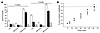Advertisement
Technical AdvanceGenetics Free access | 10.1172/JCI34316
Serotype-dependent packaging of large genes in adeno-associated viral vectors results in effective gene delivery in mice
Mariacarmela Allocca,1 Monica Doria,1 Marco Petrillo,1 Pasqualina Colella,1 Maria Garcia-Hoyos,1 Daniel Gibbs,2 So Ra Kim,3 Albert Maguire,4 Tonia S. Rex,4 Umberto Di Vicino,1 Luisa Cutillo,1 Janet R. Sparrow,3 David S. Williams,2 Jean Bennett,4 and Alberto Auricchio1,5
1Telethon Institute of Genetics and Medicine (TIGEM), Naples, Italy. 2Department of Pharmacology and Department of Neurosciences, UCSD School of Medicine, La Jolla, California, USA. 3Department of Ophthalmology, Columbia University, New York, New York, USA. 4F.M. Kirby Center for Molecular Ophthalmology, Scheie Eye Institute, University of Pennsylvania, Philadelphia, Pennsylvania, USA. 5Medical Genetics, Department of Pediatrics, University of Naples Federico II, Naples, Italy.
Address correspondence to: Alberto Auricchio, Telethon Institute of Genetics and Medicine, Via P. Castellino 111, 80131 Naples, Italy. Phone: 11-39-081-6132229; Fax: 11-39-081-5790919; E-mail: [email protected].
Find articles by Allocca, M. in: JCI | PubMed | Google Scholar
1Telethon Institute of Genetics and Medicine (TIGEM), Naples, Italy. 2Department of Pharmacology and Department of Neurosciences, UCSD School of Medicine, La Jolla, California, USA. 3Department of Ophthalmology, Columbia University, New York, New York, USA. 4F.M. Kirby Center for Molecular Ophthalmology, Scheie Eye Institute, University of Pennsylvania, Philadelphia, Pennsylvania, USA. 5Medical Genetics, Department of Pediatrics, University of Naples Federico II, Naples, Italy.
Address correspondence to: Alberto Auricchio, Telethon Institute of Genetics and Medicine, Via P. Castellino 111, 80131 Naples, Italy. Phone: 11-39-081-6132229; Fax: 11-39-081-5790919; E-mail: [email protected].
Find articles by Doria, M. in: JCI | PubMed | Google Scholar
1Telethon Institute of Genetics and Medicine (TIGEM), Naples, Italy. 2Department of Pharmacology and Department of Neurosciences, UCSD School of Medicine, La Jolla, California, USA. 3Department of Ophthalmology, Columbia University, New York, New York, USA. 4F.M. Kirby Center for Molecular Ophthalmology, Scheie Eye Institute, University of Pennsylvania, Philadelphia, Pennsylvania, USA. 5Medical Genetics, Department of Pediatrics, University of Naples Federico II, Naples, Italy.
Address correspondence to: Alberto Auricchio, Telethon Institute of Genetics and Medicine, Via P. Castellino 111, 80131 Naples, Italy. Phone: 11-39-081-6132229; Fax: 11-39-081-5790919; E-mail: [email protected].
Find articles by Petrillo, M. in: JCI | PubMed | Google Scholar
1Telethon Institute of Genetics and Medicine (TIGEM), Naples, Italy. 2Department of Pharmacology and Department of Neurosciences, UCSD School of Medicine, La Jolla, California, USA. 3Department of Ophthalmology, Columbia University, New York, New York, USA. 4F.M. Kirby Center for Molecular Ophthalmology, Scheie Eye Institute, University of Pennsylvania, Philadelphia, Pennsylvania, USA. 5Medical Genetics, Department of Pediatrics, University of Naples Federico II, Naples, Italy.
Address correspondence to: Alberto Auricchio, Telethon Institute of Genetics and Medicine, Via P. Castellino 111, 80131 Naples, Italy. Phone: 11-39-081-6132229; Fax: 11-39-081-5790919; E-mail: [email protected].
Find articles by Colella, P. in: JCI | PubMed | Google Scholar
1Telethon Institute of Genetics and Medicine (TIGEM), Naples, Italy. 2Department of Pharmacology and Department of Neurosciences, UCSD School of Medicine, La Jolla, California, USA. 3Department of Ophthalmology, Columbia University, New York, New York, USA. 4F.M. Kirby Center for Molecular Ophthalmology, Scheie Eye Institute, University of Pennsylvania, Philadelphia, Pennsylvania, USA. 5Medical Genetics, Department of Pediatrics, University of Naples Federico II, Naples, Italy.
Address correspondence to: Alberto Auricchio, Telethon Institute of Genetics and Medicine, Via P. Castellino 111, 80131 Naples, Italy. Phone: 11-39-081-6132229; Fax: 11-39-081-5790919; E-mail: [email protected].
Find articles by Garcia-Hoyos, M. in: JCI | PubMed | Google Scholar
1Telethon Institute of Genetics and Medicine (TIGEM), Naples, Italy. 2Department of Pharmacology and Department of Neurosciences, UCSD School of Medicine, La Jolla, California, USA. 3Department of Ophthalmology, Columbia University, New York, New York, USA. 4F.M. Kirby Center for Molecular Ophthalmology, Scheie Eye Institute, University of Pennsylvania, Philadelphia, Pennsylvania, USA. 5Medical Genetics, Department of Pediatrics, University of Naples Federico II, Naples, Italy.
Address correspondence to: Alberto Auricchio, Telethon Institute of Genetics and Medicine, Via P. Castellino 111, 80131 Naples, Italy. Phone: 11-39-081-6132229; Fax: 11-39-081-5790919; E-mail: [email protected].
Find articles by Gibbs, D. in: JCI | PubMed | Google Scholar
1Telethon Institute of Genetics and Medicine (TIGEM), Naples, Italy. 2Department of Pharmacology and Department of Neurosciences, UCSD School of Medicine, La Jolla, California, USA. 3Department of Ophthalmology, Columbia University, New York, New York, USA. 4F.M. Kirby Center for Molecular Ophthalmology, Scheie Eye Institute, University of Pennsylvania, Philadelphia, Pennsylvania, USA. 5Medical Genetics, Department of Pediatrics, University of Naples Federico II, Naples, Italy.
Address correspondence to: Alberto Auricchio, Telethon Institute of Genetics and Medicine, Via P. Castellino 111, 80131 Naples, Italy. Phone: 11-39-081-6132229; Fax: 11-39-081-5790919; E-mail: [email protected].
Find articles by Kim, S. in: JCI | PubMed | Google Scholar
1Telethon Institute of Genetics and Medicine (TIGEM), Naples, Italy. 2Department of Pharmacology and Department of Neurosciences, UCSD School of Medicine, La Jolla, California, USA. 3Department of Ophthalmology, Columbia University, New York, New York, USA. 4F.M. Kirby Center for Molecular Ophthalmology, Scheie Eye Institute, University of Pennsylvania, Philadelphia, Pennsylvania, USA. 5Medical Genetics, Department of Pediatrics, University of Naples Federico II, Naples, Italy.
Address correspondence to: Alberto Auricchio, Telethon Institute of Genetics and Medicine, Via P. Castellino 111, 80131 Naples, Italy. Phone: 11-39-081-6132229; Fax: 11-39-081-5790919; E-mail: [email protected].
Find articles by Maguire, A. in: JCI | PubMed | Google Scholar
1Telethon Institute of Genetics and Medicine (TIGEM), Naples, Italy. 2Department of Pharmacology and Department of Neurosciences, UCSD School of Medicine, La Jolla, California, USA. 3Department of Ophthalmology, Columbia University, New York, New York, USA. 4F.M. Kirby Center for Molecular Ophthalmology, Scheie Eye Institute, University of Pennsylvania, Philadelphia, Pennsylvania, USA. 5Medical Genetics, Department of Pediatrics, University of Naples Federico II, Naples, Italy.
Address correspondence to: Alberto Auricchio, Telethon Institute of Genetics and Medicine, Via P. Castellino 111, 80131 Naples, Italy. Phone: 11-39-081-6132229; Fax: 11-39-081-5790919; E-mail: [email protected].
Find articles by Rex, T. in: JCI | PubMed | Google Scholar
1Telethon Institute of Genetics and Medicine (TIGEM), Naples, Italy. 2Department of Pharmacology and Department of Neurosciences, UCSD School of Medicine, La Jolla, California, USA. 3Department of Ophthalmology, Columbia University, New York, New York, USA. 4F.M. Kirby Center for Molecular Ophthalmology, Scheie Eye Institute, University of Pennsylvania, Philadelphia, Pennsylvania, USA. 5Medical Genetics, Department of Pediatrics, University of Naples Federico II, Naples, Italy.
Address correspondence to: Alberto Auricchio, Telethon Institute of Genetics and Medicine, Via P. Castellino 111, 80131 Naples, Italy. Phone: 11-39-081-6132229; Fax: 11-39-081-5790919; E-mail: [email protected].
Find articles by Di Vicino, U. in: JCI | PubMed | Google Scholar
1Telethon Institute of Genetics and Medicine (TIGEM), Naples, Italy. 2Department of Pharmacology and Department of Neurosciences, UCSD School of Medicine, La Jolla, California, USA. 3Department of Ophthalmology, Columbia University, New York, New York, USA. 4F.M. Kirby Center for Molecular Ophthalmology, Scheie Eye Institute, University of Pennsylvania, Philadelphia, Pennsylvania, USA. 5Medical Genetics, Department of Pediatrics, University of Naples Federico II, Naples, Italy.
