). CD34+ CML cells display a malfunctioning mismatch repair (MMR) pathway, which can facilitate accumulation of point mutations (116) (Figure 3). BCR-ABL1 also promotes mutagenic nucleotide excision repair (NER) (117) and stimulates DSB repair, but the fidelity of the repair mechanisms (homologous recombination repair [HRR], nonhomologous end-joining [NHEJ], and single-strand annealing [SSA]) is compromised (37, 110, 118, 119). In BCR-ABL1–positive cells, point mutations were introduced during usually faithful HRR, extensive nucleobase loss was associated with NHEJ, and enhanced SSA generated large deletions. Overexpression and tyrosine phosphorylation of RAD51, a key element in HRR responsible for strand invasion and pairing, may result in aberrant HRR. Deregulation of DNA ligase IIIα, Werner helicase/exonuclease, and Artemis may contribute to excessive loss of DNA bases during NHEJ in BCR-ABL1–positive cells.
Consequences of genomic instability in CML. Genomic instability is probably responsible for two major problems in CML: TKI resistance and disease progression (13). Both phenomena could be induced by accumulation of point mutations and additional chromosomal aberrations in CML-CP cells irreversibly changing their phenotype toward that in CML-BP.
BCR-ABL1 point mutations have been detected in 50%–90% of patients displaying resistance to imatinib, including approximately 23% of imatinib-naive patients (120). Moreover, second-generation TKI treatment in imatinib-resistant cases led to selection of additional resistance mutations (121).
TKI-resistant BCR-ABL1 mutants exhibit altered kinase activity, substrate utilization, and transformation potency and are associated with clonal cytogenetic evolution, which may have an impact on disease progression (120, 122). Accordingly, BCR-ABL1 kinase mutations are associated with greater likelihood of disease progression, which suggests enhanced genomic instability in these cells (12).
Accumulation of various chromosomal aberrations and mutations is believed to be responsible for the transition of a relatively benign CP to aggressive BP (13). The frequency of additional chromosomal abnormalities is approximately 7% in CML-CP and increases to 40%–70% in the advanced phases of disease, as evaluated by standard cytogenetic analysis (23).
Numeric chromosomal changes are detected at a 50-fold higher frequency and structural changes at a 12-fold higher frequency in CML-BP, in comparison with CML-CP. More sensitive comparative genomic hybridization (CGH) and SNP analyses detect multiple genetic aberrations already in CP, but CML-BP patients have much more complex karyotypes (123, 124). This observation suggests that genomic instability is an early event in CML. Patients from the pre-imatinib and imatinib era display similar types of genetic aberrations (125).
These aberrations involve acquisition of major alterations, such as the following: (a) the acquisition of additional chromosomes (e.g., +Ph, +8, +19); (b) the acquisition of isochromosome i(17q); (c) the acquisition of t(1;17), which is associated with loss of p53; (d) the acquisition of t(1;21), which affects RUNX1 (which is also known as AML1); (e) the acquisition of t(3;21), which generates the AML-1/EVI-1 fusion protein (a negative transcriptional regulator and cell signaling modulator); (f) the acquisition of t(7;11), which produces the NUP98-HOXA9 fusion protein that causes aberrant self renewal; and (g) the acquisition of translocations and inversions associated with AML/myelodysplasia (e.g., inv[3] and t[15;17]). In addition, minor genetic aberrations such as loss of heterozygosity (LOH) at 14q32, homozygous mutations/deletions of pRB, inactivating point mutations in P53 and in interferon consensus sequence binding protein (ICSBP, which encodes an interferon regulatory transcription factor with leukemia-suppressor activity), gain-of-function mutations in GATA-2 (which regulates myelomonocytic differentiation) and RAS (small GTP-binding signal transduction protein), and mutations in a zinc finger transcription factor PR domain containing 16 (PRDM16, mutated in myelodysplastic syndrome and AML) have been also detected. Numerous SNPs have been reported in additional genes regulating cell differentiation, such as ICSBP, GATA-3, and AML1 in myeloid CML-BP (86); however, these results await confirmation.
In addition, mutations in CDKN2A/B and IKZF1 facilitate CML-CP progression to CML-lymphoid BP (24, 25). Moreover, BCR-ABL1–mediated stimulation of activation-induced cytidine deaminase (AID) leads to a hypermutator phenotype, CML-lymphoid BP, and imatinib resistance (126).
Experimental findings support the conclusion that genetic aberrations contribute to malignant progression of CML. For example, loss of p53 led to a CML-BP–like disorder in mice (127). CDKN2A gene loss enhanced oncogenicity in mouse models of BCR-ABL1–induced ALL (128). Coexpression of BCR-ABL1 and NUP98-HOXA9 caused CML-BP–like disease in mice (129). GATA-2 gain-of-function mutations, partial deletions of PMRD16 and RUNX1, and expression of RUNX1-PMRD16 detected in CML-myeloid BP may disturb myelomonocytic differentiation, strongly suggesting their involvement in acute myeloid transformation (86, 130).
Moreover, genetic aberrations associated with CML-BP progression likely play a role in TKI resistance (131), causing a high risk of treatment failure (132). For example, additional chromosomal aberrations, loss of P53, and CDKN2A and RUNX1 abnormalities may be responsible for disease persistence under imatinib treatment (128, 133–135).
