Advertisement
Research ArticleAutoimmunity Free access | 10.1172/JCI34523
Mechanisms of an autoimmunity syndrome in mice caused by a dominant mutation in Aire
Maureen A. Su,1,2 Karen Giang,1 Kristina Žumer,3 Huimin Jiang,3 Irena Oven,3 John L. Rinn,4 Jason J. DeVoss,1 Kellsey P.A. Johannes,1 Wen Lu,1 James Gardner,1 Angela Chang,1 Paula Bubulya,5 Howard Y. Chang,4 B. Matija Peterlin,3 and Mark S. Anderson1,3
1Diabetes Center, 2Department of Pediatrics, and 3Department of Medicine, UCSF, San Francisco, California, USA. 4Program in Epithelial Biology, Cancer Biology Program, Stanford University School of Medicine, Stanford, California, USA. 5Department of Biological Sciences, Wright State University, Dayton, Ohio, USA.
Address correspondence to: Mark S. Anderson, UCSF Diabetes Center, 513 Parnassus Ave., Box 0540, San Francisco, California 94143, USA. Phone: (415) 502-8052; Fax: (415) 564-5813; E-mail: [email protected].
Find articles by Su, M. in: JCI | PubMed | Google Scholar
1Diabetes Center, 2Department of Pediatrics, and 3Department of Medicine, UCSF, San Francisco, California, USA. 4Program in Epithelial Biology, Cancer Biology Program, Stanford University School of Medicine, Stanford, California, USA. 5Department of Biological Sciences, Wright State University, Dayton, Ohio, USA.
Address correspondence to: Mark S. Anderson, UCSF Diabetes Center, 513 Parnassus Ave., Box 0540, San Francisco, California 94143, USA. Phone: (415) 502-8052; Fax: (415) 564-5813; E-mail: [email protected].
Find articles by Giang, K. in: JCI | PubMed | Google Scholar
1Diabetes Center, 2Department of Pediatrics, and 3Department of Medicine, UCSF, San Francisco, California, USA. 4Program in Epithelial Biology, Cancer Biology Program, Stanford University School of Medicine, Stanford, California, USA. 5Department of Biological Sciences, Wright State University, Dayton, Ohio, USA.
Address correspondence to: Mark S. Anderson, UCSF Diabetes Center, 513 Parnassus Ave., Box 0540, San Francisco, California 94143, USA. Phone: (415) 502-8052; Fax: (415) 564-5813; E-mail: [email protected].
Find articles by Žumer, K. in: JCI | PubMed | Google Scholar
1Diabetes Center, 2Department of Pediatrics, and 3Department of Medicine, UCSF, San Francisco, California, USA. 4Program in Epithelial Biology, Cancer Biology Program, Stanford University School of Medicine, Stanford, California, USA. 5Department of Biological Sciences, Wright State University, Dayton, Ohio, USA.
Address correspondence to: Mark S. Anderson, UCSF Diabetes Center, 513 Parnassus Ave., Box 0540, San Francisco, California 94143, USA. Phone: (415) 502-8052; Fax: (415) 564-5813; E-mail: [email protected].
Find articles by Jiang, H. in: JCI | PubMed | Google Scholar
1Diabetes Center, 2Department of Pediatrics, and 3Department of Medicine, UCSF, San Francisco, California, USA. 4Program in Epithelial Biology, Cancer Biology Program, Stanford University School of Medicine, Stanford, California, USA. 5Department of Biological Sciences, Wright State University, Dayton, Ohio, USA.
Address correspondence to: Mark S. Anderson, UCSF Diabetes Center, 513 Parnassus Ave., Box 0540, San Francisco, California 94143, USA. Phone: (415) 502-8052; Fax: (415) 564-5813; E-mail: [email protected].
Find articles by Oven, I. in: JCI | PubMed | Google Scholar
1Diabetes Center, 2Department of Pediatrics, and 3Department of Medicine, UCSF, San Francisco, California, USA. 4Program in Epithelial Biology, Cancer Biology Program, Stanford University School of Medicine, Stanford, California, USA. 5Department of Biological Sciences, Wright State University, Dayton, Ohio, USA.
Address correspondence to: Mark S. Anderson, UCSF Diabetes Center, 513 Parnassus Ave., Box 0540, San Francisco, California 94143, USA. Phone: (415) 502-8052; Fax: (415) 564-5813; E-mail: [email protected].
Find articles by Rinn, J. in: JCI | PubMed | Google Scholar
1Diabetes Center, 2Department of Pediatrics, and 3Department of Medicine, UCSF, San Francisco, California, USA. 4Program in Epithelial Biology, Cancer Biology Program, Stanford University School of Medicine, Stanford, California, USA. 5Department of Biological Sciences, Wright State University, Dayton, Ohio, USA.
Address correspondence to: Mark S. Anderson, UCSF Diabetes Center, 513 Parnassus Ave., Box 0540, San Francisco, California 94143, USA. Phone: (415) 502-8052; Fax: (415) 564-5813; E-mail: [email protected].
Find articles by DeVoss, J. in: JCI | PubMed | Google Scholar
1Diabetes Center, 2Department of Pediatrics, and 3Department of Medicine, UCSF, San Francisco, California, USA. 4Program in Epithelial Biology, Cancer Biology Program, Stanford University School of Medicine, Stanford, California, USA. 5Department of Biological Sciences, Wright State University, Dayton, Ohio, USA.
Address correspondence to: Mark S. Anderson, UCSF Diabetes Center, 513 Parnassus Ave., Box 0540, San Francisco, California 94143, USA. Phone: (415) 502-8052; Fax: (415) 564-5813; E-mail: [email protected].
Find articles by Johannes, K. in: JCI | PubMed | Google Scholar
1Diabetes Center, 2Department of Pediatrics, and 3Department of Medicine, UCSF, San Francisco, California, USA. 4Program in Epithelial Biology, Cancer Biology Program, Stanford University School of Medicine, Stanford, California, USA. 5Department of Biological Sciences, Wright State University, Dayton, Ohio, USA.
Address correspondence to: Mark S. Anderson, UCSF Diabetes Center, 513 Parnassus Ave., Box 0540, San Francisco, California 94143, USA. Phone: (415) 502-8052; Fax: (415) 564-5813; E-mail: [email protected].
Find articles by Lu, W. in: JCI | PubMed | Google Scholar
1Diabetes Center, 2Department of Pediatrics, and 3Department of Medicine, UCSF, San Francisco, California, USA. 4Program in Epithelial Biology, Cancer Biology Program, Stanford University School of Medicine, Stanford, California, USA. 5Department of Biological Sciences, Wright State University, Dayton, Ohio, USA.
Address correspondence to: Mark S. Anderson, UCSF Diabetes Center, 513 Parnassus Ave., Box 0540, San Francisco, California 94143, USA. Phone: (415) 502-8052; Fax: (415) 564-5813; E-mail: [email protected].
Find articles by Gardner, J. in: JCI | PubMed | Google Scholar
1Diabetes Center, 2Department of Pediatrics, and 3Department of Medicine, UCSF, San Francisco, California, USA. 4Program in Epithelial Biology, Cancer Biology Program, Stanford University School of Medicine, Stanford, California, USA. 5Department of Biological Sciences, Wright State University, Dayton, Ohio, USA.
Address correspondence to: Mark S. Anderson, UCSF Diabetes Center, 513 Parnassus Ave., Box 0540, San Francisco, California 94143, USA. Phone: (415) 502-8052; Fax: (415) 564-5813; E-mail: [email protected].
Find articles by Chang, A. in: JCI | PubMed | Google Scholar
1Diabetes Center, 2Department of Pediatrics, and 3Department of Medicine, UCSF, San Francisco, California, USA. 4Program in Epithelial Biology, Cancer Biology Program, Stanford University School of Medicine, Stanford, California, USA. 5Department of Biological Sciences, Wright State University, Dayton, Ohio, USA.
Address correspondence to: Mark S. Anderson, UCSF Diabetes Center, 513 Parnassus Ave., Box 0540, San Francisco, California 94143, USA. Phone: (415) 502-8052; Fax: (415) 564-5813; E-mail: [email protected].
Find articles by Bubulya, P. in: JCI | PubMed | Google Scholar
1Diabetes Center, 2Department of Pediatrics, and 3Department of Medicine, UCSF, San Francisco, California, USA. 4Program in Epithelial Biology, Cancer Biology Program, Stanford University School of Medicine, Stanford, California, USA. 5Department of Biological Sciences, Wright State University, Dayton, Ohio, USA.
Address correspondence to: Mark S. Anderson, UCSF Diabetes Center, 513 Parnassus Ave., Box 0540, San Francisco, California 94143, USA. Phone: (415) 502-8052; Fax: (415) 564-5813; E-mail: [email protected].
Find articles by Chang, H. in: JCI | PubMed | Google Scholar
1Diabetes Center, 2Department of Pediatrics, and 3Department of Medicine, UCSF, San Francisco, California, USA. 4Program in Epithelial Biology, Cancer Biology Program, Stanford University School of Medicine, Stanford, California, USA. 5Department of Biological Sciences, Wright State University, Dayton, Ohio, USA.
Address correspondence to: Mark S. Anderson, UCSF Diabetes Center, 513 Parnassus Ave., Box 0540, San Francisco, California 94143, USA. Phone: (415) 502-8052; Fax: (415) 564-5813; E-mail: [email protected].
Find articles by Peterlin, B. in: JCI | PubMed | Google Scholar
1Diabetes Center, 2Department of Pediatrics, and 3Department of Medicine, UCSF, San Francisco, California, USA. 4Program in Epithelial Biology, Cancer Biology Program, Stanford University School of Medicine, Stanford, California, USA. 5Department of Biological Sciences, Wright State University, Dayton, Ohio, USA.
Address correspondence to: Mark S. Anderson, UCSF Diabetes Center, 513 Parnassus Ave., Box 0540, San Francisco, California 94143, USA. Phone: (415) 502-8052; Fax: (415) 564-5813; E-mail: [email protected].
Find articles by Anderson, M. in: JCI | PubMed | Google Scholar
Published April 15, 2008 - More info
J Clin Invest. 2008;118(5):1712–1726. https://doi.org/10.1172/JCI34523.
