Advertisement
Research ArticleOncology Free access | 10.1172/JCI33656
BRAF gene duplication constitutes a mechanism of MAPK pathway activation in low-grade astrocytomas
Stefan Pfister,1,2 Wibke G. Janzarik,3,4 Marc Remke,1 Aurélie Ernst,1 Wiebke Werft,5 Natalia Becker,1 Grischa Toedt,1 Andrea Wittmann,1 Christian Kratz,6 Heike Olbrich,4 Rezvan Ahmadi,7 Barbara Thieme,8 Stefan Joos,1 Bernhard Radlwimmer,1 Andreas Kulozik,2 Torsten Pietsch,9 Christel Herold-Mende,7 Astrid Gnekow,8 Guido Reifenberger,10 Andrey Korshunov,1,11 Wolfram Scheurlen,12 Heymut Omran,4 and Peter Lichter1
1Division Molecular Genetics, German Cancer Research Center (DKFZ), Heidelberg, Germany. 2Department of Pediatric Oncology, Hematology, and Immunology, University of Heidelberg, Heidelberg, Germany. 3Department of Neurology, University Hospital Freiburg, Freiburg, Germany. 4Department of Pediatric Neurology and Muscle Disorders, University Hospital Freiburg, Freiburg, Germany. 5Division of Biostatistics, DKFZ, Heidelberg, Germany. 6Division of Pediatric Hematology and Oncology, Department of Pediatrics and Adolescent Medicine, University of Freiburg, Freiburg, Germany. 7Department of Neurosurgery, University of Heidelberg, Heidelberg, Germany. 8Hospital for Children and Adolescents Augsburg, Augsburg, Germany. 9Department of Neuropathology, University of Bonn, Bonn, Germany. 10Department of Neuropathology, Heinrich-Heine-University, Düsseldorf, Germany. 11Department of Neuropathology, Burdenko Neurosurgical Institute, Moscow, Russia. 12Cnopf’sche Kinderklinik, Nuremberg Children’s Hospital, Nuremberg, Germany.
Address correspondence to: Peter Lichter, Division Molecular Genetics, German Cancer Research Center (DKFZ), In Neuenheimer Feld 280, 69120 Heidelberg, Germany. Phone: 49-6221-424619; Fax: 49-6221-424639; E-mail: [email protected].
Find articles by Pfister, S. in: JCI | PubMed | Google Scholar
1Division Molecular Genetics, German Cancer Research Center (DKFZ), Heidelberg, Germany. 2Department of Pediatric Oncology, Hematology, and Immunology, University of Heidelberg, Heidelberg, Germany. 3Department of Neurology, University Hospital Freiburg, Freiburg, Germany. 4Department of Pediatric Neurology and Muscle Disorders, University Hospital Freiburg, Freiburg, Germany. 5Division of Biostatistics, DKFZ, Heidelberg, Germany. 6Division of Pediatric Hematology and Oncology, Department of Pediatrics and Adolescent Medicine, University of Freiburg, Freiburg, Germany. 7Department of Neurosurgery, University of Heidelberg, Heidelberg, Germany. 8Hospital for Children and Adolescents Augsburg, Augsburg, Germany. 9Department of Neuropathology, University of Bonn, Bonn, Germany. 10Department of Neuropathology, Heinrich-Heine-University, Düsseldorf, Germany. 11Department of Neuropathology, Burdenko Neurosurgical Institute, Moscow, Russia. 12Cnopf’sche Kinderklinik, Nuremberg Children’s Hospital, Nuremberg, Germany.
Address correspondence to: Peter Lichter, Division Molecular Genetics, German Cancer Research Center (DKFZ), In Neuenheimer Feld 280, 69120 Heidelberg, Germany. Phone: 49-6221-424619; Fax: 49-6221-424639; E-mail: [email protected].
Find articles by Janzarik, W. in: JCI | PubMed | Google Scholar
1Division Molecular Genetics, German Cancer Research Center (DKFZ), Heidelberg, Germany. 2Department of Pediatric Oncology, Hematology, and Immunology, University of Heidelberg, Heidelberg, Germany. 3Department of Neurology, University Hospital Freiburg, Freiburg, Germany. 4Department of Pediatric Neurology and Muscle Disorders, University Hospital Freiburg, Freiburg, Germany. 5Division of Biostatistics, DKFZ, Heidelberg, Germany. 6Division of Pediatric Hematology and Oncology, Department of Pediatrics and Adolescent Medicine, University of Freiburg, Freiburg, Germany. 7Department of Neurosurgery, University of Heidelberg, Heidelberg, Germany. 8Hospital for Children and Adolescents Augsburg, Augsburg, Germany. 9Department of Neuropathology, University of Bonn, Bonn, Germany. 10Department of Neuropathology, Heinrich-Heine-University, Düsseldorf, Germany. 11Department of Neuropathology, Burdenko Neurosurgical Institute, Moscow, Russia. 12Cnopf’sche Kinderklinik, Nuremberg Children’s Hospital, Nuremberg, Germany.
Address correspondence to: Peter Lichter, Division Molecular Genetics, German Cancer Research Center (DKFZ), In Neuenheimer Feld 280, 69120 Heidelberg, Germany. Phone: 49-6221-424619; Fax: 49-6221-424639; E-mail: [email protected].
Find articles by Remke, M. in: JCI | PubMed | Google Scholar
1Division Molecular Genetics, German Cancer Research Center (DKFZ), Heidelberg, Germany. 2Department of Pediatric Oncology, Hematology, and Immunology, University of Heidelberg, Heidelberg, Germany. 3Department of Neurology, University Hospital Freiburg, Freiburg, Germany. 4Department of Pediatric Neurology and Muscle Disorders, University Hospital Freiburg, Freiburg, Germany. 5Division of Biostatistics, DKFZ, Heidelberg, Germany. 6Division of Pediatric Hematology and Oncology, Department of Pediatrics and Adolescent Medicine, University of Freiburg, Freiburg, Germany. 7Department of Neurosurgery, University of Heidelberg, Heidelberg, Germany. 8Hospital for Children and Adolescents Augsburg, Augsburg, Germany. 9Department of Neuropathology, University of Bonn, Bonn, Germany. 10Department of Neuropathology, Heinrich-Heine-University, Düsseldorf, Germany. 11Department of Neuropathology, Burdenko Neurosurgical Institute, Moscow, Russia. 12Cnopf’sche Kinderklinik, Nuremberg Children’s Hospital, Nuremberg, Germany.