BCR-ABL1 kinase–dependent and –independent genomic instability in CML-CP LSCs and/or LPCs. The (9;22) translocation that results in the formation of the Ph chromosome may be a random event or may result from preexisting conditions associated with genomic instability in HSCs. Therefore, additional genetic aberrations accumulated during the course of CML may be promoted by BCR-ABL1 kinase and also by a preexisting abnormality responsible for the formation of t(9;22)(q34;q11). The former statement is supported by reports that BCR-ABL1 kinase–positive cells acquire more oxidative DNA lesions than normal counterparts in response to endogenous ROS and genotoxic treatment (109, 110) and that BCR-ABL1 can inhibit some DNA repair mechanisms (MMR) and stimulate other mechanisms (NER, HRR, NHEJ, and SSA) at the cost of their fidelity (136) (Figure 3). However, the latter speculation about preexisting abnormality cannot be ruled out because chromosome abnormalities were detected in t(9;22)(q34;q11)-negative metaphases appearing during imatinib therapy in patients with newly diagnosed CML-CP (121).
Genomic instability in CML-CP most likely occurs in the LSC-enriched CD34+CD38– population and/or the LPC-rich CD34+ population because TKI-resistant BCR-ABL1 mutants and chromosomal aberrations were detected in both subpopulations (38, 41, 114). As CML-CP can progress to either myeloid or lymphoid BP (sometimes a mixed myeloid/lymphoid phenotype) and chromosomal abnormalities are documented in both phenotypes (137), this suggests that genomic instability may occur at the LSC and/or LPC level. Mutations acquired by the LSCs are likely to be passed on to successive generations of LPCs. On the other hand, genetic aberrations acquired by CML-CP LPCs may “upgrade” them to the status of CML-BP LSCs (49).
Altogether, we postulate that elevated levels of DNA damage combined with unfaithful/inefficient DNA repair may generate mutations and chromosomal aberrations in CML-CP LSCs and/or LPCs, causing resistance to TKIs and progression toward CML-BP. These mechanisms at least partially depend on BCR-ABL1 kinase. Since LSCs, in contrast to LPCs, are not sensitive to TKIs, LSCs may be “ticking time bombs,” eventually exploding to produce a TKI-resistant LPC clone that may evolve into a CML-BP clone.
Genomic instability in CML cells in the era of TKIs. BCR-ABL1 kinase induces genomic instability (13); therefore, imatinib and other TKIs should prevent accumulation of additional genetic changes in CML cells. In fact, imatinib reduced ROS, oxidative DNA damage, point mutations, and other genetic aberrations in BCR-ABL1–positive cells (109, 110, 138). Nevertheless, imatinib-treated CML patients continue to accumulate point mutations (including those causing resistance to other TKIs) and chromosomal aberrations (21, 121, 130, 139).
There are several possible explanations for persistent genomic instability during TKI treatment. First, although TKIs inhibit BCR-ABL1 kinase activity in CML-CP LPCs, their effectiveness in CML-CP LSCs is questionable. The effect of TKIs on BCR-ABL1 kinase–induced signaling may be obscured by growth factors, usually resulting in incomplete inhibition or even stimulation of signaling pathways, such as those involving STAT5, AKT, and MAPKs (140, 141). Therefore, TKIs cannot completely eliminate the effects of BCR-ABL1 kinase and may not effectively inhibit genomic instability. Second, imatinib may exert mutagenic activity to induce centrosome and chromosome aberrations (142). The appearance of cytogenetic aberrations in t(9;22)(q34;q11)-negative cells following imatinib therapy supports this hypothesis (143). Third, if CML-CP cells display an active preexisting genomic instability responsible for generation of t(9;22), this process should be BCR-ABL1 kinase independent and will continue generating errors despite treatment (121). This speculation implicates BCR-ABL1 kinase–dependent and –independent genomic instability in CML cells.
Prevention of genomic instability in CML-CP to improve therapeutic effects of TKIs and antagonize CML-BP. The majority of CML-CP patients at diagnosis do not have mutations or a “critical” combination of aberrations causing either TKI resistance or disease progression. However, a cohort of TKI-treated patients still develops mutations and chromosomal aberrations even though imatinib efficiently antagonizes genomic instability in experimental CML models. Given the fact that BCR-ABL1–negative patients, as assessed by reverse transcriptase PCR, may contain up to 106 CML cells in their body, that CML-CP patients can have approximately 5 × 107 CD34+ cells displaying innate imatinib resistance (144), and that even imatinib-sensitive CD34+ LPCs can still undergo up to 1–3 cell cycles in the presence of the drug and growth factors before eventually being eliminated (71), prevention of genomic instability may be critical for a better therapeutic effect or even eradication of CML.
ROS cause oxidative DNA damage resulting in both clinically relevant BCR-ABL1 mutations and chromosomal aberrations often detected in CML-BP (i.e., aneuploidy, translocations, and truncations) (109, 113). Antioxidants diminished ROS-mediated oxidative DNA damage and reduced the appearance of TKI-resistant mutations and chromosomal aberrations (37, 109, 110). Because the combination of imatinib and an antioxidant exerted a synergistic/additive antimutagenic effect (109), it is possible that the combination of TKI and antioxidants may prevent CML-BP by reducing the appearance of TKI-resistant clones and accumulation of a “critical” combination of genetic aberrations.