Address correspondence to: Alberto Auricchio, Telethon Institute of Genetics and Medicine, Via P. Castellino 111, 80131 Naples, Italy. Phone: 11-39-081-6132229; Fax: 11-39-081-5790919; E-mail: [email protected].
Find articles by Cutillo, L. in: JCI | PubMed | Google Scholar
1Telethon Institute of Genetics and Medicine (TIGEM), Naples, Italy. 2Department of Pharmacology and Department of Neurosciences, UCSD School of Medicine, La Jolla, California, USA. 3Department of Ophthalmology, Columbia University, New York, New York, USA. 4F.M. Kirby Center for Molecular Ophthalmology, Scheie Eye Institute, University of Pennsylvania, Philadelphia, Pennsylvania, USA. 5Medical Genetics, Department of Pediatrics, University of Naples Federico II, Naples, Italy.
Address correspondence to: Alberto Auricchio, Telethon Institute of Genetics and Medicine, Via P. Castellino 111, 80131 Naples, Italy. Phone: 11-39-081-6132229; Fax: 11-39-081-5790919; E-mail: [email protected].
Find articles by Sparrow, J. in: JCI | PubMed | Google Scholar
1Telethon Institute of Genetics and Medicine (TIGEM), Naples, Italy. 2Department of Pharmacology and Department of Neurosciences, UCSD School of Medicine, La Jolla, California, USA. 3Department of Ophthalmology, Columbia University, New York, New York, USA. 4F.M. Kirby Center for Molecular Ophthalmology, Scheie Eye Institute, University of Pennsylvania, Philadelphia, Pennsylvania, USA. 5Medical Genetics, Department of Pediatrics, University of Naples Federico II, Naples, Italy.
Address correspondence to: Alberto Auricchio, Telethon Institute of Genetics and Medicine, Via P. Castellino 111, 80131 Naples, Italy. Phone: 11-39-081-6132229; Fax: 11-39-081-5790919; E-mail: [email protected].
Find articles by Williams, D. in: JCI | PubMed | Google Scholar
1Telethon Institute of Genetics and Medicine (TIGEM), Naples, Italy. 2Department of Pharmacology and Department of Neurosciences, UCSD School of Medicine, La Jolla, California, USA. 3Department of Ophthalmology, Columbia University, New York, New York, USA. 4F.M. Kirby Center for Molecular Ophthalmology, Scheie Eye Institute, University of Pennsylvania, Philadelphia, Pennsylvania, USA. 5Medical Genetics, Department of Pediatrics, University of Naples Federico II, Naples, Italy.
Address correspondence to: Alberto Auricchio, Telethon Institute of Genetics and Medicine, Via P. Castellino 111, 80131 Naples, Italy. Phone: 11-39-081-6132229; Fax: 11-39-081-5790919; E-mail: [email protected].
Find articles by Bennett, J. in: JCI | PubMed | Google Scholar
1Telethon Institute of Genetics and Medicine (TIGEM), Naples, Italy. 2Department of Pharmacology and Department of Neurosciences, UCSD School of Medicine, La Jolla, California, USA. 3Department of Ophthalmology, Columbia University, New York, New York, USA. 4F.M. Kirby Center for Molecular Ophthalmology, Scheie Eye Institute, University of Pennsylvania, Philadelphia, Pennsylvania, USA. 5Medical Genetics, Department of Pediatrics, University of Naples Federico II, Naples, Italy.
Address correspondence to: Alberto Auricchio, Telethon Institute of Genetics and Medicine, Via P. Castellino 111, 80131 Naples, Italy. Phone: 11-39-081-6132229; Fax: 11-39-081-5790919; E-mail: [email protected].
Find articles by Auricchio, A. in: JCI | PubMed | Google Scholar
Published April 15, 2008 - More info
J Clin Invest. 2008;118(5):1955–1964. https://doi.org/10.1172/JCI34316.
© 2008 The American Society for Clinical Investigation
Received: October 24, 2007; Accepted: February 29, 2008
-
Abstract
Vectors derived from adeno-associated virus (AAV) are promising for human gene therapy, including treatment for retinal blindness. One major limitation of AAVs as vectors is that AAV cargo capacity has been considered to be restricted to 4.7 kb. Here we demonstrate that vectors with an AAV5 capsid (i.e., rAAV2/5) incorporated up to 8.9 kb of genome more efficiently than 6 other serotypes tested, independent of the efficiency of the rAAV2/5 production process. Efficient packaging of the large murine Abca4 and human MYO7A and CEP290 genes, which are mutated in common blinding diseases, was obtained, suggesting that this packaging efficiency is independent of the specific sequence packaged. Expression of proteins of the appropriate size and function was observed following transduction with rAAV2/5 carrying large genes. Intraocular administration of rAAV2/5 encoding ABCA4 resulted in protein localization to rod outer segments and significant and stable morphological and functional improvement of the retina in Abca4–/– mice. This use of rAAV2/5 may be a promising therapeutic strategy for recessive Stargardt disease, the most common form of inherited macular degeneration. The possibility of packaging large genes in AAV greatly expands the therapeutic potential of this vector system.
-
Introduction
Vectors derived from the small, icosahedral, single-stranded DNA adeno-associated virus (AAV) are very promising for gene therapy of human diseases (1). The safety and efficacy of recombinant AAV (rAAV) vectors have been successfully tested in humans in muscle, liver, lung, central nervous system (2) and are currently being tested in the retina (3). So far, the results of applications of rAAV to neurodegenerative diseases are particularly promising, as evidenced by safety and efficacy results after delivery to the subthalamic nucleus in patients with Parkinson disease (4).
Wild-type AAV contains a 4.7-kb genome made up of the rep and cap genes encoding 4 replication and 3 capsid proteins, respectively, flanked by two 145-bp inverted terminal repeats (ITRs) (5). rAAV vectors retain only the AAV ITRs, thus leaving up to 4.7 kb for packaging of therapeutic DNA (5). The availability of more than 100 different capsids derived from the same number of novel AAV serotypes allows one to exchange capsids between different serotypes and to produce dozens of rAAVs containing the same genome. For example, one can package the ITRs of the best-studied AAV serotype, AAV2, in capsids from other AAV serotypes and thus obtain rAAV2/n, where the first number defines the ITRs and the second the capsid of origin (6). Capsids are the main determinant of rAAV tropism and transduction characteristics. Therefore, the availability of such a high number of AAV serotypes allows efficient in vivo targeting of several tissues (6). In particular, gene transfer to the retina offers several advantages when compared with other tissues: the eye is small and enclosed, requiring small doses of vector for efficient targeting, thus limiting exposure to other organs (7–9). One main limitation of rAAV2 is represented by its native packaging capacity, which is considered to be restricted to 4.7 kb (10, 11), the size of the parental viral genome between the ITRs, which does not significantly vary among AAV serotypes. A recent report suggests that rAAV2/1–5 are capable of packaging and protecting recombinant genomes as large as 6 kb, although these larger genome-containing virions are preferentially degraded by the proteasome unless inhibitors are added (12).
Many common human inherited diseases are caused by mutations in genes with open reading frames largely exceeding rAAV cargo capacity. These include Duchenne muscular dystrophy, cystic fibrosis, hemophilia A, and sensorineural diseases such as recessive Stargardt disease (rSTGD) (13), Usher syndrome (USH) (14), and Leber congenital amaurosis (LCA) (15). The possibility of efficiently packaging large genomes in AAV capsids combined with the ability of rAAV to efficiently transduce the affected tissues would allow the development of rAAV-based gene therapies for these otherwise untreatable diseases. We postulated that different AAV capsids differ in their ability to tolerate large genomes. Based on the technical advantages of retinal gene transfer, we selected 3 different and common blinding diseases to test the applicability of our results: (a) rSTGD due to mutations in ABCA4 (16), which has a prevalence of 1 in 10,000 individuals and represents the most common inherited macular degeneration; (b) USH due to mutations in MYO7A (USH1B), which is a common cause of inherited severe combined deafness and blindness (14); and (c) LCA due to mutations in CEP290 (15), which is an inherited retinal degenerative disease characterized by severe loss of vision at birth.
We tested the ability of 8 different rAAV serotypes (rAAV2/1–9, with the exception of rAAV2/6, whose capsid is almost identical to that of rAAV2/1) to package a large expression cassette containing either Abca4 (8.9 kb) or MYO7A (8.1 kb). We found that rAAV2/5 yields were significantly higher than those of 6 other serotypes tested. Similar rAAV2/5 titers were obtained when packaging CEP290 or a large EGFP transgene linked to a stuffer sequence and resulting in rAAV genome sizes of similar length. rAAV2/5-mediated transduction in vitro and in vivo resulted in expression of proteins of the expected molecular weight and function and in substantial morphological and functional improvement of the ocular phenotype in the Abca4–/– mice.
