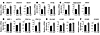Advertisement
Research Article Free access | 10.1172/JCI34090
FSP27 contributes to efficient energy storage in murine white adipocytes by promoting the formation of unilocular lipid droplets
Naonobu Nishino,1 Yoshikazu Tamori,1 Sanshiro Tateya,1 Takayuki Kawaguchi,1 Tetsuro Shibakusa,2 Wataru Mizunoya,2 Kazuo Inoue,2 Riko Kitazawa,3 Sohei Kitazawa,3 Yasushi Matsuki,4 Ryuji Hiramatsu,4 Satoru Masubuchi,5 Asako Omachi,6 Kazuhiro Kimura,6 Masayuki Saito,7 Taku Amo,8 Shigeo Ohta,8 Tomohiro Yamaguchi,9 Takashi Osumi,9 Jinglei Cheng,10 Toyoshi Fujimoto,10 Harumi Nakao,11 Kazuki Nakao,12 Atsu Aiba,11 Hitoshi Okamura,5 Tohru Fushiki,2 and Masato Kasuga1
1Department of Internal Medicine, Kobe University Graduate School of Medicine, Kobe, Japan. 2Laboratory of Nutrition Chemistry, Division of Food Science and Biotechnology, Graduate School of Agriculture, Kyoto University, Kyoto, Japan. 3Department of Biomedical Informatics, Kobe University Graduate School of Medicine, Kobe, Japan. 4Drug Research Division, Dainippon Sumitomo Pharma Co. Ltd., Takarazuka, Japan. 5Department of Brain Sciences, Kobe University Graduate School of Medicine, Kobe, Japan. 6Department of Biomedical Sciences, Graduate School of Veterinary Medicine, Hokkaido University, Sapporo, Japan. 7Department of Nutrition, School of Nursing and Nutrition, Tenshi College, Sapporo, Japan. 8Department of Biochemistry and Cell Biology, Graduate School of Medicine, Nippon Medical School, Kawasaki, Japan. 9Graduate School of Life Science, University of Hyogo, Kamigori, Japan. 10Department of Anatomy and Molecular Cell Biology, Graduate School of Medicine, Nagoya University, Nagoya, Japan. 11Department of Physiology and Cell Biology, Kobe University Graduate School of Medicine, Kobe, Japan. 12Laboratory for Animal Resources and Genetic Engineering Team, RIKEN Center for Developmental Biology (CDB), Kobe, Japan.
Address correspondence to: Masato Kasuga, Department of Internal Medicine, Division of Diabetes, Metabolism, and Endocrinology, Kobe University Graduate School of Medicine, 7-5-1 Kusunoki-cho, Chuo-ku, Kobe 650-0017, Japan. Phone: 81-78-382-5860; Fax: 81-78-382-2084; E-mail: [email protected].
Find articles by Nishino, N. in: JCI | PubMed | Google Scholar
1Department of Internal Medicine, Kobe University Graduate School of Medicine, Kobe, Japan. 2Laboratory of Nutrition Chemistry, Division of Food Science and Biotechnology, Graduate School of Agriculture, Kyoto University, Kyoto, Japan. 3Department of Biomedical Informatics, Kobe University Graduate School of Medicine, Kobe, Japan. 4Drug Research Division, Dainippon Sumitomo Pharma Co. Ltd., Takarazuka, Japan. 5Department of Brain Sciences, Kobe University Graduate School of Medicine, Kobe, Japan. 6Department of Biomedical Sciences, Graduate School of Veterinary Medicine, Hokkaido University, Sapporo, Japan. 7Department of Nutrition, School of Nursing and Nutrition, Tenshi College, Sapporo, Japan. 8Department of Biochemistry and Cell Biology, Graduate School of Medicine, Nippon Medical School, Kawasaki, Japan. 9Graduate School of Life Science, University of Hyogo, Kamigori, Japan. 10Department of Anatomy and Molecular Cell Biology, Graduate School of Medicine, Nagoya University, Nagoya, Japan. 11Department of Physiology and Cell Biology, Kobe University Graduate School of Medicine, Kobe, Japan. 12Laboratory for Animal Resources and Genetic Engineering Team, RIKEN Center for Developmental Biology (CDB), Kobe, Japan.
Address correspondence to: Masato Kasuga, Department of Internal Medicine, Division of Diabetes, Metabolism, and Endocrinology, Kobe University Graduate School of Medicine, 7-5-1 Kusunoki-cho, Chuo-ku, Kobe 650-0017, Japan. Phone: 81-78-382-5860; Fax: 81-78-382-2084; E-mail: [email protected].
Find articles by Tamori, Y. in: JCI | PubMed | Google Scholar
1Department of Internal Medicine, Kobe University Graduate School of Medicine, Kobe, Japan. 2Laboratory of Nutrition Chemistry, Division of Food Science and Biotechnology, Graduate School of Agriculture, Kyoto University, Kyoto, Japan. 3Department of Biomedical Informatics, Kobe University Graduate School of Medicine, Kobe, Japan. 4Drug Research Division, Dainippon Sumitomo Pharma Co. Ltd., Takarazuka, Japan. 5Department of Brain Sciences, Kobe University Graduate School of Medicine, Kobe, Japan. 6Department of Biomedical Sciences, Graduate School of Veterinary Medicine, Hokkaido University, Sapporo, Japan. 7Department of Nutrition, School of Nursing and Nutrition, Tenshi College, Sapporo, Japan. 8Department of Biochemistry and Cell Biology, Graduate School of Medicine, Nippon Medical School, Kawasaki, Japan. 9Graduate School of Life Science, University of Hyogo, Kamigori, Japan. 10Department of Anatomy and Molecular Cell Biology, Graduate School of Medicine, Nagoya University, Nagoya, Japan. 11Department of Physiology and Cell Biology, Kobe University Graduate School of Medicine, Kobe, Japan. 12Laboratory for Animal Resources and Genetic Engineering Team, RIKEN Center for Developmental Biology (CDB), Kobe, Japan.
Address correspondence to: Masato Kasuga, Department of Internal Medicine, Division of Diabetes, Metabolism, and Endocrinology, Kobe University Graduate School of Medicine, 7-5-1 Kusunoki-cho, Chuo-ku, Kobe 650-0017, Japan. Phone: 81-78-382-5860; Fax: 81-78-382-2084; E-mail: [email protected].
Find articles by Tateya, S. in: JCI | PubMed | Google Scholar
1Department of Internal Medicine, Kobe University Graduate School of Medicine, Kobe, Japan. 2Laboratory of Nutrition Chemistry, Division of Food Science and Biotechnology, Graduate School of Agriculture, Kyoto University, Kyoto, Japan. 3Department of Biomedical Informatics, Kobe University Graduate School of Medicine, Kobe, Japan. 4Drug Research Division, Dainippon Sumitomo Pharma Co. Ltd., Takarazuka, Japan. 5Department of Brain Sciences, Kobe University Graduate School of Medicine, Kobe, Japan. 6Department of Biomedical Sciences, Graduate School of Veterinary Medicine, Hokkaido University, Sapporo, Japan. 7Department of Nutrition, School of Nursing and Nutrition, Tenshi College, Sapporo, Japan. 8Department of Biochemistry and Cell Biology, Graduate School of Medicine, Nippon Medical School, Kawasaki, Japan. 9Graduate School of Life Science, University of Hyogo, Kamigori, Japan. 10Department of Anatomy and Molecular Cell Biology, Graduate School of Medicine, Nagoya University, Nagoya, Japan. 11Department of Physiology and Cell Biology, Kobe University Graduate School of Medicine, Kobe, Japan. 12Laboratory for Animal Resources and Genetic Engineering Team, RIKEN Center for Developmental Biology (CDB), Kobe, Japan.
Address correspondence to: Masato Kasuga, Department of Internal Medicine, Division of Diabetes, Metabolism, and Endocrinology, Kobe University Graduate School of Medicine, 7-5-1 Kusunoki-cho, Chuo-ku, Kobe 650-0017, Japan. Phone: 81-78-382-5860; Fax: 81-78-382-2084; E-mail: [email protected].
Find articles by Kawaguchi, T. in: JCI | PubMed | Google Scholar
1Department of Internal Medicine, Kobe University Graduate School of Medicine, Kobe, Japan. 2Laboratory of Nutrition Chemistry, Division of Food Science and Biotechnology, Graduate School of Agriculture, Kyoto University, Kyoto, Japan. 3Department of Biomedical Informatics, Kobe University Graduate School of Medicine, Kobe, Japan. 4Drug Research Division, Dainippon Sumitomo Pharma Co. Ltd., Takarazuka, Japan. 5Department of Brain Sciences, Kobe University Graduate School of Medicine, Kobe, Japan. 6Department of Biomedical Sciences, Graduate School of Veterinary Medicine, Hokkaido University, Sapporo, Japan. 7Department of Nutrition, School of Nursing and Nutrition, Tenshi College, Sapporo, Japan. 8Department of Biochemistry and Cell Biology, Graduate School of Medicine, Nippon Medical School, Kawasaki, Japan. 9Graduate School of Life Science, University of Hyogo, Kamigori, Japan. 10Department of Anatomy and Molecular Cell Biology, Graduate School of Medicine, Nagoya University, Nagoya, Japan. 11Department of Physiology and Cell Biology, Kobe University Graduate School of Medicine, Kobe, Japan. 12Laboratory for Animal Resources and Genetic Engineering Team, RIKEN Center for Developmental Biology (CDB), Kobe, Japan.
Address correspondence to: Masato Kasuga, Department of Internal Medicine, Division of Diabetes, Metabolism, and Endocrinology, Kobe University Graduate School of Medicine, 7-5-1 Kusunoki-cho, Chuo-ku, Kobe 650-0017, Japan. Phone: 81-78-382-5860; Fax: 81-78-382-2084; E-mail: [email protected].
Find articles by Shibakusa, T. in: JCI | PubMed | Google Scholar
1Department of Internal Medicine, Kobe University Graduate School of Medicine, Kobe, Japan. 2Laboratory of Nutrition Chemistry, Division of Food Science and Biotechnology, Graduate School of Agriculture, Kyoto University, Kyoto, Japan. 3Department of Biomedical Informatics, Kobe University Graduate School of Medicine, Kobe, Japan. 4Drug Research Division, Dainippon Sumitomo Pharma Co. Ltd., Takarazuka, Japan. 5Department of Brain Sciences, Kobe University Graduate School of Medicine, Kobe, Japan. 6Department of Biomedical Sciences, Graduate School of Veterinary Medicine, Hokkaido University, Sapporo, Japan. 7Department of Nutrition, School of Nursing and Nutrition, Tenshi College, Sapporo, Japan. 8Department of Biochemistry and Cell Biology, Graduate School of Medicine, Nippon Medical School, Kawasaki, Japan. 9Graduate School of Life Science, University of Hyogo, Kamigori, Japan. 10Department of Anatomy and Molecular Cell Biology, Graduate School of Medicine, Nagoya University, Nagoya, Japan. 11Department of Physiology and Cell Biology, Kobe University Graduate School of Medicine, Kobe, Japan. 12Laboratory for Animal Resources and Genetic Engineering Team, RIKEN Center for Developmental Biology (CDB), Kobe, Japan.
Address correspondence to: Masato Kasuga, Department of Internal Medicine, Division of Diabetes, Metabolism, and Endocrinology, Kobe University Graduate School of Medicine, 7-5-1 Kusunoki-cho, Chuo-ku, Kobe 650-0017, Japan. Phone: 81-78-382-5860; Fax: 81-78-382-2084; E-mail: [email protected].
Find articles by Mizunoya, W. in: JCI | PubMed | Google Scholar
1Department of Internal Medicine, Kobe University Graduate School of Medicine, Kobe, Japan. 2Laboratory of Nutrition Chemistry, Division of Food Science and Biotechnology, Graduate School of Agriculture, Kyoto University, Kyoto, Japan. 3Department of Biomedical Informatics, Kobe University Graduate School of Medicine, Kobe, Japan. 4Drug Research Division, Dainippon Sumitomo Pharma Co. Ltd., Takarazuka, Japan. 5Department of Brain Sciences, Kobe University Graduate School of Medicine, Kobe, Japan. 6Department of Biomedical Sciences, Graduate School of Veterinary Medicine, Hokkaido University, Sapporo, Japan. 7Department of Nutrition, School of Nursing and Nutrition, Tenshi College, Sapporo, Japan. 8Department of Biochemistry and Cell Biology, Graduate School of Medicine, Nippon Medical School, Kawasaki, Japan. 9Graduate School of Life Science, University of Hyogo, Kamigori, Japan. 10Department of Anatomy and Molecular Cell Biology, Graduate School of Medicine, Nagoya University, Nagoya, Japan. 11Department of Physiology and Cell Biology, Kobe University Graduate School of Medicine, Kobe, Japan. 12Laboratory for Animal Resources and Genetic Engineering Team, RIKEN Center for Developmental Biology (CDB), Kobe, Japan.
Address correspondence to: Masato Kasuga, Department of Internal Medicine, Division of Diabetes, Metabolism, and Endocrinology, Kobe University Graduate School of Medicine, 7-5-1 Kusunoki-cho, Chuo-ku, Kobe 650-0017, Japan. Phone: 81-78-382-5860; Fax: 81-78-382-2084; E-mail: [email protected].
Find articles by Inoue, K. in: JCI | PubMed | Google Scholar
1Department of Internal Medicine, Kobe University Graduate School of Medicine, Kobe, Japan. 2Laboratory of Nutrition Chemistry, Division of Food Science and Biotechnology, Graduate School of Agriculture, Kyoto University, Kyoto, Japan. 3Department of Biomedical Informatics, Kobe University Graduate School of Medicine, Kobe, Japan. 4Drug Research Division, Dainippon Sumitomo Pharma Co. Ltd., Takarazuka, Japan. 5Department of Brain Sciences, Kobe University Graduate School of Medicine, Kobe, Japan. 6Department of Biomedical Sciences, Graduate School of Veterinary Medicine, Hokkaido University, Sapporo, Japan. 7Department of Nutrition, School of Nursing and Nutrition, Tenshi College, Sapporo, Japan. 8Department of Biochemistry and Cell Biology, Graduate School of Medicine, Nippon Medical School, Kawasaki, Japan. 9Graduate School of Life Science, University of Hyogo, Kamigori, Japan. 10Department of Anatomy and Molecular Cell Biology, Graduate School of Medicine, Nagoya University, Nagoya, Japan. 11Department of Physiology and Cell Biology, Kobe University Graduate School of Medicine, Kobe, Japan. 12Laboratory for Animal Resources and Genetic Engineering Team, RIKEN Center for Developmental Biology (CDB), Kobe, Japan.
Address correspondence to: Masato Kasuga, Department of Internal Medicine, Division of Diabetes, Metabolism, and Endocrinology, Kobe University Graduate School of Medicine, 7-5-1 Kusunoki-cho, Chuo-ku, Kobe 650-0017, Japan. Phone: 81-78-382-5860; Fax: 81-78-382-2084; E-mail: [email protected].
Find articles by Kitazawa, R. in: JCI | PubMed | Google Scholar
1Department of Internal Medicine, Kobe University Graduate School of Medicine, Kobe, Japan. 2Laboratory of Nutrition Chemistry, Division of Food Science and Biotechnology, Graduate School of Agriculture, Kyoto University, Kyoto, Japan. 3Department of Biomedical Informatics, Kobe University Graduate School of Medicine, Kobe, Japan. 4Drug Research Division, Dainippon Sumitomo Pharma Co. Ltd., Takarazuka, Japan. 5Department of Brain Sciences, Kobe University Graduate School of Medicine, Kobe, Japan. 6Department of Biomedical Sciences, Graduate School of Veterinary Medicine, Hokkaido University, Sapporo, Japan. 7Department of Nutrition, School of Nursing and Nutrition, Tenshi College, Sapporo, Japan. 8Department of Biochemistry and Cell Biology, Graduate School of Medicine, Nippon Medical School, Kawasaki, Japan. 9Graduate School of Life Science, University of Hyogo, Kamigori, Japan. 10Department of Anatomy and Molecular Cell Biology, Graduate School of Medicine, Nagoya University, Nagoya, Japan. 11Department of Physiology and Cell Biology, Kobe University Graduate School of Medicine, Kobe, Japan. 12Laboratory for Animal Resources and Genetic Engineering Team, RIKEN Center for Developmental Biology (CDB), Kobe, Japan.