Address correspondence to: Peter Lichter, Division Molecular Genetics, German Cancer Research Center (DKFZ), In Neuenheimer Feld 280, 69120 Heidelberg, Germany. Phone: 49-6221-424619; Fax: 49-6221-424639; E-mail: [email protected].
Find articles by Ernst, A. in: JCI | PubMed | Google Scholar
1Division Molecular Genetics, German Cancer Research Center (DKFZ), Heidelberg, Germany. 2Department of Pediatric Oncology, Hematology, and Immunology, University of Heidelberg, Heidelberg, Germany. 3Department of Neurology, University Hospital Freiburg, Freiburg, Germany. 4Department of Pediatric Neurology and Muscle Disorders, University Hospital Freiburg, Freiburg, Germany. 5Division of Biostatistics, DKFZ, Heidelberg, Germany. 6Division of Pediatric Hematology and Oncology, Department of Pediatrics and Adolescent Medicine, University of Freiburg, Freiburg, Germany. 7Department of Neurosurgery, University of Heidelberg, Heidelberg, Germany. 8Hospital for Children and Adolescents Augsburg, Augsburg, Germany. 9Department of Neuropathology, University of Bonn, Bonn, Germany. 10Department of Neuropathology, Heinrich-Heine-University, Düsseldorf, Germany. 11Department of Neuropathology, Burdenko Neurosurgical Institute, Moscow, Russia. 12Cnopf’sche Kinderklinik, Nuremberg Children’s Hospital, Nuremberg, Germany.
Address correspondence to: Peter Lichter, Division Molecular Genetics, German Cancer Research Center (DKFZ), In Neuenheimer Feld 280, 69120 Heidelberg, Germany. Phone: 49-6221-424619; Fax: 49-6221-424639; E-mail: [email protected].
Find articles by Werft, W. in: JCI | PubMed | Google Scholar
1Division Molecular Genetics, German Cancer Research Center (DKFZ), Heidelberg, Germany. 2Department of Pediatric Oncology, Hematology, and Immunology, University of Heidelberg, Heidelberg, Germany. 3Department of Neurology, University Hospital Freiburg, Freiburg, Germany. 4Department of Pediatric Neurology and Muscle Disorders, University Hospital Freiburg, Freiburg, Germany. 5Division of Biostatistics, DKFZ, Heidelberg, Germany. 6Division of Pediatric Hematology and Oncology, Department of Pediatrics and Adolescent Medicine, University of Freiburg, Freiburg, Germany. 7Department of Neurosurgery, University of Heidelberg, Heidelberg, Germany. 8Hospital for Children and Adolescents Augsburg, Augsburg, Germany. 9Department of Neuropathology, University of Bonn, Bonn, Germany. 10Department of Neuropathology, Heinrich-Heine-University, Düsseldorf, Germany. 11Department of Neuropathology, Burdenko Neurosurgical Institute, Moscow, Russia. 12Cnopf’sche Kinderklinik, Nuremberg Children’s Hospital, Nuremberg, Germany.
Address correspondence to: Peter Lichter, Division Molecular Genetics, German Cancer Research Center (DKFZ), In Neuenheimer Feld 280, 69120 Heidelberg, Germany. Phone: 49-6221-424619; Fax: 49-6221-424639; E-mail: [email protected].
Find articles by Becker, N. in: JCI | PubMed | Google Scholar
1Division Molecular Genetics, German Cancer Research Center (DKFZ), Heidelberg, Germany. 2Department of Pediatric Oncology, Hematology, and Immunology, University of Heidelberg, Heidelberg, Germany. 3Department of Neurology, University Hospital Freiburg, Freiburg, Germany. 4Department of Pediatric Neurology and Muscle Disorders, University Hospital Freiburg, Freiburg, Germany. 5Division of Biostatistics, DKFZ, Heidelberg, Germany. 6Division of Pediatric Hematology and Oncology, Department of Pediatrics and Adolescent Medicine, University of Freiburg, Freiburg, Germany. 7Department of Neurosurgery, University of Heidelberg, Heidelberg, Germany. 8Hospital for Children and Adolescents Augsburg, Augsburg, Germany. 9Department of Neuropathology, University of Bonn, Bonn, Germany. 10Department of Neuropathology, Heinrich-Heine-University, Düsseldorf, Germany. 11Department of Neuropathology, Burdenko Neurosurgical Institute, Moscow, Russia. 12Cnopf’sche Kinderklinik, Nuremberg Children’s Hospital, Nuremberg, Germany.
Address correspondence to: Peter Lichter, Division Molecular Genetics, German Cancer Research Center (DKFZ), In Neuenheimer Feld 280, 69120 Heidelberg, Germany. Phone: 49-6221-424619; Fax: 49-6221-424639; E-mail: [email protected].
Find articles by Toedt, G. in: JCI | PubMed | Google Scholar
1Division Molecular Genetics, German Cancer Research Center (DKFZ), Heidelberg, Germany. 2Department of Pediatric Oncology, Hematology, and Immunology, University of Heidelberg, Heidelberg, Germany. 3Department of Neurology, University Hospital Freiburg, Freiburg, Germany. 4Department of Pediatric Neurology and Muscle Disorders, University Hospital Freiburg, Freiburg, Germany. 5Division of Biostatistics, DKFZ, Heidelberg, Germany. 6Division of Pediatric Hematology and Oncology, Department of Pediatrics and Adolescent Medicine, University of Freiburg, Freiburg, Germany. 7Department of Neurosurgery, University of Heidelberg, Heidelberg, Germany. 8Hospital for Children and Adolescents Augsburg, Augsburg, Germany. 9Department of Neuropathology, University of Bonn, Bonn, Germany. 10Department of Neuropathology, Heinrich-Heine-University, Düsseldorf, Germany. 11Department of Neuropathology, Burdenko Neurosurgical Institute, Moscow, Russia. 12Cnopf’sche Kinderklinik, Nuremberg Children’s Hospital, Nuremberg, Germany.