-
Results
Vectors with AAV5 capsids efficiently package up to 8.9 kb. To test the ability of various AAV serotypes to package large genomes and to study whether the results obtained were dependent on the nucleotide composition of the sequence packaged, murine Abca4 and human MYO7A and CEP290 cDNA sequences were separately cloned in the pAAV-CMV-EGFP plasmid (17) between the AAV2 ITRs downstream of the CMV promoter and upstream of a poly(A) signal. The resulting pAAV-CMV-Abca4, -MYO7A, and -CEP290 constructs contained 8.9, 8.1, and 8.9 kb, respectively, including the ITRs. pAAV-CMV-Abca4 was used to produce large preparations of rAAV2/1–9, with the exception of rAAV2/6, whose capsid is almost identical to that of rAAV2/1. Since the final volume is identical among large preparations, the preparation titers reflect the vector yields. Figure 1A shows the titers of the various rAAV-CMV-Abca4 serotypes. The titers of the rAAV2/5-CMV-Abca4 preparations were significantly higher (P ≤ 0.05) than those of rAAV2/2, 3, 4, 7, 8, and 9. A 1-way ANOVA analysis followed by a multiple comparison test (Bonferroni adjustment for multiplicity) confirmed that rAAV2/5 titers have population marginal means significantly different from those of rAAV2/2–9 (Supplemental Figure 1A; supplemental material available online with this article; doi: 10.1172/JCI34316DS1). Similar results were obtained using pAAV2.1-CMV-MYO7A for production of large preparations of AAV serotypes: rAAV2/5-CMV-MYO7A titers were 2.5 × 1011 ± 8.8 × 1010 genome copies (GC)/ml (n = 4). In addition, the titers of the rAAV2/5-CMV-CEP290 large preparations were also similar to those of rAAV2/5-CMV-Abca4 (5.1 × 1011 GC/ml; n = 2), suggesting that packaging is independent of the sequence tested.
 Figure 1
Figure 1Packaging capacity of rAAV serotypes. (A) Average titers of rAAV2/1, 2, 3, 4, 5, 7, 8, and 9 containing the Abca4 cDNA. The rAAV genome is composed of AAV2 ITRs, CMV promoter, and Abca4 cDNA sequence (rAAV genome size 8.9 kb). Data are shown as average ± SE. *P ≤ 0.05, compared with rAAV2/5; n, number of rAAV large preparations. The numbers above the SE bars represent the average titers. (B) Large genome to small genome titer ratio for each of the rAAV serotypes tested (average ± SE). The large genome was represented by Abca4 (rAAV genome size 8.9 kb). The small genomes (containing various transgenes) were 4.7 kb or less. n, number of rAAV preparations. (C) Average titers ± SE of rAAV2/5-CMV-Abca4 vectors with genome size of 8.9, 10, and 11.4 kb. The 10- and 11.4-kb genomes were obtained by adding 1.1 and 2.5 kb of stuffer sequence, respectively, to the rAAV2/5-CMV-Abca4 genome. The numbers above the SE bars represent the average titers. (D) Average titers (±SE) of rAAV2/5- (gray bars) and rAAV2/2-CMV-EGFP (white bars) vectors with genome sizes of 4.1, 5.1, 6.1, 7.1, and 8.9 kb. The different-size genomes were obtained by adding stuffer sequences of different lengths after the poly(A) sequence in the pAAV2.1-CMV-EGFP plasmid. The results shown are the average of titers obtained by RT-PCR from 3 small-scale vector preparations.
Next, we addressed whether this difference reflected higher vector yields during the production and purification process or the ability of AAV5 capsids to package larger genomes. For each serotype, the titer of preparations containing the large CMV-Abca4 genome was compared with that of preparations containing smaller genomes (ranging from 2.6 to 4.7 kb between the ITRs). Unlike the titers of AAV packaging a large genome (8.9 kb; Supplemental Figure 1A), significant differences were not observed among the titers of the various serotypes packaging smaller genomes (≤4.7 kb) (Supplemental Figure 1B). As a result, the large genome to small genome titer ratio was higher for AAV2/5 than it was for other serotypes tested (Figure 1B and Supplemental Table 1), thus confirming that virions with the AAV5 capsid have significantly higher packaging capacity than the 6 other serotypes tested.
To define the upper limit of rAAV2/5 packaging ability, we cloned 1.1 and 2.5 kb of stuffer sequence in pAAV-CMV-Abca4 and used the resulting plasmids to produce large preparations of rAAV2/5, the titers of which were compared with those of rAAV2/5-CMV-Abca4 (Figure 1C). A 2.6-fold drop in vector titer was observed when the rAAV-2/5-CMV-Abca4 genome contained an additional 1.1 kb.
To characterize the ability of AAV2/5 to package genomes of lengths progressively increasing from 4.1 to 8.9 kb, we separately cloned 5 stuffer sequences, resulting in genome lengths of 4.1, 5.1, 6.1, 7.1, and 8.9 kb between the AAV2 ITRs of the pAAV2.1-CMV-EGFP plasmid. The stuffer sequence was cloned downstream of the CMV-EGFP expression cassette present between the AAV2 ITRs of the plasmid. These constructs were used for small-scale vector production as described in Methods. The titers (average ± SE; n = 3) of the corresponding rAAV2/5 were compared with those of rAAV2/2, the best-studied AAV serotype (Figure 1D). When genome size was increased, rAAV2/2 titers decreased progressively from 5.6 × 1011 (4.1 kb) to 5.7 × 109 GC/ml (8.9 kb), an almost 100-fold decrease. A 50-fold decrease was observed when comparing the titers of rAAV2/2 large preparations (Supplemental Table 1). The 2-fold difference observed in these 2 sets of data may be due to differences in small- versus large-scale vector production. In contrast to rAAV2/2, rAAV2/5 titers decreased by only 8.5-fold (from 1.7 × 1012 to 2.05 × 1011), results similar to those described in Figure 1A and Supplemental Table 1. Large preparations of rAA2/5-CMV-EGFP-8.9 were produced, and the titers were similar to those of the AAV2/5 preparations containing the large Abca4, MYO7A, or CEP290 genes (6.3 × 1011 GC/ml; n = 2).
To demonstrate that the rAAV2/5-CMV-Abca4, -MYO7A, -CEP290, and -EGFP-8.9 vectors package their genome in its entire length, viral DNA was extracted from 2.5 × 1010 particles of each vector and analyzed by Southern blot following separation on alkaline agarose gel. DNase-resistant bands of the expected molecular weight (8.9 kb for CMV-Abca4, -EGFP-8.9, and -CEP290 and 8.1 kb for -MYO7A) were observed (Figure 2A, lanes 1–10). The lower intensity of some of the bands corresponding to genomes following DNase digestion may reflect the presence of unpackaged genomes that are degraded by the DNase. This was also observed following DNase treatment of viral genomes whose size is 4.7 kb or less (Figure 2A, lanes 11 and 12), suggesting that it is independent of genome size. As further evidence that rAAV2/5 particles contain intact large genomes, we analyzed by Southern blot the genomic DNA from murine skeletal muscle administered with rAAV-CMV-Abca4 (9 × 1010 GC/mouse injected in the right gastrocnemius 4 weeks before the analysis). This was digested with either 1 (NcoI) or 2 (NcoI and NotI; Figure 2B, top panel) single-cutting restriction enzymes. Digestion with NcoI releases a band higher than 8 kb (Figure 2B, bottom panel, upper blots, lane 4) corresponding to episomal monomers or head-to-tail concatemers (Figure 2B, top panel). Digestion with NcoI and NotI released a 7.8-kb band (Figure 2B, bottom panel, upper blots, lane 2), corresponding to the sequence between the AAV2 ITRs (Figure 2B, top panel), while smaller genome fragments were not detected. Translation of a protein of the appropriate size provides additional indirect evidence of the integrity of the rAAV2/5 genomes. Western blot analysis of Cos cells transduced (1 × 105 GC/cell; Figure 2C, upper blot) with either rAAV2/5-CMV-Abca4 (lane 2) or -EGFP (lane 3) showed a band of 220 kDa recognized by the anti-ABCA4 antibody only in lane 2 or in the control wild-type retina (lane 1).
 Figure 2
Figure 2Genome integrity of rAAV2/5-CMV-Abca4, -MYO7A, -CEP290, and -EGFP-8.9. (A) Southern blot analysis of vector DNA isolated directly from rAAV large preparations (2.5 × 1010 GC/lane) and separated on alkaline agarose gels. Lane 1 contains a marker DNA fragment obtained by restriction digestion from the pAAV2.1-CMV-Abca4 plasmid; lane 2 contains the same DNA fragment as in lane 1 digested with DNaseI, as control of DNaseI activity; lanes 3 and 4: genomes isolated from rAAV2/5-CMV-Abca4; lanes 5 and 6: genomes isolated from rAAV2/5-CMV-MYO7A; lanes 7 and 8: genomes isolated from rAAV2/5-CMV-CEP290; lanes 9 and 10: genomes isolated from rAAV2/5-CMV-EGFP-8.9 (lanes 1–10 belong to the same gel but were noncontiguous); lanes 11 and 12: genomes isolated from rAAV2/5-CMV-EGFP (2.6 kb). Samples in lanes 3, 5, 7, 9, and 11 were treated with DNaseI. (B) Assessment of rAAV2/5-CMV-Abca4 genome length following in vivo delivery. Top panel: Schematic representation of the rAAV2/5-CMV-Abca4 genome with the 2 probes used for the Southern blot analysis. Bottom panel, upper blots: Southern blot analysis of genomic DNA from uninjected muscles (lanes 1 and 3) and an equivalent amount of genomic DNA from murine muscle injected with rAAV2/5-CMV-Abca4 (lanes 2 and 4) digested with NcoI and NotI (lanes 1 and 2) or NcoI alone (lanes 3 and 4). Lanes belong to the same gel but were noncontiguous. The arrows indicate the bands of the expected size. Lower blots: Southern blot analysis with a probe specific for the PDE6B gene used as loading control. Molecular weights are indicated on the left. (C) Western blot analysis with anti-ABCA4 (upper blot) or anti–α-tubulin (lower blot) antibodies of lysates from Cos cells transduced with rAAV2/5. Lane 1: retina from wild-type mouse; lane 2: samples transduced with rAAV2/5-CMV-Abca4; lane 3: samples transduced with rAAV2/5-CMV-EGFP. Anti–α-tubulin was used as loading control. The amounts (μg) of protein loaded are indicated under the respective lanes.