Address correspondence to: Masato Kasuga, Department of Internal Medicine, Division of Diabetes, Metabolism, and Endocrinology, Kobe University Graduate School of Medicine, 7-5-1 Kusunoki-cho, Chuo-ku, Kobe 650-0017, Japan. Phone: 81-78-382-5860; Fax: 81-78-382-2084; E-mail: [email protected].
Find articles by Kitazawa, S. in: JCI | PubMed | Google Scholar
1Department of Internal Medicine, Kobe University Graduate School of Medicine, Kobe, Japan. 2Laboratory of Nutrition Chemistry, Division of Food Science and Biotechnology, Graduate School of Agriculture, Kyoto University, Kyoto, Japan. 3Department of Biomedical Informatics, Kobe University Graduate School of Medicine, Kobe, Japan. 4Drug Research Division, Dainippon Sumitomo Pharma Co. Ltd., Takarazuka, Japan. 5Department of Brain Sciences, Kobe University Graduate School of Medicine, Kobe, Japan. 6Department of Biomedical Sciences, Graduate School of Veterinary Medicine, Hokkaido University, Sapporo, Japan. 7Department of Nutrition, School of Nursing and Nutrition, Tenshi College, Sapporo, Japan. 8Department of Biochemistry and Cell Biology, Graduate School of Medicine, Nippon Medical School, Kawasaki, Japan. 9Graduate School of Life Science, University of Hyogo, Kamigori, Japan. 10Department of Anatomy and Molecular Cell Biology, Graduate School of Medicine, Nagoya University, Nagoya, Japan. 11Department of Physiology and Cell Biology, Kobe University Graduate School of Medicine, Kobe, Japan. 12Laboratory for Animal Resources and Genetic Engineering Team, RIKEN Center for Developmental Biology (CDB), Kobe, Japan.
Address correspondence to: Masato Kasuga, Department of Internal Medicine, Division of Diabetes, Metabolism, and Endocrinology, Kobe University Graduate School of Medicine, 7-5-1 Kusunoki-cho, Chuo-ku, Kobe 650-0017, Japan. Phone: 81-78-382-5860; Fax: 81-78-382-2084; E-mail: [email protected].
Find articles by Matsuki, Y. in: JCI | PubMed | Google Scholar
1Department of Internal Medicine, Kobe University Graduate School of Medicine, Kobe, Japan. 2Laboratory of Nutrition Chemistry, Division of Food Science and Biotechnology, Graduate School of Agriculture, Kyoto University, Kyoto, Japan. 3Department of Biomedical Informatics, Kobe University Graduate School of Medicine, Kobe, Japan. 4Drug Research Division, Dainippon Sumitomo Pharma Co. Ltd., Takarazuka, Japan. 5Department of Brain Sciences, Kobe University Graduate School of Medicine, Kobe, Japan. 6Department of Biomedical Sciences, Graduate School of Veterinary Medicine, Hokkaido University, Sapporo, Japan. 7Department of Nutrition, School of Nursing and Nutrition, Tenshi College, Sapporo, Japan. 8Department of Biochemistry and Cell Biology, Graduate School of Medicine, Nippon Medical School, Kawasaki, Japan. 9Graduate School of Life Science, University of Hyogo, Kamigori, Japan. 10Department of Anatomy and Molecular Cell Biology, Graduate School of Medicine, Nagoya University, Nagoya, Japan. 11Department of Physiology and Cell Biology, Kobe University Graduate School of Medicine, Kobe, Japan. 12Laboratory for Animal Resources and Genetic Engineering Team, RIKEN Center for Developmental Biology (CDB), Kobe, Japan.
Address correspondence to: Masato Kasuga, Department of Internal Medicine, Division of Diabetes, Metabolism, and Endocrinology, Kobe University Graduate School of Medicine, 7-5-1 Kusunoki-cho, Chuo-ku, Kobe 650-0017, Japan. Phone: 81-78-382-5860; Fax: 81-78-382-2084; E-mail: [email protected].
Find articles by Hiramatsu, R. in: JCI | PubMed | Google Scholar
1Department of Internal Medicine, Kobe University Graduate School of Medicine, Kobe, Japan. 2Laboratory of Nutrition Chemistry, Division of Food Science and Biotechnology, Graduate School of Agriculture, Kyoto University, Kyoto, Japan. 3Department of Biomedical Informatics, Kobe University Graduate School of Medicine, Kobe, Japan. 4Drug Research Division, Dainippon Sumitomo Pharma Co. Ltd., Takarazuka, Japan. 5Department of Brain Sciences, Kobe University Graduate School of Medicine, Kobe, Japan. 6Department of Biomedical Sciences, Graduate School of Veterinary Medicine, Hokkaido University, Sapporo, Japan. 7Department of Nutrition, School of Nursing and Nutrition, Tenshi College, Sapporo, Japan. 8Department of Biochemistry and Cell Biology, Graduate School of Medicine, Nippon Medical School, Kawasaki, Japan. 9Graduate School of Life Science, University of Hyogo, Kamigori, Japan. 10Department of Anatomy and Molecular Cell Biology, Graduate School of Medicine, Nagoya University, Nagoya, Japan. 11Department of Physiology and Cell Biology, Kobe University Graduate School of Medicine, Kobe, Japan. 12Laboratory for Animal Resources and Genetic Engineering Team, RIKEN Center for Developmental Biology (CDB), Kobe, Japan.
Address correspondence to: Masato Kasuga, Department of Internal Medicine, Division of Diabetes, Metabolism, and Endocrinology, Kobe University Graduate School of Medicine, 7-5-1 Kusunoki-cho, Chuo-ku, Kobe 650-0017, Japan. Phone: 81-78-382-5860; Fax: 81-78-382-2084; E-mail: [email protected].
Find articles by Masubuchi, S. in: JCI | PubMed | Google Scholar
1Department of Internal Medicine, Kobe University Graduate School of Medicine, Kobe, Japan. 2Laboratory of Nutrition Chemistry, Division of Food Science and Biotechnology, Graduate School of Agriculture, Kyoto University, Kyoto, Japan. 3Department of Biomedical Informatics, Kobe University Graduate School of Medicine, Kobe, Japan. 4Drug Research Division, Dainippon Sumitomo Pharma Co. Ltd., Takarazuka, Japan. 5Department of Brain Sciences, Kobe University Graduate School of Medicine, Kobe, Japan. 6Department of Biomedical Sciences, Graduate School of Veterinary Medicine, Hokkaido University, Sapporo, Japan. 7Department of Nutrition, School of Nursing and Nutrition, Tenshi College, Sapporo, Japan. 8Department of Biochemistry and Cell Biology, Graduate School of Medicine, Nippon Medical School, Kawasaki, Japan. 9Graduate School of Life Science, University of Hyogo, Kamigori, Japan. 10Department of Anatomy and Molecular Cell Biology, Graduate School of Medicine, Nagoya University, Nagoya, Japan. 11Department of Physiology and Cell Biology, Kobe University Graduate School of Medicine, Kobe, Japan. 12Laboratory for Animal Resources and Genetic Engineering Team, RIKEN Center for Developmental Biology (CDB), Kobe, Japan.
Address correspondence to: Masato Kasuga, Department of Internal Medicine, Division of Diabetes, Metabolism, and Endocrinology, Kobe University Graduate School of Medicine, 7-5-1 Kusunoki-cho, Chuo-ku, Kobe 650-0017, Japan. Phone: 81-78-382-5860; Fax: 81-78-382-2084; E-mail: [email protected].
Find articles by Omachi, A. in: JCI | PubMed | Google Scholar
1Department of Internal Medicine, Kobe University Graduate School of Medicine, Kobe, Japan. 2Laboratory of Nutrition Chemistry, Division of Food Science and Biotechnology, Graduate School of Agriculture, Kyoto University, Kyoto, Japan. 3Department of Biomedical Informatics, Kobe University Graduate School of Medicine, Kobe, Japan. 4Drug Research Division, Dainippon Sumitomo Pharma Co. Ltd., Takarazuka, Japan. 5Department of Brain Sciences, Kobe University Graduate School of Medicine, Kobe, Japan. 6Department of Biomedical Sciences, Graduate School of Veterinary Medicine, Hokkaido University, Sapporo, Japan. 7Department of Nutrition, School of Nursing and Nutrition, Tenshi College, Sapporo, Japan. 8Department of Biochemistry and Cell Biology, Graduate School of Medicine, Nippon Medical School, Kawasaki, Japan. 9Graduate School of Life Science, University of Hyogo, Kamigori, Japan. 10Department of Anatomy and Molecular Cell Biology, Graduate School of Medicine, Nagoya University, Nagoya, Japan. 11Department of Physiology and Cell Biology, Kobe University Graduate School of Medicine, Kobe, Japan. 12Laboratory for Animal Resources and Genetic Engineering Team, RIKEN Center for Developmental Biology (CDB), Kobe, Japan.
Address correspondence to: Masato Kasuga, Department of Internal Medicine, Division of Diabetes, Metabolism, and Endocrinology, Kobe University Graduate School of Medicine, 7-5-1 Kusunoki-cho, Chuo-ku, Kobe 650-0017, Japan. Phone: 81-78-382-5860; Fax: 81-78-382-2084; E-mail: [email protected].
Find articles by Kimura, K. in: JCI | PubMed | Google Scholar
1Department of Internal Medicine, Kobe University Graduate School of Medicine, Kobe, Japan. 2Laboratory of Nutrition Chemistry, Division of Food Science and Biotechnology, Graduate School of Agriculture, Kyoto University, Kyoto, Japan. 3Department of Biomedical Informatics, Kobe University Graduate School of Medicine, Kobe, Japan. 4Drug Research Division, Dainippon Sumitomo Pharma Co. Ltd., Takarazuka, Japan. 5Department of Brain Sciences, Kobe University Graduate School of Medicine, Kobe, Japan. 6Department of Biomedical Sciences, Graduate School of Veterinary Medicine, Hokkaido University, Sapporo, Japan. 7Department of Nutrition, School of Nursing and Nutrition, Tenshi College, Sapporo, Japan. 8Department of Biochemistry and Cell Biology, Graduate School of Medicine, Nippon Medical School, Kawasaki, Japan. 9Graduate School of Life Science, University of Hyogo, Kamigori, Japan. 10Department of Anatomy and Molecular Cell Biology, Graduate School of Medicine, Nagoya University, Nagoya, Japan. 11Department of Physiology and Cell Biology, Kobe University Graduate School of Medicine, Kobe, Japan. 12Laboratory for Animal Resources and Genetic Engineering Team, RIKEN Center for Developmental Biology (CDB), Kobe, Japan.
Address correspondence to: Masato Kasuga, Department of Internal Medicine, Division of Diabetes, Metabolism, and Endocrinology, Kobe University Graduate School of Medicine, 7-5-1 Kusunoki-cho, Chuo-ku, Kobe 650-0017, Japan. Phone: 81-78-382-5860; Fax: 81-78-382-2084; E-mail: [email protected].
Find articles by Saito, M. in: JCI | PubMed | Google Scholar
1Department of Internal Medicine, Kobe University Graduate School of Medicine, Kobe, Japan. 2Laboratory of Nutrition Chemistry, Division of Food Science and Biotechnology, Graduate School of Agriculture, Kyoto University, Kyoto, Japan. 3Department of Biomedical Informatics, Kobe University Graduate School of Medicine, Kobe, Japan. 4Drug Research Division, Dainippon Sumitomo Pharma Co. Ltd., Takarazuka, Japan. 5Department of Brain Sciences, Kobe University Graduate School of Medicine, Kobe, Japan. 6Department of Biomedical Sciences, Graduate School of Veterinary Medicine, Hokkaido University, Sapporo, Japan. 7Department of Nutrition, School of Nursing and Nutrition, Tenshi College, Sapporo, Japan. 8Department of Biochemistry and Cell Biology, Graduate School of Medicine, Nippon Medical School, Kawasaki, Japan. 9Graduate School of Life Science, University of Hyogo, Kamigori, Japan. 10Department of Anatomy and Molecular Cell Biology, Graduate School of Medicine, Nagoya University, Nagoya, Japan. 11Department of Physiology and Cell Biology, Kobe University Graduate School of Medicine, Kobe, Japan. 12Laboratory for Animal Resources and Genetic Engineering Team, RIKEN Center for Developmental Biology (CDB), Kobe, Japan.
Address correspondence to: Masato Kasuga, Department of Internal Medicine, Division of Diabetes, Metabolism, and Endocrinology, Kobe University Graduate School of Medicine, 7-5-1 Kusunoki-cho, Chuo-ku, Kobe 650-0017, Japan. Phone: 81-78-382-5860; Fax: 81-78-382-2084; E-mail: [email protected].
Find articles by Amo, T. in: JCI | PubMed | Google Scholar
1Department of Internal Medicine, Kobe University Graduate School of Medicine, Kobe, Japan. 2Laboratory of Nutrition Chemistry, Division of Food Science and Biotechnology, Graduate School of Agriculture, Kyoto University, Kyoto, Japan. 3Department of Biomedical Informatics, Kobe University Graduate School of Medicine, Kobe, Japan. 4Drug Research Division, Dainippon Sumitomo Pharma Co. Ltd., Takarazuka, Japan. 5Department of Brain Sciences, Kobe University Graduate School of Medicine, Kobe, Japan. 6Department of Biomedical Sciences, Graduate School of Veterinary Medicine, Hokkaido University, Sapporo, Japan. 7Department of Nutrition, School of Nursing and Nutrition, Tenshi College, Sapporo, Japan. 8Department of Biochemistry and Cell Biology, Graduate School of Medicine, Nippon Medical School, Kawasaki, Japan. 9Graduate School of Life Science, University of Hyogo, Kamigori, Japan. 10Department of Anatomy and Molecular Cell Biology, Graduate School of Medicine, Nagoya University, Nagoya, Japan. 11Department of Physiology and Cell Biology, Kobe University Graduate School of Medicine, Kobe, Japan. 12Laboratory for Animal Resources and Genetic Engineering Team, RIKEN Center for Developmental Biology (CDB), Kobe, Japan.
Address correspondence to: Masato Kasuga, Department of Internal Medicine, Division of Diabetes, Metabolism, and Endocrinology, Kobe University Graduate School of Medicine, 7-5-1 Kusunoki-cho, Chuo-ku, Kobe 650-0017, Japan. Phone: 81-78-382-5860; Fax: 81-78-382-2084; E-mail: [email protected].
Find articles by Ohta, S. in: JCI | PubMed | Google Scholar
1Department of Internal Medicine, Kobe University Graduate School of Medicine, Kobe, Japan. 2Laboratory of Nutrition Chemistry, Division of Food Science and Biotechnology, Graduate School of Agriculture, Kyoto University, Kyoto, Japan. 3Department of Biomedical Informatics, Kobe University Graduate School of Medicine, Kobe, Japan. 4Drug Research Division, Dainippon Sumitomo Pharma Co. Ltd., Takarazuka, Japan. 5Department of Brain Sciences, Kobe University Graduate School of Medicine, Kobe, Japan. 6Department of Biomedical Sciences, Graduate School of Veterinary Medicine, Hokkaido University, Sapporo, Japan. 7Department of Nutrition, School of Nursing and Nutrition, Tenshi College, Sapporo, Japan. 8Department of Biochemistry and Cell Biology, Graduate School of Medicine, Nippon Medical School, Kawasaki, Japan. 9Graduate School of Life Science, University of Hyogo, Kamigori, Japan. 10Department of Anatomy and Molecular Cell Biology, Graduate School of Medicine, Nagoya University, Nagoya, Japan. 11Department of Physiology and Cell Biology, Kobe University Graduate School of Medicine, Kobe, Japan. 12Laboratory for Animal Resources and Genetic Engineering Team, RIKEN Center for Developmental Biology (CDB), Kobe, Japan.