Address correspondence to: Peter Lichter, Division Molecular Genetics, German Cancer Research Center (DKFZ), In Neuenheimer Feld 280, 69120 Heidelberg, Germany. Phone: 49-6221-424619; Fax: 49-6221-424639; E-mail: [email protected].
Find articles by Wittmann, A. in: JCI | PubMed | Google Scholar
1Division Molecular Genetics, German Cancer Research Center (DKFZ), Heidelberg, Germany. 2Department of Pediatric Oncology, Hematology, and Immunology, University of Heidelberg, Heidelberg, Germany. 3Department of Neurology, University Hospital Freiburg, Freiburg, Germany. 4Department of Pediatric Neurology and Muscle Disorders, University Hospital Freiburg, Freiburg, Germany. 5Division of Biostatistics, DKFZ, Heidelberg, Germany. 6Division of Pediatric Hematology and Oncology, Department of Pediatrics and Adolescent Medicine, University of Freiburg, Freiburg, Germany. 7Department of Neurosurgery, University of Heidelberg, Heidelberg, Germany. 8Hospital for Children and Adolescents Augsburg, Augsburg, Germany. 9Department of Neuropathology, University of Bonn, Bonn, Germany. 10Department of Neuropathology, Heinrich-Heine-University, Düsseldorf, Germany. 11Department of Neuropathology, Burdenko Neurosurgical Institute, Moscow, Russia. 12Cnopf’sche Kinderklinik, Nuremberg Children’s Hospital, Nuremberg, Germany.
Address correspondence to: Peter Lichter, Division Molecular Genetics, German Cancer Research Center (DKFZ), In Neuenheimer Feld 280, 69120 Heidelberg, Germany. Phone: 49-6221-424619; Fax: 49-6221-424639; E-mail: [email protected].
Find articles by Kratz, C. in: JCI | PubMed | Google Scholar
1Division Molecular Genetics, German Cancer Research Center (DKFZ), Heidelberg, Germany. 2Department of Pediatric Oncology, Hematology, and Immunology, University of Heidelberg, Heidelberg, Germany. 3Department of Neurology, University Hospital Freiburg, Freiburg, Germany. 4Department of Pediatric Neurology and Muscle Disorders, University Hospital Freiburg, Freiburg, Germany. 5Division of Biostatistics, DKFZ, Heidelberg, Germany. 6Division of Pediatric Hematology and Oncology, Department of Pediatrics and Adolescent Medicine, University of Freiburg, Freiburg, Germany. 7Department of Neurosurgery, University of Heidelberg, Heidelberg, Germany. 8Hospital for Children and Adolescents Augsburg, Augsburg, Germany. 9Department of Neuropathology, University of Bonn, Bonn, Germany. 10Department of Neuropathology, Heinrich-Heine-University, Düsseldorf, Germany. 11Department of Neuropathology, Burdenko Neurosurgical Institute, Moscow, Russia. 12Cnopf’sche Kinderklinik, Nuremberg Children’s Hospital, Nuremberg, Germany.
Address correspondence to: Peter Lichter, Division Molecular Genetics, German Cancer Research Center (DKFZ), In Neuenheimer Feld 280, 69120 Heidelberg, Germany. Phone: 49-6221-424619; Fax: 49-6221-424639; E-mail: [email protected].
Find articles by Olbrich, H. in: JCI | PubMed | Google Scholar
1Division Molecular Genetics, German Cancer Research Center (DKFZ), Heidelberg, Germany. 2Department of Pediatric Oncology, Hematology, and Immunology, University of Heidelberg, Heidelberg, Germany. 3Department of Neurology, University Hospital Freiburg, Freiburg, Germany. 4Department of Pediatric Neurology and Muscle Disorders, University Hospital Freiburg, Freiburg, Germany. 5Division of Biostatistics, DKFZ, Heidelberg, Germany. 6Division of Pediatric Hematology and Oncology, Department of Pediatrics and Adolescent Medicine, University of Freiburg, Freiburg, Germany. 7Department of Neurosurgery, University of Heidelberg, Heidelberg, Germany. 8Hospital for Children and Adolescents Augsburg, Augsburg, Germany. 9Department of Neuropathology, University of Bonn, Bonn, Germany. 10Department of Neuropathology, Heinrich-Heine-University, Düsseldorf, Germany. 11Department of Neuropathology, Burdenko Neurosurgical Institute, Moscow, Russia. 12Cnopf’sche Kinderklinik, Nuremberg Children’s Hospital, Nuremberg, Germany.
Address correspondence to: Peter Lichter, Division Molecular Genetics, German Cancer Research Center (DKFZ), In Neuenheimer Feld 280, 69120 Heidelberg, Germany. Phone: 49-6221-424619; Fax: 49-6221-424639; E-mail: [email protected].
Find articles by Ahmadi, R. in: JCI | PubMed | Google Scholar
1Division Molecular Genetics, German Cancer Research Center (DKFZ), Heidelberg, Germany. 2Department of Pediatric Oncology, Hematology, and Immunology, University of Heidelberg, Heidelberg, Germany. 3Department of Neurology, University Hospital Freiburg, Freiburg, Germany. 4Department of Pediatric Neurology and Muscle Disorders, University Hospital Freiburg, Freiburg, Germany. 5Division of Biostatistics, DKFZ, Heidelberg, Germany. 6Division of Pediatric Hematology and Oncology, Department of Pediatrics and Adolescent Medicine, University of Freiburg, Freiburg, Germany. 7Department of Neurosurgery, University of Heidelberg, Heidelberg, Germany. 8Hospital for Children and Adolescents Augsburg, Augsburg, Germany. 9Department of Neuropathology, University of Bonn, Bonn, Germany. 10Department of Neuropathology, Heinrich-Heine-University, Düsseldorf, Germany. 11Department of Neuropathology, Burdenko Neurosurgical Institute, Moscow, Russia. 12Cnopf’sche Kinderklinik, Nuremberg Children’s Hospital, Nuremberg, Germany.