-
Concluding remarks
To date, there is strong evidence supporting the idea that the level of BCR-ABL1 kinase activity plays a pivotal role in almost all CML patients undergoing progression and that BCR-ABL1–induced genetic/chromosomal abnormalities can predispose to transformation and/or markedly influence the aggressiveness of the blast crisis progenitor cell clone. However, there are several crucial and burning questions that remain to be answered. What controls BCR-ABL1 expression and activity during progression? Does malignant progression originate from CML-CP LSCs and/or LPCs? Is the acquisition of self renewal, impaired differentiation, and increased genomic instability of CML-BP stem and/or progenitor cells solely a BCR-ABL1–dependent effect? A possible scenario might envision a BCR-ABL1 autoregulatory loop that amplifies signals that positively influence BCR-ABL1 gene transcription and enhance its protein stability. Likewise, it is highly plausible that, in CML-CP, BCR-ABL1–induced genomic aberrations and/or BCR-ABL1–independent preexisting genetic lesions function as “amplifiers” of a genetically unstable phenotype and thereby predispose CML to blastic transformation by affecting stemness, survival, proliferation, differentiation, and/or genome stability of the Ph-positive bone marrow stem and progenitor cells.
-
Acknowledgments
This work was supported in part by grants from the National Cancer Institute (CA095512 to D. Perrotti; CA123014 and CA133646 to T. Skorski) and the US Army and CML Research Program (W81XWH-07-1-0270 to D. Perrotti). D. Perrotti is a Scholar of The Leukemia and Lymphoma Society. C. Jamieson is funded by California Institute for Regenerative Medicine (CIRM) grants. We thank Paolo Neviani and Stephen Lee (The Ohio State University) and Elisabeth Bolton (Temple University) for editorial and/or graphical assistance.
Address correspondence to: Danilo Perrotti, Department of Molecular Virology, Immunology and Medical Genetics and Comprehensive Cancer Center, The Ohio State University, 892 Biomedical Research Tower, 460 West 12th Avenue, Columbus, Ohio 41230, USA. Phone: 614.292.3255; Fax: 614.688.4181; E-mail: [email protected]. Or to: Tomasz Skorski, Department of Microbiology and Immunology, School of Medicine, Temple University, 3400 N. Broad Street, MRB 548, Philadelphia, Pennsylvania 19140, USA. Phone: 215.707.9157; Fax: 215.707.9160; E-mail: [email protected].
-
Footnotes
Conflict of interest: Catriona Jamieson has research grants from Pfizer, Coronado Biosciences, and Celgene and has also consulted for Bristol-Myers Squibb and Wintherix.
Citation for this article: J Clin Invest. 2010;120(7):2254–2264. doi:10.1172/JCI41246.
-
References
- Cross NC, et al. BCR-ABL1-positive CML and BCR-ABL1-negative chronic myeloproliferative disorders: some common and contrasting features. Leukemia. 2008;22(11):1975–1989.
- Bruns I, et al. The hematopoietic stem cell in chronic phase CML is characterized by a transcriptional profile resembling normal myeloid progenitor cells and reflecting loss of quiescence. Leukemia. 2009;23(5):892–899.
- Hochhaus A, et al. Six-year follow-up of patients receiving imatinib for the first-line treatment of chronic myeloid leukemia. Leukemia. 2009;23(6):1054–1061.
- Druker BJ, et al. Five-year follow-up of patients receiving imatinib for chronic myeloid leukemia. N Engl J Med. 2006;355(23):2408–2417.
- Barnes DJ, Melo JV. Primitive, quiescent and difficult to kill: the role of non-proliferating stem cells in chronic myeloid leukemia. Cell Cycle. 2006;5(24):2862–2866.View this article via: PubMed Google Scholar
- Apperley JF. Part I: mechanisms of resistance to imatinib in chronic myeloid leukaemia. Lancet Oncol. 2007;8(11):1018–1029.
- Baccarani M, et al. Evolving concepts in the management of chronic myeloid leukemia: recommendations from an expert panel on behalf of the European LeukemiaNet. Blood. 2006;108(6):1809–1820.
- Rosenthal S, Canellos GP, Whang-Peng J, Gralnick HR. Blast crisis of chronic granulocytic leukemia. Morphologic variants and therapeutic implications. Am J Med. 1977;63(4):542–547.
- Atallah E, et al. Chronic myelogenous leukemia in T cell lymphoid blastic phase achieving durable complete cytogenetic and molecular remission with imatinib mesylate (STI571; Gleevec) therapy. Cancer. 2002;94(11):2996–2999.
- Gratwohl A, et al. Allogeneic hematopoietic stem cell transplantation for chronic myeloid leukemia in Europe 2006: transplant activity, long-term data and current results. An analysis by the Chronic Leukemia Working Party of the European Group for Blood and Marrow Transplantation (EBMT). Haematologica. 2006;91(4):513–521.View this article via: PubMed Google Scholar
- Foroni L, et al. Technical aspects and clinical applications of measuring BCR-ABL1 transcripts number in chronic myeloid leukemia. Am J Hematol. 2009;84(8):517–522.
- Soverini S, et al. ABL mutations in late chronic phase chronic myeloid leukemia patients with up-front cytogenetic resistance to imatinib are associated with a greater likelihood of progression to blast crisis and shorter survival: a study by the GIMEMA Working Party on Chronic Myeloid Leukemia. J Clin Oncol. 2005;23(18):4100–4109.
- Skorski T. BCR/ABL, DNA damage and DNA repair: implications for new treatment concepts. Leuk Lymphoma. 2008;49(4):610–614.
- Copland M, et al. BMS-214662 potently induces apoptosis of chronic myeloid leukemia stem and progenitor cells and synergizes with tyrosine kinase inhibitors. Blood. 2008;111(5):2843–2853.
- Bellodi C, et al. Targeting autophagy potentiates tyrosine kinase inhibitor-induced cell death in Philadelphia chromosome-positive cells, including primary CML stem cells. J Clin Invest. 2009;119(5):1109–1123.
- Dierks C, et al. Expansion of Bcr-Abl-positive leukemic stem cells is dependent on Hedgehog pathway activation. Cancer Cell. 2008;14(3):238–249.