Address correspondence to: Masato Kasuga, Department of Internal Medicine, Division of Diabetes, Metabolism, and Endocrinology, Kobe University Graduate School of Medicine, 7-5-1 Kusunoki-cho, Chuo-ku, Kobe 650-0017, Japan. Phone: 81-78-382-5860; Fax: 81-78-382-2084; E-mail: [email protected].
Find articles by Yamaguchi, T. in: JCI | PubMed | Google Scholar
1Department of Internal Medicine, Kobe University Graduate School of Medicine, Kobe, Japan. 2Laboratory of Nutrition Chemistry, Division of Food Science and Biotechnology, Graduate School of Agriculture, Kyoto University, Kyoto, Japan. 3Department of Biomedical Informatics, Kobe University Graduate School of Medicine, Kobe, Japan. 4Drug Research Division, Dainippon Sumitomo Pharma Co. Ltd., Takarazuka, Japan. 5Department of Brain Sciences, Kobe University Graduate School of Medicine, Kobe, Japan. 6Department of Biomedical Sciences, Graduate School of Veterinary Medicine, Hokkaido University, Sapporo, Japan. 7Department of Nutrition, School of Nursing and Nutrition, Tenshi College, Sapporo, Japan. 8Department of Biochemistry and Cell Biology, Graduate School of Medicine, Nippon Medical School, Kawasaki, Japan. 9Graduate School of Life Science, University of Hyogo, Kamigori, Japan. 10Department of Anatomy and Molecular Cell Biology, Graduate School of Medicine, Nagoya University, Nagoya, Japan. 11Department of Physiology and Cell Biology, Kobe University Graduate School of Medicine, Kobe, Japan. 12Laboratory for Animal Resources and Genetic Engineering Team, RIKEN Center for Developmental Biology (CDB), Kobe, Japan.
Address correspondence to: Masato Kasuga, Department of Internal Medicine, Division of Diabetes, Metabolism, and Endocrinology, Kobe University Graduate School of Medicine, 7-5-1 Kusunoki-cho, Chuo-ku, Kobe 650-0017, Japan. Phone: 81-78-382-5860; Fax: 81-78-382-2084; E-mail: [email protected].
Find articles by Osumi, T. in: JCI | PubMed | Google Scholar
1Department of Internal Medicine, Kobe University Graduate School of Medicine, Kobe, Japan. 2Laboratory of Nutrition Chemistry, Division of Food Science and Biotechnology, Graduate School of Agriculture, Kyoto University, Kyoto, Japan. 3Department of Biomedical Informatics, Kobe University Graduate School of Medicine, Kobe, Japan. 4Drug Research Division, Dainippon Sumitomo Pharma Co. Ltd., Takarazuka, Japan. 5Department of Brain Sciences, Kobe University Graduate School of Medicine, Kobe, Japan. 6Department of Biomedical Sciences, Graduate School of Veterinary Medicine, Hokkaido University, Sapporo, Japan. 7Department of Nutrition, School of Nursing and Nutrition, Tenshi College, Sapporo, Japan. 8Department of Biochemistry and Cell Biology, Graduate School of Medicine, Nippon Medical School, Kawasaki, Japan. 9Graduate School of Life Science, University of Hyogo, Kamigori, Japan. 10Department of Anatomy and Molecular Cell Biology, Graduate School of Medicine, Nagoya University, Nagoya, Japan. 11Department of Physiology and Cell Biology, Kobe University Graduate School of Medicine, Kobe, Japan. 12Laboratory for Animal Resources and Genetic Engineering Team, RIKEN Center for Developmental Biology (CDB), Kobe, Japan.
Address correspondence to: Masato Kasuga, Department of Internal Medicine, Division of Diabetes, Metabolism, and Endocrinology, Kobe University Graduate School of Medicine, 7-5-1 Kusunoki-cho, Chuo-ku, Kobe 650-0017, Japan. Phone: 81-78-382-5860; Fax: 81-78-382-2084; E-mail: [email protected].
Find articles by Cheng, J. in: JCI | PubMed | Google Scholar
1Department of Internal Medicine, Kobe University Graduate School of Medicine, Kobe, Japan. 2Laboratory of Nutrition Chemistry, Division of Food Science and Biotechnology, Graduate School of Agriculture, Kyoto University, Kyoto, Japan. 3Department of Biomedical Informatics, Kobe University Graduate School of Medicine, Kobe, Japan. 4Drug Research Division, Dainippon Sumitomo Pharma Co. Ltd., Takarazuka, Japan. 5Department of Brain Sciences, Kobe University Graduate School of Medicine, Kobe, Japan. 6Department of Biomedical Sciences, Graduate School of Veterinary Medicine, Hokkaido University, Sapporo, Japan. 7Department of Nutrition, School of Nursing and Nutrition, Tenshi College, Sapporo, Japan. 8Department of Biochemistry and Cell Biology, Graduate School of Medicine, Nippon Medical School, Kawasaki, Japan. 9Graduate School of Life Science, University of Hyogo, Kamigori, Japan. 10Department of Anatomy and Molecular Cell Biology, Graduate School of Medicine, Nagoya University, Nagoya, Japan. 11Department of Physiology and Cell Biology, Kobe University Graduate School of Medicine, Kobe, Japan. 12Laboratory for Animal Resources and Genetic Engineering Team, RIKEN Center for Developmental Biology (CDB), Kobe, Japan.
Address correspondence to: Masato Kasuga, Department of Internal Medicine, Division of Diabetes, Metabolism, and Endocrinology, Kobe University Graduate School of Medicine, 7-5-1 Kusunoki-cho, Chuo-ku, Kobe 650-0017, Japan. Phone: 81-78-382-5860; Fax: 81-78-382-2084; E-mail: [email protected].
Find articles by Fujimoto, T. in: JCI | PubMed | Google Scholar
1Department of Internal Medicine, Kobe University Graduate School of Medicine, Kobe, Japan. 2Laboratory of Nutrition Chemistry, Division of Food Science and Biotechnology, Graduate School of Agriculture, Kyoto University, Kyoto, Japan. 3Department of Biomedical Informatics, Kobe University Graduate School of Medicine, Kobe, Japan. 4Drug Research Division, Dainippon Sumitomo Pharma Co. Ltd., Takarazuka, Japan. 5Department of Brain Sciences, Kobe University Graduate School of Medicine, Kobe, Japan. 6Department of Biomedical Sciences, Graduate School of Veterinary Medicine, Hokkaido University, Sapporo, Japan. 7Department of Nutrition, School of Nursing and Nutrition, Tenshi College, Sapporo, Japan. 8Department of Biochemistry and Cell Biology, Graduate School of Medicine, Nippon Medical School, Kawasaki, Japan. 9Graduate School of Life Science, University of Hyogo, Kamigori, Japan. 10Department of Anatomy and Molecular Cell Biology, Graduate School of Medicine, Nagoya University, Nagoya, Japan. 11Department of Physiology and Cell Biology, Kobe University Graduate School of Medicine, Kobe, Japan. 12Laboratory for Animal Resources and Genetic Engineering Team, RIKEN Center for Developmental Biology (CDB), Kobe, Japan.
Address correspondence to: Masato Kasuga, Department of Internal Medicine, Division of Diabetes, Metabolism, and Endocrinology, Kobe University Graduate School of Medicine, 7-5-1 Kusunoki-cho, Chuo-ku, Kobe 650-0017, Japan. Phone: 81-78-382-5860; Fax: 81-78-382-2084; E-mail: [email protected].
Find articles by Nakao, H. in: JCI | PubMed | Google Scholar
1Department of Internal Medicine, Kobe University Graduate School of Medicine, Kobe, Japan. 2Laboratory of Nutrition Chemistry, Division of Food Science and Biotechnology, Graduate School of Agriculture, Kyoto University, Kyoto, Japan. 3Department of Biomedical Informatics, Kobe University Graduate School of Medicine, Kobe, Japan. 4Drug Research Division, Dainippon Sumitomo Pharma Co. Ltd., Takarazuka, Japan. 5Department of Brain Sciences, Kobe University Graduate School of Medicine, Kobe, Japan. 6Department of Biomedical Sciences, Graduate School of Veterinary Medicine, Hokkaido University, Sapporo, Japan. 7Department of Nutrition, School of Nursing and Nutrition, Tenshi College, Sapporo, Japan. 8Department of Biochemistry and Cell Biology, Graduate School of Medicine, Nippon Medical School, Kawasaki, Japan. 9Graduate School of Life Science, University of Hyogo, Kamigori, Japan. 10Department of Anatomy and Molecular Cell Biology, Graduate School of Medicine, Nagoya University, Nagoya, Japan. 11Department of Physiology and Cell Biology, Kobe University Graduate School of Medicine, Kobe, Japan. 12Laboratory for Animal Resources and Genetic Engineering Team, RIKEN Center for Developmental Biology (CDB), Kobe, Japan.
Address correspondence to: Masato Kasuga, Department of Internal Medicine, Division of Diabetes, Metabolism, and Endocrinology, Kobe University Graduate School of Medicine, 7-5-1 Kusunoki-cho, Chuo-ku, Kobe 650-0017, Japan. Phone: 81-78-382-5860; Fax: 81-78-382-2084; E-mail: [email protected].
Find articles by Nakao, K. in: JCI | PubMed | Google Scholar
1Department of Internal Medicine, Kobe University Graduate School of Medicine, Kobe, Japan. 2Laboratory of Nutrition Chemistry, Division of Food Science and Biotechnology, Graduate School of Agriculture, Kyoto University, Kyoto, Japan. 3Department of Biomedical Informatics, Kobe University Graduate School of Medicine, Kobe, Japan. 4Drug Research Division, Dainippon Sumitomo Pharma Co. Ltd., Takarazuka, Japan. 5Department of Brain Sciences, Kobe University Graduate School of Medicine, Kobe, Japan. 6Department of Biomedical Sciences, Graduate School of Veterinary Medicine, Hokkaido University, Sapporo, Japan. 7Department of Nutrition, School of Nursing and Nutrition, Tenshi College, Sapporo, Japan. 8Department of Biochemistry and Cell Biology, Graduate School of Medicine, Nippon Medical School, Kawasaki, Japan. 9Graduate School of Life Science, University of Hyogo, Kamigori, Japan. 10Department of Anatomy and Molecular Cell Biology, Graduate School of Medicine, Nagoya University, Nagoya, Japan. 11Department of Physiology and Cell Biology, Kobe University Graduate School of Medicine, Kobe, Japan. 12Laboratory for Animal Resources and Genetic Engineering Team, RIKEN Center for Developmental Biology (CDB), Kobe, Japan.
Address correspondence to: Masato Kasuga, Department of Internal Medicine, Division of Diabetes, Metabolism, and Endocrinology, Kobe University Graduate School of Medicine, 7-5-1 Kusunoki-cho, Chuo-ku, Kobe 650-0017, Japan. Phone: 81-78-382-5860; Fax: 81-78-382-2084; E-mail: [email protected].
Find articles by Aiba, A. in: JCI | PubMed | Google Scholar
1Department of Internal Medicine, Kobe University Graduate School of Medicine, Kobe, Japan. 2Laboratory of Nutrition Chemistry, Division of Food Science and Biotechnology, Graduate School of Agriculture, Kyoto University, Kyoto, Japan. 3Department of Biomedical Informatics, Kobe University Graduate School of Medicine, Kobe, Japan. 4Drug Research Division, Dainippon Sumitomo Pharma Co. Ltd., Takarazuka, Japan. 5Department of Brain Sciences, Kobe University Graduate School of Medicine, Kobe, Japan. 6Department of Biomedical Sciences, Graduate School of Veterinary Medicine, Hokkaido University, Sapporo, Japan. 7Department of Nutrition, School of Nursing and Nutrition, Tenshi College, Sapporo, Japan. 8Department of Biochemistry and Cell Biology, Graduate School of Medicine, Nippon Medical School, Kawasaki, Japan. 9Graduate School of Life Science, University of Hyogo, Kamigori, Japan. 10Department of Anatomy and Molecular Cell Biology, Graduate School of Medicine, Nagoya University, Nagoya, Japan. 11Department of Physiology and Cell Biology, Kobe University Graduate School of Medicine, Kobe, Japan. 12Laboratory for Animal Resources and Genetic Engineering Team, RIKEN Center for Developmental Biology (CDB), Kobe, Japan.
Address correspondence to: Masato Kasuga, Department of Internal Medicine, Division of Diabetes, Metabolism, and Endocrinology, Kobe University Graduate School of Medicine, 7-5-1 Kusunoki-cho, Chuo-ku, Kobe 650-0017, Japan. Phone: 81-78-382-5860; Fax: 81-78-382-2084; E-mail: [email protected].
Find articles by Okamura, H. in: JCI | PubMed | Google Scholar
1Department of Internal Medicine, Kobe University Graduate School of Medicine, Kobe, Japan. 2Laboratory of Nutrition Chemistry, Division of Food Science and Biotechnology, Graduate School of Agriculture, Kyoto University, Kyoto, Japan. 3Department of Biomedical Informatics, Kobe University Graduate School of Medicine, Kobe, Japan. 4Drug Research Division, Dainippon Sumitomo Pharma Co. Ltd., Takarazuka, Japan. 5Department of Brain Sciences, Kobe University Graduate School of Medicine, Kobe, Japan. 6Department of Biomedical Sciences, Graduate School of Veterinary Medicine, Hokkaido University, Sapporo, Japan. 7Department of Nutrition, School of Nursing and Nutrition, Tenshi College, Sapporo, Japan. 8Department of Biochemistry and Cell Biology, Graduate School of Medicine, Nippon Medical School, Kawasaki, Japan. 9Graduate School of Life Science, University of Hyogo, Kamigori, Japan. 10Department of Anatomy and Molecular Cell Biology, Graduate School of Medicine, Nagoya University, Nagoya, Japan. 11Department of Physiology and Cell Biology, Kobe University Graduate School of Medicine, Kobe, Japan. 12Laboratory for Animal Resources and Genetic Engineering Team, RIKEN Center for Developmental Biology (CDB), Kobe, Japan.
Address correspondence to: Masato Kasuga, Department of Internal Medicine, Division of Diabetes, Metabolism, and Endocrinology, Kobe University Graduate School of Medicine, 7-5-1 Kusunoki-cho, Chuo-ku, Kobe 650-0017, Japan. Phone: 81-78-382-5860; Fax: 81-78-382-2084; E-mail: [email protected].
Find articles by Fushiki, T. in: JCI | PubMed | Google Scholar
1Department of Internal Medicine, Kobe University Graduate School of Medicine, Kobe, Japan. 2Laboratory of Nutrition Chemistry, Division of Food Science and Biotechnology, Graduate School of Agriculture, Kyoto University, Kyoto, Japan. 3Department of Biomedical Informatics, Kobe University Graduate School of Medicine, Kobe, Japan. 4Drug Research Division, Dainippon Sumitomo Pharma Co. Ltd., Takarazuka, Japan. 5Department of Brain Sciences, Kobe University Graduate School of Medicine, Kobe, Japan. 6Department of Biomedical Sciences, Graduate School of Veterinary Medicine, Hokkaido University, Sapporo, Japan. 7Department of Nutrition, School of Nursing and Nutrition, Tenshi College, Sapporo, Japan. 8Department of Biochemistry and Cell Biology, Graduate School of Medicine, Nippon Medical School, Kawasaki, Japan. 9Graduate School of Life Science, University of Hyogo, Kamigori, Japan. 10Department of Anatomy and Molecular Cell Biology, Graduate School of Medicine, Nagoya University, Nagoya, Japan. 11Department of Physiology and Cell Biology, Kobe University Graduate School of Medicine, Kobe, Japan. 12Laboratory for Animal Resources and Genetic Engineering Team, RIKEN Center for Developmental Biology (CDB), Kobe, Japan.