Address correspondence to: Peter Lichter, Division Molecular Genetics, German Cancer Research Center (DKFZ), In Neuenheimer Feld 280, 69120 Heidelberg, Germany. Phone: 49-6221-424619; Fax: 49-6221-424639; E-mail: [email protected].
Find articles by Thieme, B. in: JCI | PubMed | Google Scholar
1Division Molecular Genetics, German Cancer Research Center (DKFZ), Heidelberg, Germany. 2Department of Pediatric Oncology, Hematology, and Immunology, University of Heidelberg, Heidelberg, Germany. 3Department of Neurology, University Hospital Freiburg, Freiburg, Germany. 4Department of Pediatric Neurology and Muscle Disorders, University Hospital Freiburg, Freiburg, Germany. 5Division of Biostatistics, DKFZ, Heidelberg, Germany. 6Division of Pediatric Hematology and Oncology, Department of Pediatrics and Adolescent Medicine, University of Freiburg, Freiburg, Germany. 7Department of Neurosurgery, University of Heidelberg, Heidelberg, Germany. 8Hospital for Children and Adolescents Augsburg, Augsburg, Germany. 9Department of Neuropathology, University of Bonn, Bonn, Germany. 10Department of Neuropathology, Heinrich-Heine-University, Düsseldorf, Germany. 11Department of Neuropathology, Burdenko Neurosurgical Institute, Moscow, Russia. 12Cnopf’sche Kinderklinik, Nuremberg Children’s Hospital, Nuremberg, Germany.
Address correspondence to: Peter Lichter, Division Molecular Genetics, German Cancer Research Center (DKFZ), In Neuenheimer Feld 280, 69120 Heidelberg, Germany. Phone: 49-6221-424619; Fax: 49-6221-424639; E-mail: [email protected].
Find articles by Joos, S. in: JCI | PubMed | Google Scholar
1Division Molecular Genetics, German Cancer Research Center (DKFZ), Heidelberg, Germany. 2Department of Pediatric Oncology, Hematology, and Immunology, University of Heidelberg, Heidelberg, Germany. 3Department of Neurology, University Hospital Freiburg, Freiburg, Germany. 4Department of Pediatric Neurology and Muscle Disorders, University Hospital Freiburg, Freiburg, Germany. 5Division of Biostatistics, DKFZ, Heidelberg, Germany. 6Division of Pediatric Hematology and Oncology, Department of Pediatrics and Adolescent Medicine, University of Freiburg, Freiburg, Germany. 7Department of Neurosurgery, University of Heidelberg, Heidelberg, Germany. 8Hospital for Children and Adolescents Augsburg, Augsburg, Germany. 9Department of Neuropathology, University of Bonn, Bonn, Germany. 10Department of Neuropathology, Heinrich-Heine-University, Düsseldorf, Germany. 11Department of Neuropathology, Burdenko Neurosurgical Institute, Moscow, Russia. 12Cnopf’sche Kinderklinik, Nuremberg Children’s Hospital, Nuremberg, Germany.
Address correspondence to: Peter Lichter, Division Molecular Genetics, German Cancer Research Center (DKFZ), In Neuenheimer Feld 280, 69120 Heidelberg, Germany. Phone: 49-6221-424619; Fax: 49-6221-424639; E-mail: [email protected].
Find articles by Radlwimmer, B. in: JCI | PubMed | Google Scholar
1Division Molecular Genetics, German Cancer Research Center (DKFZ), Heidelberg, Germany. 2Department of Pediatric Oncology, Hematology, and Immunology, University of Heidelberg, Heidelberg, Germany. 3Department of Neurology, University Hospital Freiburg, Freiburg, Germany. 4Department of Pediatric Neurology and Muscle Disorders, University Hospital Freiburg, Freiburg, Germany. 5Division of Biostatistics, DKFZ, Heidelberg, Germany. 6Division of Pediatric Hematology and Oncology, Department of Pediatrics and Adolescent Medicine, University of Freiburg, Freiburg, Germany. 7Department of Neurosurgery, University of Heidelberg, Heidelberg, Germany. 8Hospital for Children and Adolescents Augsburg, Augsburg, Germany. 9Department of Neuropathology, University of Bonn, Bonn, Germany. 10Department of Neuropathology, Heinrich-Heine-University, Düsseldorf, Germany. 11Department of Neuropathology, Burdenko Neurosurgical Institute, Moscow, Russia. 12Cnopf’sche Kinderklinik, Nuremberg Children’s Hospital, Nuremberg, Germany.
Address correspondence to: Peter Lichter, Division Molecular Genetics, German Cancer Research Center (DKFZ), In Neuenheimer Feld 280, 69120 Heidelberg, Germany. Phone: 49-6221-424619; Fax: 49-6221-424639; E-mail: [email protected].
Find articles by Kulozik, A. in: JCI | PubMed | Google Scholar
1Division Molecular Genetics, German Cancer Research Center (DKFZ), Heidelberg, Germany. 2Department of Pediatric Oncology, Hematology, and Immunology, University of Heidelberg, Heidelberg, Germany. 3Department of Neurology, University Hospital Freiburg, Freiburg, Germany. 4Department of Pediatric Neurology and Muscle Disorders, University Hospital Freiburg, Freiburg, Germany. 5Division of Biostatistics, DKFZ, Heidelberg, Germany. 6Division of Pediatric Hematology and Oncology, Department of Pediatrics and Adolescent Medicine, University of Freiburg, Freiburg, Germany. 7Department of Neurosurgery, University of Heidelberg, Heidelberg, Germany. 8Hospital for Children and Adolescents Augsburg, Augsburg, Germany. 9Department of Neuropathology, University of Bonn, Bonn, Germany. 10Department of Neuropathology, Heinrich-Heine-University, Düsseldorf, Germany. 11Department of Neuropathology, Burdenko Neurosurgical Institute, Moscow, Russia. 12Cnopf’sche Kinderklinik, Nuremberg Children’s Hospital, Nuremberg, Germany.