© 2008 The American Society for Clinical Investigation
Received: November 19, 2007; Accepted: February 27, 2008
-
Abstract
Homozygous loss-of-function mutations in AIRE cause autoimmune polyglandular syndrome type 1 (APS 1), which manifests in a classic triad of hypoparathyroidism, adrenal insufficiency, and candidiasis. Interestingly, a kindred with a specific G228W AIRE variant presented with an autosomal dominant autoimmune phenotype distinct from APS 1. We utilized a novel G228W-knockin mouse model to show that this variant acted in a dominant-negative manner to cause a unique autoimmunity syndrome. In addition, the expression of a large number of Aire-regulated thymic antigens was partially inhibited in these animals, demonstrating the importance of quantitative changes in thymic antigen expression in determining organ-specific autoimmunity. Furthermore, the dominant-negative effect of the G228W variant was exerted through recruitment of WT Aire away from active sites of transcription in the nucleus of medullary thymic epithelial cells in vivo. Together, these results may demonstrate a mechanism by which autoimmune predisposition to phenotypes distinct from APS 1 can be mediated in a dominant-negative fashion by Aire.
-
Introduction
Autoimmune diseases arise from a breakdown in immune tolerance to self. Recently, new insights into how immune tolerance is controlled have come from the detailed study of the monogenic disorder autoimmune polyglandular syndrome type 1 (APS 1). This syndrome is due to homozygous loss of AIRE gene function (1, 2), which is important in promoting immune tolerance within the thymus (3–5). Patients with APS 1 develop a classic triad of hypoparathyroidism, adrenal insufficiency, and mucocutaneous candidiasis, along with other autoimmune manifestations at lower incidences (6). Like patients with homozygous loss-of-function mutations in AIRE, Aireo/o (previously referred to as Aire–/–; ref. 4) mice develop autoantibodies and immune infiltrates in multiple organs (4). Through the use of Aireo/o mice, Aire has been shown to promote the promiscuous gene expression (PGE) of a number of tissue-restricted antigens (TRAs) within medullary thymic epithelial cells (mTECs). Loss of Aire results in failure to delete T cells reactive against these TRAs in the thymus, resulting in autoimmunity (3–5).
Within the AIRE gene, a large number of mutations have been described that result in APS 1 when present in homozygosity or in compound heterozygosity with another mutation (6, 7). It remains unclear, however, whether carriers of 1 copy of these AIRE mutations are predisposed to autoimmunity (8–10). Surprisingly, an Italian kindred has been described with apparent autosomal dominant autoimmunity manifesting predominantly as thyroiditis that segregates with a single copy of an AIRE G228W variant (11).
The G228W substitution described in this family occurs in the SAND domain, a highly conserved domain that has been proposed to be a DNA-binding domain (12–14). The Aire protein, however, does not contain the KWDK motif predicted to be essential for DNA binding in other SAND domain–containing proteins (15, 16). Aire has also been demonstrated to form large multimers in biochemical studies of cell transfectants (14, 17). Given the data indicating that Aire multimerizes and that the G228W mutation is outside of the multimerization domain, we reasoned that the G228W mutation in this family could be behaving in a dominant-negative fashion. We therefore chose to study this human mutation to understand further how Aire acts to promote immune tolerance.
Here we describe what we believe to be a novel murine model with a knockin G228W mutation in Aire. Like their human counterparts, these mice develop autosomal dominant autoimmunity that has a disease spectrum different from that observed in Aireo/o mice. Analysis of PGE within the thymus of these mice revealed a global suppression but not complete block of this activity. G228W Aire appears to prevent WT Aire from reaching sites of active transcription and localizes Aire to nuclear inclusion bodies within mTECs. Taken together, these results demonstrate how quantitative changes in the level of thymic self-antigen expression can determine autoimmunity and the mechanism by which a dominant-negative protein can interfere with Aire’s function to cause what we believe to be a novel autoimmunity syndrome.
-
Results
Generation of a murine model of the G228W mutation. A family has been described in which autoimmune thyroiditis appears to be inherited in an autosomal dominant fashion (11). Autoimmunity in this family associates with a point mutation in the SAND domain of AIRE at position 228 that converts a glycine to a tryptophan. This glycine is highly conserved in the Aire gene across a large number of vertebrate species (data not shown). To further unravel how this mutation may be causing an autosomal dominant effect, we developed a murine model using homologous recombination. Briefly, we used Cre-loxP technology to introduce 2 nucleic acid changes in exon 6 of the murine Aire gene (Figure 1, A and B). DNA sequence analysis of mice generated using this approach confirmed the presence of the point mutation in exon 6 (Figure 1C). To detect the presence of Aire protein in this G228W-knockin line, immunohistochemistry was performed using an Aire-specific monoclonal antibody on thymic sections. Consistent with previous studies (18, 19), a subset of cytokeratin 5–positive mTECs were Aire positive. These cells were seen in WT (Aire+/+) mice but not in Aireo/o controls (Figure 1D, top panels). Like WT Aire mice, both G228W heterozygous (AireGW/+) and G228W homozygous (AireGW/GW) mice have detectable Aire protein in the thymic medulla (Figure 1D, bottom panels). On morphometric analysis, the density of Aire-positive cells per thymic medullary area was not different in Aire+/+, null heterozygote (Aire+/o), AireGW/+, and AireGW/GW thymi (Figure 1E, top). Furthermore, the total thymic medullary areas were comparable in Aire+/+ and AireGW/+ mice (Figure 1E, bottom). Consistent with these observations, we also found that the total number of mTECs per thymus as determined by flow cytometry (Supplemental Figure 1A; supplemental material available online with this article; doi: 10.1172/JCI34523DS1) was not different in Aire+/+ and AireGW/+ mice.
 Figure 1
Figure 1The G228W-knockin mouse expresses Aire protein. (A) Schematic of the first 8 exons of targeted G228W allele. The asterisk marks the site of the missense mutation. Arrowheads flanking the neo cassette represent loxP sites. The targeting construct is indicated by the dotted line, and the 5′ and 3′ ends are marked with vertical dotted lines. A single loxP remained after Cre-mediated neo excision in ES cells (bottom). (B) Southern blot of electroporated ES cell clones using a probe external to the construct (shown in A) to ascertain homologous recombination. Banding pattern of genomic DNA cut with KpnI from WT (left) and a targeted ES cell clone (right) are shown. Lanes were run on the same gel but were noncontiguous. (C) DNA (first line) and amino acid (second line) sequences around G228W point mutation. The mutated codon is outlined with a rectangle. (D) Immunohistochemical staining of thymi for cytokeratin 5 (red) and Aire (green). Scale bar: 100 μm. (E) Top: Quantitation of number of Aire-positive cells per area of thymic medulla in thymic sections. Bottom: Quantitation of the thymic medullary areas per section of Aire+/+ (n = 17) and AireGW/+ (n = 13) thymi. Sections were randomly selected from at least 3 different mice per genotype. Averages ± SD are shown. (F) Histogram of Aire expression by flow cytometry in mTEChi (red) cells, cortical thymic epithelial cells (cTECs) (green) cells, and isotype control (blue). Numbers represent average ± SD within gated region of mTEChi cells; MFI is presented as mean ± SD (n = 3 for each genotype).
To more closely determine Aire protein levels, we also analyzed Aire expression in the thymus of AireGW/+ mice by flow cytometry using a method recently described by Gray et al. (19). In this analysis, we found evidence for Aire expression in the MHC class IIhi subset of mTECs (mTEChi) except in Aireo/o controls (Figure 1F). Within the mTEChi population, the percent of Aire-positive cells was similar in Aire+/+, Aire+/o, and AireGW/+ mice. The intensity of Aire staining (as reflected by MFI) was similar in Aire+/+ and Aire+/o mice and increased in AireGW/+ compared with Aire+/+ mice (P = 0.04). Furthermore, consistent with our morphometric analysis, the total number of Aire+ mTECs was not significantly different in Aire+/+, Aire+/o, and AireGW/+ mice (Supplemental Figure 1B). Additionally, AireGW/+ mice demonstrated normal-appearing thymic architecture (Supplemental Figure 1C). Analysis of thymocytes showed that the percentages and numbers of CD4/CD8 double-negative, double-positive, and single-positive thymocytes were similar in Aire+/+ and AireGW/+ mice (Supplemental Figure 1D and data not shown). In addition, the absolute number and percentages of CD4+FoxP3+ cells was not altered in either the thymus or peripheral lymphoid organs in AireGW/+ or AireGW/GW mice (Supplemental Figure 1D and data not shown).
Heterozygous G228W mice develop spontaneous organ-specific autoimmunity that is thymus dependent. To determine whether the presence of the G228W allele predisposes to autoimmunity, 15- to 25-week-old Aire+/+, AireGW/+, and AireGW/GW mice in a mixed C57BL/6-129 genetic background were analyzed for evidence of lymphocytic infiltrates in selected organs. As shown in Figure 2, histological sections of lacrimal and salivary glands from AireGW/+ mice showed areas of lymphocytic infiltration (Figure 2A and Table 1) similar in pattern and severity to that seen previously in Aireo/o mice (4). Interestingly, AireGW/+ mice did not show evidence of retinal lymphocytic infiltration in the eye, a lesion that has been described previously in Aireo/o mice in this background (Figure 2B). Retinal infiltration, however, was seen in AireGW/GW homozygous mice. Furthermore, unlike Aireo/o mice in this mixed background (4), AireGW/+ mice do not have histological infiltration in the stomach, liver, or gonad (data not shown). Thus, in this mixed background, AireGW/+ mice develop autoimmunity in a subset of organs affected in Aireo/o mice.
 Figure 2
Figure 2AireGW/+ mice (mixed C57BL/6-129 background) develop spontaneous autoimmune disease that is thymus dependent. (A) Representative H&E-stained sections of lacrimal (top row) and salivary glands (bottom row) in Aire+/+ (left column) and AireGW/+ (right column) mice at 20 weeks of age. Arrows indicate areas of lymphocytic infiltration seen in AireGW/+ mice. Images were taken at ×20 magnification. (B) Infiltration scores for lacrimal gland in bone marrow chimeras aged 10 weeks after bone marrow transplantation. The genotypes of the bone marrow recipients (either Aire+/+ or AireGW/+) and donors (either Aire+/+ or AireGW/+) are shown for each cohort. Bars represent average infiltrate score for each group. *P < 0.003 between Aire+/+ recipients and AireGW/+ recipients. Each circle represents an individual mouse. (C) Infiltration scores for salivary gland of thymic transplants into nude C57BL/6 mice aged 12 weeks after transplantation. The genotypes of thymic donors (either Aire+/+ or AireGW/+) are shown. Bars represent average infiltrate score for each group. *P = 0.016 between Aire+/+ and AireGW/+ thymic donors. Each circle represents an individual mouse.