Address correspondence to: Masato Kasuga, Department of Internal Medicine, Division of Diabetes, Metabolism, and Endocrinology, Kobe University Graduate School of Medicine, 7-5-1 Kusunoki-cho, Chuo-ku, Kobe 650-0017, Japan. Phone: 81-78-382-5860; Fax: 81-78-382-2084; E-mail: [email protected].
Find articles by Kasuga, M. in: JCI | PubMed | Google Scholar
Published July 24, 2008 - More info
J Clin Invest. 2008;118(8):2808–2821. https://doi.org/10.1172/JCI34090.
© 2008 The American Society for Clinical Investigation
Received: September 28, 2007; Accepted: May 21, 2008
Related article:
Abstract
Fat-specific protein of 27 kDa (FSP27) is a highly expressed adipocyte protein that promotes triglyceride accumulation within lipid droplets. In this issue of the JCI, Nishino et al. show that FSP27 also helps to maintain the characteristically large unilocular lipid droplet structure within each white adipocyte (see the related article beginning on page 2808). Fragmentation of lipid droplets in white adipocytes from FSP27-KO mice caused both increased lipolysis and upregulation of genes enhancing mitochondrial oxidative metabolism. This increased energy expenditure in turn protected the mice from diet-induced obesity and insulin resistance. These new results highlight powerful mechanisms that tightly coordinate rates of triglyceride storage in lipid droplets with mitochondrial fatty acid oxidation in white adipocytes.
Authors
Vishwajeet Puri, Michael P. Czech
-
Abstract
White adipocytes are unique in that they contain large unilocular lipid droplets that occupy most of the cytoplasm. To identify genes involved in the maintenance of mature adipocytes, we expressed dominant-negative PPARγ in 3T3-L1 cells and performed a microarray screen. The fat-specific protein of 27 kDa (FSP27) was strongly downregulated in this context. FSP27 expression correlated with induction of differentiation in cultured preadipocytes, and the protein localized to lipid droplets in murine white adipocytes in vivo. Ablation of FSP27 in mice resulted in the formation of multilocular lipid droplets in these cells. Furthermore, FSP27-deficient mice were protected from diet-induced obesity and insulin resistance and displayed an increased metabolic rate due to increased mitochondrial biogenesis in white adipose tissue (WAT). Depletion of FSP27 by siRNA in murine cultured white adipocytes resulted in the formation of numerous small lipid droplets, increased lipolysis, and decreased triacylglycerol storage, while expression of FSP27 in COS cells promoted the formation of large lipid droplets. Our results suggest that FSP27 contributes to efficient energy storage in WAT by promoting the formation of unilocular lipid droplets, thereby restricting lipolysis. In addition, we found that the nature of lipid accumulation in WAT appears to be associated with maintenance of energy balance and insulin sensitivity.
-
Introduction
Obesity is associated with various complications such as insulin resistance, type 2 diabetes, dyslipidemia, and atherosclerosis (1, 2). It results from an imbalance between energy intake and energy expenditure (3, 4). In general, obesity is characterized by the excessive accumulation of triacylglycerol (TAG) in adipose tissue, especially in white adipose tissue (WAT). In the fed state, WAT synthesizes TAG from FFAs and glucose and efficiently stores it in large lipid droplets. In the fasted state, WAT generates FFAs and glycerol as a result of the hydrolysis of stored TAG and releases them into the circulation as energy sources for other tissues, suggesting that WAT is an energy reservoir. Almost the entire cytoplasm of white adipocytes is occupied with a large (10–200 μm in diameter), unilocular, TAG-rich lipid droplet. Whereas most mammalian cells, including those of the liver, muscle, heart, kidney, intestine, and mammary gland, have the capacity to store TAG as lipid droplets surrounded by a phospholipid monolayer in the cytoplasm (5), these cells, in contrast to white adipocytes, accumulate multiple small lipid droplets (6). Brown adipocytes, which mediate thermogenesis by oxidizing FFAs in their abundant mitochondria, also possess small multilocular lipid droplets in their cytoplasm. These differences in the pattern of lipid accumulation are thought to markedly affect the functional characteristics of the respective cells. The unilocular lipid droplets of white adipocytes may represent an efficient structure for lipid storage. Although several proteins, including perilipin, adipose differentiation–related protein (ADRP, or adipophilin), tail-interacting protein of 47 kDa (TIP47), and S3-12, have been shown to associate with the surface of intracellular lipid droplets and to regulate their formation and mobilization (7, 8), the mechanism responsible for the formation of unilocular lipid droplets in WAT has remained unknown (6, 9). Fat-specific protein of 27 kDa (FSP27), a protein whose function has been unclear (10), was also recently shown to localize to lipid droplets and to promote lipid accumulation in cultured adipocytes (11). By generating FSP27-knockout mice, we have now shown that FSP27 plays a key role not only in efficient energy storage but also in whole-body energy metabolism by promoting the packaging of TAG in the unilocular lipid droplets of white adipocytes.
-
Results
Expression of FSP27 is largely restricted to WAT. PPARγ is important for adipogenesis and for maintenance of the characteristics of mature adipocytes (12). To identify molecules that mediate this latter function of PPARγ, we performed microarray analysis with total RNA isolated from mature 3T3-L1 adipocytes infected either with an adenovirus encoding a dominant-negative mutant of PPARγ (PPARγ-ΔC) that attenuates mature adipocyte functions (12) or with a control adenovirus. This analysis revealed FSP27 mRNA as one of the transcripts whose abundance was most decreased by expression of PPARγ-ΔC. FSP27 was originally identified as a fat-specific molecule (10) and was later shown to be associated with intracellular lipid droplets (13), but its function has remained unknown. We were not able to detect FSP27 mRNA in 3T3-L1 preadipocytes by northern blot analysis, but expression of Fsp27 was induced in these cells in association with their differentiation into adipocytes (Figure 1A). We also confirmed that expression of PPARγ-ΔC resulted in a decrease in the amount of FSP27 mRNA in 3T3-L1 adipocytes (Figure 1B). Furthermore, the thiazolidinedione BRL49653, an agonist of PPARγ that promotes adipocyte differentiation (14), increased the abundance of FSP27 mRNA in 3T3-L1 adipocytes, whereas TNF-α, which inhibits adipocyte differentiation (15), induced a marked decrease in Fsp27 expression in these cells (Figure 1B and Supplemental Figure 1; supplemental material available online with this article; doi: 10.1172/JCI34090DS1). These results suggested that the expression of Fsp27 is associated with the differentiation or maturation of adipocytes.
 Figure 1
Figure 1Adipocyte-specific expression of Fsp27. (A) Northern blot analysis of FSP27 mRNA in 3T3-L1 cells at the indicated times after the onset of induction of adipocyte differentiation. The region of the ethidium bromide–stained gel containing 28S rRNA is also shown. (B) Northern blot analysis of FSP27 mRNA in 3T3-L1 adipocytes, either after incubation for 48 hours with 5 μM BRL49653 or TNF-α (10 ng/ml) or 48 hours after infection with adenoviral vectors encoding wild-type mouse PPARγ (adex-PPARγ-WT) or PPARγ-ΔC (adex-PPARγ-ΔC) or with the corresponding empty vector (adex-empty), at an MOI of 60 PFU per cell. (C) Northern blot analysis of FSP27 mRNA in various organs and tissues of C57BL/6J mice at 4 weeks of age. Sub, subcutaneous; Epi, epididymal. (D) Immunoblot analysis of FSP27 in total lysates of WAT (subcutaneous or epididymal) and BAT isolated from C57BL/6J mice at 4 weeks of age. Both α-tubulin (loading control) and UCP-1 (BAT marker) were also examined. (E) Northern blot analysis of FSP27 mRNA in HB2 and HW cells at the indicated times after the onset of induction of adipocyte differentiation. (F) Immunoblot analysis of FSP27 in HB2 and HW cells at the indicated times after the onset of induction of adipocyte differentiation using the antibodies to a COOH-terminal peptide of FSP27. Both α-tubulin and β-actin (loading controls) as well as COXIV (mitochondrial marker) were also examined.
We next investigated the tissue distribution of FSP27 mRNA in mice and found that Fsp27 is expressed almost exclusively in adipose tissue, including subcutaneous WAT, epididymal WAT, and brown adipose tissue (BAT) (Figure 1C). In contrast, we found that FSP27 protein was abundant in WAT but almost undetectable in BAT by immunoblot analysis, using the antibodies generated in response to a COOH-terminal peptide of FSP27 (Figure 1D). To further examine the expression of FSP27, we isolated mature brown adipose cells and white adipose cells by collagenase digestion and subjected them to immunoblot analysis with 2 different types of antibodies to FSP27. The antibodies to the COOH-terminal peptide did not yield a major band corresponding to FSP27 in isolated brown adipose cells, although a faint band that migrated slightly slower than the major band in isolated white adipose cells was observed (Supplemental Figure 2). The antibodies to the GST fusion protein of FSP27 also did not yield an immunoreactive band corresponding to FSP27 in isolated brown adipose cells (Supplemental Figure 2). In addition, we further examined the expression of FSP27 in brown adipocyte (HB2) and white adipocyte (HW) cell lines that were established from preadipocytes of BAT and WAT, from mice deficient in the tumor suppressor p53 (16). After induction of adipogenesis, HB2 and HW cells show morphological characteristics of brown and white adipocytes, including the formation of small and large lipid droplets in the cytoplasm, respectively (Supplemental Figure 3). Northern blot analysis revealed that expression of Fsp27 was induced in both HB2 and HW cells during their differentiation into mature adipocytes (Figure 1E). However, expression of FSP27 protein was induced only in HW adipocytes (Figure 1F). We next attempted to overexpress FSP27 in HW white adipocytes and HB2 brown adipocytes with the use of an adenoviral vector. In HW adipocytes infected with an adenovirus encoding FSP27, the expressed protein appeared by immunofluorescence microscopy to be localized both surrounding the lipid droplets and in the cytoplasm (Supplemental Figure 4). In contrast, FSP27 was not detected in HB2 adipocytes infected with the adenovirus for this protein, even though it was apparent in preadipocytes scattered among the HB2 adipocytes (Supplemental Figure 4). These results suggest that FSP27 may be unstable in brown adipocytes even if overexpressed exogenously. Together, these results indicated that FSP27 is expressed at the protein level mainly in WAT, and they suggested that FSP27 plays an important role in mature WAT.
Protection from diet-induced obesity and insulin resistance and increased energy expenditure in FSP27-knockout mice. To investigate the physiological role of FSP27 in vivo, we generated FSP27-knockout mice by homologous recombination (Supplemental Figure 5). FSP27 homozygous knockout (FSP27-KO) mice were born in a Mendelian ratio and appeared physically normal at birth. Food intake did not differ between wild-type and FSP27-KO mice (Table 1). The body weights of wild-type, FSP27 heterozygous knockout, and FSP27-KO mice fed a standard diet also did not differ up to 14 weeks of age (Figure 2A). However, the body weight of FSP27-KO mice was significantly decreased compared with that of wild-type mice from 16 weeks of age (Supplemental Figure 6). For animals fed a high-fat diet, the gain in body weight was significantly smaller for FSP27-KO mice than for wild-type or FSP27 heterozygous knockout mice from 11 or 10 weeks of age, respectively (Figure 2A). These results suggested that FSP27-KO mice dissipate more energy than wild-type mice.
 Figure 2
Figure 2Glucose and energy metabolism in FSP27-knockout mice. (A) Body weight of wild-type, FSP27 heterozygous knockout (+/–), and FSP27-KO (–/–) mice maintained on a standard diet (SD) or fed a high-fat diet (HFD). Data are mean ± SEM (n = 7–11). §P < 0.05 versus corresponding values for wild-type or FSP27 heterozygous knockout mice on the high-fat diet; #P < 0.05 versus corresponding value for FSP27 heterozygous knockout mice on the high-fat diet. (B) Plasma glucose concentrations during an intraperitoneal glucose tolerance test (left panel) or an insulin tolerance test (right panel) in 12-week-old wild-type and FSP27-KO (KO) mice, either maintained on a standard diet or fed a high-fat diet from 4 weeks of age. Data are mean ± SEM (n = 11 for standard diet; n = 8 for high-fat diet.). *P < 0.05, **P < 0.01 versus corresponding value for wild-type animals on the standard diet; †P < 0.05, ††P < 0.01 versus corresponding value for wild-type animals on the high-fat diet. (C) Hyperinsulinemic-euglycemic clamp analysis in 10- to 12-week-old wild-type and FSP27-KO mice fed a high-fat diet. GIR, glucose infusion rate; Rd, rate of glucose disappearance; BHGP, basal hepatic glucose production; CHGP, hepatic glucose production during the clamp period. Data are mean ± SEM (n = 6). §§P < 0.05, ##P < 0.01 versus corresponding value for wild type. (D and E) Whole-body oxygen consumption rate (VO2, expressed in milliliters per minute per gram of body weight) during a 12-hour dark/12-hour light cycle in 12-week-old mice fed a standard diet (n = 3 for wild type; n = 5 for FSP27-KO) (D) or a high-fat diet (n = 6) (E) is shown in the left panels. The average values for the 24-hour period are shown in the right panels. Data are mean ± SEM. ‡P < 0.05, ‡‡P < 0.01 versus wild type. (F and G) Average values of respiratory quotient (VCO2/VO2) (F) and energy expenditure (G) for the 24-hour period in the experiments shown in D and E. Data are mean ± SEM. ‡P < 0.05, ‡‡P < 0.01 versus wild type.
We next examined the effects of loss of FSP27 on metabolic parameters. FSP27-KO mice maintained on a standard diet showed both a significantly lower plasma glucose level in the fed state (Table 1) and a significantly higher glucose tolerance in an intraperitoneal glucose tolerance test (Figure 2B) compared with wild-type mice. Plasma glucose levels in both the fasting and fed states were significantly lower in FSP27-KO mice maintained on a high-fat diet than in wild-type animals on the same diet (Table 1). In addition, FSP27-KO mice appeared to be largely protected from the glucose intolerance and insulin resistance induced by the high-fat diet in wild-type mice (Figure 2B). Hyperinsulinemic-euglycemic clamp analysis also revealed that the glucose infusion rate was significantly increased and that hepatic glucose production during the clamp period was significantly decreased in FSP27-KO mice fed a high-fat diet compared with those in wild-type mice (Figure 2C), confirming that FSP27-KO mice were indeed protected from insulin resistance induced by such a diet. Furthermore, FSP27-KO mice were also protected from high-fat diet–induced accumulation of fat in the liver (Table 1). With regard to energy metabolism, body temperature did not differ significantly between FSP27-KO and wild-type mice (36.3 ± 0.08°C versus 36.4 ± 0.08°C, respectively; mean ± SEM, n = 4 and 3, respectively) maintained at room temperature. However, the basal rate of oxygen consumption was significantly increased in FSP27-KO mice maintained on a standard or high-fat diet compared with the corresponding values for wild-type animals (Figure 2, D and E). The respiratory quotient was slightly but significantly higher for FSP27-KO mice maintained on a high-fat diet than for wild-type animals (Figure 2F). Energy expenditure was thus significantly increased in FSP27-KO mice maintained on either a standard or high-fat diet (Figure 2G). Monitoring of the frequency of voluntary movements revealed that locomotor activity tended to be reduced in FSP27-KO mice compared with that in wild-type animals, although this difference was not statistically significant (Supplemental Figure 7), suggesting that the increased energy expenditure in FSP27-KO mice was not attributable to hyperactivity.