Address correspondence to: Peter Lichter, Division Molecular Genetics, German Cancer Research Center (DKFZ), In Neuenheimer Feld 280, 69120 Heidelberg, Germany. Phone: 49-6221-424619; Fax: 49-6221-424639; E-mail: [email protected].
Find articles by Pietsch, T. in: JCI | PubMed | Google Scholar
1Division Molecular Genetics, German Cancer Research Center (DKFZ), Heidelberg, Germany. 2Department of Pediatric Oncology, Hematology, and Immunology, University of Heidelberg, Heidelberg, Germany. 3Department of Neurology, University Hospital Freiburg, Freiburg, Germany. 4Department of Pediatric Neurology and Muscle Disorders, University Hospital Freiburg, Freiburg, Germany. 5Division of Biostatistics, DKFZ, Heidelberg, Germany. 6Division of Pediatric Hematology and Oncology, Department of Pediatrics and Adolescent Medicine, University of Freiburg, Freiburg, Germany. 7Department of Neurosurgery, University of Heidelberg, Heidelberg, Germany. 8Hospital for Children and Adolescents Augsburg, Augsburg, Germany. 9Department of Neuropathology, University of Bonn, Bonn, Germany. 10Department of Neuropathology, Heinrich-Heine-University, Düsseldorf, Germany. 11Department of Neuropathology, Burdenko Neurosurgical Institute, Moscow, Russia. 12Cnopf’sche Kinderklinik, Nuremberg Children’s Hospital, Nuremberg, Germany.
Address correspondence to: Peter Lichter, Division Molecular Genetics, German Cancer Research Center (DKFZ), In Neuenheimer Feld 280, 69120 Heidelberg, Germany. Phone: 49-6221-424619; Fax: 49-6221-424639; E-mail: [email protected].
Find articles by Herold-Mende, C. in: JCI | PubMed | Google Scholar
1Division Molecular Genetics, German Cancer Research Center (DKFZ), Heidelberg, Germany. 2Department of Pediatric Oncology, Hematology, and Immunology, University of Heidelberg, Heidelberg, Germany. 3Department of Neurology, University Hospital Freiburg, Freiburg, Germany. 4Department of Pediatric Neurology and Muscle Disorders, University Hospital Freiburg, Freiburg, Germany. 5Division of Biostatistics, DKFZ, Heidelberg, Germany. 6Division of Pediatric Hematology and Oncology, Department of Pediatrics and Adolescent Medicine, University of Freiburg, Freiburg, Germany. 7Department of Neurosurgery, University of Heidelberg, Heidelberg, Germany. 8Hospital for Children and Adolescents Augsburg, Augsburg, Germany. 9Department of Neuropathology, University of Bonn, Bonn, Germany. 10Department of Neuropathology, Heinrich-Heine-University, Düsseldorf, Germany. 11Department of Neuropathology, Burdenko Neurosurgical Institute, Moscow, Russia. 12Cnopf’sche Kinderklinik, Nuremberg Children’s Hospital, Nuremberg, Germany.
Address correspondence to: Peter Lichter, Division Molecular Genetics, German Cancer Research Center (DKFZ), In Neuenheimer Feld 280, 69120 Heidelberg, Germany. Phone: 49-6221-424619; Fax: 49-6221-424639; E-mail: [email protected].
Find articles by Gnekow, A. in: JCI | PubMed | Google Scholar
1Division Molecular Genetics, German Cancer Research Center (DKFZ), Heidelberg, Germany. 2Department of Pediatric Oncology, Hematology, and Immunology, University of Heidelberg, Heidelberg, Germany. 3Department of Neurology, University Hospital Freiburg, Freiburg, Germany. 4Department of Pediatric Neurology and Muscle Disorders, University Hospital Freiburg, Freiburg, Germany. 5Division of Biostatistics, DKFZ, Heidelberg, Germany. 6Division of Pediatric Hematology and Oncology, Department of Pediatrics and Adolescent Medicine, University of Freiburg, Freiburg, Germany. 7Department of Neurosurgery, University of Heidelberg, Heidelberg, Germany. 8Hospital for Children and Adolescents Augsburg, Augsburg, Germany. 9Department of Neuropathology, University of Bonn, Bonn, Germany. 10Department of Neuropathology, Heinrich-Heine-University, Düsseldorf, Germany. 11Department of Neuropathology, Burdenko Neurosurgical Institute, Moscow, Russia. 12Cnopf’sche Kinderklinik, Nuremberg Children’s Hospital, Nuremberg, Germany.
Address correspondence to: Peter Lichter, Division Molecular Genetics, German Cancer Research Center (DKFZ), In Neuenheimer Feld 280, 69120 Heidelberg, Germany. Phone: 49-6221-424619; Fax: 49-6221-424639; E-mail: [email protected].
Find articles by Reifenberger, G. in: JCI | PubMed | Google Scholar
1Division Molecular Genetics, German Cancer Research Center (DKFZ), Heidelberg, Germany. 2Department of Pediatric Oncology, Hematology, and Immunology, University of Heidelberg, Heidelberg, Germany. 3Department of Neurology, University Hospital Freiburg, Freiburg, Germany. 4Department of Pediatric Neurology and Muscle Disorders, University Hospital Freiburg, Freiburg, Germany. 5Division of Biostatistics, DKFZ, Heidelberg, Germany. 6Division of Pediatric Hematology and Oncology, Department of Pediatrics and Adolescent Medicine, University of Freiburg, Freiburg, Germany. 7Department of Neurosurgery, University of Heidelberg, Heidelberg, Germany. 8Hospital for Children and Adolescents Augsburg, Augsburg, Germany. 9Department of Neuropathology, University of Bonn, Bonn, Germany. 10Department of Neuropathology, Heinrich-Heine-University, Düsseldorf, Germany. 11Department of Neuropathology, Burdenko Neurosurgical Institute, Moscow, Russia. 12Cnopf’sche Kinderklinik, Nuremberg Children’s Hospital, Nuremberg, Germany.