- Zhao C, et al. Hedgehog signalling is essential for maintenance of cancer stem cells in myeloid leukaemia. Nature. 2009;458(7239):776–779.
- Perrotti D, Neviani P. Protein phosphatase 2A (PP2A), a drugable tumor suppressor in Ph1(+) leukemias. Cancer Metastasis Rev. 2008;27(2):159–168.
- Calabretta B, Perrotti D. The biology of CML blast crisis. Blood. 2004;103(11):4010–4022.
- Yamamoto M, Kakihana K, Kurosu T, Murakami N, Miura O. Clonal evolution with inv(11)(p15q22) and NUP98/DDX10 fusion gene in imatinib-resistant chronic myelogenous leukemia. Cancer Genet Cytogenet. 2005;157(2):104–108.
- Fabarius A, et al. Dynamics of cytogenetic aberrations in Philadelphia chromosome positive and negative hematopoiesis during dasatinib therapy of chronic myeloid leukemia patients after imatinib failure. Haematologica. 2007;92(6):834–837.
- Roche-Lestienne C, et al. RUNX1 DNA-binding mutations and RUNX1-PRDM16 cryptic fusions in BCR-ABL+ leukemias are frequently associated with secondary trisomy 21 and may contribute to clonal evolution and imatinib resistance. Blood. 2008;111(7):3735-3741.View this article via: PubMed Google Scholar
- Bacher U, Haferlach T, Hiddemann W, Schnittger S, Kern W, Schoch C. Additional clonal abnormalities in Philadelphia-positive ALL and CML demonstrate a different cytogenetic pattern at diagnosis and follow different pathways at progression. Cancer Genet Cytogenet. 2005;157(1):53–61.
- Mullighan CG, Williams RT, Downing JR, Sherr CJ. Failure of CDKN2A/B (INK4A/B-ARF)-mediated tumor suppression and resistance to targeted therapy in acute lymphoblastic leukemia induced by BCR-ABL. Genes Dev. 2008;22(11):1411–1415.
- Mullighan CG, et al. BCR-ABL1 lymphoblastic leukaemia is characterized by the deletion of Ikaros. Nature. 2008;453(7191):110–114.
- Nakayama H, et al. Decreases in Ikaros activity correlate with blast crisis in patients with chronic myelogenous leukemia. Cancer Res. 1999;59(16):3931–3934.View this article via: PubMed Google Scholar
- Sill H, Goldman JM, Cross NC. Homozygous deletions of the p16 tumor-suppressor gene are associated with lymphoid transformation of chronic myeloid leukemia. Blood. 1995;85(8):2013–2016.View this article via: PubMed Google Scholar
- Lanza F, Bi S. Role of p53 in leukemogenesis of chronic myeloid leukemia. Stem Cells. 1995;13(4):445–452.
- Melo JV, Barnes DJ. Chronic myeloid leukaemia as a model of disease evolution in human cancer. Nat Rev Cancer. 2007;7(6):441–453.
- Gaiger A, et al. Increase of bcr-abl chimeric mRNA expression in tumor cells of patients with chronic myeloid leukemia precedes disease progression. Blood. 1995;86(6):2371–2378.View this article via: PubMed Google Scholar
- Yong AS, Melo JV. The impact of gene profiling in chronic myeloid leukaemia. Best Pract Res Clin Haematol. 2009;22(2):181–190.
- Radich JP, et al. Gene expression changes associated with progression and response in chronic myeloid leukemia. Proc Natl Acad Sci U S A. 2006;103(8):2794–2799.
- Terragna C, et al. Gene expression profile (GEP) of chronic myeloid leukemia (CML) patients at diagnosis: Two distinguished subgroups of CML patients identified, based on a molecular signature, irrespective of their sokal risk score. ASH Annual Meeting. Blood. 2008;112:1095.(Abstract 3190).
- Oehler VG, et al. The preferentially expressed antigen in melanoma (PRAME) inhibits myeloid differentiation in normal hematopoietic and leukemic progenitor cells. Blood. 2009;114(15):3299–3308.
- Jamieson CH. Chronic myeloid leukemia stem cells. Hematology Am Soc Hematol Educ Program. 2008;2008:436–442.View this article via: PubMed Google Scholar
- Oehler VG, Yeung KY, Choi YE, Bumgarner RE, Raftery AE, Radich JP. The derivation of diagnostic markers of chronic myeloid leukemia progression from microarray data. Blood. 2009;114(15):3292–3298.
- Cramer K, et al. BCR/ABL and other kinases from chronic myeloproliferative disorders stimulate single-strand annealing, an unfaithful DNA double-strand break repair. Cancer Res. 2008;68(17):6884–6888.
- Jiang X, Saw KM, Eaves A, Eaves C. Instability of BCR-ABL gene in primary and cultured chronic myeloid leukemia stem cells. J Natl Cancer Inst. 2007;99(9):680–693.View this article via: PubMed Google Scholar
- Brazma D, et al. Genomic profile of chronic myelogenous leukemia: Imbalances associated with disease progression. Genes Chromosomes Cancer. 2007;46(11):1039–1050.
- Schultheis B, Szydlo R, Mahon FX, Apperley JF, Melo JV. Analysis of total phosphotyrosine levels in CD34+ cells from CML patients to predict the response to imatinib mesylate treatment. Blood. 2005;105(12):4893–4894.
- Barnes DJ, et al. Bcr-Abl expression levels determine the rate of development of resistance to imatinib mesylate in chronic myeloid leukemia. Cancer Res. 2005;65(19):8912–8919.