Multilocular lipid droplet formation as well as enhanced mitochondrial biogenesis and glucose and FFA oxidation in WAT of FSP27-KO mice. To investigate the mechanism responsible for the increased oxygen consumption and energy expenditure of FSP27-KO mice, we examined adipose tissue of these animals. The amounts of epididymal and subcutaneous WAT, but not those of BAT, were reduced in FSP27-KO mice compared with those in wild-type mice (Figure 3A). Furthermore, epididymal WAT of FSP27-KO mice was a brownish color similar to that of BAT (Figure 3A). A high-fat diet increased the amounts of epididymal and subcutaneous WAT to a greater extent in wild-type mice than in FSP27-KO mice (Supplemental Figure 8). The weight and appearance of adipose tissue of FSP27 heterozygous knockout mice were similar to those for wild-type mice (data not shown). Both subcutaneous and epididymal white adipocytes of FSP27-KO mice accumulated multilocular fat droplets (Figure 3B and Supplemental Figure 9). The size of lipid droplets in BAT of FSP27-KO mice also appeared to be increased (Figure 3B and Supplemental Figure 9). Transmission electron microscopy confirmed that lipid droplets in subcutaneous white adipocytes of FSP27-KO mice are multilocular and smaller than those of wild-type mice (Figure 3C). It also revealed that the number of mitochondria per cell in WAT of FSP27-KO mice was twice that in wild-type mice, although the size of these organelles did not differ between the 2 genotypes (Figure 3, C and D). In addition, Southern blot analysis with a probe specific for the mitochondrial gene for subunit I of cytochrome c oxidase (COXI) showed that the amount of mitochondrial DNA was increased in WAT of FSP27-KO mice (Figure 3E). No difference in mitochondrial number or size in BAT was apparent between FSP27-KO and wild-type mice (Figure 3, C and D). These results thus showed that mitochondrial biogenesis is substantially increased in WAT of FSP27-KO mice.
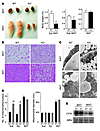 Figure 3
Figure 3Characterization of adipose tissue in FSP27 knockout mice. (A) Comparison of interscapular brown fat pads (BAT) and epididymal white fat pads (WAT) of 14-week-old wild-type and FSP27-KO mice (left panel). Weights of epididymal WAT, subcutaneous WAT, and BAT isolated from 14-week-old wild-type and FSP27-KO mice (right panels). Data are mean ± SEM (n = 4–8). †P < 0.05 versus the corresponding value for wild type. (B) Sections of subcutaneous WAT and interscapular BAT from 14-week-old wild-type and FSP27-KO mice were stained with hematoxylin-eosin and examined by light microscopy. Scale bar: 20 μm. (C) Transmission electron microscopy of subcutaneous WAT and interscapular BAT from 6-week-old wild-type and FSP27-KO mice. L, lipid droplet; *, mitochondria. Scale bar: 500 nm. (D) Mitochondrial number (left panel) and size (right panel) in subcutaneous WAT, epididymal WAT, and interscapular BAT determined from electron micrographs similar to those in C. Mitochondrial number is expressed per nucleus and was determined for 25 wild-type and 28 FSP27-KO cells for subcutaneous WAT, 33 cells for epididymal WAT, and 56 wild-type and 64 FSP27-KO cells for BAT. Mitochondrial diameter was measured with a scale of 25 nm in 120 cells. Data are mean ± SEM. ††P < 0.01 versus the corresponding value for wild type. (E) Southern blot analysis of total cellular DNA from BAT or WAT of 9-week-old wild-type or FSP27-KO mice with probes specific for the mitochondrial COXI gene and the nuclear 36B4 gene.
We next examined the abundance in adipose tissue of mRNAs for nuclear respiratory factor 1 (NRF1) and mitochondrial transcription factor A (mtTFA), both of which are transcription factors that play an important role in mitochondrial biogenesis (17, 18). RT-PCR analysis revealed that the amount of NRF1 mRNA in WAT was significantly increased in FSP27-KO mice, reaching a level similar to that apparent in BAT of wild-type animals (Figure 4A). The amount of mtTFA mRNA in WAT was also increased significantly in FSP27-KO mice (Figure 4A). In addition, the amounts of mRNAs for PPARγ coactivator 1α (PGC1a) and PGC1b, important regulators of mitochondrial thermogenesis and biogenesis (19), were significantly increased in WAT of FSP27-KO mice, but these increased levels remained much smaller than those in BAT (Figure 4A). We also examined the abundance of mRNAs for acyl-CoA dehydrogenases, which are important for fatty acid oxidation in mitochondria. The amounts of mRNAs for very long-chain acyl-CoA dehydrogenase (VLCAD), long-chain acyl-CoA dehydrogenase (LCAD), and medium-chain acyl-CoA dehydrogenase (MCAD) were increased in WAT of FSP27-KO mice (Figure 4B). Furthermore, the amounts of mRNAs for COXI, COXII, and COXIV, all of which are components of COX and important for electron transport in mitochondria, were substantially increased in WAT of FSP27-KO mice (Figure 4C). However, the mRNA for uncoupling protein–1 (UCP1) was almost undetectable in WAT of FSP27-KO mice in contrast to its abundance in BAT (Figure 4C). These results thus suggested that the increase in mitochondrial mass in WAT of FSP27-KO mice is accompanied by an increase in FFA oxidation and subsequent oxidative phosphorylation but not by increased uncoupling or thermogenesis. To exclude a possible contribution of the liver and skeletal muscle to the increased oxygen consumption observed in FSP27-KO mice, we examined the abundance of mRNAs for carnitine palmitoyltransferase–1 (CPT1) and acyl-CoA dehydrogenases (VLCAD, LCAD, and MCAD) in these tissues by RT-PCR analysis and found no significant differences between FSP27-KO and wild-type mice (data not shown). In addition, there were no substantial differences between wild-type and FSP27-KO mice in the extent of lipid droplet accumulation as revealed by staining of TAG with oil red O in the liver or skeletal muscle (Supplemental Figure 10).
 Figure 4
Figure 4Mitochondrion-related gene expression and metabolic rate in WAT of FSP27-knockout mice. (A and B) Quantitative RT-PCR analysis of the expression of genes related to mitochondrial biogenesis (A) or to FFA oxidation (B) in WAT and BAT of 20-week-old wild-type and FSP27-KO mice. Data were normalized by the amount of 36B4 mRNA and expressed relative to the corresponding value for WAT of wild-type mice; data are mean ± SEM (n = 4). *P < 0.05, **P < 0.01, ***P < 0.001 versus the corresponding value for wild-type mice. (C) Northern blot analysis of mRNAs for COXI, COXII, COXIV, and UCP1 in WAT and BAT of 14-week-old wild-type and FSP27-KO mice. (D) Oxygen consumption by adipocytes isolated from inguinal WAT (IWAT; left panel) or interscapular BAT (right panel) of wild-type and FSP27-KO mice. Arrows indicate the addition of the β3-adrenergic agonist CL316,243 or vehicle (Ca2+- and Mg2+-free PBS). Data are mean ± SEM of values from 4 independent experiments. (E and F) Glucose (E) and oleic acid (F) oxidation in adipocytes isolated from epididymal WAT or interscapular BAT of wild-type and FSP27-KO mice. Data are expressed relative to the corresponding value for WAT of wild-type mice and are mean ± SEM of values from 4 WT or 3 KO independent experiments. †P < 0.05, ††P < 0.01 versus the corresponding value for wild-type cells.
We also examined urinary catecholamines in FSP27-KO mice in order to evaluate the extent of systemic sympathetic activation, given that systemic β-adrenergic stimulation in mice and rats results in fragmentation of large lipid storage droplets and induction of mitochondrial biogenesis in WAT as well as upregulation of genes involved in fatty acid oxidation and mitochondrial electron transport in this tissue (20, 21). However, we found no significant differences in the amounts of urinary catecholamines (mean ± SEM) collected over a period of 72 hours between wild-type (n = 3) and FSP27-KO (n = 4) mice (adrenaline, 84.7 ± 10.3 versus 73.5 ± 6.5 ng, respectively; noradrenaline, 1,275.6 ± 36.1 versus 1,234.4 ± 168.5 ng, respectively; dopamine, 2,966.3 ± 212.3 versus 2,063.8 ± 299.4 ng, respectively), suggesting that the changes in WAT of FSP27-KO mice are not attributable to sympathetic β-adrenergic stimulation.
To examine directly the possible role of FSP27 in mitochondrial function in adipocytes, we first measured oxygen consumption of such cells isolated from FSP27-KO and wild-type mice. Consistent with the increased number of mitochondria in white adipocytes of FSP27-KO mice, the basal oxygen consumption of these cells was about twice that of cells from wild-type mice (Figure 4D). However, the β3-adrenergic agonist CL316,243 did not substantially increase oxygen consumption in white adipocytes of wild-type or FSP27-KO mice (Figure 4D). In addition, the uncoupled respiration ratio for white adipocytes in the basal state did not differ between wild-type and FSP27-KO mice (Supplemental Figure 11), possibly reflecting the low abundance of UCP1 mRNA in white adipocytes of both genotypes (Figure 4C). These results showed that mitochondrial function is upregulated in WAT of FSP27-KO mice, suggesting that the increased energy expenditure of these animals is attributable to increased mitochondrial biogenesis and function in WAT. In contrast to its lack of effect in WAT, CL316,243 induced a marked increase in oxygen consumption by adipocytes isolated from BAT of FSP27-KO or wild-type mice (Figure 4D). However, both basal and CL316,243-stimulated oxygen consumption were reduced by approximately 50% in brown adipocytes of FSP27-KO mice compared with those apparent for wild-type cells (Figure 4D). To examine further the role of FSP27 in mitochondrial function, we measured glucose and FFA oxidation in isolated adipocytes. Glucose and oleic acid oxidation were both increased in white adipocytes of FSP27-KO mice (Figure 4, E and F), consistent with the notion that mitochondrial function is upregulated in these cells. In contrast, oleic acid oxidation was decreased in brown adipocytes of FSP27-KO mice (Figure 4F). The findings that oxygen consumption and FFA oxidation were both decreased in brown adipocytes of FSP27-KO mice may be related to the increased size of lipid droplets in BAT of these animals (Figure 3B). Given that FSP27 was virtually undetectable in BAT of wild-type mice (Figure 1D), these changes observed in BAT of FSP27-KO mice may represent a compensatory response to the increased energy expenditure in WAT.
Ablation of FSP27 results in formation of multiple small lipid droplets, enhanced lipolysis, and depletion of TAG in HW adipocytes. Immunofluorescence microscopy with antibodies to FSP27 revealed that FSP27 was distributed in a ring around the lipid droplets of HW adipocytes (Figure 5A), suggesting that FSP27 localizes to the surface of these droplets. We also examined the distribution of perilipin, which is localized almost exclusively at the surface of lipid droplets in fat cells and steroidogenic cells and which inhibits lipolysis by blocking the access of lipases to the droplets (22–24). Perilipin was also localized to the surface of lipid droplets in HW adipocytes (Figure 5B), showing a distribution pattern similar to that of FSP27.
 Figure 5
Figure 5Localization of FSP27 to lipid droplets and its role in the pattern of lipid droplet accumulation in HW adipocytes. (A and B) Immunofluorescence localization of FSP27 (A) and perilipin (B) in HW adipocytes by confocal laser microscopy. TAG was stained with Bodipy 493/503. (C) Immunoblot analysis of FSP27, perilipin, and β-actin (control) in HW adipocytes 2 days after the introduction of FSP27 or perilipin siRNAs as indicated. (D) Pattern of lipid droplet accumulation in HW adipocytes 2 days after siRNA introduction as in C. TAG was stained with Bodipy 493/503. (E) Glycerol release of HW adipocytes during incubation for 4 or 8 hours, 2 days after introduction of siRNAs as in C. Data are mean ± SEM (n = 6) and are expressed relative to the value for control cells incubated for 4 hours. *P < 0.01 versus corresponding value for control siRNA; **P < 0.01 versus corresponding value for FSP27 or perilipin siRNA. (F) Glycerol release of HW adipocytes during incubation for 40 minutes with 10 μM isoproterenol 2 days after introduction of siRNAs as in C. Data are mean ± SEM (n = 5) and are expressed relative to the value for control cells. *P < 0.01 versus corresponding value for control siRNA; †P < 0.01 versus corresponding value for perilipin siRNA. (G) Isolated white adipocytes of wild-type or FSP27-KO mice were incubated for 60 minutes in the absence (basal) or presence of 10 μM isoproterenol, after which concentrations of FFAs (left panel) and glycerol (right panel) in the culture medium were measured. Data are mean ± SEM (n = 5 WT or 4 KO for FFAs; n = 4 for glycerol) and are expressed relative to the corresponding basal value for wild-type cells. §P < 0.05, §§P < 0.01 versus corresponding value for wild type. (H) Serum FFA (left panel) and glycerol (right panel) concentrations in wild-type and FSP27-KO mice measured 20 minutes after intraperitoneal injection of CL316,243 (0.1 mg/kg) or vehicle. Data are mean ± SEM (n = 4 WT or 3 KO) and are expressed relative to the corresponding basal value for wild-type mice. Original magnification, ×630.
We next examined the effect of depletion of FSP27 by RNAi on lipid droplet formation in HW adipocytes. Introduction of an siRNA specific for FSP27 mRNA into differentiated HW adipocytes resulted in the almost complete loss of FSP27 within 2 days (Figure 5C). This depletion of FSP27 resulted in the formation of numerous small lipid droplets in HW adipocytes, in contrast to the fewer and larger lipid droplets observed in control cells (Figure 5D). Quantitative analysis confirmed that depletion of FSP27 in HW adipocytes for 2 days resulted in an increase in lipid droplet number and a decrease in the mean area of individual droplets per cell (Supplemental Figure 12). Cells depleted of perilipin (Figure 5C) still manifested large lipid droplets, although all lipid droplets in such cells were generally smaller than those in control cells (Figure 5D). Cells depleted of both FSP27 and perilipin (Figure 5C) exhibited a pattern of lipid droplet formation similar to that observed in cells depleted of FSP27 alone (Figure 5D). The intracellular TAG content of HW adipocytes was significantly reduced 3 days after the introduction of FSP27 or perilipin siRNAs, and the effects of the 2 siRNAs appeared to be additive (Supplemental Figure 13). Measurement of glycerol released into the medium during culture of cells under basal conditions for 4 or 8 hours revealed that the extent of lipolysis was significantly increased in FSP27-depleted HW adipocytes compared with that apparent in control adipocytes at both time points (Figure 5E). Consistent with previous observations (25, 26), depletion of perilipin also increased the extent of basal lipolysis in HW adipocytes (Figure 5E). Depletion of both FSP27 and perilipin again increased basal lipolysis to a greater extent than did that of either protein alone. Examination of lipolysis in response to β-adrenergic stimulation revealed that depletion of FSP27 and perilipin also potentiated isoproterenol-induced lipolysis in an approximately additive manner (Figure 5F). These results suggested that FSP27 is important for the formation of large lipid droplets and that the loss of FSP27 promotes lipolysis and the consequent depletion of intracellular TAG by a mechanism distinct from that by which loss of perilipin induces this same effect.