Address correspondence to: Peter Lichter, Division Molecular Genetics, German Cancer Research Center (DKFZ), In Neuenheimer Feld 280, 69120 Heidelberg, Germany. Phone: 49-6221-424619; Fax: 49-6221-424639; E-mail: [email protected].
Find articles by Korshunov, A. in: JCI | PubMed | Google Scholar
1Division Molecular Genetics, German Cancer Research Center (DKFZ), Heidelberg, Germany. 2Department of Pediatric Oncology, Hematology, and Immunology, University of Heidelberg, Heidelberg, Germany. 3Department of Neurology, University Hospital Freiburg, Freiburg, Germany. 4Department of Pediatric Neurology and Muscle Disorders, University Hospital Freiburg, Freiburg, Germany. 5Division of Biostatistics, DKFZ, Heidelberg, Germany. 6Division of Pediatric Hematology and Oncology, Department of Pediatrics and Adolescent Medicine, University of Freiburg, Freiburg, Germany. 7Department of Neurosurgery, University of Heidelberg, Heidelberg, Germany. 8Hospital for Children and Adolescents Augsburg, Augsburg, Germany. 9Department of Neuropathology, University of Bonn, Bonn, Germany. 10Department of Neuropathology, Heinrich-Heine-University, Düsseldorf, Germany. 11Department of Neuropathology, Burdenko Neurosurgical Institute, Moscow, Russia. 12Cnopf’sche Kinderklinik, Nuremberg Children’s Hospital, Nuremberg, Germany.
Address correspondence to: Peter Lichter, Division Molecular Genetics, German Cancer Research Center (DKFZ), In Neuenheimer Feld 280, 69120 Heidelberg, Germany. Phone: 49-6221-424619; Fax: 49-6221-424639; E-mail: [email protected].
Find articles by Scheurlen, W. in: JCI | PubMed | Google Scholar
1Division Molecular Genetics, German Cancer Research Center (DKFZ), Heidelberg, Germany. 2Department of Pediatric Oncology, Hematology, and Immunology, University of Heidelberg, Heidelberg, Germany. 3Department of Neurology, University Hospital Freiburg, Freiburg, Germany. 4Department of Pediatric Neurology and Muscle Disorders, University Hospital Freiburg, Freiburg, Germany. 5Division of Biostatistics, DKFZ, Heidelberg, Germany. 6Division of Pediatric Hematology and Oncology, Department of Pediatrics and Adolescent Medicine, University of Freiburg, Freiburg, Germany. 7Department of Neurosurgery, University of Heidelberg, Heidelberg, Germany. 8Hospital for Children and Adolescents Augsburg, Augsburg, Germany. 9Department of Neuropathology, University of Bonn, Bonn, Germany. 10Department of Neuropathology, Heinrich-Heine-University, Düsseldorf, Germany. 11Department of Neuropathology, Burdenko Neurosurgical Institute, Moscow, Russia. 12Cnopf’sche Kinderklinik, Nuremberg Children’s Hospital, Nuremberg, Germany.
Address correspondence to: Peter Lichter, Division Molecular Genetics, German Cancer Research Center (DKFZ), In Neuenheimer Feld 280, 69120 Heidelberg, Germany. Phone: 49-6221-424619; Fax: 49-6221-424639; E-mail: [email protected].
Find articles by Omran, H. in: JCI | PubMed | Google Scholar
1Division Molecular Genetics, German Cancer Research Center (DKFZ), Heidelberg, Germany. 2Department of Pediatric Oncology, Hematology, and Immunology, University of Heidelberg, Heidelberg, Germany. 3Department of Neurology, University Hospital Freiburg, Freiburg, Germany. 4Department of Pediatric Neurology and Muscle Disorders, University Hospital Freiburg, Freiburg, Germany. 5Division of Biostatistics, DKFZ, Heidelberg, Germany. 6Division of Pediatric Hematology and Oncology, Department of Pediatrics and Adolescent Medicine, University of Freiburg, Freiburg, Germany. 7Department of Neurosurgery, University of Heidelberg, Heidelberg, Germany. 8Hospital for Children and Adolescents Augsburg, Augsburg, Germany. 9Department of Neuropathology, University of Bonn, Bonn, Germany. 10Department of Neuropathology, Heinrich-Heine-University, Düsseldorf, Germany. 11Department of Neuropathology, Burdenko Neurosurgical Institute, Moscow, Russia. 12Cnopf’sche Kinderklinik, Nuremberg Children’s Hospital, Nuremberg, Germany.
Address correspondence to: Peter Lichter, Division Molecular Genetics, German Cancer Research Center (DKFZ), In Neuenheimer Feld 280, 69120 Heidelberg, Germany. Phone: 49-6221-424619; Fax: 49-6221-424639; E-mail: [email protected].
Find articles by Lichter, P. in: JCI | PubMed | Google Scholar
Published April 8, 2008 - More info
J Clin Invest. 2008;118(5):1739–1749. https://doi.org/10.1172/JCI33656.
© 2008 The American Society for Clinical Investigation
Received: August 20, 2007; Accepted: February 13, 2008
-
Abstract
The molecular pathogenesis of pediatric astrocytomas is still poorly understood. To further understand the genetic abnormalities associated with these tumors, we performed a genome-wide analysis of DNA copy number aberrations in pediatric low-grade astrocytomas by using array-based comparative genomic hybridization. Duplication of the BRAF protooncogene was the most frequent genomic aberration, and tumors with BRAF duplication showed significantly increased mRNA levels of BRAF and a downstream target, CCND1, as compared with tumors without duplication. Furthermore, denaturing HPLC showed that activating BRAF mutations were detected in some of the tumors without BRAF duplication. Similarly, a marked proportion of low-grade astrocytomas from adult patients also had BRAF duplication. Both the stable silencing of BRAF through shRNA lentiviral transduction and pharmacological inhibition of MEK1/2, the immediate downstream phosphorylation target of BRAF, blocked the proliferation and arrested the growth of cultured tumor cells derived from low-grade gliomas. Our findings implicate aberrant activation of the MAPK pathway due to gene duplication or mutation of BRAF as a molecular mechanism of pathogenesis in low-grade astrocytomas and suggest inhibition of the MAPK pathway as a potential treatment.