Efficient in vitro and in vivo transduction with rAAV2/5 vectors packaging large genes. The next step was to test whether transduction mediated by rAAV2/5 vectors packaging large genomes results in efficient expression in vitro and in vivo. Cos cells were infected with rAAV2/5-CMV-EGFP containing genomes of 4.1, 5.1, or 8.9 kb at various MOIs (Figure 3A). No significant differences in transduction efficiencies 72 hours after infection were observed among the various vectors packaging genomes of different lengths. Seeking additional evidence of the transduction efficiency of the AAV2/5 vectors packaging large genomes, we compared ABCA4 and MYO7A expression levels following AAV-mediated transduction of Abca4–/– retinas or retinal pigment epithelial (RPE) cells of Shaker1 mice (a Myo7a-null model; ref. 18) with those of wild-type murine retina or RPE cell cultures (Figure 3, B and C). rAAV2/5-CMV-Abca4– or -MYO7A–mediated transduction results in efficient expression in vivo or in vitro. Western blot analysis of Abca4–/– murine retinas transduced (1.2 × 109 GC/retina; Figure 3B, top panel) with either rAAV2/5-CMV-Abca4 (lane 2) or -EGFP (lane 3) showed a band only in lane 2. The level of ABCA4 expression in Abca4–/– retinas transduced with rAAV2/5-CMV-Abca4 was approximately 45% of that in wild-type retina (as assessed by densitometric analysis), which is consistent with the levels of transduction (40%–50%) of the whole murine retina following subretinal administration of AAV (19). Similarly, primary RPE cells from Shaker1 mice were transduced with either rAAV2/5-CMV-MYO7A or -EGFP (104 GC/cell; Figure 3C). Western blot analysis with anti-MYO7A antibodies showed expression of the 250-kDa MYO7A band only in RPE cells infected with rAAV2/5-CMV-MYO7A (lanes 1–4) but not with -EGFP (lane 5). rAAV2/5-CMV-MYO7A–mediated MYO7A expression persisted for at least 2 weeks after infection (the duration of the study) in these nondividing RPE cell cultures (lanes 3–4). At 7 days after infection, the level of MYO7A expression in Myo7a-null RPE cells transduced with rAAV2/5-CMV-MYO7A was approximately 70% of that in wild-type RPE cells (as assessed by densitometric analysis). This level of expression was slightly above that found in Myo7a+/– tissues (~50% of wild-type levels; D. Gibbs and D.S. Williams, unpublished data).
 Figure 3
Figure 3EGFP, ABCA4, and MYO7A expression following rAAV2/5 delivery. (A) Transduction efficiency (assessed as green forming units [GFU]) of rAAV2/5-CMV-EGFP with genome size of 4.1 (black bars), 5.1 (gray bars), and 8.9 kb (white bars) at various MOI (GC/cells) on Cos cells 72 hours after the infection. Results are shown as the average of 3 independent experiments ± SE. (B) Western blot analysis with anti-ABCA4 (top panel) and anti–α-tubulin (middle panel) antibodies and 8-azido-[α-32P]-ATP labeling of ABCA4 (bottom panel) of lysates from Abca4–/– retinas transduced with rAAV2/5. Lane 1: retina from wild-type mouse; lane 2: samples transduced with rAAV2/5-CMV-Abca4; lane 3: samples transduced with rAAV2/5-CMV-EGFP. Anti–α-tubulin was used as loading control. The amounts of protein (μg) loaded are indicated under the respective lanes. (C) Western blot analysis with anti-MYO7A (top panel) and anti–retinal G protein–coupled receptor (anti-RGR; bottom panel) antibodies of lysates from primary RPE cell cultures. The cells were from Myo7a-null mice and were transduced with rAAV2/5-CMV-MYO7A. Lane 1 and 2: 7 days after infection with rAAV2/5-CMV-MYO7A; lanes 3 and 4: 17 days after infection with rAAV2/5-CMV-MYO7A; lane 5: 17 days after infection with rAAV2/5-CMV-EGFP; lane 6: uninfected RPE lysates from wild-type C57BL/6 mice 7 days after plating. Anti-RGR antibody was used as loading control. Molecular weights are indicated on the left. The amounts of protein (μg) loaded are indicated below each lane.
Abca4 encodes a member of the ABC transporter family. It is expressed in photoreceptors and has been localized to the rim of outer segment discs (Rim protein) (20, 21). To test whether the recombinant ABCA4 protein produced following rAAV2/5-mediated |gene transfer is functional, its ability to bind ATP was evaluated. Photoaffinity labeling of Abca4–/– retinas (Figure 3B, bottom panel) transduced with either rAAV2/5-CMV-Abca4 (lane 2) or -EGFP (lane 3) showed a 220-kDa radioactive band containing 8-azido-[α-32P]-ATP only in lane 2 or in the control wild-type retina (lane 1). These results demonstrate that rAAV2/5 vectors express ABCA4 correctly in vitro and in vivo and that the recombinant ABCA4 properly binds ATP. In addition, we show proper and sustained MYO7A expression in Myo7a-null RPE cells infected with rAAV2/5 vectors.
rAAV2/5 administration in a mouse model of rSTGD significantly improves retinal morphology and function. Based on the results above, we tested the efficacy of AAV2/5-mediated retinal gene transfer in a murine model of rSTGD. Targeted disruption of the Abca4 locus in pigmented (22) and albino (23) mice (Abca4–/–) results in a phenotype that recapitulates some rSTGD characteristics: accumulation of lipofuscin in the RPE, thicker RPE cells, slow photoreceptor degeneration, and delayed dark adaptation (22–24). To test whether rAAV2/5-mediated gene delivery results in correction of the Abca4–/– mutant phenotype, 1-month-old mice were injected subretinally with 2 μl of rAAV2/5-CMV-Abca4 (corresponding to 1.2 × 109 GC) in one eye and with the same dose of rAAV2/5-CMV-EGFP in the contralateral eye. The impact of gene transfer on Abca4–/– retinas was evaluated 3 months later (age of the animals: 4 months) unless otherwise noted. We initially analyzed recombinant ABCA4 expression by immunohistochemistry on retinal sections and found that it properly localized to photoreceptor outer segments (Figure 4A) as the endogenous ABCA4 does and as expected according to the described rAAV2/5 tropism (25, 26).
 Figure 4
Figure 4Morphological analysis of Abca4–/– retinas following rAAV-mediated gene transfer. (A) Immunohistochemical analysis with anti-ABCA4 (Rim 3F4) antibody of retinal sections from 4-month-old Abca4+/+ mice and Abca4–/– pigmented mice injected subretinally at 1 month of age with rAAV2/5-CMV-EGFP and the contralateral eye with rAAV2/5-CMV-Abca4. OS, outer segment (photoreceptors); ONL, outer nuclear layer; INL, inner nuclear layer; GCL, ganglion cell layer. Original magnification, ×20. (B) Electron microscopic analysis of RPE from pigmented 5-month-old Abca4–/– mice. RPE from 1 eye injected subretinally at 1 month of age with rAAV2/5-CMV-EGFP (left) and the contralateral eye with rAAV2/5-CMV-Abca4 (right). Ch, choroid; BrM, Bruch’s membrane. White arrows indicate the irregularly shaped lipofuscin pigment granules to be distinguished from the larger oval melanosomes. Micrographs were obtained at the same magnification (×6,000). (C) Number of lipofuscin granules (left) and RPE thickness (right) in the RPE of Abca4+/+ or Abca4–/– mice injected subretinally with rAAV2/5-CMV-EGFP or rAAV2/5-CMV-Abca4 (n = 2 eyes/group).
We then evaluated the impact of rAAV2/5-mediated gene transfer on Abca4–/– RPE abnormalities such as presence of lipofuscin granules and thicker RPE (22, 24). Electron microscopic analysis of RPE cells located in the region of injection revealed a reduced number of lipofuscin granules and decreased RPE thickness (both similar to those seen in Abca4+/+ RPE) in the Abca4–/– retinas treated with rAAV2/5-CMV-Abca4 compared with those treated with rAAV2/5-CMV-EGFP (Figure 4, B and C). This suggests that rAAV2/5-mediated Abca4 gene transfer ameliorates the RPE ultrastructural abnormalities associated with the Abca4–/– phenotype.
Consistent with a role for ABCA4 in the transport of N-retinylidene-phosphatidylethanolamine across photoreceptor disk membranes (27, 28), the lipofuscin granules present in the RPE of Abca4–/– mice contain the bisretinoid fluorophores A2E, all-trans-retinal-dimer-ethanolamine (atRALdi-E), and all-trans-retinal-dimer-phosphatidylethanolamine (atRALdi-PE) (29). The levels of the fluorophores A2E, atRALdi-E, and atRALdi-PE were significantly reduced in both albino (age: 4 months) and pigmented (age: 6 months) Abca4–/– retinas treated with rAAV2/5-CMV-Abca4, when compared with the rAAV2/5-CMV-EGFP–treated contralateral eyes (Figure 5A). In addition, the ability of Abca4–/– photoreceptors to recover from light desensitization was significantly improved in the retinas treated with the therapeutic vector when compared with control rAAV2/5-CMV-EGFP–treated retinas (Figure 5B). H&E staining of retinal sections did not reveal any inflammatory infiltrate or a reduction in the outer nuclear layer thickness in either rAAV2/5-CMV-Abca4– or -EGFP–treated eyes (data not shown).