To address further the function of FSP27 in lipolysis, we investigated the effect of FSP27 depletion on lipolysis both in isolated mouse adipocytes and in mice. We evaluated lipolysis in isolated adipocytes by measuring FFA and glycerol concentrations of the culture medium. Basal lipolysis was increased in isolated white adipocytes from FSP27-KO mice, although isoproterenol-stimulated lipolysis was impaired in these cells compared with that in white adipocytes from wild-type mice (Figure 5G). We next administered CL316,243 to mice and measured the products of lipolysis, FFAs and glycerol, in serum. Serum FFA and glycerol concentrations were similar in wild-type and FSP27-KO mice under basal or CL316,243-stimulated conditions (Figure 5H). These results differed from those previously obtained with perilipin-knockout mice, in which lipolysis stimulated by a β-adrenergic agonist in vivo was decreased compared with that in wild-type mice (25). The physiological role of FSP27 in mice may thus differ from that of perilipin.
We next examined the effects of FSP27 ablation on gene expression in HW adipocytes at both 2 and 4 days after the introduction of FSP27 siRNA. We confirmed that FSP27 remained almost completely depleted 4 days after siRNA introduction (data not shown). As shown above, depletion of FSP27 for 2 days was sufficient to induce multilocularization of lipid droplets (Figure 5D) and to increase lipolysis (Figure 5, E and F). RT-PCR analysis performed 2 days after introduction of siRNA revealed that the amounts of mRNAs for diacylglycerol acyltransferase type 1 (DGAT1), DGAT2, and stearoyl-CoA desaturase 1 (SCD1), all of which participate in lipid synthesis (27–31), did not differ between FSP27-depleted and control HW adipocytes (data not shown). In addition, the amounts of mRNAs for hormone-sensitive lipase (HSL) and adipose triglyceride lipase (ATGL), both of which contribute to TAG hydrolysis in adipocytes (32), were not significantly different between the FSP27-depleted and control cells (data not shown). However, 4 days after the introduction of siRNA, the amounts of these various mRNAs, with the exception of that of SCD1 mRNA, were all increased in the FSP27-depleted cells (Figure 6, A and B). The abundance of mRNAs for perilipin, adipophilin, and TIP47, all of which coat lipid droplets and are important in lipid metabolism (7, 33), was not affected by depletion of FSP27 for 2 days (data not shown), but the amounts of perilipin and adipophilin mRNAs were increased significantly after 4 days (Figure 6C). Finally, we examined the expression of genes related to mitochondrial biogenesis or oxidative phosphorylation. Depletion of FSP27 for 2 days did not affect the abundance of mRNAs for NRF1, mtTFA, PGC1a, PGC1b, or the acyl-CoA dehydrogenases (VLCAD, LCAD, MCAD) in HW adipocytes (data not shown). The abundance of COXII mRNA was significantly increased (~1.87-fold; P < 0.008) by FSP27 depletion for 2 days, whereas that of COXIV mRNA was unaffected (data not shown). However, depletion of FSP27 for 4 days resulted in increases in the amounts of these various mRNAs, with the exception of those of PGC1a and MCAD (Figure 6, D–F). These results thus suggested that the increased expression of genes involved in lipid metabolism, lipid coating, or mitochondrial biogenesis or function is not the cause of multilocularization of lipid droplets but may be the result of it.
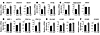 Figure 6
Figure 6Effects of FSP27 depletion for 4 days on gene expression in HW adipocytes. Quantitative RT-PCR analysis of the expression of genes whose products are associated with lipid synthesis (A), lipolysis (B), lipid droplets (C), mitochondrial biogenesis (D), fatty acid oxidation (E), or oxidative phosphorylation (F) in HW adipocytes 4 days after the introduction of FSP27 siRNA (KD). Data were normalized by the amount of 36B4 mRNA and expressed relative to the corresponding value for control cells; they are mean ± SEM from 4 independent experiments. Cont, control. *P < 0.01 versus control.
Forced expression of FSP27 promotes formation of large lipid droplets in COS cells. To confirm the function of FSP27 in WAT, we transfected COS cells, which do not express endogenous FSP27, with a vector that encodes both FSP27 and the fluorescence marker DsRed2. The expression of FSP27 in the transfected cells was confirmed by immunoblot analysis (Figure 7A). Control COS cells accumulated multiple small lipid droplets in the cytoplasm during culture for 24 hours in the presence of 400 μM oleate. In contrast, COS cells expressing FSP27, which were identified by monitoring the fluorescence of DsRed2, accumulated large lipid droplets (Figure 7B). Quantitative analysis revealed that the mean area of individual droplets per cell was significantly increased (Figure 7C), whereas the droplet number per cell was significantly decreased (Figure 7D) in cells expressing FSP27 compared with that in control cells. In addition, the total amount of lipid per cell was significantly increased in cells expressing FSP27 (Figure 7E). These results thus indicated that FSP27 promotes the formation of large lipid droplets and, consequently, increases cellular lipid content.
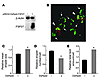 Figure 7
Figure 7Formation of large lipid droplets induced by forced expression of FSP27 in COS cells. (A) Immunoblot analysis of FSP27 and β-actin (loading control) in COS cells 2 days after transfection with an expression plasmid encoding both FSP27 and DsRed2 (pIRES2-DsRed2-FSP27). (B) COS cells transfected with pIRES2-DsRed2-FSP27 as in A were incubated with 400 μM oleic acid for 24 hours and then stained with Bodipy 493/503 (for TAG). A merged image of Bodipy 493/503 and DsRed2 fluorescence is shown. Arrowheads and arrows indicate cells positive or negative for FSP27 expression, respectively. Original magnification, ×630. (C–E) Quantitation of mean droplet size (C), droplet number (D), and total lipid amount (E) per cell for COS cells treated as in B but also stained with Hoechst 33258 (for nuclei). Total lipid amount was calculated as the product of the area and green fluorescence density of each droplet per cell. Data are mean ± SEM of values from 1,502 and 444 cells negative or positive for DsRed2 fluorescence, respectively, and are expressed relative to the corresponding value for DsRed2-negative cells. *P < 0.001 versus DsRed2-negative cells.
-
Discussion
Obesity is a disorder of energy balance, in which excess energy accumulates in WAT in the form of TAG. In white adipocytes, TAG is stored in large, often solitary cytoplasmic organelles, known as lipid droplets. These droplets are composed of a core of TAG surrounded by a phospholipid monolayer, in which numerous proteins are embedded (5). To understand the pathogenesis of obesity, it is therefore essential to characterize the mechanism of lipid droplet formation and its regulation. We have now shown that FSP27 is expressed at the protein level mainly in WAT, is associated with the large unilocular lipid droplets of white adipocytes, and is indispensable for the formation or maintenance of these droplets. In contrast to WAT, BAT accumulates small multilocular lipid droplets, likely because FSP27 is almost undetectable in this tissue, even though its mRNA is present in amounts similar to those in WAT.
Mammalian lipid droplets are associated with a specific set of proteins that are members of the PAT (perilipin-ADRP-TIP47) domain family, which include perilipin, ADRP (or adipophilin), TIP47, and S3-12 (7, 8). Loss of perilipin results in constitutive lipolysis in adipocytes under basal conditions (25, 26). We also found that perilipin depletion increased lipolysis in HW adipocytes, but this effect was not accompanied by a pronounced change in the pattern of lipid droplet accumulation in contrast to the multilocularization of lipid droplets induced in these cells by depletion of FSP27. These results suggest that the lipolysis induced in HW adipocytes by depletion of FSP27 is mediated by a mechanism distinct from that by which perilipin depletion elicits this effect. The change in the pattern of lipid droplet accumulation induced by loss of FSP27 suggests that the accompanying increase in lipolysis may result from the multilocularization of large lipid droplets. Such multilocularization thus increases the total surface area of lipid droplets in a cell and thereby likely increases the access of lipases to stored lipid. Thus, multilocularization of lipid droplets can be the cause of the increased lipolytic rate in WAT lacking FSP27. Conversely, unilocular lipid droplets are thought to impede lipolysis by minimizing the accessibility of stored lipid to lipases and thereby to be important for efficient TAG storage in WAT. Acute and robust lipolysis of multilocular lipid droplets may be necessary for the efficient supply of FFAs to mitochondria in energy-consuming tissues such as BAT. Indeed, the small lipid droplets of BAT are surrounded by a large number of mitochondria, which are responsible for the uncoupled respiration of FFAs derived from lipid droplets (34). The pattern of intracellular lipid formation may therefore be closely related to the metabolic characteristics of cells.
FSP27-KO mice show a lean phenotype, increased metabolic rate, and resistance to diet-induced obesity. These characteristics likely result from the decreased TAG storage function, the increased mitochondrial biogenesis, and the increased energy expenditure in WAT of these animals. Depletion of FSP27 in HW adipocytes for 2 days resulted in the formation of multiple small lipid droplets and enhanced lipolysis, without an increase in the expression of genes involved in lipid metabolism or mitochondrial biogenesis or function. However, depletion of FSP27 for 4 days led to upregulation of the expression of such genes. These results suggest that the primary event induced by FSP27 depletion may indeed be the multilocularization of lipid droplets and the consequent increase in lipolysis. The increased mitochondrial biogenesis observed in WAT of FSP27-KO mice hence may be secondary to the increased lipolysis. The increased availability of FFAs and their metabolites may stimulate members of the PPAR family in WAT of FSP27-KO mice and thereby induce the secondary mitochondrial biogenesis. Indeed, several FFAs have been shown to stimulate PPARα and PPARδ (35). In addition, activation of PPARα and PPARγ has been shown to increase mitochondrial copy number as well as the expression of UCP-1, MCAD, and CPT-1 (36). Activation of PPARδ in adipose tissue also induces expression of genes required for FFA oxidation and energy expenditure (37). The increase in intracellular lipolysis in adipocytes can thus lead to increased mitochondrial biogenesis and FFA oxidation.
Perilipin-knockout mice are characterized by constitutive lipolysis. They have reduced adipose mass and are also resistant to diet-induced obesity (25, 26). These phenotypes are similar to those of FSP27-KO mice. The most prominent difference between these 2 mouse lines is the pattern of lipid droplet formation in WAT. The multilocular pattern of lipid droplet formation observed in WAT of FSP27-KO mice is thus not apparent in perilipin-knockout mice (25, 26). In addition, increased mitochondrial biogenesis was not apparent in WAT of the perilipin-knockout mice, at least not in histological analysis. Furthermore, perilipin-knockout mice show a tendency to develop insulin resistance (26, 38), despite increased β-oxidation in skeletal muscle, heart, and WAT (38, 39), that is thought to be an adaptive response to consume the FFAs generated by the increased rate of lipolysis in WAT. In contrast, FSP27-KO mice showed improved glucose tolerance and insulin sensitivity, without an apparent increase in β-oxidation in the liver or skeletal muscle. These results suggest that the FFAs generated by the increased lipolysis in WAT of FSP27-KO mice are efficiently oxidized by the increased number of mitochondria in WAT itself, without the need for compensation by the liver or skeletal muscle. The multilocular pattern of lipid droplet accumulation in WAT of FSP27-KO mice likely facilitates the efficient transfer of FFAs to the abundant neighboring mitochondria. These data suggest the mechanism by which increased lipolysis in WAT does not result in insulin resistance in FSP27-KO mice.
FSP27 belongs to the CIDE (cell death–inducing DFF45-like effector) family of proteins, which show sequence similarity to the NH2-terminal region of the proapoptotic DNA fragmentation factor 40 (DFF40) and its inhibitor DFF45 (40). This family also includes CIDE-A and CIDE-B in addition to FSP27 (40). CIDE-A is expressed specifically in BAT in mice, and its deficiency in these animals results in resistance to diet-induced obesity as well as in upregulation of thermogenesis and metabolic rate through release of direct suppression of UCP-1 activity (41). CIDE-B is expressed predominantly in liver, and CIDE-B–knockout mice are resistant to diet-induced obesity and liver steatosis (42). FSP27 and CIDE-A may therefore play regulatory roles in mice for lipid storage in WAT and energy expenditure in BAT, respectively. In humans, however, CIDEA mRNA is present in WAT, and a low level of CIDEA gene expression in adipose tissue was associated with several features of metabolic syndrome (43). In addition, there are several similarities between human CIDE-A and mouse FSP27. Culture of human primary adipocytes with TNF-α thus resulted in a decrease in the amount of CIDEA mRNA, and depletion of CIDE-A from these cells by RNAi induced an increase in lipolysis (43). The human ortholog of mouse FSP27 has been identified, but the characteristics of this protein, including its expression in adipose tissue, were not examined (44). During the preparation of this manuscript, FSP27 was shown to enhance TAG storage in cultured cells (11). Further studies are necessary to characterize the functions of CIDE family proteins, especially in humans.
In summary, we have shown that FSP27 plays a physiologically important role in efficient energy storage in WAT, by promoting the formation of unilocular lipid droplets in this tissue. Such regulation of lipid droplet formation by FSP27 has pronounced effects on energy metabolism of the whole body. Our data suggest that alteration of the pattern of lipid droplet accumulation in white adipocytes by modulation of FSP27 function is a potential therapeutic strategy for treatment of obesity and its related disorders.
-
Methods
Oligonucleotide microarray analysis. Total RNA was prepared from 3T3-L1 cells that had been induced to differentiate into adipocytes as previously described (12) and then infected for 48 hours with an adenovirus encoding PPARγ-ΔC (12) or a control adenovirus at an MOI of 60 PFU per cell. RT, second-strand cDNA synthesis, and production of biotin-labeled cRNA were performed with 10 μg of total RNA as the initial template. The labeled cRNA was purified, fragmented, and subjected to hybridization with GeneChip mouse microarrays (Mu11K; Affymetrix). After washing and staining, the arrays were scanned with a Hewlett Packard confocal laser scanner and visualized with Affymetrix GeneChip 3.1 software. The data were analyzed with GeneChip Analysis Suite software version 4.0 (Affymetrix), and the fold differences in hybridization intensity between the two 3T3-L1 adipocyte samples were determined.
Generation of FSP27-knockout mice. We isolated Fsp27 genomic clones from the female mouse 129s6/SvEvTAC Taconic BAC Library (Children’s Hospital Oakland Research Institute, Oakland, California, USA) and prepared a targeting construct in which the Fsp27 sequence between ApaI and XhoI sites, including the first and second exons, was replaced with the neomycin resistance gene (Supplemental Figure 5A). The targeting plasmid was linearized with NotI and introduced by electroporation into EB3/5 embryonic stem cells. Cell clones resistant to G418 and gancyclovir were isolated, and genomic DNA was extracted from these cells, digested with EcoRV, and subjected to Southern blot analysis in order to confirm homologous recombination (Supplemental Figure 5B). Two lines of chimeric mice were obtained by injection of cells from these clones into C57BL/6N blastocysts. Crossing of the chimeric mice with C57BL/6J mice yielded offspring that harbored the disrupted Fsp27 allele in the germ line. FSP27-KO mice were obtained by breeding of heterozygotes. Immunoblot analysis did not detect FSP27 in WAT of FSP27-KO mice and revealed that the amount of this protein was reduced by approximately 50% in WAT of FSP27 heterozygous-knockout mice (Supplemental Figure 5C). For the experiments in the present study, we used male mice that had been backcrossed to the C57BL/6J background for more than 6 generations. Mice were maintained under a 12-hour light/12-hour dark cycle and had free access to food and water. In experiments examining the effects of a high-fat diet, mice were fed chow in which fat constituted 56% of calories (Oriental Yeast), beginning at 4 weeks of age. Mice were obtained from Japan SLC. Experimental protocols with mice were performed in accordance with the guidelines of the and with approval of the animal ethics committee of Kobe University Graduate School of Medicine.