-
Introduction
Pilocytic astrocytomas of WHO grade I are the most common primary brain tumors in children and are usually associated with a favorable prognosis, as indicated by a 10-year survival rate of 96% in the population-based study of Ohgaki and Kleihues (1). Diffuse astrocytomas of WHO grade II constitute a biologically and clinically distinct group of “low-grade” astrocytomas that are far less frequent in the pediatric age group. In contrast to pilocytic astrocytomas, diffuse astrocytomas are more common in the cerebral hemispheres than in the cerebellum, demonstrate a diffusely infiltrative growth behavior, and have a higher risk for recurrence and malignant progression. Therefore, the overall prognosis of diffuse astrocytomas is less favorable, with a median survival time of 5.6 years (1). Interestingly, diffuse astrocytomas in children show malignant transformation in only around 10% of cases (2), while the majority (60%–70%) of these tumors in adults eventually progress to anaplastic astrocytoma or secondary glioblastoma (1, 3). In addition to the WHO grade, the prognosis of low-grade astrocytoma patients depends on the tumor localization, with survival being particularly poor for diffuse brain stem astrocytomas (4). The usual first-line treatment of pilocytic and diffuse astrocytomas consists of surgical resection. Although children with these tumors often survive for many years after operation, a significant number will experience tumor recurrence, especially when surgical tumor resection is incomplete. Radiation and/or chemotherapy are largely reserved for patients with recurrent and/or progressive disease. To date, knowledge about the genetic abnormalities involved in the pathogenesis of pediatric low-grade astrocytomas is limited, and even less is known about genetic changes associated with tumor recurrence or malignant transformation. Genetic alterations frequently observed in adult anaplastic astrocytomas and/or glioblastomas, such as mutations of TP53 or PTEN, homozygous deletion of CDKN2A, amplification of CDK4 or EGFR, and losses of chromosome 10, are only rarely encountered in pediatric pilocytic and low-grade diffuse astrocytomas (5). Previous studies investigating copy number changes in these tumors have shown normal karyotypes in the vast majority of cases (4, 6). Analyses of copy number changes in a total of more than 160 cases examined in different studies employing a variety of technologies revealed only a few consistent findings. The most frequent aberrations detected in these studies involved trisomy of chromosome 5 and of chromosome 7 or gains of 7q (6–10).
Neurofibromatosis type 1 (NF1) is an autosomal, dominantly inherited disorder known to be associated with a predisposition to astrocytoma development. In particular, pilocytic astrocytoma of the optic nerve and chiasm but also diffuse astrocytoma occur with increased frequency (11). Neurofibromin, the gene product of the NF1 gene, physiologically contributes to growth arrest of astrocytic cells and neuronal differentiation by downregulation of the MAPK signaling pathway via its GTPase-activating domain. Loss of neurofibromin expression conversely leads to increased Ras activity and astrocyte proliferation (12). Mutations of other components of the MAPK pathway cause several other inherited diseases, such as Noonan syndrome, LEOPARD syndrome, cardio-facio-cutaneous (CFC) syndrome, and Costello syndrome (13–18). Somatic mutations in some of the genes responsible for these syndromes (PTPN11, KRAS, BRAF, HRAS) have been implicated in the pathogenesis of various tumor types, including brain tumors (19–21). We and others identified activating mutations in KRAS, which result in activation of MAPK signaling, in a minor fraction of sporadic pilocytic astrocytomas (22, 23). In one study, it was shown that the MAPK pathway is activated in virtually all sporadic pilocytic astrocytomas (22). Gene expression profiles of NF1-associated pilocytic astrocytomas and sporadic pilocytic astrocytomas showed similar activation of MAPK pathway target genes, further indicating that the MAPK pathway is commonly involved in the pathogenesis of sporadic astrocytomas as well (24, 25). However, the underlying genetic events leading to MAPK pathway activation in sporadic low-grade astrocytomas of childhood have only been identified in a small proportion of tumors. Therefore, it is of great interest to search for further mechanisms leading to MAPK pathway activation in these tumors. In the present study, we used array-based comparative genomic hybridization (array-CGH) or matrix-CGH to study a large series of 66 pediatric low-grade astrocytomas for genome-wide copy number changes. Array-CGH allows for a much better resolution than conventional CGH on metaphase chromosomes and therefore can identify much smaller imbalances (26).