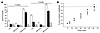 Figure 5
Figure 5Reduction of lipofuscin levels and improved recovery from photoreceptor desensitization in Abca4–/– mice injected with rAAV2/5-CMV-Abca4. (A) Effect of rAAV2/5-mediated Abca4 gene transfer on lipofuscin accumulation in the retina of Abca4–/– mice. A2E (combined A2E and iso-A2E), atRALdi-E, and atRALdi-PE levels in eyecups of 4- and 6-month-old albino and pigmented Abca4–/– mice, respectively, injected at postnatal day 30 in 1 eye with rAAV2/5-CMV-Abca4 (black bars) and in the contralateral eye with rAAV2/5-CMV-EGFP (white bars). Age-matched albino BALB/c and pigmented Abca4+/+ mice are represented in gray bars. Values are the average of 2 independent samples containing 4 eye cups each. (B) Rescue from delayed recovery from photoreceptor desensitization in Abca4–/– mice treated with rAAV2/5-CMV-Abca4. Progressive recovery after bleaching of the b-wave amplitude in 4-month-old Abca4–/– mice injected subretinally with either rAAV2/5-CMV-Abca4 (red triangles; n = 4 eyes) or rAAV2/5-CMV-EGFP (green squares; n = 4 eyes) and in age-matched wild-type BALB/c mice (black circles; n = 10 eyes). Data are shown as average ± SE. *P ≤ 0.05.
-
Discussion
According to current dogma, AAV can efficiently package cargo up to 4.7 kb. However, a previous report suggested that AAV2/1–5 are capable of packaging up to 6 kb (12). Here we show that rAAV vectors with AAV5 capsids are capable of packaging up to 8.9 kb of single-stranded DNA more efficiently than 6 other serotypes tested. Efficient packaging occurred whether the genome included the Abca4, MYO7A, CEP290, or EGFP-8.9 kb cDNA, suggesting that it is independent of the sequence packaged. We have also shown that this is the result of the intrinsic ability of the AAV5 capsids to package large genomes rather than higher vector yields of rAAV2/5 compared with those of other serotypes. These results open new perspectives for AAV vector development: testing of additional naturally occurring AAV serotypes or systematic mutagenesis of existing AAV capsid sequences may provide additional rAAV vectors with packaging ability similar to or better than that of rAAV2/5. Why AAV5 capsids have this capacity in contrast to the other AAV capsids tested remains to be elucidated. Amino acid sequence comparison allowed us to identify 2 amino acid stretches that vary between the AAV5 VP1 capsid protein and the VP1 from the other serotypes. We have exchanged these sequences, separately or together, between the AAV5 (GenBank accession number AAY00268, sequence 1: aa 140–184; sequence 2: aa 489–547) and AAV2 (GenBank accession number AA095537, aa 141–195; aa 482–558) capsids to test whether rAAV2/5 large genome packaging capacity can be transferred to other serotypes. We could not detect improvements in rAAV2/2-CMV-Abca4 titers despite production of capsids of the expected size as assessed by Western blot analysis (data not shown). This suggests that AAV5 capsid sequences 1 and 2 are not sufficient for large genome encapsidation. Notably, the amino acid sequence of the entire AAV5 capsid is significantly different from that of other AAV capsids identified so far (30). Therefore, many single amino acid residues other than the 2 stretches that we have tested are divergent between AAV5 and the other AAV serotypes. We are currently using a systematic approach to mutagenize AAV capsids and test whether single amino acid(s) play a role in packaging of large genomes in AAV.
We show that rAAV2/5-CMV-Abca4 vector contains an intact genome and that transduction of Cos cells as well as subretinal delivery of the vector in Abca4–/– mice result in production of ABCA4 of the appropriate molecular weight and biological activity as shown by the photoaffinity labeling assay. We obtained similar results following transduction of Shaker1 RPE cells with rAAV2/5-CMV-MYO7A. The possibility of including large genomes in a single rAAV vector that efficiently expresses a therapeutic gene is advantageous compared with strategies that are used to overcome limited rAAV cargo capacity, such as rAAV genome intermolecular recombination. In this latter case, a large open reading frame is split into 2 different rAAV vectors. Reconstitution of a single protein depends on efficient infection of the same cell by the 2 rAAVs, intermolecular rejoining of their genomes, and expression of a single protein either by homologous recombination through overlapping sequences (31) or by trans-splicing of a common reconstituted transcript (32).
Our results have important implications for gene therapy of human diseases due to mutations in large genes, such as cystic fibrosis, Duchenne muscular dystrophy, and hemophilia A. The efficacy of rAAV2/5-mediated gene transfer in animal models of these diseases remains to be assessed. Although modifications of rAAV vector production strategies may increase the yield of rAAV2/5 carrying large cargo, the yields we report here would certainly be sufficient to treat the diseases of sensorineural origin that we describe. An advantage of retinal (or cochlear; ref. 33) gene delivery is the target size of the tissue, which is orders of magnitude smaller than that of other organs, such as liver or lung. We and others have previously demonstrated that vectors with AAV5 capsids efficiently transduce RPE and photoreceptors of several species (25, 26, 34, 35) as well as murine cochlear inner hair cells (33), the appropriate targets for gene therapy of rSTGD and USH. The possibility of efficiently packaging MYO7A or CEP290 in a single rAAV2/5 vector opens new therapeutic perspectives for USH1B, the most frequent cause of hereditary deafness and blindness in humans (14), and LCA, the most common inherited cause of blindness in infancy. While formal proof of principle of the efficacy of rAAV2/5-mediated MYO7A or CEP290 gene transfer in Shaker1 (18) or rd16 (36) mice, respectively, remains to be provided, here we show that subretinal delivery of rAAV2/5-CMV-Abca4 in a murine model of STGD results in significant correction of lipofuscin levels, RPE abnormalities, and retinal function. The treatment was stable at least through 6 months, the longest time period after which treatment effects were evaluated in vivo. This is the first evidence to our knowledge of rescue in an animal model of rSTGD, thus making rAAV2/5 an attractive candidate for gene therapy of this disease as well as of others associated with mutations in ABCA4, such as cone-rod dystrophy (37) and retinitis pigmentosa (38, 39). Importantly, heterozygous ABCA4 mutations in humans (40) have been associated with age-related macular degeneration (AMD), the most common blinding disease in the elderly (41).
In conclusion, our discovery greatly expands the therapeutic potential of rAAV as a gene therapy vector for diseases due to mutations in large genes. In addition it provides the first proof of principle of gene therapy for the currently untreatable and common blinding macular degeneration found in Stargardt disease. This comes at a very exciting time in the promising field of rAAV-mediated retinal gene transfer, where the first evaluations of rAAV-mediated retinal gene transfer efficacy and safety in humans are being performed (3).
-
Methods
Generation of the plasmid constructs. For the production of rAAV encoding EGFP and ABCA4, the pAAV2.1-CMV-EGFP (17) and pZac2.1-CMV-Abca4 plasmids were used (CMV sequence from NC001347.3 nt 174,661 to 175,243). The pZac2.1-CMV-Abca4 was obtained by cloning the murine Abca4 cDNA (7,268 bp, including the coding sequence as well as some 5ι and 3ι UTR region) between the EcoRI and SalI sites in the pZac2.1 plasmid (42). The Abca4 cDNA was obtained from the pBluescript SK(-)Abca4 plasmid (a kind gift from G.H. Travis, University of Texas, Dallas, Texas, USA; ref. 21) by digestion with EcoRI and XhoI enzymes.
For the production of rAAV encoding MYO7A, the pAAV2.1-CMV-MYO7A was constructed as follows. Human MYO7A cDNA (6,648 bp, consisting of the MYO7A coding sequence without UTR regions) was cloned in the pAAV2.1-CMV-EGFP plasmid between the NotI and SacII sites (complete rAAV genome size: 8,107 bp). For this purpose, the MYO7A coding sequence (GenBank accession number NM_000260) was divided into 3 fragments of 2,853 bp, 2,275 bp, and 1,890 bp, respectively and amplified by PCR from cDNA of human retina (BD Biosciences) with the following oligonucleotides: F1 (NotI): ATTTGCGGCCGCATGGTGATTCTTCAGCAGGGG; R1: CCCCAGGAAGCCAAACATCT; F2: AGGGCTGAGTATCTGTGG; R2: CGGGGTTGGGGTTATCCT; F3: GCTGAGGACATTCGTGAC; R3(SacII): TCCCCGCGGTCACTTGCCGCTCCTGGAG (underlining indicates the restriction site sequences inserted). Then, the 3 fragments, designated F1, F2, and F3, were separately cloned in pZero Blunt Vector (Invitrogen), sequenced, and then cloned via triple ligation reaction in pAAV2.1-CMV-EGFP. For the production of rAAV encoding CEP290, the pAAV2.1-CMV-CEP290 plasmid was produced as follows. The human CEP290 cDNA (7,440 bp, consisting of the CEP290 coding sequence; GenBank accession number NM025114) was PCR amplified from a human osteosarcoma cell line (U2OS) cDNA and then cloned in the pAAV2.1-CMV-EGFP plasmid between the NotI and SacII sites (complete rAAV genome size, 8,900 bp). To determine the upper limit of genome size packaged by rAAV, the pZac2.1-CMV-Abca4+1,300 and the pZac2.1-CMV-Abca4+2,522 plasmid were produced. Stuffer regions of 1,300 and 2,522 bp were designed and cloned (NheI-EcoRI) in the expression cassette between the AAV2 ITRs of pZac2.1-CMV-Abca4. The total vector genome sizes in the pZac2.1-CMV-Abca4, pZac2.1-CMV-Abca4+1,300, and pZac2.1-CMV-Abca4+2,522 plasmids are 8,916 bp, 10,000 bp and 11,400 bp, respectively.