- Perrotti D, et al. BCR-ABL suppresses C/EBPalpha expression through inhibitory action of hnRNP E2. Nat Genet. 2002;30(1):48–58.
- Lapidot T, et al. A cell initiating human acute myeloid leukaemia after transplantation into SCID mice. Nature. 1994;367(6464):645–648.
- Bonnet D, Dick JE. Human acute myeloid leukemia is organized as a hierarchy that originates from a primitive hematopoietic cell. Nat Med. 1997;3(7):730–737.
- Jorgensen HG, Holyoake TL. Characterization of cancer stem cells in chronic myeloid leukaemia. Biochem Soc Trans. 2007;35(pt 5):1347–1351.
- Huntly BJ, et al. MOZ-TIF2, but not BCR-ABL, confers properties of leukemic stem cells to committed murine hematopoietic progenitors. Cancer Cell. 2004;6(6):587–596.
- Cozzio A, Passegue E, Ayton PM, Karsunky H, Cleary ML, Weissman IL. Similar MLL-associated leukemias arising from self-renewing stem cells and short-lived myeloid progenitors. Genes Dev. 2003;17(24):3029–3035.
- Lemoli RM, et al. Molecular and functional analysis of the stem cell compartment of chronic myelogenous leukemia reveals the presence of a CD34- cell population with intrinsic resistance to imatinib. Blood. 2009;114(25):5191–5200.
- Jamieson CH, et al. Granulocyte-macrophage progenitors as candidate leukemic stem cells in blast-crisis CML. N Engl J Med. 2004;351(7):657–667.
- Salesse S, Dylla SJ, Verfaillie CM. p210BCR/ABL-induced alteration of pre-mRNA splicing in primary human CD34+ hematopoietic progenitor cells. Leukemia. 2004;18(4):727–733.
- Perrotti D, Neviani P. From mRNA metabolism to cancer therapy: chronic myelogenous leukemia shows the way. Clin Cancer Res. 2007;13(6):1638–1642.
- Klein F, et al. BCR-ABL1 induces aberrant splicing of IKAROS and lineage infidelity in pre-B lymphoblastic leukemia cells. Oncogene. 2006;25(7):1118–1124.
- Iacobucci I, et al. Expression of spliced oncogenic Ikaros isoforms in Philadelphia-positive acute lymphoblastic leukemia patients treated with tyrosine kinase inhibitors: implications for a new mechanism of resistance. Blood. 2008;112(9):3847–3855.
- Lee TS, et al. BCR-ABL alternative splicing as a common mechanism for imatinib resistance: evidence from molecular dynamics simulations. Mol Cancer Ther. 2008;7(12):3834–3841.
- Abrahamsson AE, et al. Glycogen synthase kinase 3beta missplicing contributes to leukemia stem cell generation. Proc Natl Acad Sci U S A. 2009;106(10):3925–3929.
- Santaguida M, et al. JunB protects against myeloid malignancies by limiting hematopoietic stem cell proliferation and differentiation without affecting self-renewal. Cancer Cell. 2009;15(4):341–352.
- Neviani P, et al. Activation of PP2A by FTY720 inhibits survival and self-renewal of the Ph(+) chronic myelogenous leukemia (CML) CD34+/CD38– stem cell through the simultaneous suppression of BCR/ABL and BCR/ABL– independent signals. ASH Annual Meeting. Blood. 2008;112:189.
- Coluccia AM, et al. Bcr-Abl stabilizes beta-catenin in chronic myeloid leukemia through its tyrosine phosphorylation. EMBO J. 2007;26(5):1456–1466.
- Minami Y, et al. BCR-ABL-transformed GMP as myeloid leukemic stem cells. Proc Natl Acad Sci U S A. 2008;105(46):17967–17972.
- Zhao C, et al. Loss of beta-catenin impairs the renewal of normal and CML stem cells in vivo. Cancer Cell. 2007;12(6):528–541.
- Hofmann I, et al. Hedgehog signaling is dispensable for adult murine hematopoietic stem cell function and hematopoiesis. Cell Stem Cell. 2009;4(6):559–567.
- Niki M, et al. Role of Dok-1 and Dok-2 in leukemia suppression. J Exp Med. 2004;200(12):1689–1695.
- Ito K, et al. PML targeting eradicates quiescent leukaemia-initiating cells. Nature. 2008;453(7198):1072–1078.
- Jaiswal S, Traver D, Miyamoto T, Akashi K, Lagasse E, Weissman IL. Expression of BCR/ABL and BCL-2 in myeloid progenitors leads to myeloid leukemias. Proc Natl Acad Sci U S A. 2003;100(17):10002–10007.
- Kim DH, et al. Genetic variants in the candidate genes of the apoptosis pathway and susceptibility to chronic myeloid leukemia. Blood. 2009;113(11):2517–2525.
- Kuribara R, et al. Roles of Bim in apoptosis of normal and Bcr-Abl-expressing hematopoietic progenitors. Mol Cell Biol. 2004;24(14):6172–6183.
- Shah NP, et al. Transient potent BCR-ABL inhibition is sufficient to commit chronic myeloid leukemia cells irreversibly to apoptosis. Cancer Cell. 2008;14(6):485–493.
- Samanta AK, et al. Jak2 inhibition deactivates Lyn kinase through the SET-PP2A-SHP1 pathway, causing apoptosis in drug-resistant cells from chronic myelogenous leukemia patients. Oncogene. 2009;28(14):1669–1681.
- Chen Y, Hu Y, Zhang H, Peng C, Li S. Loss of the Alox5 gene impairs leukemia stem cells and prevents chronic myeloid leukemia. Nat Genet. 2009;41(7):783–792.