 Table 1
Table 1
Lacrimal, salivary, and eye infiltration scores of cohorts of 15- to 25-week-old Aire+/+, AireGW/+, and AireGW/GW mice in the mixed C57BL/6-129 background
Previous work with Aireo/o mice has demonstrated thymic dependency for the generation of autoimmunity (3, 4). To formally test this in AireGW/+ mice, we performed bone marrow chimera and thymic transfer experiments. For bone marrow chimeras, T cell–depleted bone marrow derived from either AireGW/+ or WT mice was transplanted into 4- to 6-week-old lethally irradiated AireGW/+ or WT recipient mice. Autoimmune infiltration in the lacrimal (Figure 2B) and salivary (data not shown) glands occurred at a higher incidence and with greater severity in bone marrow chimeras in which recipient mice were AireGW/+, consistent with a stromal defect.
To more specifically map the G228W defect to the thymus, thymic stroma from AireGW/+ or Aire+/+ mice were transplanted into athymic C57BL/6 nude mice. Infiltration of the lacrimal gland (data not shown) and salivary gland (Figure 2C) was seen at a higher incidence and greater severity in mice that received thymi from AireGW/+ donors. Taken together, these experiments demonstrate that the autoimmune-inducing disease defect in AireGW/+ mice tracks with thymic stroma.
In the NOD background, AireGW/+ mice develop spontaneous autoimmunity that is distinct from that in both Aireo/o and Aire+/o mice. Previous reports have suggested a gene dosage effect of Aire on immune tolerance (18). We therefore tested whether AireGW/+ mice were distinguishable from Aire+/o mice in the development of spontaneous autoimmunity. Because the null allele in our colony had been backcrossed into the NOD Lt/J and C57BL/6 backgrounds, it was impossible for us to have genetically matched Aire+/o mice in the mixed C57BL/6-129 genetic background. The G228W allele was therefore backcrossed into the NOD background using a speed congenic approach. We also chose this background because of the propensity for the NOD background to cause a more severe autoimmune phenotype in Aireo/o mice (20, 21). Age-matched female cohorts of Aire+/+, Aire+/o, AireGW/+, and AireGW/o mice were analyzed at an early time point (10 weeks) and at a late time point (25–30 weeks).
By 10 weeks, Aireo/o mice in the NOD background (NOD.o/o mice) develop autoimmunity in a number of organs, including the eye, salivary gland, lacrimal gland, exocrine pancreas, stomach, and lung (20, 21) (Figure 3A and data not shown). As shown in the top panel of Figure 3A, 10-week-old AireGW/+ mice in the NOD background (NOD.GW/+ mice) also had increased salivary and lacrimal infiltration when compared with age-matched WT (NOD.+/+) and null heterozygote (NOD.+/o) controls. Other organs affected in 10-week-old NOD.o/o mice did not show infiltration in 10-week-old NOD.GW/+ mice (Figure 3A and data not shown). Thus, at 10 weeks of age, NOD.GW/+ mice seem to develop autoimmunity in a subset of organs seen in NOD.o/o mice, and, importantly, the severity of autoimmunity seen in NOD.GW/+ mice was significantly greater than that in either NOD.+/+ or NOD.+/o mice.
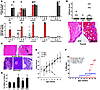 Figure 3
Figure 3NOD.GW/+ mice develop dominant autoimmune disease that is distinct from that of NOD.+/o and NOD.o/o. (A) Infiltration scores of NOD.+/+, NOD.+/o, NOD.GW/+, NOD.GW/o, and NOD.o/o cohorts at 10 (top) and 25–30 weeks (bottom) for organs indicated. Each circle represents an individual mouse. Bars indicate average infiltrate score for each cohort. **P ≤ 0.001 compared with both Aire+/+ and Aire+/o. NA, not available due to death. (B) Top: Thyroid infiltrate scores of 15- to 25-week-old mice in the NOD background of given genotype. Each circle represents an individual mouse. Bottom: H&E-stained sections (original magnification, ×5) of thyroid gland from 20-week-old NOD.+/+ (left) and NOD.GW/+ (right) mice. (C) Top: H&E-stained sections (original magnification, ×20) of sciatic nerve from 25-week-old NOD.+/+ (left) and NOD.GW/+ (right) littermates. Bottom: H&E-stained pancreas sections (original magnification, ×5) from NOD.o/o (left), NOD.GW/+ (middle), and NOD.GW/o (right). Arrows point to islets; arrowhead points to intra-islet infiltration. (D) Percentage of activated CD4+ splenocytes (CD62Llo, CD44hi) in 6- to 10-week-old mice of given genotype in the NOD background by flow cytometry. Averages ± SD of 5 independent experiments are shown. **P ≤ 0.03 compared with both NOD.+/+ and NOD.+/o. (E) Weight curves of NOD.+/+ (n = 10), NOD.GW/+ (n = 11), and NOD.GW/o (n = 4) cohorts, showing average weight ± SD. (F) Neuropathy (red squares) and diabetes (blue circles) incidence curves of NOD.+/+ (n = 10, open symbols) and NOD.GW/+ (n = 11, filled symbols) littermates. *P = 0.0035, difference in neuropathy between NOD.+/+ and NOD.GW/+ mice.
Previous work on Aireo/o mice demonstrated increased numbers of CD4- and CD8-positive T cells with an activated/memory phenotype in the peripheral lymphoid organs (4). Consistent with the observed difference in salivary and lacrimal gland infiltrates, we also found that increases in these populations were significantly greater in 10-week-old NOD.GW/+ mice than in NOD.+/+ or NOD.+/o mice (Figure 3D).
In the 10-week cohort, we also analyzed a group of mice that had 1 G228W allele and 1 null allele (NOD.GW/o). Like NOD.o/o mice, NOD.GW/o mice developed severe infiltrates in the exocrine pancreas, eye, salivary gland, lacrimal gland, stomach, and lung (Figure 3A and data not shown) (20, 21). NOD.GW/o mice also developed severe thyroiditis at an incidence similar to that seen in NOD.o/o mice (data not shown and refs. 20, 21). Furthermore, these NOD.GW/o mice developed weight loss by 6 weeks of age that was not seen in NOD.GW/+ mice (Figure 3E). These results show that NOD.GW/o mice closely phenocopy NOD.o/o mice and support the hypothesis that the G228W allele is nonfunctional.
A second cohort of NOD.GW/+ mice aged to 25–30 weeks showed additional autoimmunity effects. Like humans heterozygous for the G288W mutation, NOD.GW/+ mice develop lymphocytic infiltration in the thyroid gland, with some mice demonstrating complete obliteration of the thyroid follicles (Figure 3B, bottom panels). Four of the 16 NOD.GW/+ mice had at least 50% of their thyroid gland effaced, whereas none of the 21 NOD.+/+ or NOD.+/o mice showed this severe destruction.
NOD.GW/+ mice also develop progressive peripheral neuropathy that resulted in paralysis of the hind limbs beginning at 17 weeks of age (Figure 3F and Supplemental Figure 2A). By 22 weeks of age, 9 of 11 mice had developed neuropathy. Importantly, neuropathy was not observed in any NOD.+/+ littermate controls (Figure 3F). Consistent with these findings, sciatic nerve lymphocytic infiltration was seen on histology in all neuropathic mice (Figure 3, A and C, top row) and not in any of the NOD.+/+ or NOD.+/o age-matched controls.
Diabetes mellitus was noted in NOD.GW/+ females at an incidence similar to that seen in NOD.+/+ mice (Figure 3F) and insulitis scores were similar between NOD.GW/+, NOD.+/+, and NOD.+/o mice at 10 weeks of age (Supplemental Figure 2B). Additionally, at 10 weeks of age, histological sections of pancreas revealed only insulitis, without any evidence of the severe exocrine pancreatitis seen in AireGW/o and Aireo/o mice (Figure 3, A and C, bottom row). Exocrine pancreatitis was never observed in NOD.GW/+ mice on histology, even when they were aged to 30 weeks (Figure 3A). All NOD.GW/+ mice (11 of 11) developed either diabetes or neuropathy by 22 weeks. Similar to what was seen in other experimental systems (22, 23), diabetes and neuropathy were never observed in the same mouse. Because of this, it is difficult to determine whether the AireGW/+ allele predisposes to diabetes, as the neuropathy phenotype may be protective against diabetes.
Taken together, these data demonstrate that the G228W allele exerts an autosomal dominant effect and AireGW/+ mice develop spontaneous autoimmunity at an increased incidence and severity compared with Aire+/o mice. This effect is not complete, as these mice did not develop some of the clinical phenotypes observed in NOD.o/o mice (i.e., exocrine pancreatitis or lung infiltrates). Finally, data with the NOD.GW/o mice support the notion that the G228W allele is nonfunctional, in that these mice phenocopy NOD.o/o mice for autoimmunity.
Heterozygous G228W mice have a defect in negative selection. Previous work has directly demonstrated a defect in negative selection of autoreactive T cells in Aireo/o mice through the use of TCR-Tg models (3, 5). To determine the effect of the G228W Aire mutation on thymic negative selection, we crossed AireGW/+ mice to the OT-II × Rip-mOVA TCR-Tg model, which has previously been shown to be Aire dependent (3). OT-II is a TCR-Tg line derived from a CD4+ T cell clone that reacts against ovalbumin (24) in the context of the I-Ab class II molecule. RIP-mOVA transgenic Tg mice express a membrane-bound version of ovalbumin under the control of the rat insulin promoter. In addition to pancreatic islet, this transgene is expressed within the thymus (3), and large-scale deletion of OT-II TCR-Tg thymocytes is seen when both the OT-II TCR transgene and the RIP-mOVA transgene are present. As shown in Figure 4, strong positive selection of the OT-II TCR is observed in the thymus of OT-II single-Tg mice (Figure 4A, first column) as evidenced by the presence of a large number of CD4 single-positive cells expressing the Vα2/Vβ5 clonotype TCR (93.3% of CD4+CD8– thymocytes). Consistent with a previous study (3), we found no evidence of an effect by Aire on positive selection in OT-II single-Tg mice on the Aireo/o or AireGW/+ backgrounds (data not shown). In OT-II × Rip-mOVA Tg animals, we observed negative selection of OT-II T cells, as evidenced by a significant decrease in CD4+CD8– thymocytes that were clonotype positive (52.5% of CD4+CD8– thymocytes; Figure 4A, second column). This deletion process was dependent on Aire, as Aireo/o mice showed a large number of CD4+CD8– thymocytes that were clonotype positive (85.0% of CD4+CD8– thymocytes; Figure 4A, third column). AireGW/+ mice also showed an abrogation of negative selection that was quantitatively similar to that in Aireo/o mice (93.2% of CD4+CD8– thymocytes; Figure 4A, far-right column), with similar percentages of CD4+CD8– thymocytes that were clonotype positive.