In order to systematically characterize the packaging capacity and transduction efficacy of rAAV vectors with increasing genome size, 5 different constructs encoding for EGFP were generated. Five stuffers of different sizes (1,400 bp, 2,400 bp, 3,400 bp, 4,400 bp, and 6,100 bp) were inserted in the pAAV2.1-CMV-EGFP plasmid downstream of the poly(A) sequence. The stuffer sequences were PCR amplified from the same DNA sequence (Abca4) with XhoI cloning sites at both ends and cloned in pAAV2.1-CMV-EGFP. Therefore, the following plasmids were generated: pAAV2.1-CMV-EGFP-4.1, pAAV2.1-CMV-EGFP-5.1, pAAV2.1-CMV-EGFP-6.1, pAAV2.1-CMV-EGFP-7.1, and pAAV2.1-CMV-EGFP-8.9, where the last number indicates the length of the genome including the ITRs.
rAAV vector production. Large preparations of rAAV vectors were produced by the TIGEM AAV Vector Core using the pAAV2.1-CMV-EGFP, pZac2.1-CMV-Abca4, pZac2.1-CMV-Abca4+1,300, pZac2.1-CMV-Abca4+2,522, pAAV2.1-CMV-CEP290, pAAV-CMV-MYO7A, pAAV2.1-CMV-EGFP-4.1, pAAV2.1-CMV-EGFP-5.1, and pAAV2.1-CMV-EGFP-8.9 plasmids. In addition, 101 large preparations of rAAV with genome size ranging from 2.7 to 4.6 kb were produced by the TIGEM Vector Core and their titers included in Figure 1B, Supplemental Table 1, and Supplemental Figure 1. These rAAVs contain 70 different expression cassettes with various transgenes. rAAV2/1, 2, 3, 4, 5, 7, 8, and 9 viruses were produced by triple transfection of 293 cells followed by 2 rounds of CsCl2 purification (25). For each viral preparation, physical titers (GC/ml) were determined by dot blot analysis (43) by the TIGEM AAV Vector Core and by PCR quantification using TaqMan (Applied Biosystems) using 2 different probes (42) by the University of Pennsylvania Vector Core. For each large preparation, the titer was averaged from the dot blot and the TaqMan PCR quantification analyses. For Figure 1D, rAAV2/5 and rAAV2/2 vectors were produced in small scale (single 15-cm dish) in 293 cells triple transfected and lysed by freeze-and-thaw in a volume that was 2% of the final volume of a large preparation (made from 50 dishes). Titers were determined by PCR quantification using TaqMan following DNase and RNase treatment of crude lysates.
Animal models and vector administration. All procedures on animals were performed under a protocol approved by the Hospital Cardarelli Ethics Committee, Naples, Italy, and by the Italian Ministry of Health. Pigmented (22) and albino Abca4–/– (23) mice generated through successive crosses and backcrosses with BALB/c mice (homozygous for Rpe65 Leu450; ref. 44), Shaker 1 mice (carrying the 4626SB allele, an effective null mutation on a C57BL/6HNSD background; ref. 17), and wild-type C57BL/6 and BALB/c mice (Harlan Italy) were used. Either subretinal or intramuscular injections were performed. Subretinal vector administration was performed in 1-month-old Abca4–/– mice as described previously (45). Subretinal administration and intramuscular injections were supplemented with 40 μM of proteasome inhibitors (LnLL; Sigma-Aldrich) to increase rAAV transduction for the experiments depicted in Figure 2B and Figure 3B (12). For the in vivo experiments aimed at assessing AAV-mediated morphological and functional rescue (Figures 4 and 5), proteasome inhibitors were not used. Before vector administration, mice were anesthetized with an intraperitoneal injection of avertin at 2 ml/100 g body weight (46). Then, mice were injected with 2 μl of rAAV2/5-CMV-Abca4 (1.2 × 109 GC) in the right eye. The same dose of rAAV2/5-CMV-EGFP was delivered to the left eye, as negative control. Intramuscular injections were performed in the right gastrocnemius of C57BL/6 mice with 150 μl of rAAV2/5-CMV-Abca4 (9 × 1010 GC).
Statistics. Data are presented as average ± SE. Two-tailed Student’s t test, ANOVA, and a multiple comparison test with a Bonferroni adjustment for multiplicity were used to determine statistical significance where indicated. P ≤ 0.05 was considered significant.
Southern blot analyses of rAAV vector DNA. DNA was extracted from 2.5 × 1010 viral particles (measured as GC). To digest unpackaged genomes, the vector solution was incubated with 11 μl of DNase (Roche) in a total volume of 250 μl, containing 50 mM Tris, pH 7.5, and 1 mM MgCl2 for 1 hour at 37°C. The DNase was then inactivated with 50 mM EDTA, followed by incubation at 50°C for 45 minutes with proteinase K and 2.5% N-lauryl-sarcosil solution to lyse the capsids. The DNA was extracted twice with phenol-chloroform and precipitated with 2 volumes of ethanol and 10% sodium acetate (3 M). Alkaline agarose gel electrophoresis was performed as previously described (47). Markers were produced by double digestion of the pZac2.1-CMV-Abca4 with NcoI and NotI, to produce a band of 7,835 bp. Probe 2 was used to identify rAAV2/5-CMV-Abca4 (Figure 2B, top panel), while to identify all the other rAAV vector DNA, a probe specific for the poly(A) sequence was used. All probe sequences are provided in the supplemental material.
Southern blot analysis of muscle genomic DNA following transduction with rAAV. DNA was isolated from mouse gastrocnemius 21 days after intramuscular injections by the Hirt extraction method (26, 48). The DNA (30 μg) was digested with NcoI and NotI or NcoI alone, separated on a 0.8% agarose gel, and detected with probes 1 and 2 (Figure 2B, top panel) or with a probe specific for the PDE6B gene (used as loading control) radiolabeled using the Rediprime II Random prime labeling system (Amersham) and [α-32]-CTP according to the manufacturer’s instructions.
rAAV infection of Cos and Shaker1 primary RPE cells. For the experiments depicted in Figure 3A, Cos cells were plated in MW-96 (7 × 104 cell/well) and infected with 102, 103, 104, or 105 GC/cell of rAAV2/5-CMV-EGFP-4.1, -5.1, -8.9. The medium was supplemented with 10 μM proteasome inhibitors. After 72 hours, the number of cells expressing EGFP (green forming units [GFU]) was determined.
Cos cells were plated in 6-well plates to a concentration of 3 × 105 cells/well. Forty-four hours later, the cells were incubated with 105 GC/cell of rAAV2/5-CMV-EGFP or rAAV2/5-CMV-Abca4 in serum-free DMEM with 10 μM proteasome inhibitors. Forty-eight hours later, the cells were harvested by scraping for Western blot analyses (see below).
Primary mouse RPE cells were isolated from 12-day-old Shaker1 mice and cultured on 24-well Transwell polycarbonate membrane inserts (Fisher Scientific), as described previously (49). The primary RPE cells were infected 48 hours later with either rAAV2/5-CMV-MYO7A or rAAV2/5-CMV-EGFP (104 GC/cell) as described above. Infected RPE cells were maintained in culture until MYO7A expression was assayed by Western blot at either 7 or 17 days after infection.
Analysis of ABCA4 and MYO7A expression by Western blot. Western blot analysis was performed on retinas and on Cos and Shaker 1 primary RPE cells infected with rAAV. Retinas were harvested as described previously (50). Samples were lysed in SIE buffer (250 mM sucrose, 3 mM imidazoles [pH 7.4], 1% ethanol, and 1% NP-40) on ice for 30 minutes, and proteins were denatured by heating at 37°C for 30 minutes in sample buffer with 8 M urea and separated by 6% SDS-PAGE. After blotting, specific proteins were labeled using anti-ABCA4 (Santa Cruz Biotechnology Inc.), anti–α tubulin (Sigma-Aldrich), anti-MYO7A (pAb2.2), and anti–retinal G protein–coupled receptor (RGR; mcDE5; a kind gift from Henry K.W. Fong, University of Southern California, Los Angeles, California, USA; RGR was used as loading control) antibodies.
Photoaffinity labeling assay on infected Cos cells and retinas. Protein extraction from Cos membranes was performed 48 hours after infection with rAAV. Cells were harvested in hypotonic buffer (10 mM Tris-HCl [pH 7.4] and 0.5 mM EDTA). After 1 hour at 4°C, the samples were passed through a 28-G needle to disrupt the cells and centrifuged for 1 hour at 16,000 g. The resulting membrane pellet was dissolved in the resuspension buffer (25 mM HEPES [pH 7.5], 150 mM NaCl, and 5 mM MgCl2).
Proteins were extracted from rod outer segments by vortexing retinas in 100 μl of 45% sucrose, 20 mM Tris-HCl (pH 7.4), 1 mM EDTA, 2 mM MgCl2, 20 μM leupeptin, and 2 mM PMSF (22). Then, retinas were centrifuged for 10 minutes at 4,000 g, and the supernatants were collected, diluted with an equal volume of 150 mM NaCl, 20 mM Tris-HCl (pH 7.4), 1 mM EDTA, and 2 mM MgCl2, and recentrifuged for 1 hour at 16,000 g. The outer segment pellet was dissolved in 30 μl of resuspension buffer.