Isolation of total RNA, northern blot analysis, and quantitation of mitochondrial DNA. Total RNA was extracted from various tissues and cells with the use of an RNeasy kit (QIAGEN) or the TRIzol reagent (Invitrogen), and portions (10 μg) of the isolated RNA were subjected to northern blot analysis. Probes were prepared by RT-PCR from total RNA extracted from 3T3-L1 adipocytes (for FSP27 and 36B4) or from mouse BAT (for COXI, COXII, COXIV, and UCP-1). The probes were labeled with 32P with the use of a Rediprime II Random-Primer Labeling system (GE Healthcare). Autoradiograms of blots were visualized with a BAS2500 image analyzer (Fuji Film). Measurement of mitochondrial DNA was performed as described previously (45). In brief, cDNA probes for COXI and 36B4 genes were generated by RT-PCR from total RNA extracted from mouse BAT and were labeled with 32P with the use of a Rediprime Random-Primer Labeling kit. Total DNA was extracted from mouse epididymal WAT or BAT, and portions (10 μg) of the DNA were digested with EcoRV and BglII. The resulting DNA fragments were fractionated by electrophoresis on a 0.7% agarose gel and then transferred overnight to a nylon membrane (HyBond N+; GE Healthcare). The membrane was incubated for 30 minutes at 65°C with Rapid-Hyb Hybridization buffer (GE Healthcare), subjected to hybridization with the radioactive probes overnight at 65°C, and then washed at 65°C twice for 30 minutes with 0.1× SSC containing 0.1% SDS. Autoradiograms of blots were visualized with a BAS2500 image analyzer.
Preparation of HB2 and HW cells and measurement of glycerol release and cellular TAG. HB2 and HW cells were prepared from interscapular BAT and epididymal WAT, respectively, of p53 homozygous knockout mice as described previously (16). The cells were maintained in DMEM (Sigma-Aldrich) supplemented with 10% FBS (JRH Biosciences), streptomycin (50 μg/ml), and penicillin (50 U/ml). At confluence, fresh medium supplemented with 0.5 mM 3-isobutyl-1-methylxanthine and 1 nM dexamethasone was added to the cells for induction of adipocyte differentiation. After 2 days, the medium was changed to DMEM supplemented with 50 nM triiodothyronine and insulin (10 μg/ml) and was refreshed every 2 days. The amount of glycerol released into culture medium by HW cells was determined with the use of a colorimetric assay (Sigma-Aldrich). The TAG content of isopropanol extracts prepared from HW cells grown in 12-well plates was determined with an acetyl acetone–based colorimetric assay kit (Wako)
Quantitative RT-PCR. Complementary DNA synthesized from total RNA was analyzed in a Sequence Detector (model 7900; PE Applied Biosystems) with specific primers and SYBR Green PCR Master reagents (Perkin Elmer Life Sciences). The relative abundance of mRNAs was calculated with 36B4 mRNA as the invariant control. The primers (sense and antisense, respectively) were as follows: FSP27, 5′-CTGGAGGAAGATGGCACAATCGTG-3′ and 5′-CAGCCAATAAAGTCCTGAGGGTTCA-3′; mtTFA, 5′-AGTTCCCACGCTGGTAGTGT-3′ and 5′-GCGCACATCTCGACCC-3′; NRF1, 5′-CAGCAACCCTGATGGCACCGTGTCG-3′ and 5′-GGCCTCTGATGCTTGCGTCGTCTGG-3′; PGC1a, 5′-GGAGCTGGATGGCTTGGGACAT-3′ and 5′-TTCGCAGGCTCATTGTTGTACTGGT-3′; PGC1b, 5′-GGCTCTTCCGCTCACA-3′ and 5′-ACCTGGCAGTAGTCGTGGTC-3′; CPT1, 5′-GCTGCTTCCCCTCACAAGTTCC-3′ and 5′-GCTTTGGCTGCCTGTGTCAGTATGC-3′; VLCAD, 5′-GAATGACCCTGCCAAGAACGA-3′ and 5′-ATGCCCACAATCTCTGCCAAG-3′; LCAD, 5′-GGACTCCGGTTCTGCTTCCA-3′ and 5′-TGCAATCGGGTACTCCCACA-3′; MCAD, 5′-CAACACTCGAAAGCGGCTCA-3′ and 5′-ACTTGCGGGCAGTTGCTTG-3′; DGAT1, 5′-ACCGCGAGTTCTACA-3′ and 5′-AGGGGAACGCTCACTAGGTA-3′; DGAT2, 5′-GCGCTACTTCCGAGACTACTTT-3′ and 5′-GCACAGGCATCCGGAAGTTACC-3′; SCD1, 5′-TCTCAGAAACACACGCCGACC-3′ and 5′-AGGCCGGGCTTGTAGTACCTC-3′; HSL, 5′-TGTGGCACAGACCTCTAAAT-3′ and 5′-GGCATATCCGCTCTC-3′; ATGL, 5′-GGAGACCAAGTGGAACATCTCA-3′ and 5′-AATAATGTTGGCACCTGCTTCA-3′; perilipin, 5′-TGCTGGATGGAGACCTC-3′ and 5′-ACCGGCTCCATGCTCCA-3′; adipophilin, 5′-AAGCATCGGCTACGACGACAC-3′ and 5′-GGACAGTCTGGCATGTAGTCTGGA-3′; TIP47, 5′-AGGACTTTGCAACTGCTGCTGA-3′ and 5′-AGGACTTTGCAACTGCTGCTGA-3′; COXII, 5′-GCCGACTAAATCAAGCAACAG-3′ and 5′-TCTAGGACAATGGGCATAAAGCTATTAA-3′; COXIV, 5′-AGCCATTTCTACTTCGGTGTG-3′ and 5′-GCAGACAGCATCGTGACAT-3′; and 36B4, 5′-GAGGAATCAGATGAGGATATGGGA-3′ and 5′-AAGCAGGCTGACTTGGTTGC-3′.
Measurement of metabolic parameters. Plasma insulin concentration was measured with an insulin assay kit (Morinaga Institute of Biological Science). FFA and TAG concentrations in serum were measured with NEFA-C and Triglyceride E tests (Wako), respectively. Serum adiponectin and leptin levels were measured with ELISA kits from Otsuka Pharmaceutical and R&D Systems, respectively. Liver lipids were extracted from tissue homogenates with isopropanol, and the TAG content was determined with a Triglyceride E test. For intraperitoneal glucose tolerance tests, mice were deprived of food for 16 to 18 hours and then injected with glucose (2 g per kilogram of body mass). For intraperitoneal insulin tolerance tests, mice were injected with human regular insulin (0.75 U/kg; Eli Lilly). Blood samples were collected before and after injections, and plasma glucose concentration was measured with a Glutestsensor (Sanwa Chemical). Food intake was measured by weighing the food given at the beginning and that remaining at the end of each day for 1 week. For catecholamine measurements, urine was collected in bottles containing HCl and subjected to HPLC.
Hyperinsulinemic-euglycemic clamp analysis was performed as described (46), with minor modifications. In brief, 5–7 days before the clamp, mice were anesthetized by intraperitoneal injection of sodium pentobarbital (80–100 mg/kg) and a catheter was inserted into the right internal jugular vein for infusion. The analysis was performed under nonstressful conditions with conscious mice that had been deprived of food overnight for 4 hours. [3-3H]Glucose (NEN Life Science) was infused for 2 hours at a rate of 0.05 μCi/min before initiation of the clamp, and a blood sample was collected at the end of this period to estimate basal glucose turnover. After a bolus injection of [3-3H]glucose (10 μCi) and the onset of subsequent continuous infusion of [3-3H]glucose (0.1 μCi/min), a hyperinsulinemic-euglycemic clamp was applied for 120 minutes with continuous infusion of insulin at a rate of 2.5 mU/kg per minute. Plasma glucose concentration was monitored every 10 minutes, and 30% glucose was infused at a variable rate to maintain plasma glucose at basal concentrations. Blood samples were collected 80, 90, 100, 110, and 120 minutes after the onset of the clamp for determination of the plasma concentrations of [3-3H]glucose and 3H2O. The rates of glucose disposal and hepatic glucose production were calculated as described (47).
Measurement of body temperature, locomotor activity, and oxygen consumption in mice. Mice were housed individually at 22° to 23°C in transparent plastic cages (31 × 20 × 13 cm) placed in a light-controlled (12-hour light/12-hour dark cycle; fluorescent light, 200–300 lux) monitoring chamber (40 × 33 × 33 cm) that was equipped with a radioreceiver of a telemetry system (RPC-1; Data Sciences International) for retrieval of body temperature (48). A temperature probe (TA10TA-F20; Data Sciences International) was introduced into the abdominal cavity of mice anesthetized with a mixture of halothane and O2 (Fluothane; Takeda; 3%–4% for induction and 1% for maintenance). After recovery for more than 10 days, the mice were monitored for body temperature every 1 hour for 3 days. Locomotor activity was detected by passive (pyroelectric) infrared sensors (FA-05 F5B; Omron). Data were collected and analyzed with the use of a Chronobiology kit (Stanford Software Systems) (49). Oxygen consumption of mice in the fed condition was measured with an indirect calorimetric system every 6.5 minutes for 24 hours at 28°C as described previously (50). In brief, room air was pumped through an acrylic metabolic chamber, and the expired gas was filtered through thin cotton, dried, and subjected to gas analysis with an Alco System model RL-600 instrument. Energy expenditure was calculated as the product of the calorific value of oxygen (3.815 + [1.232 × respiratory quotient]) and the volume of O2 consumed.
Immunoblot analysis. Total cell lysates were prepared with a lysis buffer containing 25 mM Tris-HCl (pH 7.4), 150 mM NaCl, 1 mM EDTA, 1% Triton X-100, 50 mM NaF, 10 mM sodium pyrophosphate, 1 mM sodium vanadate, and 1 mM PMSF. Tissue homogenates prepared in the same lysis buffer were rotated for 1 hour at 4°C, in the additional presence of 0.2% sarkosyl and 10% glycerol before analysis. Protein samples were subjected to immunoblot analysis with antibodies to FSP27, perilipin (Sigma-Aldrich), UCP-1 (51), COXIV (Molecular Probes), α-tubulin (Sigma-Aldrich), or β-actin (Sigma-Aldrich). Immune complexes were detected with HRP-conjugated secondary antibodies and enhanced chemiluminescence reagents (GE Healthcare). The polyclonal antibodies to FSP27 were generated by injection of rabbits with a synthetic peptide (CTEEEQPAKSSLL) corresponding to the COOH-terminal portion of mouse FSP27 or with a glutathione-S-transferase fusion protein of mouse FSP27 (amino acid residues 45–127) that was expressed in and purified from Esherichia coli. The antibodies generated in response to the fusion protein were purified by affinity chromatography with Hi-Trap NHS-activated columns (GE Healthcare) coupled with glutathione-S-transferase or the fusion protein.
Microscopic analyses. For light microscopy, the tissue was fixed with formalin, embedded in paraffin, sectioned at a thickness of 6 μm, and mounted on glass slides by standard procedures. The sections were stained with hematoxylin-eosin. For electron microscopy, tissue was fixed for at least 4 hours at 4°C with 2.5% glutaraldehyde in 0.1 M sodium cacodylate buffer and then cut into pieces of approximately 1 mm3. The tissue fragments were then fixed with 1% osmium tetroxide in 0.2 M sodium cacodylate buffer, washed in the buffer alone, dehydrated with a series of alcohol solutions, and embedded in resin. After polymerization of the resin at 60°C for 48 hours, sections were cut with a microtome and observed with an electron microscope (JEM-1200EX; JEOL) (52).
For oil red O staining, cells were fixed for 1 hour at room temperature with 10% formalin in PBS, washed 3 times with distilled water, and then stained for 1 hour with filtered 0.5% oil red O in 60% isopropanol. After washing 3 times with distilled water, the cells were examined with a light microscope. For immunofluorescence microscopy, cells were washed twice with PBS, fixed for 20 minutes at room temperature with 4% formaldehyde in PBS, washed 3 times with PBS, permeabilized with 0.2% Triton X-100 for 5 minutes, and washed an additional 3 times with PBS. They were then incubated for 60 minutes with 2% BSA in PBS, before exposure for 60 minutes at room temperature to rabbit polyclonal antibodies to FSP27 or to perilipin (Sigma-Aldrich). The cells were washed 3 times with PBS and then incubated for 1 hour with Alexa Fluor 555–conjugated goat antibodies to rabbit IgG (Molecular Probes) as well as with Bodipy 493/503 (Molecular Probes) for staining of neutral lipid. Cover slips were applied to the slides with Fluoro Guard Antifade Reagent (Bio-Rad), and the cells were examined with a laser-scanning microscope (LSM5 PASCAL, version 3; Carl Zeiss).
Measurement of oxygen consumption in isolated adipocytes. Mice were anesthetized with ether and perfused, first with Ca2+- and Mg2+-free PBS containing 1 mM EDTA and heparin sodium (10 U/ml) and then with Krebs-Ringer bicarbonate HEPES buffer (KRBH, pH 7.4) containing collagenase (1 mg/ml), through a cannula inserted in an open-chest procedure into the left ventricle, with draining from the right atrium. BAT and inguinal WAT were then rapidly removed and placed in PBS. After the removal of extraneous tissue, the remaining adipose tissue was minced, and WAT or BAT from 3 mice was combined, transferred to an incubator, and incubated with slow shaking at 37°C in KRBH containing fatty acid–free BSA (10 mg/ml), 2.5 mM glucose, and collagenase (1 mg/ml). BAT was incubated for 30 minutes, with additional shaking by hand every 5 minutes, after which the mixture was passed through a 200-mm nylon filter and centrifuged at 200 g for 1 minute at room temperature. The floating adipocytes were collected. The pieces of BAT remaining on the nylon filter were incubated for an additional 30 minutes with the collagenase solution, and the released cells were collected and combined with the initial batch. The pieces of WAT were incubated for 1 hour, and the released cells were collected as for BAT. All adipocytes were washed 3 times with KRBH to eliminate collagenase and were counted with a Thoma’s hemacytometer. The cells were diluted to a density of 1 × 105 to 5 × 105 cells/ml for BAT and 2 × 105 to 15 × 105 cells/ml for WAT with KRBH containing fatty acid–free BSA (40 mg/ml) and 2.7 mM glucose. The cell suspensions were maintained for 1 hour at room temperature before analysis.
Oxygen consumption by the isolated adipocytes was measured with a Clark-style oxygen electrode (782 2-Channel Oxygen System, version 1.0; Strathkelvin Instruments). The cell suspension (200 μl) was added to a magnetically stirred chamber set with a thermostat to 37°C and was adjusted to a final volume of 1 ml with KRBH containing fatty acid–free BSA (40 mg/ml) and 2.7 mM glucose. The chamber was closed, and the cells were incubated for 5 minutes to determine the basal respiratory rate. CL316,243 (Sigma-Aldrich) was then injected to give a final concentration of 100 nM with a Hamilton syringe through a small hole in the cover of the chamber. Alternatively, after measurement of basal oxygen consumption rate, oligomycin, a specific inhibitor of mitochondrial ATP synthase, was added to a concentration of 0.1 mg/ml for measurement of respiration devoted to ATP production. Cyanide, an inhibitor of the mitochondrial respiratory chain, was next added at a concentration of 3 mM to estimate respiration associated with proton leak. Uncoupled respiration was calculated by subtracting the oxygen consumption devoted to ATP production from total mitochondrial oxygen consumption.
Measurement of glucose and FFA oxidation in isolated adipocytes. Glucose and FFA oxidation were measured as described previously (53), with some modifications. In brief, for measurement of glucose oxidation, isolated adipocytes (2 × 105 cells) were incubated in a polypropylene tube with 1 ml of KRBH (pH 7.4) containing 2 mM glucose, 2% BSA, and 0.5 μCi of [1-14C]-d-glucose (52 mCi/mmol). For measurement of FFA oxidation, isolated adipocytes (2 × 105 cells) were washed twice with KRBH containing 2% BSA and were then incubated with 1 ml of the same solution containing 0.4 mM oleic acid and 1.0 μCi of [1-14C]oleic acid (53 mCi/mmol). The cells were incubated for 3 hours at 37°C, after which 0.5 ml of 4 M H2SO4 was added to the tube, and the generated 14CO2 was trapped by filter paper treated with hyamine hydroxide and measured by scintillation spectrometry.