 Figure 4
Figure 4AireGW/+ mice have a defect in negative selection in the OTII/RIP-mOVA system. (A) Representative flow cytometric plots of thymocytes. CD4 versus CD8 plots (top row) and Vα2 versus Vβ5 plots (bottom row) are shown. Numbers indicate percentage of lymphocytes in each gate. DP, double positive. Plots of OTII single-Tg mice are shown in the left column. Plots of OTII × Rip-mOVA double-Tg mice are shown in the second column. Plots of OTII × RIP-mOVA double-Tg mice in the Aireo/o setting are shown in the third column. Plots of OTII × RIP-mOVA double-Tg mice in the G228W heterozygous (AireGW/+) setting are shown in the right column. (B) Average ± SD thymocyte numbers of CD4+CD8+ clonotype-positive for each genotype. n = 7 for OTII alone; n = 9 for Aire+/+ OTII × RIP-mOVA; Aire+/o OTII × RIP-mOVA; n = 6 for Aireo/o OTII × RIP-mOVA; n = 9 for AireGW/+ OTII × RIP-mOVA. (C) Relative expression of thymic mOVA as determined by real-time RT-PCR on whole thymic stromal preparations of WT (Aire+/+) and AireGW/+ mice carrying the RIP-mOVA transgene. Data shown are representative of 2 experiments.
Cumulative data from these experiments are shown in Figure 4B. The total number of CD4+CD8– clonotype-positive thymocytes is shown for each genotype. Defects in negative selection were seen in both Aireo/o (P < 0.001) and AireGW/+ mice (P < 0.002) when compared with Aire+/+ mice. Importantly, heterozygous Aire-null (Aire+/o) mice maintained the negative selection phenotype of WT mice and the total number of clonotype-positive thymocytes was significantly greater in Aire+/o mice than in AireGW/+ mice, ruling out the possibility that Aire haploinsufficiency is causing the negative selection defect in AireGW/+ mice.
Although Aireo/o mice have been shown to have a defect in thymic negative selection in the OTII/RIP-mOVA double-Tg system, the level of RIP-mOVA expression was unexpectedly similar in WT and Aireo/o mice carrying this transgene (3). This suggested that a mechanism other than decreased thymic antigen expression in the Aireo/o mouse may contribute to the negative selection defect. One possibility is that Aire can also regulate genes involved in antigen processing, antigen presentation, and mTEC-thymocyte interactions. Indeed, a number of such genes were found to be differentially regulated in Aireo/o mice (3). We tested whether the G228W Aire mutation also resulted in these findings. Indeed, mOVA expression was also similar in Aire+/+ and AireGW/+ mice (Figure 4C). Finally, a number of genes involved in antigen presentation and processing were found to be differentially regulated in AireGW/+ mice (Table 2). Thus, like Aireo/o mice, AireGW/+ mice also harbor a defect in thymic negative selection that is independent of Aire-control of self-antigen expression.
 Table 2
Table 2
Genes differentially regulated between WT and AireGW/+ mTECs according to gene chip analysis
Heterozygous G228W mice have partial inhibition of thymic self-antigen expression. Aireo/o mice have decreased thymic expression of certain TRAs (4, 25). Like Aireo/o mice, AireGW/+ mice develop spontaneous organ-specific autoimmunity; however, the specificity of the organs targeted differs in Aireo/o and AireGW/+ in both mice and humans. To test the hypothesis that AireGW/+ mice have a distinct thymic antigen expression profile to account for these differences, we performed microarray studies comparing the gene expression pattern in mTECs from Aire+/+ and AireGW/+ mice.
For this analysis, RNA was prepared from purified mTECs pooled from 6- to 8-week-old Aire+/+ and AireGW/+ mice, and labeled cDNA was hybridized to an Affymetrix U430 2.0 microarray. A total of 343 genes were upregulated and 511 genes were downregulated in AireGW/+ compared with WT control mTECs (P < 0.01) (Supplemental Figure 3). We further tested whether AireGW/+ mTECs downregulated a subset of TRAs that has previously been shown to be downregulated by Aireo/o mTECs in similar analyses (4, 26). Strikingly, a large number of known Aire-regulated genes were also downregulated in AireGW/+ mTECs (Figure 5A). Thus, on a global level, there is a significant overlap in the genes whose expression is downregulated in AireGW/+ mTECs and those downregulated in Aireo/o mTECs.
 Figure 5
Figure 5Aire-dependent organ-specific antigen expression is partially inhibited in AireGW/+ thymi. (A) Heat map of genes previously shown to be downregulated in Aireo/o mTECs (26), comparing 3 replicates of Aire+/+ versus AireGW/+ mTECs. Scale bar: log2 ratio of a given sample relative to the average abundance across all 6 samples. (B) Twenty-one most downregulated genes in AireGW/+ compared with Aire+/+ as determined by fold change (FC, defined as expression in AireGW/+ divided by that in WT). All genes shown were differentially expressed between AireGW/+ and Aire+/+ mTECs (P ≤ 0.01). Genes are highlighted in yellow if they were expressed in only 1 tissue; in orange if expressed in several tissues; and in blue if ubiquitously expressed. (C and D) Relative expression levels by real-time RT-PCR of genes known to be downregulated in Aireo/o mice (C) or newly discovered to be downregulated in AireGW/+ by microarray analysis (D). Representative results for insulin 2 (Ins 2), spermine binding protein (Sbp), IRBP, hemoglobin Y (Hbby), salivary protein 2 (Spt2), and Spt1 are shown in C. Representative results for prostaglandin D2 synthase (Ptdgs), similar to common salivary protein 1 (Dcpp), G protein–coupled receptor 50 (Gpr50), and cellular retinoic acid binding protein I (Crabp1) are shown in D. Relative expression levels in Aire+/o, Aireo/o, and AireGW/+ compared with Aire+/+ thymi are shown for each gene. A representative experiment is shown, with each measurement done in quadruplicate. At least 2 independent experiments were performed.
Previous analysis of downregulated genes in Aireo/o mTECs revealed that the majority could be classified as TRAs (4). We therefore analyzed the top 21 AireGW/+ downregulated genes (P < 0.01 and >2-fold decrease) in a similar fashion. Twenty of these 21 downregulated genes fit the criteria of a TRA (Figure 5B). Thus, genes downregulated in mTECs in AireGW/+ mice are also highly enriched for TRAs.
We next postulated that the distinct autoimmune phenotype observed in AireGW/+ mice could be explained by quantitative changes in thymic TRA expression. We therefore tested thymic expression of 2 groups of thymic TRAs using real-time RT-PCR. First, we tested 6 transcripts (Ins2, SBP, IRBP, Hbby, Spt2, Spt1) that have previously been shown to be Aire regulated (4, 26, 27). In all transcripts tested, expression was repressed in both Aireo/o and AireGW/+ compared with Aire+/+ thymic stroma (Figure 5C). Remarkably, the level of expression for each individual TRA was incrementally higher in AireGW/+ when compared with Aireo/o mice (Figure 5C). TRA expression levels in AireGW/GW were similar to those in Aireo/o (data not shown).
Second, we tested 7 transcripts (Ptdgs, Dcpp, Gpr50, Crabp1, Nts, elastase, and NPY) that were identified as being downregulated (P < 0.01 and >2-fold downregulated) in the AireGW/+ mTECs microarray analysis and had not previously been identified as being downregulated in Aireo/o mTECs (4, 26). Once again, these 7 transcripts were repressed in both Aireo/o and AireGW/+ thymi (Figure 5D and data not shown). Of note, expression levels in Aire+/+ thymi compared with Aire+/o thymi were similar in all TRAs evaluated, suggesting that there is not a gene dosage effect (Figure 5, C and D). These data thus support a model in which the presence of one G228W allele leads to a partial repression of all Aire-dependent TRAs rather than a repression of a subset of Aire-dependent TRAs.
A quantitative change in thymic self-antigen expression is important in preventing autoimmunity. The expression of a single thymic TRA, interphotoreceptor retinoid-binding protein (IRBP) has been shown to be sufficient to prevent autoimmune uveitis (27), and thymic IRBP expression has been correlated with uveitis resistance (28). This antigen is of particular interest because 15- to 25-week-old AireGW/+ mice in the mixed C57BL/6-129 background do not develop uveitis, unlike Aireo/o mice in this mixed background (Table 1; ref. 4). Given the clear link of uveitis to IRBP, we further investigated uveitis and thymic IRBP expression in AireGW/+ mice. To eliminate the effects of the mixed genetic background, AireGW/+ mice were further backcrossed into the C57BL/6 (B6.GW/+) background (more than 5 generations). These B6.GW/+ mice were aged to 15–25 weeks and again showed no signs of eye autoimmune infiltration or eye-specific autoantibodies (Figure 6A). Consistent with previous observations, B6.o/o mice at this age did develop eye infiltrates at a high penetrance (Figure 6A and ref. 27). Like other TRAs tested, IRBP appears to be partially downregulated in AireGW/+ mice when compared with Aireo/o mice (Figure 5C). Thus, the incremental increase in IRBP expression in AireGW/+ thymi appears to be sufficient to prevent uveitis in this background.
 Figure 6
Figure 6Partial IRBP expression in AireGW/+ thymi protects against uveitis. (A and B) Eye infiltration scores for Aire+/+, AireGW/+, and Aireo/o mice in the C57BL/6 (A) and NOD (B) backgrounds. Mice were aged to 15–25 weeks, except for NOD.o/o mice (rightmost cohort in B), which were sacrificed when requiring euthanasia (≤10 weeks). *P ≤ 0.01 compared with Aire+/+ littermate controls. (C) Immunoblot of whole eye extract using sera from Aireo/o in the C57BL/6 background (B6.o/o); WT mice in the C57BL/6 background (B6.+/+); G228W heterozygous mice in the C57BL/6 background (B6.GW/+); Aireo/o in the NOD background (NOD.o/o); WT mice in the NOD background (NOD.+/+), and G228W heterozygous mice in the NOD background (NOD.GW/+). NOD.o/o mice were 6–12 weeks of age; all others were 15–25 weeks of age. The arrow indicates band of the expected size for IRBP. (D) Results of real-time RT-PCR for IRBP in AireGW/+ compared with Aire+/+ mice are shown. Mice from the C57BL/6 background (left) and the NOD background (right) were tested.