For photoaffinity labeling assays, protein extracts from Cos membranes or rod outer segments were incubated at room temperature with 4 μM 8-azido-[α-32P]-ATP (Affinity Labeling Technologies Inc.) for 1 minute under ultraviolet light (320 nm) at a distance of 10 cm (51). Samples were then mixed with SDS-PAGE sample buffer without heating, and the proteins were resolved by SDS-PAGE. 8-Azido-[α-32P]-ATP–labeled proteins were detected with a PhosphorImager (Amersham) by autoradiography.
Extraction and HPLC analysis of RPE lipofuscin pigments. HPLC analysis was performed on eyecups from 4-month-old albino and 6-month-old pigmented Abca4–/– mice injected with rAAV2/5-CMV-Abca4 in one eye and rAAV2/5-CMV-EGFP in the contralateral eye. Eyecups from age-matched Abca4+/+ and BALB/c mice were used as control. Posterior eyecups of dark adapted mice were pooled (4 eyecups per sample), homogenized, and extracted 3 times in chloroform/methanol (1:1) (44). After centrifugation (1,000 g for 2 minutes), the organic extract was filtered through cotton and a reversed-phase (C8 Sep-Pak; Millipore) cartridge with 0.1% TFA in methanol. The extract was subsequently concentrated by evaporation of solvent under argon gas, redissolved in 50% methanolic chloroform (1 or 2 eyes/10 μl solvent), and analyzed by reverse-phase HPLC using an Alliance System (Waters) equipped with 2695 Separation Module, 2996 Photodiode Array Detector, and a 2475 Multi λ Fluorescence Detector. For chromatographic separation, an analytical scale Atlantis dC18 (3 μm, 4.6 × 150 mm; Waters) column was utilized with an acetonitrile and water gradient and 0.1% trifluoroacetic acid (90%–100%, 0–10 minutes; 100% acetonitrile, 10–20 minutes; monitoring at 430 nm; 10 μl injection volume). Extraction and injection for HPLC were performed under dim red light. Integrated peak areas were determined using Waters Empower software, and picomolar concentrations per eyecup were calculated by reference to an external standard of synthesized compound and by normalizing to the ratio of the HPLC injection volume to total extract volume. The structures of synthesized standards of A2E, atRALdi-E, and atRALdi-PE have been confirmed (29, 52–54).
Electrophysiological recordings. Electroretinographical (ERG) analysis was performed in 4-month-old albino Abca4–/– and wild-type, age-matched BALB/c mice. Flash ERG was evoked by 10-ms flashes of light generated through a Ganzfeld stimulator (Lace). The electrophysiological signals were recorded through gold-plated electrodes inserted under the lower eyelids in contact with the cornea previously anesthetized with oxybuprocaine (Novartis Pharma). The electrode in each eye was referenced to a needle electrode inserted subcutaneously at the level of corresponding frontal region. The different electrodes were connected to a 2-channel amplifier. After 180 minutes of dark adaptation, mice were anesthetized and loosely mounted in a stereotaxic apparatus under dim red light with body temperature maintained at 37.5°C. Mice were then exposed to a constant light, the intensity of which was set at 300 candelas (cd)/m2 for 80 seconds (preadapting light, bleaching condition). Recovery of b-wave was monitored at fixed intervals after preadapting light (0, 5, 15, 30, 45, 60 minutes). The amplitude of b-wave in response to a flash of 1 cd/m2/s1 after the preadapting light was measured and expressed as a relative value with respect to that measured before the preadapting light.
Electron microscopic and histological analyses and immunohistochemistry. Mice were perfused through the heart with 2% paraformaldehyde and 1% glutaraldehyde in PBS (pH 7.4). Then the eyeballs were removed and fixed overnight in 0.1 M sodium cacodylate buffer (pH 7.4) containing 2% paraformaldehyde and 2% glutaraldehyde. The fixed eyeballs were cut so that the lens and vitreous could be removed while leaving the eyecup. The eyecups were treated with 1% osmium tetroxide and stained with 1% aqueous uranyl acetate. The specimens were then dehydrated and embedded in EPON-812. Thin sections from the temporal side of each eye, which corresponded to the injected side, were prepared on an Ultracut microtome (Leica Microsystems). Electron microscopic images were acquired from thin sections under a FEI Philips Tecnai-12 electron microscope (Philips) using an ULTRA VIEW CCD digital camera. Micrographs were obtained at ×6,000 magnification. Quantitative analysis of numbers of lipofuscin granules was made by counting in 3 different optical fields for each eye the smaller structures of variable density representing lipofuscin granules distinct from the large oval structures of high electron density representing melanosomes. RPE thickness measurements were done in 20 different places per specimen (10 measurements across the nuclear area, where the cell is thicker, and 10 across the cell-to-cell border, where the cell is thinner). Then, the counts were averaged.
For histological analysis, mouse eyecups were harvested, fixed by immersion in 4% paraformaldehyde, and embedded in OCT (kaltek). For each eye, serial sections (11 μm thick) were cut along the horizontal meridian and distributed on 10 slides so that each slide contained representative sections of the whole eye at different levels. The sections were stained with H&E (Sigma-Aldrich), and retinal histology was analyzed by light microscopy. For the ABCA4 staining, the tissue sections were incubated for 1 hour with blocking solution (1× PBS, 0.5% Tween-20, 0.1% BSA) and 10% FBS (GIBCO BRL; Invitrogen) before incubation overnight with the Rim 3F4 antibody (a kind gift from Robert S. Molday, University of British Columbia, Vancouver, British Columbia, Canada). After washing, sections were incubated for 1 hour with secondary anti-mouse IgG conjugated to HRP (Vector Laboratories) followed by 30 minutes DAB staining (Vector Laboratories). The counterstaining was performed for 1 minute with hematoxylin (Sigma-Aldrich). Stained sections were mounted with Eukitt (kaltek).
-
Acknowledgments
We thank Graciana Diez Roux (TIGEM) and Andrea Ballabio (TIGEM) for the critical reading of the manuscript; Diego di Bernardo (TIGEM) and Maria Lanzillo (TIGEM) for help with statistical analysis and subcloning, respectively; the TIGEM AAV Vector Core and the Telethon Electron Microscopy Core. Gabriel Travis (UCLA School of Medicine) generously provided the pigmented Abca4–/– mice and the pBluescript SK(-)Abca4 plasmid, and Robert S. Molday (University of British Columbia) the Rim 3F4 antibody. M. Allocca is the recipient of a fellowship from the European School of Molecular Medicine. This work was supported by Telethon grant TIGEM P21; Italian Ministry of Agriculture grant D.M.589/7303/04; EC-FP6 projects LSHB-CT-2005-512146 “DiMI” and 018933 “Clinigene”; NIH grants EY015136, EY10820, EY07042, and EY12951; Foundation Fighting Blindness; Research to Prevent Blindness; the Paul and Evanina Bell Mackall Foundation Trust; the F.M. Kirby Foundation; the Ruth and Milton Steinbach Fund; and the Kaplen Foundation.
Address correspondence to: Alberto Auricchio, Telethon Institute of Genetics and Medicine, Via P. Castellino 111, 80131 Naples, Italy. Phone: 11-39-081-6132229; Fax: 11-39-081-5790919; E-mail: [email protected].
-
Footnotes
Nonstandard abbreviations used: AAV, adeno-associated virus; atRALdi-E, all-trans-retinal-dimer-ethanolamine; atRALdi-PE, all-trans-retinal-dimer-phosphatidylethanolamine; GC, genome copies; ITR, inverted terminal repeat; LCA, Leber congenital amaurosis; rAAV, recombinant AAV; RPE, retinal pigment epithelium; rSTGD, recessive Stargardt disease; USH, Usher syndrome.
Conflict of interest: The authors have declared that no conflict of interest exists.
Reference information: J. Clin. Invest.118:1955–1964 (2008). doi:10.1172/JCI34316
-
References
- Warrington (Jr.), K.H., Herzog, R.W. 2006. Treatment of human disease by adeno-associated viral gene transfer. Hum. Genet. 119:571-603.
- Carter, B.J. 2005. Adeno-associated virus vectors in clinical trials. Hum. Gene Ther. 16:541-550.
- Bennett, J. 2006. Commentary: an aye for eye gene therapy. Hum. Gene Ther. 17:177-179.
- Kaplitt, M.G., et al. 2007. Safety and tolerability of gene therapy with an adeno-associated virus (AAV) borne GAD gene for Parkinson’s disease: an open label, phase I trial. Lancet. 369:2097-2105.
- Muzyczka, N., and Berns, K.I. 2001. Parvoviridae: the viruses and their replication. Lippincott Williams & Wilkins. Philadelphia, Pennsylvania, USA. 1089–1122.
- Gao, G., Vandenberghe, L.H., Wilson, J.M. 2005. New recombinant serotypes of AAV vectors. Curr. Gene Ther. 5:285-297.
- Bennett, J., Maguire, A.M. 2000. Gene therapy for ocular disease. Mol. Ther. 1:501-505.
- Dinculescu, A., Glushakova, L., Min, S.H., Hauswirth, W.W. 2005. Adeno-associated virus-vectored gene therapy for retinal disease. Hum. Gene Ther. 16:649-663.
- Bainbridge, J.W., Tan, M.H., Ali, R.R. 2006. Gene therapy progress and prospects: the eye. Gene Ther. 13:1191-1197.
- Dong, J.Y., Fan, P.D., Frizzell, R.A. 1996. Quantitative analysis of the packaging capacity of recombinant adeno-associated virus. Hum Gene Ther 7:2101-2112.