- Jiang X, et al. Chronic myeloid leukemia stem cells possess multiple unique features of resistance to BCR-ABL targeted therapies. Leukemia. 2007;21(5):926–935.View this article via: PubMed Google Scholar
- Graham SM, et al. Primitive, quiescent, Philadelphia-positive stem cells from patients with chronic myeloid leukemia are insensitive to STI571 in vitro. Blood. 2002;99(1):319–325.
- Barnes DJ, Schultheis B, Adedeji S, Melo JV. Dose-dependent effects of Bcr-Abl in cell line models of different stages of chronic myeloid leukemia. Oncogene. 2005;24(42):6432–6440.View this article via: PubMed Google Scholar
- Trotta R, et al. BCR/ABL activates mdm2 mRNA translation via the La antigen. Cancer Cell. 2003;3(2):145–160.
- Neviani P, et al. The tumor suppressor PP2A is functionally inactivated in blast crisis CML through the inhibitory activity of the BCR/ABL-regulated SET protein. Cancer Cell. 2005;8(5):355–368.
- Donato NJ, et al. BCR-ABL independence and LYN kinase overexpression in chronic myelogenous leukemia cells selected for resistance to STI571. Blood. 2003;101(2):690–698.
- Mahon FX, et al. Lyn kinase overexpression is one of the mechanisms of resistance to nilotinib in chronic myeloid leukemia. Blood. 2008;112(1):3181.
- Samanta A, et al. Disruption of the Bcr-Abl/Jak2/HSP90 network complex leading to induction of apoptosis in drug-resistant CML cells by a new compound WP1193. Blood. 2008;112:3199.
- Veselovska J, et al. Bcr-Abl-independent activation of Src kinases associated with development of dasatinib resistance in a CML patient. Blood. 2008;112:1624.
- Li M, Damuni Z. I1PP2A and I2PP2A. Two potent protein phosphatase 2A-specific inhibitor proteins. Methods Mol Biol. 1998;93:59–66.View this article via: PubMed Google Scholar
- Iervolino A, et al. hnRNP A1 nucleocytoplasmic shuttling activity is required for normal myelopoiesis and BCR/ABL leukemogenesis. Mol Cell Biol. 2002;22(7):2255–2266.
- Junttila MR, et al. CIP2A inhibits PP2A in human malignancies. Cell. 2007;130(1):51–62.
- Neviani P, et al. FTY720, a new alternative for treating blast crisis chronic myelogenous leukemia and Philadelphia chromosome-positive acute lymphocytic leukemia. J Clin Invest. 2007;117(9):2408–2421.
- Liedtke M, Pandey P, Kumar S, Kharbanda S, Kufe D. Regulation of Bcr-Abl-induced SAP kinase activity and transformation by the SHPTP1 protein tyrosine phosphatase. Oncogene. 1998;17(15):1889–1892.
- Chen P, Levis M, Brown P, Kim KT, Allebach J, Small D. FLT3/ITD mutation signaling includes suppression of SHP-1. J Biol Chem. 2005;280(7):5361–5369.
- Wu C, Sun M, Liu L, Zhou GW. The function of the protein tyrosine phosphatase SHP-1 in cancer. Gene. 2003;306:1–12.
- Zhang SJ, et al. Gain-of-function mutation of GATA-2 in acute myeloid transformation of chronic myeloid leukemia. Proc Natl Acad Sci U S A. 2008;105(6):2076–2081.
- Mitani K, et al. Generation of the AML1-EVI-1 fusion gene in the t(3;21)(q26;q22) causes blastic crisis in chronic myelocytic leukemia. EMBO J. 1994;13(3):504–510.View this article via: PubMed Google Scholar
- Nakamura T, et al. Fusion of the nucleoporin gene NUP98 to HOXA9 by the chromosome translocation t(7;11)(p15;p15) in human myeloid leukaemia. Nat Genet. 1996;12(2):154–158.View this article via: PubMed Google Scholar
- Chang JS, et al. High levels of the BCR/ABL oncoprotein are required for the MAPK-hnRNP-E2 dependent suppression of C/EBPalpha-driven myeloid differentiation. Blood. 2007;110(3):994–1003.
- Gorre ME, et al. Clinical resistance to STI-571 cancer therapy caused by BCR-ABL gene mutation or amplification. Science. 2001;293(5531):876–880.
- le Coutre P, et al. Induction of resistance to the Abelson inhibitor STI571 in human leukemic cells through gene amplification. Blood. 2000;95(5):1758–1766.View this article via: PubMed Google Scholar
- Marega M, Piazza R, Meneghetti I, Redaelli S, Mogavero A, Gambacorti C. BCR and BCR/ABL regulation during myeloid differentiation in healthy donors and in chronic phase/blast crisis CML patients. Blood. 2008;112:3204.
- Bueno MJ, et al. Genetic and epigenetic silencing of microRNA-203 enhances ABL1 and BCR-ABL1 oncogene expression. Cancer Cell. 2008;13(6):496–506.
- Amin HM, Hoshino K, Yang H, Lin Q, Lai R, Garcia-Manero G. Decreased expression level of SH2 domain-containing protein tyrosine phosphatase-1 (Shp1) is associated with progression of chronic myeloid leukaemia. J Pathol. 2007;212(4):402–410.
- Schuster C, et al. The effects of Bcr-Abl on C/EBP transcription-factor regulation and neutrophilic differentiation are reversed by the Abl kinase inhibitor imatinib mesylate. Blood. 2003;101(2):655–663.