NOD.GW/+ mice developed retinal infiltrates in a delayed fashion when compared with NOD.o/o mice. At 10 weeks, NOD.o/o mice developed retinal infiltration, while NOD.GW/+ mice did not (Figure 3A). All 15- to 25-week-old NOD.GW/+ mice, however, had retinal infiltration (Figure 3A and Figure 6B) and, like Aireo/o mice in both the C56BL/6 and NOD backgrounds, demonstrated autoantibodies to eye extracts that were predominantly reactive against the 130-kDa IRBP eye antigen (Figure 6C and ref. 27). Of note, in both the C57BL/6 and NOD backgrounds, similar decreases in IRBP expression in AireGW/+ compared with Aire+/+ thymi were seen (Figure 6D). Thus, this partial thymic IRBP expression in AireGW/+ mice appears to delay uveitis onset in the NOD background.
The G228W Aire protein acts in a dominant-negative fashion to inhibit the localization of Aire to active sites of transcription. Given that the G228W Aire allele results in autosomal dominant autoimmunity, we investigated potential mechanisms by which this allele could exert a dominant-negative effect. G228W Aire transfected into COS-1 or Caco2 cells has been reported to remain in the cytosol (29, 30), suggesting that this point mutation may interfere with nuclear localization (29, 30). To test this possibility, we stained thymic sections from Aire+/+ and AireGW/+ mice with an anti-Aire antibody and performed confocal microscopy to test whether this is the case in vivo. In contrast to previous studies, we found that Aire localized to the nucleus and not the cytoplasm in AireGW/+ mTECs (Figure 7B, center column). Aire localized in a dot-like pattern in the euchromatin (DAPI light staining) regions of the nucleus of mTECs in both Aire+/+ and AireGW/+ mice. Of note, Aire was also seen in larger aggregate structures in the nucleus of AireGW/+ mTECs (Figure 7B, center column). These aggregates were seen in 60% of Aire-positive cells in AireGW/+ mice, compared with less than 1% of Aire-positive cells in Aire+/+ mice (Supplemental Figure 4). These aggregate structures were also seen in AireGW/GW mTECs (data not shown). It should also be noted that these aggregates may also explain the higher intensity of Aire staining in mTEChi cells on flow cytometry in AireGW/+ mice in Figure 1F.
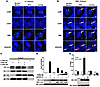 Figure 7
Figure 7The G228W protein acts in a dominant-negative fashion to inhibit localization of Aire to sites of active transcription. (A and B) Immunohistochemistry of Aire+/+ (A) and AireGW/+ (B) thymi stained with the indicated nuclear marker (green; left columns), anti-Aire antibody (red; middle columns) and DAPI (blue). Arrows indicate nuclear inclusion bodies. Scale bar: 3.17 μm. (C) Coimmunoprecipitation of G228W and WT AIRE. myc-tagged WT AIRE (m.AIRE WT), G228W AIRE (m.AIRE GW), and/or FLAG-tagged G228W AIRE (f.AIRE GW) were transfected into 1C6 cells, immunoprecipitated with anti-FLAG antibody, and subjected to Western blotting with either anti-myc (top row) or FLAG antibody (second row). To ensure protein expression, 10% of lysates were blotted with anti-myc (third row) or anti-FLAG (fourth row) antibody. (D) Insulin promoter activation by G228W and WT AIRE. m.AIRE WT, f.AIRE GW, or both (GW/WT of 1:5, 1:2, and 1:1 in columns 3–5, respectively) were transfected with the HIP339Luc insulin promoter–luciferase reporter into 1C6 cells. Protein expression using anti-FLAG (top row), anti-myc (middle row), or anti-GAPDH (bottom row) is shown by Western blot. (E) Chromatin immunoprecipitation of 1C6 cells transfected with empty vector (vector), WT AIRE (WT), G228W AIRE (GW), or the latter 2 in combination (WT+GW) with anti-AIRE antibody (black bars) or isotype control (white bars). Average amount (±SD) of mouse insulin 2 promoter (normalized as percentage of input) in the immunoprecipitates is shown. Relative amounts of AIRE input and GAPDH (loading control) are shown by Western blot (bottom panel). Measurements were done in triplicate in 2 independent experiments, with results of a representative experiment shown.
Staining of WT Aire in this dot-like pattern is consistent with previous studies (18), but colocalization with subnuclear structures in vivo has not been described. We therefore sought to investigate this and whether the subnuclear localization of Aire is altered in AireGW/+ mTECs. Thymic sections from Aire+/+ and AireGW/+ mice were stained with a panel of antibody markers specific for subnuclear structures. Markers for nuclear speckles (anti–small nuclear ribonuclear protein [anti-snRP]; refs. 31, 32), cajal bodies (anti-coilin; ref. 33), and promyelocytic leukemia protein (PML) bodies (anti-PML; ref. 33) were used. In Aire+/+ mice, Aire was distributed in subnuclear structures that were immediately adjacent to nuclear speckles (Figure 7A, top row). Aire appeared to partially colocalize with cajal bodies (Figure 7A, second row). Additionally, consistent with previous in vitro studies (34), Aire did not colocalize with PML bodies (Figure 7A, third row).
In AireGW/+ mTECs, Aire was also found adjacent to nuclear speckles (Figure 7B, top row) and in the larger Aire-containing subnuclear aggregates that also costained for snRP, coilin, and PML (Figure 7B, top 3 rows). These larger structures are reminiscent of ubiquitin-containing nuclear inclusion bodies, which have previously been described in neurodegenerative disease models (35). To test whether the Aire-containing aggregates were structurally similar to nuclear inclusion bodies, thymic sections from AireGW/+ mice were stained with an anti-ubiquitin antibody. Positive staining for ubiquitin in the large Aire-containing subnuclear structures was seen in AireGW/+ mTECs (Figure 7B, bottom row). Thus, in AireGW/+ mTECs, G228W Aire can also localize next to nuclear speckles. In addition, we observes the formation of Aire-containing inclusion bodies in the nucleus of AireGW/+ mTECs that may be a result of or directly involved in the inhibition of Aire function.
We next sought to determine how G228W Aire acts as a dominant-negative protein by utilizing the 1C6 mTEC cell line (which does not express detectable levels of Aire; ref. 36). 1C6 cells transfected with WT and G228W Aire were stained with anti-Aire antibody to ensure nuclear localization of both WT and G228W Aire protein (Supplemental Figure 5). We first tested whether G228W Aire multimerizes with WT Aire protein in this system. As shown in Figure 7C, immunoprecipitation studies using 1C6 cells cotransfected with WT and G228W human AIRE demonstrate that G228W and WT AIRE interact with each other. We next tested the ability of the G228W protein to activate transcription of a reporter under the control of the insulin promoter. WT AIRE promoted the transcriptional activity of the insulin promoter by 60-fold in a luciferase reporter assay (Figure 7D), consistent with a previous report (37). G228W AIRE, however, did not increase insulin promoter transcription and inhibited WT AIRE activity in a dose-dependent manner when cotransfected, again consistent with a dominant-negative effect.
To determine whether G228W Aire prevents Aire-containing complexes from gaining access to transcriptional target sites, we performed chromatin immunoprecipitation experiments using 1C6 cells transfected with WT AIRE, G228W AIRE, or a 1:1 mixture of both. Crosslinked chromatin was immunoprecipitated with either an anti-Aire antibody or an isotype control antibody, and the presence of the insulin 2 promoter (a known TRA target) in the precipitate was determined by real-time PCR. As shown in Figure 7E, WT AIRE was enriched more than 6-fold at the insulin 2 promoter when transfected alone, as previously observed (37). G228W AIRE, however, was not detectable when transfected alone, and transfection of a mixture of WT and G228W AIRE abolished binding of AIRE to the insulin 2 locus. These data suggest that G228W Aire acts as a dominant negative by preventing the recruitment of Aire-containing complexes to the active targets of Aire transcription in mTECs. In sum, in AireGW/+ mTECs, G228W Aire appears to localize to the nucleus, where it multimerizes with WT Aire and prevents Aire from gaining access to target sites of transcription.
-
Discussion
We have demonstrated that a naturally occurring human variant in Aire can act in an autosomal dominant fashion and provide mechanistic insights into this activity. An important implication of this study is that genetic variants in Aire can confer a dominant-negative effect with a disease phenotype altered from that seen in Aire-deficient individuals. This suggests that patients can develop autoimmune susceptibility through Aire without the classic triad of hypoparathyroidism, adrenalitis, and candidiasis. In support of this notion is the original patient kindred that harbored the G228W variant in AIRE. Certain heterozygous members of this kindred manifested only autoimmune thyroiditis, which is not a major clinical feature of APS 1 (11). Although some studies have demonstrated that Aire mutation carriers may be found in autoimmune disease cohorts (38), further study is needed to determine the exact contribution of Aire variants to autoimmune diseases with features distinct from APS 1. In addition, it may be possible that isolated kindreds with evidence of autosomal dominant autoimmunity harbor an Aire allele similar to G228W.
Aire promotes self tolerance by helping to drive expression of TRAs in mTECs (4). We find that the G228W Aire variant has a quantitative effect on thymic PGE, decreasing Aire-dependent TRA expression to approximately 10% of that seen in WT control individuals. To study how this quantitative change in thymic TRA expression affects disease, we performed an in-depth analysis of the TRA IRBP and uveitis because of the clear relationship between the loss of thymic IRBP expression and uveitis (27, 28). Here, we show that partial thymic expression of IRBP in AireGW/+ mice is sufficient to prevent uveitis in the C57BL/6 background, demonstrating that subtle changes in thymic PGE can determine autoimmune susceptibility. Within the autoimmune-prone NOD background, partial thymic expression of IRBP delayed uveitis onset, again supporting a quantitative relationship between thymic IRBP expression levels and uveitis susceptibility. These findings are consistent with reports in the NOD mouse that suggest that quantitative changes in thymic insulin expression can determine predisposition to diabetes (39, 40). Similarly, quantitative changes in the thymic expression of the P0 gene were correlated with susceptibility to experimental autoimmune neuritis (EAN) (41).
These findings are also consistent with previous studies in humans suggesting that quantitative changes in thymic PGE can determine autoimmunity. Polymorphisms in the variable number of tandem repeats (VNTR) region upstream of the insulin promoter result in variation in thymic insulin expression levels (42, 43), and alleles that cause higher levels of thymic insulin expression are associated with protection against type 1 diabetes (44). Notably, these VNTR polymorphisms appear to change thymic insulin expression levels by only 2- to 3-fold (42, 43). Additionally, a variant in the α chain of the muscle acetylcholine receptor (CHRNA1) promoter appears to modulate expression levels of CHRNA1 in human mTECs (45), and higher thymic expression levels of CHRNA1 are associated with later-onset myasthenia gravis. Once again, alleles of the CHRNA1 gene that protect against myasthenia gravis increase mean thymic expression levels by only 1.8-fold (45). These examples suggest that incremental changes in thymic antigens play an important role in the pathogenesis of autoimmunity and point to subtle modulations in thymic antigen expression levels as possible targets for therapeutic interventions.