- Hermonat, P.L., Quirk, J.G., Bishop, B.M., Han, L. 1997. The packaging capacity of adeno-associated virus (AAV) and the potential for wild-type-plus AAV gene therapy vectors. FEBS Lett. 407:78-84.
- Grieger, J.C., Samulski, R.J. 2005. Packaging capacity of adeno-associated virus serotypes: impact of larger genomes on infectivity and postentry steps. J. Virol. 79:9933-9944.
- Deutman, A.F., and Hyong, C.B. 2001. Macular dystrophies. InRetina . S.J. Ryan, T.E. Ogden, D.R. Hinton, and A.P. Schachat, editors. Mosby. St. Louis, Missouri, USA. 1210–1257.
- Keats, B.J., Corey, D.P. 1999. The usher syndromes. Am. J. Med. Genet. 89:158-166.
- Kaplan, J., Rozet, J.M., Perrault, I., and Munnich, A. 2000. Leber congenital amaurosis. InThe metabolic and molecular bases of inherited disease. 8th edition. C.R. Scriver, A.L. Beaudet, W. Sly, and D.M. Valle, editors. McGraw-Hill. New York, New York, USA. 5947–5945.
- Allikmets, R., et al. 1997. A photoreceptor cell-specific ATP-binding transporter gene (ABCR) is mutated in recessive Stargardt macular dystrophy. Nat. Genet. 15:236-246.
- Auricchio, A., Hildinger, M., O’Connor, E., Gao, G.P., Wilson, J.M. 2001. Isolation of highly infectious and pure adeno-associated virus type 2 vectors with a single-step gravity-flow column. Hum. Gene Ther. 12:71-76.
- Gibson, F., et al. 1995. A type VII myosin encoded by the mouse deafness gene shaker-1. Nature. 374:62-64.
- Allocca, M., et al. 2007. Novel adeno-associated virus serotypes efficiently transduce murine photoreceptors. J. Virol. 81:11372-11380.
- Illing, M., Molday, L.L., Molday, R.S. 1997. The 220-kDa rim protein of retinal rod outer segments is a member of the ABC transporter superfamily. J. Biol. Chem. 272:10303-10310.
- Azarian, S.M., Travis, G.H. 1997. The photoreceptor rim protein is an ABC transporter encoded by the gene for recessive Stargardt’s disease (ABCR). FEBS Lett 409:247-252.
- Weng, J., et al. 1999. Insights into the function of Rim protein in photoreceptors and etiology of Stargardt’s disease from the phenotype in abcr knockout mice. Cell 98:13-23.
- Radu, R.A., Mata, N.L., Bagla, A., Travis, G.H. 2004. Light exposure stimulates formation of A2E oxiranes in a mouse model of Stargardt’s macular degeneration. Proc. Natl. Acad. Sci. U. S. A. 101:5928-5933.
- Mata, N.L., et al. 2001. Delayed dark-adaptation and lipofuscin accumulation in abcr+/– mice: implications for involvement of ABCR in age-related macular degeneration. Invest. Ophthalmol. Vis. Sci. 42:1685-1690. View this article via: PubMed Google Scholar
- Auricchio, A., et al. 2001. Exchange of surface proteins impacts on viral vector cellular specificity and transduction characteristics: the retina as a model. Hum. Mol. Genet. 10:3075-3081.
- Yang, G.S., et al. 2002. Virus-mediated transduction of murine retina with adeno-associated virus: effects of viral capsid and genome size. J. Virol. 76:7651-7660.
- Sun, H., Molday, R.S., Nathans, J. 1999. Retinal stimulates ATP hydrolysis by purified and reconstituted ABCR, the photoreceptor-specific ATP-binding cassette transporter responsible for Stargardt disease. J. Biol. Chem. 274:8269-8281.
- Beharry, S., Zhong, M., Molday, R.S. 2004. N-retinylidene-phosphatidylethanolamine is the preferred retinoid substrate for the photoreceptor-specific ABC transporter ABCA4 (ABCR). J. Biol. Chem. 279:53972-53979.
- Fishkin, N.E., Sparrow, J.R., Allikmets, R., Nakanishi, K. 2005. Isolation and characterization of a retinal pigment epithelial cell fluorophore: an all-trans-retinal dimer conjugate. Proc. Natl. Acad. Sci. U. S. A. 102:7091-7096.
- Gao, G., et al. 2004. Clades of Adeno-associated viruses are widely disseminated in human tissues. J. Virol. 78:6381-6388.
- Ghosh, A., Yue, Y., Duan, D. 2006. Viral serotype and the transgene sequence influence overlapping adeno-associated viral (AAV) vector-mediated gene transfer in skeletal muscle. J. Gene Med. 8:298-305.
- Reich, S.J., et al. 2003. Efficient trans-splicing in the retina expands the utility of adeno-associated virus as a vector for gene therapy. Hum. Gene Ther. 14:37-44.
- Liu, Y., et al. 2005. Specific and efficient transduction of cochlear inner hair cells with recombinant adeno-associated virus type 3 vector. Mol. Ther. 12:725-733.
- Rabinowitz, J.E., et al. 2002. Cross-packaging of a single adeno-associated virus (AAV) type 2 vector genome into multiple AAV serotypes enables transduction with broad specificity. J. Virol. 76:791-801.
- Lotery, A.J., et al. 2003. Adeno-associated virus type 5: transduction efficiency and cell-type specificity in the primate retina. Hum. Gene Ther. 14:1663-1671.
- Chang, B., et al. 2006. In-frame deletion in a novel centrosomal/ciliary protein CEP290/NPHP6 perturbs its interaction with RPGR and results in early-onset retinal degeneration in the rd16 mouse. Hum. Mol. Genet. 15:1847-1857.
- Maugeri, A., et al. 2000. Mutations in the ABCA4 (ABCR) gene are the major cause of autosomal recessive cone-rod dystrophy. Am. J. Hum. Genet. 67:960-966.
- Cremers, F.P., et al. 1998. Autosomal recessive retinitis pigmentosa and cone-rod dystrophy caused by splice site mutations in the Stargardt’s disease gene ABCR. Hum. Mol. Genet. 7:355-362.
- Martinez-Mir, A., et al. 1998. Retinitis pigmentosa caused by a homozygous mutation in the Stargardt disease gene ABCR. Nat. Genet. 18:11-12.
- Allikmets, R., et al. 1997. Mutation of the Stargardt disease gene (ABCR) in age-related macular degeneration. Science. 277:1805-1807.
- Seddon, J.M. 2001. Epidemiology of age-related macular degeneration. In Retina. S.J. Ryan, T.E. Ogden, D.R. Hinton, and A.P. Schachat, editors. Mosby. St. Louis, Missouri, USA. 1039–1050.
- Gao, G., et al. 2000. Purification of recombinant adeno-associated virus vectors by column chromatography and its performance in vivo. Hum. Gene Ther. 11:2079-2091.
- Drittanti, L., Rivet, C., Manceau, P., Danos, O., Vega, M. 2000. High throughput production, screening and analysis of adeno-associated viral vectors. Gene Ther. 7:924-929.
- Kim, S.R., et al. 2004. Rpe65 Leu450Met variant is associated with reduced levels of the retinal pigment epithelium lipofuscin fluorophores A2E and iso-A2E. Proc. Natl. Acad. Sci. U. S. A. 101:11668-11672.
- Liang, F.Q., Anand, V., Maguire, A., Bennett, J. 2001. Intraocular delivery of recombinant virus. Methods Mol. Med. 47:125-139.
- Papaioannou, V.E., Fox, J.G. 1993. Efficacy of tribromoethanol anesthesia in mice. Lab. Anim. Sci. 43:189-192. View this article via: PubMed Google Scholar
- Sambrook, J., and Russell, D.W. 2001. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press. Cold Spring Harbor, New York, USA. 999 pp.
- Hirt, B. 1967. Selective extraction of polyoma DNA from infected mouse cell cultures. J. Mol. Biol. 26:365-369.
- Gibbs, D., Kitamoto, J., Williams, D.S. 2003. Abnormal phagocytosis by retinal pigmented epithelium that lacks myosin VIIa, the Usher syndrome 1B protein. Proc. Natl. Acad. Sci. U. S. A. 100:6481-6486.
- Auricchio, A., et al. 2002. Pharmacological regulation of protein expression from adeno-associated viral vectors in the eye. Mol. Ther. 6:238. View this article via: PubMed Google Scholar
- Sun, H., Smallwood, P.M., Nathans, J. 2000. Biochemical defects in ABCR protein variants associated with human retinopathies. Nat. Genet. 26:242-246.
- Sakai, N., Decatur, J., Nakanishi, K., Eldred, G.E. 1996. Ocular age pigment “A2E”: an unprecedented pyridinium bisretinoid. J. Am. Chem. Soc. 118:1559-1560. View this article via: CrossRef Google Scholar
- Parish, C.A., Hashimoto, M., Nakanishi, K., Dillon, J., Sparrow, J. 1998. Isolation and one-step preparation of A2E and iso-A2E, fluorophores from human retinal pigment epithelium. Proc. Natl. Acad. Sci. U. S. A. 95:14609-14613.
- Fishkin, N., Pescitelli, G., Sparrow, J.R., Nakanishi, K., Berova, N. 2004. Absolute configurational determination of an all-trans-retinal dimer isolated from photoreceptor outer segments. Chirality. 16:637-641.
-
Version history
- Version 1 (April 15, 2008): No description
- Version 2 (May 1, 2008): No description



Copyright © 2024 American Society for Clinical Investigation
ISSN: 0021-9738 (print), 1558-8238 (online)