- Ferrari-Amorotti G, et al. Leukemogenesis induced by wild-type and STI571-resistant BCR/ABL is potently suppressed by C/EBPalpha. Blood. 2006;108(4):1353–1362.
- Wagner K, et al. Absence of the transcription factor CCAAT enhancer binding protein alpha results in loss of myeloid identity in bcr/abl-induced malignancy. Proc Natl Acad Sci U S A. 2006;103(16):6338–6343.
- Pabst T, et al. Mutations of the myeloid transcription factor CEBPA are not associated with the blast crisis of chronic myeloid leukaemia. Br J Haematol. 2006;133(4):400–402.
- Cuenco GM, Ren R. Cooperation of BCR-ABL and AML1/MDS1/EVI1 in blocking myeloid differentiation and rapid induction of an acute myelogenous leukemia. Oncogene. 2001;20(57):8236–8248.
- Helbling D, et al. The leukemic fusion gene AML1-MDS1-EVI1 suppresses CEBPA in acute myeloid leukemia by activation of Calreticulin. Proc Natl Acad Sci U S A. 2004;101(36):13312–13317.
- Guerzoni C, et al. Inducible activation of C/EBP{beta}, a gene negatively regulated by BCR/ABL, inhibits proliferation and promotes differentiation of BCR/ABL-expressing cells. Blood. 2006;107(10):4080–4089.
- Notari M, et al. A MAPK/HNRPK pathway controls BCR/ABL oncogenic potential by regulating MYC mRNA translation. Blood. 2006;107(6):2507–2516.
- Chu S, Holtz M, Gupta M, Bhatia R. BCR/ABL kinase inhibition by imatinib mesylate enhances MAP kinase activity in chronic myelogenous leukemia CD34+ cells. Blood. 2004;103(8):3167–3174.
- Ross SE, et al. Phosphorylation of C/EBPalpha inhibits granulopoiesis. Mol Cell Biol. 2004;24(2):675–686.
- Fan AC, et al. Nanofluidic proteomic assay for serial analysis of oncoprotein activation in clinical specimens. Nat Med. 2009;15(5):566–571.
- Agirre X, et al. Down-regulation of hsa-miR-10a in chronic myeloid leukemia CD34+ cells increases USF2-mediated cell growth. Mol Cancer Res. 2008;6(12):1830–1840.
- Venturini L, et al. Expression of the miR-17-92 polycistron in chronic myeloid leukemia (CML) CD34+ cells. Blood. 2007;109(10):4399–4405.
- Eiring AM, et al. miR-328 functions as an RNA decoy to modulate hnRNP E2 regulation of mRNA translation in leukemic blasts. Cell. 2010;140(5):652–665.
- Koptyra M, et al. BCR/ABL kinase induces self-mutagenesis via reactive oxygen species to encode imatinib resistance. Blood. 2006;108(1):319–327.
- Nowicki MO, et al. BCR/ABL oncogenic kinase promotes unfaithful repair of the reactive oxygen species-dependent DNA double-strand breaks. Blood. 2004;104(12):3746–3753.
- Rodrigues MS, Reddy MM, Sattler M. Cell cycle regulation by oncogenic tyrosine kinases in myeloid neoplasias: from molecular redox mechanisms to health implications. Antioxid Redox Signal. 2008;10(10):1813–1848.View this article via: PubMed Google Scholar
- Beckman KB, Ames BN. Oxidative decay of DNA. J Biol Chem. 1997;272(32):19633–19636.
- Koptyra M, Cramer K, Slupianek A, Richardson C, Skorski T. BCR/ABL promotes accumulation of chromosomal aberrations induced by oxidative and genotoxic stress. Leukemia. 2008;22(10):1969–1972.
- Dierov J, et al. BCR/ABL induces chromosomal instability after genotoxic stress and alters the cell death threshold. Leukemia. 2009;23(2):279–286.
- Bernstein C, Bernstein H, Payne CM, Garewal H. DNA repair/pro-apoptotic dual-role proteins in five major DNA repair pathways: fail-safe protection against carcinogenesis. Mutat Res. 2002;511(2):145–178.View this article via: PubMed Google Scholar
- Stoklosa T, et al. BCR/ABL inhibits mismatch repair to protect from apoptosis and induce point mutations. Cancer Res. 2008;68(8):2576–2580.
- Canitrot Y, et al. p210 BCR/ABL kinase regulates nucleotide excision repair (NER) and resistance to UV radiation. Blood. 2003;102(7):2632–2637.
- Slupianek A, et al. BCR/ABL regulates mammalian RecA homologs, resulting in drug resistance. Mol Cell. 2001;8(4):795–806.
- Sallmyr A, Tomkinson AE, Rassool FV. Up-regulation of WRN and DNA ligase IIIalpha in chronic myeloid leukemia: consequences for the repair of DNA double-strand breaks. Blood. 2008;112(4):1413–1423.
- Willis SG, et al. High–sensitivity detection of BCR–ABL kinase domain mutations in imatinib–naive patients: correlation with clonal cytogenetic evolution but not response to therapy. Blood. 2005;106(6):2128–2137.
- Cortes J, et al. Dynamics of BCR-ABL kinase domain mutations in chronic myeloid leukemia after sequential treatment with multiple tyrosine kinase inhibitors. Blood. 2007;110(12):4005–4011.
- Griswold IJ, et al. Kinase domain mutants of Bcr-Abl exhibit altered transformation potency, kinase activity, and substrate utilization, irrespective of sensitivity to imatinib. Mol Cell Biol. 2006;26(16):6082–6093.
- Brazma D, et al. Genomic profile of chronic myelogenous leukemia: Imbalances associated with disease progression. Genes Chromosomes Cancer. 2007;46(11):1039–1050.