Interestingly, despite the relatively uniform decrease (approximately 90%) in Aire-dependent thymic PGE in AireGW/+ mice, there are some phenotypes observed in Aireo/o mice that did not emerge in AireGW/+ mice even in the autoimmune-prone NOD background. An example is the severe exocrine pancreatitis that was observed in NOD.o/o or in NOD.GW/o mice that was not detected in NOD.GW/+ mice. This suggests that thymic TRAs that protect against this phenotype can adequately tolerize developing thymocytes in AireGW/+ mice, even though their expression level has been significantly reduced. However, complete analysis of this model will require the identification of the exocrine pancreatic antigens that fail to be expressed in the Aireo/o model. Thus, some thymic TRAs with reduced expression in the AireGW/+ model may still be at expression levels adequate to induce thymic tolerance, while others are not.
In the OT-II × RIP-mOVA double-Tg model, AireGW/+ mice share with Aireo/o mice the same thymic negative selection defect. One might expect, given the partial inhibition of TRAs and the differences in organs affected in AireGW/+ mice, that this defect might be partial in AireGW/+ mice. Surprisingly, the degree of defective negative selection was quantitatively similar in AireGW/+ and Aireo/o mice. The mechanism of this defect still remains to be elucidated but may be due to Aire-dependent changes in antigen presentation activity by mTECs (3) or in subtle changes in thymic structure that have been suggested in a previous study (46). Why this negative selection defect is not partial in AireGW/+ mice, however, remains to be explained and brings into question the contribution of this mechanism, if any, to the autoimmune syndrome in Aire-defective mice.
We demonstrate that within the nucleus of mTECs, a portion of WT Aire is immediately adjacent to nuclear speckles and not other subnuclear compartments such as PML bodies. Nuclear speckles are believed to be in close physical proximity to sites of active gene transcription (47). Although transcription does not occur in nuclear speckles themselves, a number of proteins involved in transcription (for example, RNA polymerase II) and splicing are enriched in these structures (31). Thus, nuclear speckles have been suggested to be storage or assembly compartments of transcription and RNA processing factors. Given their localization adjacent to nuclear speckles, Aire-positive structures in mTECs also may not be active sites of transcription, but may be depots for factors important in transcription. Clearly, further work will need to be done to understand why Aire is distributed in this interesting subnuclear pattern.
In vivo analysis of the G228W mutant protein revealed that the mutant protein localizes to the nucleus but also forms nuclear inclusion bodies. This is different from what was found in previous in vitro transfection studies, which suggested that the mutant form of the protein is trapped in the cytoplasm and cannot enter the nucleus (29, 30). In our in vitro transfection system of 1C6 cells, G228W Aire did enter the nucleus but was not observed to form the inclusion bodies seen in endogenous mTECs. These differences can be explained by expression level differences of transfected Aire, highlighting the importance of determining the autosomal dominant mechanism in vivo. Coupling our transcriptional, biochemical, and colocalization studies, we have developed a model in which the G228W mutant gains access to the nucleus and multimerizes with WT Aire. These mixed complexes then are unable to gain access to transcriptional targets of Aire and ultimately end up in inclusion body structures within mTECs. Further study will be needed, however, to demonstrate whether these inclusion bodies directly form as a result of the G228W mutation or form downstream of inhibition of another interaction.
The G228W mutation occurs in the Aire SAND domain, a known DNA-binding domain for other SAND domain–containing proteins (15, 48, 49). However, Aire does not contain the KWDK DNA-binding motif in the SAND domain that the other SAND domain–containing family members harbor. Despite this, there has been a report of the ability of the Aire SAND domain to bind DNA (13). Interestingly, the NMR structure information for the SAND domain of Sp100 and GMEB suggests that the G228W mutation likely occurs in a turn between 2 β-pleated sheets in the Aire SAND domain (15, 16). These β-pleated sheets lie on the opposite face of the DNA-binding surface in these 2 proteins, and there is some suggestion that the β-pleated sheet face may be involved in protein-protein interactions (15). We are therefore currently determining whether the G228W mutation may be interfering with DNA binding directly or may be disrupting binding to other protein(s).
Previous studies have suggested that there may be a gene dosage effect of Aire in which mice heterozygous for the null allele (Aire+/o) are predisposed to autoimmunity (18). We were unable to demonstrate a clear effect of the null allele on PGE in mTECs or on autoimmune infiltrates when present in heterozygosity in the autoimmune-prone NOD background. These differences may be explained by a different null allele, studied by Liston et al. (18), and/or by our model utilizing a polyclonal T cell system rather than a TCR-Tg system. Importantly, heterozygous carriers of autosomal recessive mutations in AIRE have not been clearly shown as a population to have an increased predisposition to autoimmunity (8–10), which again suggests that gene dosage effects of Aire may have a limited role in autoimmune-disease risk when compared with the dominant-negative allele under study in this report.
We also observed several interesting phenotypes in AireGW/+ mice. Importantly, we have demonstrated that the allele predisposes to autoimmune thyroiditis, which mirrors the original observations in the affected human kindred (11). Additionally, we observed that NOD.GW/+ mice develop a spontaneous, autoimmune peripheral neuropathy that has not been described in NOD.o/o mice. One possible explanation is that neuropathy is specific to the AireGW/+ genotype. Alternatively, because most NOD.o/o mice die by 10 weeks of age (20), they may not live long enough to manifest this disease. Given the role of Aire in the thymus, these data suggest that there may be thyroid and peripheral nerve antigens that are expressed in the thymus under the control of Aire, but further study will be needed to show this potential connection.
In conclusion, we have demonstrated that a human variant in the SAND domain of Aire can confer a dominant-negative effect on PGE in the thymus and predispose to autoimmunity. We show that quantitative changes in thymic PGE can predispose to autoimmunity, and thus “tuning” of PGE in the thymus to appropriate levels appears to be crucial in promoting self tolerance. In addition, this study demonstrates the importance of the SAND domain in conferring proper activity of Aire, and it will be important to further determine the mechanism(s) by which the G228W mutation interferes with this activity.
-
Methods
Generation of G228W Aire knockin mice. The murine Aire gene was previously cloned from a bacteriophage mouse Sv/129 genomic library (4). Site-directed mutagenesis was performed using the QuikChange II XL (Stratagene) kit. Primers used for site-directed mutagenesis were 5′-AAGTGCATTCAGGTTGGGTGGGAGTTTTATACACCCAACAA-3′ and its complement. Mutagenized nucleotides are underlined. A cassette containing Pgk-neo flanked by loxP recombination sites that had previously been introduced into genomic Aire between exons 2 and 3 was introduced between 2 BsmBI sites in exons 2 and 3 of the mutagenized construct. The construct was sequenced in its entirety to ensure that no additional mutations were introduced. The linearized 13.4-kb targeting construct was injected into E14Tg2A.4 ES cells from the 129/ola mouse strain. Clones were screened by PCR using the primers 5′-CTCAACTGTCAACACTCATGTTAG-3′ (found external to the targeting construct) and 5′-CGGAGTAGCATAGGGTTTGC-3′ (in the Pgk-neo cassette). Clones that were positive by PCR were subjected to Southern blot analysis as described in Figure 1 and as previously described (4).
Clones carrying the homologous recombination event were transfected with Cre to excise the neo locus and injected into C57BL/6 blastocysts. Chimeras were bred onto the C57BL/6 and NOD backgrounds, and mice carrying the germline mutation were confirmed to have the knockin point mutation by Southern blotting and sequencing. For genotyping, PCR was performed with the following primers: set 1, 5′-GTCATGTTGACGGATCCAGGGTAGAAAGT-3′ and 5′-AGACTAGGTGTTCCCTCCCAACCTCAG-3′. The WT allele gives a product of 1,150 bp, while the G228W allele gives a product of 1,184 bp (due to a single remaining loxP site).
Mice. Mice in the mixed C57BL/6-129 background were generated from F2 × F2, F2 × F3, or F3 × F3 crosses of a backcross to C57BL/6 mice. Mice in the C57BL/6 background were generated from at least a N5 backcross into C57BL/6 mice. Mice in the NOD background were generated using a speed congenics approach. In brief, mice at the N2 and N4 generations of a NOD Lt/J backcross were typed for markers linked to 17 Idd loci (Idd 1–7, 9, 10, 12–15, and 17–19) that were polymorphic between NOD and 129. Mice in the N4 generation that were homozygous NOD for all loci were selected for further backcrossing. All mice used in NOD experiments were from at least the N5 backcross. In the NOD background, only female mice were analyzed. NOD.o/o and NOD.+/o mice have previously been described (20). RIP-mOVA and OTII mice were obtained from the Jackson Laboratory. All mice were maintained under barrier conditions at UCSF, and all animal studies were approved by the UCSF Institutional Animal Care and Use Committee.
Neuropathy was detected and scored as previously described (50). Diabetes incidence was determined as described in ref. 51. Weight curves were generated by weekly measurements.
Histology. Organs from mice were harvested and fixed overnight in 10% formalin at 4 degrees, washed in 30% ethanol for 30 minutes, then stored in 70% ethanol. Organs were embedded in paraffin, sectioned, and stained with H&E. Scoring of salivary, lacrimal, and eye infiltration was done as previously described in a blinded fashion (20). In brief, scores of 0, 0.5, 1, 2, 3, and 4 indicate no, trace, mild, moderate, and severe lymphocytic infiltration and complete destruction, respectively. Insulitis scoring was performed as described in ref. 51.
Immunohistochemistry. Frozen thymic sections (8 μm) were stained for Aire using a mouse monoclonal antibody (18) kindly provided by H.S. Scott at the Walter and Eliza Hall Institute of Medical Research, Melbourne, Victoria, Australia. Subnuclear markers used were anti-coilin (H-300; Santa Cruz Biotechnology Inc.), anti-EFTUD2/SNRP116 (Bethyl Laboratories Inc.), and anti-PML (clone 36.1-104; Millipore). Ubiquitin staining was performed using anti-ubiquitin (Dako) antibody. Controls using secondary antibodies alone or nonspecific primary antibodies of the same isotype were used to ascertain the specificity of the staining. Thymic medulla was labeled with anti-cytokeratin 5 (ab24647; Abcam). Cytokeratin and Aire costaining was visualized under fluorescence microscopy. All other Aire costainings were visualized using confocal microscopy. Areas of cytokeratin 5 staining and Aire were measured using MetaMorph analysis software (Universal Imaging Corp.; Molecular Devices). Staining of transfected 1C6 cells was performed as previously described (37).