Depletion of FSP27 or perilipin in HW adipocytes. Five days after the onset of induction of differentiation, HW adipocytes were washed twice with PBS, detached from the culture dish by exposure to 0.25% trypsin and collagenase (0.5 mg/ml) in PBS, and resuspended in PBS. The cells (~3 × 106) were then mixed with siRNA duplexes and subjected to electroporation with a Bio-Rad Gene Pulser II system at a setting of 0.18 kV and 0.975 μF. The FSP27 siRNA was targeted to the mRNA sequence 5′-GCACAAUCGUGGAGACAGAAGAAUA-3′. The perilipin siRNA was obtained from Invitrogen. Immediately after electroporation, the cells were mixed with fresh DMEM supplemented with 10% FBS, and 10 minutes later they were seeded onto culture plates. They were subjected to assays at the indicated times (see the legends for Figures 5 and 6) after electroporation.
Forced expression of FSP27 in COS cells and cultured adipocytes. The coding sequence of mouse FSP27 was subcloned into the pIRES2-DsRed2 vector (Clontech), and the resulting construct was introduced into COS cells with the use of the FuGENE 6 reagent (Roche). Cells were incubated with Bodipy 493/503 (for detection of lipid droplets) and Hoechst 33258 (for detection of nuclei) for 30 minutes at 37°C in a cell incubator. They were then washed twice with PBS, fixed for 20 minutes at room temperature with 4% formaldehyde in PBS, and washed 3 times with PBS. For quantitative analysis, images were acquired with an IN Cell Analyzer 1000 (GE Healthcare). The number of lipid droplets per nucleus and their mean area were then measured with the use of Developer Toolbox software. For the overexpression of mouse FSP27 tagged at its COOH-terminus with the Flag epitope in cultured HB2 and HW adipocytes, an Adenovirus Cre/loxP kit was used (Takara).
Statistics. Quantitative data are presented as mean ± SEM. Differences between groups were examined for statistical significance with 2-tailed Student’s t test. A P value of less than 0.05 was considered statistically significant.
-
Acknowledgments
We thank H. Niwa (RIKEN Center for Developmental Biology, Kobe, Japan) for EB3/5 embryonic stem cells; S. Shigeta and R. Kumagai for technical assistance; and M.P. Czech (University of Massachusetts Medical School, Worcester, Massachusetts, USA) for helpful discussion. This work was supported by a grant for the Intellectual Cluster Formation Project from the Ministry of Education, Culture, Sports, Science, and Technology of Japan (MEXT) to M. Kasuga; a grant for the 21st Century COE Program “Center of Excellence for Signal Transduction Disease: Diabetes Mellitus as a Model” from MEXT to M. Kasuga; and a Grant-in-Aid for Creative Scientific Research from MEXT to M. Kasuga.
Address correspondence to: Masato Kasuga, Department of Internal Medicine, Division of Diabetes, Metabolism, and Endocrinology, Kobe University Graduate School of Medicine, 7-5-1 Kusunoki-cho, Chuo-ku, Kobe 650-0017, Japan. Phone: 81-78-382-5860; Fax: 81-78-382-2084; E-mail: [email protected].
-
Footnotes
Nonstandard abbreviations used: BAT, brown adipose tissue; CIDE, cell death–inducing DNA fragmentation factor 45-like effector; COXI, subunit I of cytochrome c oxidase; FSP27, fat-specific protein of 27 kDa; FSP27-KO, FSP27 homozygous knockout; KRBH, Krebs-Ringer bicarbonate HEPES buffer; LCAD, long-chain acyl-CoA dehydrogenase; MCAD, medium-chain acyl-CoA dehydrogenase; mtTFA, mitochondrial transcription factor A; NRF1, nuclear respiratory factor 1; PGC1a, PPARγ coactivator 1α; PPARγ-ΔC, adenovirus encoding a dominant-negative mutant of PPARγ; TAG, triacylglycerol; TIP47, tail-interacting protein of 47 kDa; UCP1, uncoupling protein–1; VLCAD, very LCAD; WAT, white adipose tissue.
Conflict of interest: The authors have declared that no conflict of interest exists.
Reference information: J. Clin. Invest.118:2808–2821 (2008). doi:10.1172/JCI34090
See the related article at Lipid droplets: FSP27 knockout enhances their sizzle.
-
References
- Kahn, B.B., Flier, J.S. 2000. Obesity and insulin resistance. J. Clin. Invest. 106:473-481.
- Unger, R.H. 2003. Weapons of lean body mass destruction: the role of ectopic lipids in the metabolic syndrome. Endocrinology. 144:5159-5165.
- Shulman, G.I. 2000. Cellular mechanisms of insulin resistance. J. Clin. Invest. 106:171-176.
- Rosen, E.D., Spiegelman, B.M. 2006. Adipocytes as regulators of energy balance and glucose homeostasis. Nature. 444:847-853.
- Tauchi-Sato, K., Ozeki, S., Houjou, T., Taguchi, R., Fujimoto, T. 2002. The surface of lipid droplets is a phospholipid monolayer with a unique fatty acid composition. J. Biol. Chem. 277:44507-44512.
- Zweytick, D., Athenstaedt, K., Daum, G. 2000. Intracellular lipid particles of eukaryotic cells. Biochim. Biophys. Acta. 1469:101-120. View this article via: PubMed Google Scholar
- Miura, S., et al. 2002. Functional conservation for lipid storage droplet association among perilipin, ADRP, and TIP47 (PAT)-related proteins in mammals, Drosophila, and Dictyostelium. J. Biol. Chem. 277:32253-32257.
- Wolins, N.E., Brasaemle, D.L., Bickel, P.E. 2006. A proposed model of fat packaging by exchangeable lipid droplet proteins. FEBS Lett. 580:5484-5491.
- Murphy, D.J., Vance, J. 1999. Mechanisms of lipid body formation. Trends Biochem. Sci. 24:109-115.
- Danesch, U., Hoeck, W., Ringold, G.M. 1992. Cloning and transcriptional regulation of a novel adipocyte-specific gene, FSP27. CAAT-enhancer-binding protein (C/EBP) and C/EBP-like proteins interact with sequences required for differentiation-dependent expressio.
n
. J. Biol. Chem. 267:7185-7193. View this article via: PubMed Google Scholar
- Puri, V., et al. 2007. Fat specific protein 27: A novel lipid droplet protein that enhances triglyceride storage. J. Biol. Chem. 282:34213-34218.
- Tamori, Y., Masugi, J., Nishino, N., Kasuga, M. 2002. Role of peroxisome proliferator-activated receptor-gamma in maintenance of the characteristics of mature 3T3-L1 adipocytes. Diabetes. 51:2045-2055.
- Brasaemle, D.L., Dolios, G., Shapiro, L., Wang, R. 2004. Proteomic analysis of proteins associated with lipid droplets of basal and lipolytically stimulated 3T3-L1 adipocytes. J. Biol. Chem. 279:46835-46842.
- Lehmann, J.M., et al. 1995. An antidiabetic thiazolidinedione is a high affinity ligand for peroxisome proliferator-activated receptor γ (PPARγ). J. Biol. Chem. 270:12953-12956.
- Petruschke, T., Hauner, H. 1993. Tumor necrosis factor-alpha prevents the differentiation of human adipocyte precursor cells and causes delipidation of newly developed fat cells. J. Clin. Endocrinol. Metab. 76:742-747.
- Irie, Y., et al. 1999. Immortal brown adipocytes from p53-knockout mice: differentiation and expression of uncoupling proteins. Biochem. Biophys. Res. Commun. 255:221-225.
- Evans, M.J., Scarpulla, R.C. 1990. NRF-1: a trans-activator of nuclear-encoded respiratory genes in animal cells. Genes Dev. 4:1023-1034.
- Virbasius, J.V., Scarpulla, R.C. 1994. Activation of the human mitochondrial transcription factor A gene by nuclear respiratory factors: a potential regulatory link between nuclear and mitochondrial gene expression in organelle biogenesis. Proc. Natl. Acad. Sci. U. S. A. 91:1309-1313.
- Uldry, M., et al. 2006. Complementary action of the PGC-1 coactivators in mitochondrial biogenesis and brown fat differentiation. Cell Metab. 3:333-341.
- Granneman, J.G., Li, P., Zhu, Z., Lu, Y. 2005. Metabolic and cellular plasticity in white adipose tissue I: effects of β3-adrenergic receptor activation. Am. J. Physiol. Endocrinol. Metab. 289:E608-E616.
- Himms-Hagen, J., et al. 2000. Multilocular fat cells in WAT of CL-316243-treated rats derive directly from white adipocytes. Am. J. Physiol. Cell Physiol. 279:C670-C681. View this article via: PubMed Google Scholar
- Greenberg, A.S., et al. 1991. Perilipin, a major hormonally regulated adipocyte-specific phosphoprotein associated with the periphery of lipid storage droplets. J. Biol. Chem. 266:11341-11346. View this article via: PubMed Google Scholar
- Servetnick, D.A., et al. 1995. Perilipins are associated with cholesteryl ester droplets in steroidogenic adrenal cortical and Leydig cells. J. Biol. Chem. 270:16970-16973.
- Brasaemle, D.L., et al. 2000. Perilipin A increases triacylglycerol storage by decreasing the rate of triacylglycerol hydrolysis. J. Biol. Chem. 275:38486-38493.
- Martinez-Botas, J., et al. 2000. Absence of perilipin results in leanness and reverses obesity in Lepr (db/db) mice. Nat. Genet. 26:474-479.
- Tansey, J.T., et al. 2001. Perilipin ablation results in a lean mouse with aberrant adipocyte lipolysis, enhanced leptin production, and resistance to diet-induced obesity. Proc. Natl. Acad. Sci. U. S. A. 98:6494-6499.
- Cases, S., et al. 1998. Identification of a gene encoding an acyl CoA:diacylglycerol acyltransferase, a key enzyme in triacylglycerol synthesis. Proc. Natl. Acad. Sci. U. S. A. 95:13018-13023.
- Lardizabal, K.D., et al. 2001. DGAT2 is a new diacylglycerol acyltransferase gene family: purification, cloning, and expression in insect cells of two polypeptides from Mortierella ramanniana with diacylglycerol acyltransferase activity. J. Biol. Chem. 276:38862-38869.
- Cases, S., et al. 2001. Cloning of DGAT2, a second mammalian diacylglycerol acyltransferase, and related family members. J. Biol. Chem. 276:38870-38876.
- Ntambi, J.M., Miyazaki, M. 2004. Regulation of stearoyl-CoA desaturases and role in metabolism. Prog. Lipid Res. 43:91-104.
- Coleman, R.A., Lee, D.P. 2004. Enzymes of triacylglycerol synthesis and their regulation. Prog. Lipid Res. 43:134-176.
- Schweiger, M., et al. 2006. Adipose triglyceride lipase and hormone-sensitive lipase are the major enzymes in adipose tissue triacylglycerol catabolism. J. Biol. Chem. 281:40236-40241.
- Londos, C., Sztalryd, C., Tansey, J.T., Kimmel, A.R. 2005. Role of PAT proteins in lipid metabolism. Biochimie. 87:45-49.
- Murphy, D.J. 2001. The biogenesis and functions of lipid bodies in animals, plants and microorganisms. Prog. Lipid Res. 40:325-438.
- Forman, B.M., Chen, J., Evans, R.M. 1997. Hypolipidemic drugs, polyunsaturated fatty acids, and eicosanoids are ligands for peroxisome proliferator-activated receptors alpha and delta. Proc. Natl. Acad. Sci. U. S. A. 94:4312-4317.
- Bogacka, I., Ukropcova, B., McNeil, M., Gimble, J.M., Smith, S.R. 2005. Structural and functional consequences of mitochondrial biogenesis in human adipocytes in vitro. J. Clin. Endocrinol. Metab. 90:6650-6656.
- Wang, Y.X., et al. 2003. Peroxisome-proliferator-activated receptor δ activates fat metabolism to prevent obesity. Cell. 113:159-170.
- Saha, P.K., Kojima, H., Martinez-Botas, J., Sunehag, A.L., Chan, L. 2004. Metabolic adaptations in the absence of perilipin. J. Biol. Chem. 279:35150-35158.
- Castro-Chavez, F., et al. 2003. Coordinated upregulation of oxidative pathways and downregulation of lipid biosynthesis underlie obesity resistance in perilipin knockout mice: a microarray gene expression profile. Diabetes. 52:2666-2674.
- Inohara, N., Koseki, T., Chen, S., Wu, X., Nunez, G. 1998. CIDE, a novel family of cell death activators with homology to the 45 kDa subunit of the DNA fragmentation factor. EMBO J. 17:2526-2533.
- Zhou, Z., et al. 2003. Cidea-deficient mice have lean phenotype and are resistant to obesity.
Nat. Genet. 35:49-56. View this article via: PubMed Google Scholar
- Li, J.Z., et al. 2007. Cideb regulates diet-induced obesity, liver steatosis and insulin sensitivity by controlling lipogenesis and fatty acid oxidation. Diabetes. 56:2523-2540.
- Nordström, E.A., et al. 2005. A human-specific role of cell death-inducing DFFA (DNA fragmentation factor-alpha)-like effector A (CIDEA) in adipocyte lipolysis and obesity. Diabetes. 54:1726-1734.
- Liang, L., Zhao, M., Xu, Z., Yokoyama, K.K., Li, T. 2003. Molecular cloning and characterization of CIDE-3, a novel member of the cell-death-inducing DNA-fragmentation-factor (DFF45)-like effector family. Biochem. J. 370:195-203.
- Barazzoni, R., Short, K.R., Nair, K.S. 2000. Effects of aging on mitochondrial DNA copy number and cytochrome c oxidase gene expression in rat skeletal muscle, liver, and heart. J. Biol. Chem. 275:3343-3347.
- Kanda, H., et al. 2006. MCP-1 contributes to macrophage infiltration into adipose tissue, insulin resistance, and hepatic steatosis in obesity. J. Clin. Invest. 116:1494-1505.
- Kim, J.K., Kotani, K., Ciaraldi, T.P., Henry, R.R., Kahn, B.B. 2001. Glucose toxicity and the development of diabetes in mice with muscle-specific inactivation of GLUT4. J. Clin. Invest. 108:153-160.
- Mohri, T., et al. 2003. Alterations of circadian expressions of clock genes in Dahl salt-sensitive rats fed a high-salt diet. Hypertension. 42:189-194.
- Masubuchi, S., Kataoka, N., Sassone-Corsi, P., Okamura, H. 2005. Mouse Period1 (mPER1) acts as a circadian adaptor to entrain the oscillator to environmental light/dark cycles by regulating mPER2 protein. J. Neurosci. 25:4719-4724.
- Ohnuki, K., Haramizu, S., Oki, K., Ishihara, K., Fushiki, T. 2001. A single oral administration of conjugated linoleic acid enhanced energy metabolism in mice. Lipids. 36:583-587.
- Nagase, I., et al. 1996. Expression of uncoupling protein in skeletal muscle and white fat of obese mice treated with thermogenic beta 3-adrenergic agonist. J. Clin. Invest. 97:2898-2904.
- Kitazawa, S., Kitazawa, R. 2006. In situ detection of specific gene expression during and immediately after transcription at electron microscopic level. J. Struct. Biol. 153:64-72.
- Vessal, M., Mishra, S., Moulik, S., Murphy, L.J. 2006. Prohibitin attenuates insulin-stimulated glucose and fatty acid oxidation in adipose tissue by inhibition of pyruvate carboxylase. FEBS J. 273:568-576.
-
Version history
- Version 1 (July 24, 2008): No description
- Version 2 (August 1, 2008): No description



Copyright © 2024 American Society for Clinical Investigation
ISSN: 0021-9738 (print), 1558-8238 (online)