- Khorashad JS, et al. Multiple sub-microscopic genomic lesions are a universal feature of chronic myeloid leukaemia at diagnosis. Leukemia. 2008;22(9):1806–1807.
- Haferlach C, Bacher U, Schnittger S, Weiss T, Kern W, Haferlach T. Similar patterns of chromosome abnormalities in CML occur in addition to the Philadelphia chromosome with or without tyrosine kinase inhibitor treatment. Leukemia. 2009;24(3):638–640.
- Klemm L, et al. The B cell mutator AID promotes B lymphoid blast crisis and drug resistance in chronic myeloid leukemia. Cancer Cell. 2009;16(3):232–245.
- Skorski T, et al. Blastic transformation of p53-deficient bone marrow cells by p210bcr/abl tyrosine kinase. Proc Natl Acad Sci U S A. 1996;93(23):13137–13142.
- Williams RT, Roussel MF, Sherr CJ. Arf gene loss enhances oncogenicity and limits imatinib response in mouse models of Bcr-Abl-induced acute lymphoblastic leukemia. Proc Natl Acad Sci U S A. 2006;103(17):6688–6693.
- Dash AB, et al. A murine model of CML blast crisis induced by cooperation between BCR/ABL and NUP98/HOXA9. Proc Natl Acad Sci U S A. 2002;99(11):7622–7627.
- Deluche L, et al. Cryptic and partial deletions of PRDM16 and RUNX1 without t(1;21)(p36;q22) and/or RUNX1-PRDM16 fusion in a case of progressive chronic myeloid leukemia: a complex chromosomal rearrangement of underestimated frequency in disease progression? Genes Chromosomes Cancer. 2008;47(12):1110–1117.
- Lahaye T, et al. Response and resistance in 300 patients with BCR-ABL-positive leukemias treated with imatinib in a single center: a 4.5-year follow-up. Cancer. 2005;103(8):1659–1669.
- Cortes J, O’Dwyer ME. Clonal evolution in chronic myelogenous leukemia. Hematol Oncol Clin North Am. 2004;18(3):671–684, x.
- Wendel HG, et al. Loss of p53 impedes the antileukemic response to BCR-ABL inhibition. Proc Natl Acad Sci U S A. 2006;103(19):7444–7449.
- Miething C, et al. Retroviral insertional mutagenesis identifies RUNX genes involved in chronic myeloid leukemia disease persistence under imatinib treatment. Proc Natl Acad Sci U S A. 2007;104(11):4594–4599.
- Kim TD, et al. Impact of additional chromosomal aberrations and BCR-ABL kinase domain mutations on the response to nilotinib in Philadelphia chromosome-positive chronic myeloid leukemia. Haematologica. 2010;95(4):582–588.
- Nieborowska-Skorska M, et al. ATR-Chk1 axis protects BCR/ABL leukemia cells from the lethal effect of DNA double-strand breaks. Cell Cycle. 2006;5(9):994–1000.View this article via: PubMed Google Scholar
- Johansson B, Fioretos T, Mitelman F. Cytogenetic and molecular genetic evolution of chronic myeloid leukemia. Acta Haematol. 2002;107(2):76–94.
- Brain J, Saksena A, Laneuville P. The kinase inhibitor STI571 reverses the Bcr-Abl induced point mutation frequencies observed in pre-leukemic P190(Bcr-Abl) transgenic mice. Leuk Res. 2002;26(11):1011–1016.
- Nowak D, et al. SNP array analysis of tyrosine kinase inhibitor-resistant chronic myeloid leukemia identifies heterogeneous secondary genomic alterations. Blood. 1049;115(5):1049–1053.
- Jorgensen HG, Allan EK, Jordanides NE, Mountford JC, Holyoake TL. Nilotinib exerts equipotent antiproliferative effects to imatinib and does not induce apoptosis in CD34+ CML cells. Blood. 2007;109(9):4016–4019.
- Konig H, Copland M, Chu S, Jove R, Holyoake TL, Bhatia R. Effects of dasatinib on SRC kinase activity and downstream intracellular signaling in primitive chronic myelogenous leukemia hematopoietic cells. Cancer Res. 2008;68(23):9624–9633.
- Fabarius A, et al. Induction of centrosome and chromosome aberrations by imatinib in vitro. Leukemia. 2005;19(9):1573–1578.
- Jabbour E, et al. Chromosomal abnormalities in Philadelphia chromosome negative metaphases appearing during imatinib mesylate therapy in patients with newly diagnosed chronic myeloid leukemia in chronic phase. Blood. 2007;110(8):2991–2995.
- Elrick LJ, Jorgensen HG, Mountford JC, Holyoake TL. Punish the parent not the progeny. Blood. 2005;105(5):1862–1866.
-
Version history
- Version 1 (July 1, 2010): No description
Article tools
-
View PDF

- Download citation information
- Send a comment
- Terms of use
- Standard abbreviations
- Need help? Email the journal
Metrics
Go to
- Top
- Abstract
- Introduction
- CML-BP patients: a therapeutic challenge
- Biological complexity of CML-BP
- CML-BP LSCs: BCR-ABL1 overexpression, self renewal, and survival
- Global interference with BCR-ABL1–dependent and –independent signals
- Impaired myeloid differentiation in CML-BP: a true BCR-ABL1 dosage effect
- Genomic instability facilitates blastic transformation
- Concluding remarks
- Acknowledgments
- Footnotes
- References
- Version history



Copyright © 2024 American Society for Clinical Investigation
ISSN: 0021-9738 (print), 1558-8238 (online)