Bone marrow chimeras/thymic transplants. Bone marrow chimeras and thymic transplants were performed as previously described (4). For bone marrow chimeras, bone marrow was harvested from the femurs of 6-week-old mice in the C57BL/6-129 mixed background. T cells were removed from the bone marrow by complement depletion using antibodies against CD4 and CD8. Recipient mice received 2 doses of radiation (11 Gy each time) 12 hours apart. Cells (1.5 × 106) were injected into each recipient mouse by tail vein injection. Chimeras were aged for 12 weeks prior to analysis. For thymic transplants, thymi from neonates were removed and cultured in Transwell plates for 7 days in 1.35 mM 2′-deoxyguanosine (2-dG) (Sigma-Aldrich) in complete DMEM-10 to deplete the thymi of hematopoietic cells. Thymi were washed in complete DMEM-10 (without 2-dG) 2 hours prior to transplantation. Nude mice in the C57BL/6J background (Jackson Laboratory) were anesthetized, and thymi were transplanted under the kidney capsule. T cell reconstitution was confirmed by the presence of CD4+ and CD8+ cells in the peripheral blood. Transplanted mice were aged for 10 weeks prior to analysis.
Antibodies and flow cytometry. Cells were isolated from thymus, cervical lymph node, and spleen and stained with fluorescent antibody cocktails. For immunological analyses of lymphoid populations, these reagents were: anti-CD4 Alexa Fluor 700, anti-CD8 PerCP (BD Biosciences), anti-CD3 FITC (BD Biosciences), anti-CD44 PE (BD Biosciences), anti-CD25 allophycocyanin (APC; BD Bioscience), and anti-CD62L FITC (BD Biosciences). Anti-Foxp3 APC (eBioscience) staining was performed according to the manufacturers’ instructions.
For quantitation of total mTEC numbers, stromal fractions of thymi from four 4- to 6-week-old mice were stained with the following fluorescent antibody cocktails: CD45-PE, PI, G8.8-biotin, and Ly51–Alexa Fluor 488. Cells were then incubated with streptavidin-APC. Total mTEC cell numbers (defined as CD45–, PI–, G8.8+, Ly51int, as originally described in ref. 52) were obtained from the MoFlo cell sorter (Dako) and divided by 4 to obtain the mTEC cell numbers for 1 mouse. Quantitation of total Aire-positive mTECs was performed in 4- to 6-week-old mice as described in ref. 19.
Analysis of OT-II Tg mice was performed as described in ref. 3 using the following fluorescent antibodies: CD4–Alexa Fluor 700, CD8-PerCP, Vα 2-FITC, and Vβ5-PE.
Real-time RT-PCR. Thymic stromal preparations and real-time RT-PCR were performed as previously described (27). Sequences for primer and probe sets for insulin, Aire, and IRBP have been previously published (4, 27). Primer and probe sets for Hbby, NPY, Ptdgs, Spt1, and Spt2 were gifts from C. Wang (Stanford University, Palo Alto, California, USA). Sequences for these primer and probe sets and for Dcpp are listed in Supplemental Table 1. All other primer and probe sets were purchased from Applied Biosystems.
Microarray probe preparation and hybridization. Thymic stromal preparations were performed as previously described (27). mTECs were isolated from these stromal preparations based on the following surface marker profile: CD45–, PI–, G8.8+, Ly51int as originally described in ref. 52. RNA was isolated using Absolutely RNA MicroPrep Kit (Stratagene). RNA was amplified using the NuGEN WT-Ovation Pico RNA Amplification kit. The quality of the resulting cDNA was verified by Bioanalyzer chip (Agilent Technologies) and by hybridization to test chips. The cDNA was then fragmented and labeled in preparation for hybridization. The UCSF Affymetrix GeneChip Core was utilized for hybridization of probe to the Affymetrix U430 2.0 gene chips.
Microarray data analysis. Raw microarray data (.CEL files) were processed using the robust multichip analysis probe level normalization algorithm. Data were analyzed in both an unsupervised and supervised fashion. First, to assess the diversity of mTEC gene expression relative to Aire genotype in an unbiased fashion, we used a method to group samples and genes based on similarity, termed hierarchical clustering (53). This analysis revealed the extent to which Aire genotype (WT, Aireo/o, AireGW/+) contributes to the diversity of mTEC gene expression. Second, to specifically identify genes that are directly or indirectly controlled in their expression by Aire, nonparametric t test was used to identify genes that were differentially expressed in a manner dependent on Aire genotype. Genes that demonstrated statistically significant differences (P < 0.01) were selected and accounted for multiple hypothesis testing by false discovery rate (FDR) calculations (FDR <0.05). Genes were categorized as TRA using previously described criteria (4).
Western blots. Eye extracts were made from immunodeficient SCID mice, and Western blots were performed as previously described (27).
Coimmunoprecipitation. 1C6 mTECs were cotransfected with differentially tagged WT AIRE or G228W AIRE with FuGENE 6 (Roche Diagnostics). FLAG-tagged AIRE (a gift from M. Matsumoto, University of Tokushima, Tokushima, Japan) was cloned in the pCR3 vector as previously described (34), and myc-tagged AIRE (a gift from P. Peterson, University of Tartu, Tartu, Estonia) was cloned in pCDNA3.1 as previously described (54). The G228W mutation was introduced using the QuikChange II XL (Stratagene) kit. Primers used were: 5′-CATCCAGGTTGGTTGGGAGTTCTACACTC-3′ and its complement. Twenty-four hours after transfection, cells were harvested and lysed in 1% NP-40, 150 mM NaCl, 2 mM EDTA, 10 mM Tris HCl pH 7.4 with protease inhibitor. AIRE was immunoprecipitated with anti-FLAG M2 antibody conjugated to agarose (Sigma-Aldrich). Input and immunoprecipitated AIRE were analyzed by Western blotting with anti-FLAG M2 antibodies (Sigma-Aldrich) or anti–c-myc antibody 9E10 (Santa Cruz Biotechnology Inc.) and anti-mouse HRP-conjugated secondary antibody (Amersham Biosciences; GE Healthcare).
Luciferase assay. 1C6 mTECs were cotransfected with AIRE and/or G228W AIRE with a firefly luciferase reporter HIP339Luc. A Renilla luciferase control vector (10 ng) was cotransfected. The HIP339Luc reporter is driven by the human insulin promoter (–339 to +1) cloned upstream of the luciferase gene (the reporter was a gift from M. German, UCSF). To determine dose dependence, an increasing amount of G228W AIRE was transfected with the same amount of WT AIRE (ratio of WT to G228W: 5:1, 2:1, and 1:1). The amount of plasmid DNA transfected (1.5 μg of plasmid DNA per transfection) was kept constant for individual transfections by addition of empty vector. The cells were harvested 24 hours after transfection. The Dual-Luciferase kit (Promega) was used for the assay, which was done according to the manufacturer’s instructions. The firefly luciferase activities were normalized to the measured Renilla luciferase activity, and fold activation over empty vector activity was calculated for each transfection. Each data point represents an average of 3 independent transfections with calculated SEM.
Chromatin immunoprecipitation. 1C6 cells were transfected with AIRE cloned into the pcDNA3.1 vector using FuGENE 6 (Roche Diagnostics) according to the manufacturer’s instructions. Cells were harvested by trypsinization the next day. A portion of the cells was used for Western blot analysis to determine comparable expression levels of AIRE proteins. Cross-linking was performed with 1% formaldehyde at room temperature for 10 minutes and stopped with 0.125 M glycine. Nuclei from 1 × 107 cells were isolated and sonicated to generate DNA fragments of 200–500 bp. Chromatin immunoprecipitation assays were performed with rabbit normal serum (negative control) or 2 μg anti-AIRE antibodies (sc-33188; Santa Cruz Biotechnology Inc.) with protein A beads (GE Healthcare). The amount of mouse insulin 2 promoter in the immunoprecipitates was detected by real-time PCR and normalized to inputs.
Statistics. Statistical analysis was performed using Prism 4 (GraphPad) software. Rank-sum testing was performed for histological analysis, with P ≤ 0.05 considered significant. Log-rank testing was used to compare disease incidence curves, with P ≤ 0.05 considered significant.
-
Acknowledgments
We thank Jeffrey Bluestone, Abul Abbas, and members of the Anderson laboratory for helpful discussions and comments on the manuscript. We thank Nigel Killeen and the UCSF Transgenic Core for technical assistance in the production of knockin mice. We thank Hamish S. Scott at the Walter and Eliza Hall Institute of Medical Research and Clifford Wang for providing reagents. This work was supported by the NIH (AI035297, EY016408, and P30 DK063720), the Pew Scholars Program in Biomedical Sciences, the Sandler Foundation, and core support from NIH grant P30 DK063720. M.A. Su is a National Institute of Child Health and Human Development (NICHD) Fellow of the Pediatric Scientist Development Program (NICHD grant award K12-HD00850).
Address correspondence to: Mark S. Anderson, UCSF Diabetes Center, 513 Parnassus Ave., Box 0540, San Francisco, California 94143, USA. Phone: (415) 502-8052; Fax: (415) 564-5813; E-mail: [email protected].
-
Footnotes
Nonstandard abbreviations used: APS 1, autoimmune polyglandular syndrome type 1; IRBP, interphotoreceptor retinoid-binding protein; mTEC, medullary thymic epithelial cell; mTEChi, MHC class IIhi subset of mTECs; PGE, promiscuous gene expression; PML, promyelocytic leukemia protein; snRP, small nuclear ribonuclear protein; TRA, tissue-restricted antigen.
Conflict of interest: The authors have declared that no conflict of interest exists.
Reference information: J. Clin. Invest.118:1712–1726 (2008). doi:10.1172/JCI34523
-
References
- Nagamine, K., et al. 1997. Positional cloning of the APECED gene. Nat. Genet. 17:393-398.
- Finnish-German APECED Consortium. 1997. An autoimmune disease, APECED, caused by mutations in a novel gene featuring two PHD-type zinc-finger domains. Nat. Genet. 17:399-403.
- Anderson, M.S., et al. 2005. The cellular mechanism of Aire control of T cell tolerance. Immunity. 23:227-239.
- Anderson, M.S., et al. 2002. Projection of an immunological self shadow within the thymus by the aire protein. Science. 298:1395-1401.
- Liston, A., Lesage, S., Wilson, J., Peltonen, L., Goodnow, C.C. 2003. Aire regulates negative selection of organ-specific T cells. Nat. Immunol. 4:350-354.
- Peterson, P., Peltonen, L. 2005. Autoimmune polyendocrinopathy syndrome type 1 (APS1) and AIRE gene: new views on molecular basis of autoimmunity. J.Autoimmun. 25(Suppl.):49-55.
- Ruan, Q.G., She, J.X. 2004. Autoimmune polyglandular syndrome type 1 and the autoimmune regulator. Clin. Lab. Med. 24:305-317.
- Meyer, G., et al. 2001. Screening for an AIRE-1 mutation in patients with Addison’s disease, type 1 diabetes, Graves’ disease and Hashimoto’s thyroiditis as well as in APECED syndrome. Clin. Endocrinol. (Oxf.) 54:335-338.
- Torok, H.P., Tonenchi, L., Glas, J., Schiemann, U., Folwaczny, C. 2004. No significant association between mutations in exons 6 and 8 of the autoimmune regulator (AIRE) gene and inflammatory bowel disease. Eur. J. Immunogenet. 31:83-86.
- Ahonen, P. 1985. Autoimmune polyendocrinopathy--candidosis — ectodermal dystrophy (APECED): autosomal recessive inheritance. Clin. Genet. 27:535-542. View this article via: PubMed Google Scholar
- Cetani, F., et al. 2001. A novel mutation of the autoimmune regulator gene in an Italian kindred with autoimmune polyendocrinopathy-candidiasis-ectodermal dystrophy, acting in a dominant fashion and strongly cosegregating with hypothyroid autoimmune thyroiditis. J. Clin. Endocrinol. Metab. 86:4747-4752.
- Blechschmidt, K., et al. 1999. The mouse Aire gene: comparative genomic sequencing, gene organization, and expression. Genome Res. 9:158-166. View this article via: PubMed Google Scholar
- Purohit, S., Kumar, P.G., Laloraya, M., She, J.X. 2005. Mapping DNA-binding domains of the autoimmune regulator protein. Biochem. Biophys. Res. Commun. 327:939-944.
- Kumar, P.G., et al. 2001. The autoimmune regulator (AIRE) is a DNA-binding protein. J. Biol. Chem. 276:41357-41364.
- Bottomley, M.J., et al. 2001. The SAND domain structure defines a novel DNA-binding fold in transcriptional regulation. Nat. Struct. Biol. 8:626-633.
- Surdo, P.L., Bottomley, M.J., Sattler, M., Scheffzek, K. 2003. Crystal structure and nuclear magnetic resonance analyses of the SAND domain from glucocorticoid modulatory element binding protein-1 reveals deoxyribonucleic acid and zinc binding regions. Mol. Endocrinol. 17:1283-1295.
- Pitkanen, J., et al. 2000. The autoimmune regulator protein has transcriptional transactivating properties and interacts with the common coactivator CREB-binding protein. J. Biol. Chem. 275:16802-16809.
- Liston, A., et al. 2004. Gene dosage–limiting role of Aire in thymic expression, clonal deletion, and organ-specific autoimmunity. J. Exp. Med. 200:1015-1026.
- Gray, D., Abramson, J., Benoist, C., Mathis, D. 2007. Proliferative arrest and rapid turnover of thymic epithelial cells expressing Aire. J. Exp. Med. 204:2521-2528.
- Jiang, W., Anderson, M.S., Bronson, R., Mathis, D., Benoist, C. 2005. Modifier loci condition autoimmunity provoked by Aire deficiency. J. Exp. Med. 202:805-815.
- Niki, S., et al. 2006. Alteration of intra-pancreatic target-organ specificity by abrogation of Aire in NOD mice. J. Clin. Invest. 116:1292-1301.
- Salomon, B., et al. 2001. Development of spontaneous autoimmune peripheral polyneuropathy in B7-2-deficient NOD mice. J. Exp. Med. 194:677-684.
- Setoguchi, R., Hori, S., Takahashi, T., Sakaguchi, S. 2005. Homeostatic maintenance of natural Foxp3(+) CD25(+) CD4(+) regulatory T cells by interleukin (IL)-2 and induction of autoimmune disease by IL-2 neutralization. J. Exp. Med. 201:723-735.
- Barnden, M.J., Allison, J., Heath, W.R., Carbone, F.R. 1998. Defective TCR expression in transgenic mice constructed using cDNA-based alpha- and beta-chain genes under the control of heterologous regulatory elements. Immunol. Cell Biol. 76:34-40.
- Derbinski, J., et al. 2005. Promiscuous gene expression in thymic epithelial cells is regulated at multiple levels. J. Exp. Med. 202:33-45.
- Johnnidis, J.B., et al. 2005. Chromosomal clustering of genes controlled by the aire transcription factor. Proc. Natl. Acad. Sci. U. S. A. 102:7233-7238.
- Devoss, J., et al. 2006. Spontaneous autoimmunity prevented by thymic expression of a single self-antigen. J. Exp. Med. 203:2727-2735.
- Egwuagu, C.E., Charukamnoetkanok, P., Gery, I. 1997. Thymic expression of autoantigens correlates with resistance to autoimmune disease. J. Immunol. 159:3109-3112. View this article via: PubMed Google Scholar
- Halonen, M., et al. 2004. APECED-causing mutations in AIRE reveal the functional domains of the protein. Hum. Mutat. 23:245-257.
- Ilmarinen, T., et al. 2005. Functional analysis of SAND mutations in AIRE supports dominant inheritance of the G228W mutation. Hum. Mutat. 26:322-331.
- Saitoh, N., et al. 2004. Proteomic analysis of interchromatin granule clusters. Mol. Biol. Cell. 15:3876-3890.
- Fabrizio, P., Laggerbauer, B., Lauber, J., Lane, W.S., Luhrmann, R. 1997. An evolutionarily conserved U5 snRNP-specific protein is a GTP-binding factor closely related to the ribosomal translocase EF-2. EMBO J. 16:4092-4106.
- Spector, D.L. 2006. Snapshot: cellular bodies. Cell. 127:1071.e1071-e1072. View this article via: PubMed Google Scholar
- Akiyoshi, H., et al. 2004. Subcellular expression of autoimmune regulator (AIRE) is organized in a spatiotemporal manner. J. Biol. Chem. 279:33984-33991.
- Woulfe, J.M. 2007. Abnormalities of the nucleus and nuclear inclusions in neurodegenerative disease: a work in progress. Neuropathol. Appl. Neurobiol. 33:2-42. View this article via: PubMed Google Scholar
- Uchida, D., et al. 2004. AIRE functions as an E3 ubiquitin ligase. J. Exp. Med. 199:167-172.
- Oven, I., et al. 2007. AIRE recruits P-TEFb for transcriptional elongation of target genes in medullary thymic epithelial cells. Mol. Cell Biol. 27:8815-8823.
- Ferrera, F., et al. 2007. AIRE gene polymorphisms in systemic sclerosis associated with autoimmune thyroiditis. Clin. Immunol. 122:13-17.
- Thebault-Baumont, K., et al. 2003. Acceleration of type 1 diabetes mellitus in proinsulin 2-deficient NOD mice. J. Clin. Invest. 111:851-857.
- Chentoufi, A.A., Polychronakos, C. 2002. Insulin expression levels in the thymus modulate insulin-specific autoreactive T-cell tolerance: the mechanism by which the IDDM2 locus may predispose to diabetes. Diabetes. 51:1383-1390.
- Miyamoto, K., Miyake, S., Schachner, M., Yamamura, T. 2003. Heterozygous null mutation of myelin P0 protein enhances susceptibility to autoimmune neuritis targeting P0 peptide. Eur. J. Immunol. 33:656-665.
- Pugliese, A., et al. 1997. The insulin gene is transcribed in the human thymus and transcription levels correlated with allelic variation at the INS VNTR-IDDM2 susceptibility locus for type 1 diabetes. Nat. Genet. 15:293-297.
- Vafiadis, P., et al. 1997. Insulin expression in human thymus is modulated by INS VNTR alleles at the IDDM2 locus. Nat. Genet. 15:289-292.
- Bennett, S.T., et al. 1995. Susceptibility to human type 1 diabetes at IDDM2 is determined by tandem repeat variation at the insulin gene minisatellite locus. Nat. Genet. 9:284-292.
- Giraud, M., et al. 2007. An IRF8-binding promoter variant and AIRE control CHRNA1 promiscuous expression in thymus. Nature. 448:934-937.
- Gillard, G.O., Dooley, J., Erickson, M., Peltonen, L., Farr, A.G. 2007. Aire-dependent alterations in medullary thymic epithelium indicate a role for Aire in thymic epithelial differentiation. J. Immunol. 178:3007-3015. View this article via: PubMed Google Scholar
- Lamond, A.I., Spector, D.L. 2003. Nuclear speckles: a model for nuclear organelles. Nat. Rev. Mol. Cell. Biol. 4:605-612.
- Gibson, T.J., Ramu, C., Gemund, C., Aasland, R. 1998. The APECED polyglandular autoimmune syndrome protein, AIRE-1, contains the SAND domain and is probably a transcription factor. Trends Biochem. Sci. 23:242-244.
- Michelson, R.J., et al. 1999. Nuclear DEAF-1-related (NUDR) protein contains a novel DNA binding domain and represses transcription of the heterogeneous nuclear ribonucleoprotein A2/B1 promoter. J. Biol. Chem. 274:30510-30519.
- Bour-Jordan, H., Thompson, H.L., Bluestone, J.A. 2005. Distinct effector mechanisms in the development of autoimmune neuropathy versus diabetes in nonobese diabetic mice. J. Immunol. 175:5649-5655. View this article via: PubMed Google Scholar
- Coligan, J.E., et al. 2001.Current protocols in immunology. Wiley. New York, New York, USA. 5500 pp.
- Gray, D.H., Chidgey, A.P., Boyd, R.L. 2002. Analysis of thymic stromal cell populations using flow cytometry. J. Immunol. Methods. 260:15-28.
- Eisen, M.B., Spellman, P.T., Brown, P.O., Botstein, D. 1998. Cluster analysis and display of genome-wide expression patterns. Proc. Natl. Acad. Sci. U. S. A. 95:14863-14868.
- Heino, M., et al. 1999. Autoimmune regulator is expressed in the cells regulating immune tolerance in thymus medulla. Biochem. Biophys. Res. Commun. 257:821-825.
-
Version history
- Version 1 (April 15, 2008): No description
- Version 2 (May 1, 2008): No description



Copyright © 2024 American Society for Clinical Investigation
ISSN: 0021-9738 (print), 1558-8238 (online)