-
Results
Copy number changes in pediatric astrocytomas as assessed by array-CGH. A total of 66 tumors from children with low-grade astrocytomas (53 pilocytic astrocytomas, 13 diffuse astrocytomas) were analyzed by array-CGH. Two of the pilocytic astrocytomas were derived from NF1 patients; all other tumors were apparently sporadic. No detectable DNA copy number imbalances were observed in 21 tumors (32%). The overall frequency of copy number changes plotted against the chromosome position is depicted in Figure 1A. A summary of minimally altered regions occurring at a frequency of 5% or above is given in Supplemental Table 2 (supplemental material available online with this article; doi:10.1172/JCI33656DS1). Recurrent large chromosomal imbalances involving entire chromosomes were trisomies of chromosomes 5 (6/66) and chromosome 7 (4/66). The frequencies of trisomies 5 and 7 differed significantly neither between pilocytic and diffuse astrocytomas nor between infratentorial and supratentorial tumors. The most frequent aberration in 30 of the 66 astrocytomas (45%) was a copy number gain of 0.97 Mb pairs represented by 2 adjacent BAC clones (RP5-886O8 and RP4-726N20) at chromosome band 7q34 spanning the BRAF locus, among others. A detailed list of all genes contained in the minimally gained region is given in Supplemental Table 3. This aberration was present as the only detectable chromosomal aberration in 13 tumors (20%), all of which were pilocytic astrocytomas (Figure 1, B and C). The first flanking BAC clones with normal copy numbers were RP11-269N18 and RP5-1173P7, respectively. To confirm the chromosomal annotation of the BAC clone covering BRAF, FISH was performed on normal metaphase chromosomes and paraffin sections of 5 pilocytic astrocytomas with known copy number status (Figure 1D). Adjacent normal tissue did not show the copy number gain, excluding a germ-line segmental duplication. Furthermore, duplications of the BRAF locus in addition to a trisomy of chromosome 7 or a gain of chromosome arm 7q were not observed. Remarkably, duplications of the BRAF locus were detected in 28 of 53 (53%) pilocytic astrocytomas, whereas they were detected in only 2 of 13 diffuse astrocytomas (15%). The copy number gain at 7q34 was more frequent in astrocytomas with noncerebellar localization (62% versus 43% in cerebellar tumors, χ2 test; P = 0.31), patients with residual tumor after surgery (70% versus 42%, χ2 test; P = 0.15), patients with tumor recurrence (71% versus 52%, χ2 test; P = 0.59), and consequently, patients who required adjuvant therapy (75% versus 50%, χ2 test: P = 0.40; Table 1). Of the 2 NF1-associated pilocytic astrocytomas, one showed a duplication of the BRAF locus. In contrast, BRAF gains (if not part of a large chromosomal gain) were not detected in any of 28 gangliogliomas and 10 pleomorphic xanthoastrocytomas investigated by array-CGH on the same microarray (refs. 27, 28, and our unpublished data).
 Figure 1
Figure 1DNA copy-number aberrations in pediatric low-grade astrocytomas. (A) Frequencies of DNA copy number gains and losses of tumors (n = 66) were plotted against their chromosomal position. (B) Complete array-CGH trace of a pilocytic astrocytoma, with a BRAF duplication at 7q34 representing the only detectable genomic aberration in this tumor. This constellation was found in 13 of 66 (20%) tumors (arrow indicates clones with copy number gain). (C) Array-CGH profile of chromosome 7 in another pilocytic astrocytoma with duplication of the BRAF locus at 7q34 (arrow indicates clones with copy number gain). (D) Duplication of the BRAF locus in a pilocytic astrocytoma as assessed by FISH. The probe covering the BRAF locus (3 copies) was labeled in green, and the centromeric control probe (2 copies) in red. Original magnification, ×100.
Activating mutations in the BRAF gene in pediatric astrocytomas. Since activating mutations of the BRAF gene have been described in different tumor entities, we investigated all tumors enrolled in the array-CGH study for activating mutations in exons known to carry such mutations in malignant (exons 11 and 15) and benign (exons 6, 12, and 14 in CFC syndrome) conditions by using denaturing HPLC (DHPLC). Tumors showing an abnormal DHPLC profile were subsequently analyzed by direct DNA sequencing. We detected point mutations affecting the hot spot codon 600 in exon 15 of BRAF in 4 of the 66 pediatric astrocytomas (6%; data not shown), all carrying the same mutation, V600E. This well-established activating mutation was found in 3 pilocytic astrocytomas and 1 diffuse astrocytoma. Lymphocyte DNA obtained from the same patients lacked these mutations, thus excluding the presence of germ-line mutations (data not shown). Interestingly, none of the 4 BRAF-mutant tumors carried a duplication of the BRAF locus, but 1 of the mutant pilocytic tumors displayed a trisomy of chromosome 7. In this particular case, we additionally investigated a recurrent tumor that still carried the BRAF mutation but had lost the trisomy of chromosome 7 (data not shown).
Expression of BRAF and its downstream target CCND1 in pediatric astrocytomas. Levels of BRAF mRNA expression were measured by quantitative real-time PCR in those pilocytic astrocytomas from which high-quality RNA was available. In total, 5 tumors with a balanced BRAF locus and 8 tumors with small copy number gains of the BRAF locus were studied. Normalized BRAF transcript levels were 1.4- to 7.3-fold higher in pilocytic astrocytomas when compared with nonneoplastic brain tissue pooled from 5 different individuals. The median BRAF mRNA expression in tumors with BRAF duplications was 2-fold higher as compared with tumors with a balanced BRAF status (Figure 2A; P = 0.01). In the same tumors, expression levels of CCND1 transcripts, which encode cyclin D1, a well-established target gene of MAPK signaling, were measured. CCND1 mRNA levels varied between 0.78- and 55-fold in the astrocytomas when compared with nonneoplastic brain tissue. The median CCND1 mRNA expression level was 3.7-fold higher in tumors with BRAF duplication than in tumors without this aberration (Figure 2B; P = 0.008). Six tumor samples with BRAF duplications were investigated by Western blot analysis, and all of them showed higher total expression of BRAF protein and increased phosphorylation of ERK1/2 when compared with nonneoplastic brain tissue (Figure 2C).
 Figure 2
Figure 2BRAF and CCND1 expression in primary pilocytic astrocytomas. (A) BRAF mRNA expression levels in 5 pilocytic astrocytomas with 2 copies of BRAF (left) and 8 pilocytic astrocytomas with 3 copies of BRAF due to gene duplications (right) in relation to the BRAF transcript levels in nonneoplastic brain tissue. Note that the median BRAF mRNA expression level is significantly higher in tumors with BRAF duplications as compared with tumors without this aberration (P = 0.01). (B) CCND1 mRNA expression in the same tumors. Median CCND1 mRNA expression levels are significantly higher in tumors with BRAF duplications than in tumors with 2 gene copies (P = 0.008). (C) Western blot analysis demonstrating BRAF protein expression (upper panel) and ERK1/2 phosphorylation (pERK1/2, middle panel) in 6 tumors with BRAF duplication (lanes 2–7) and in nonneoplastic brain tissue (lane 1, pooled protein fractions from 5 brain tissue samples from 5 different individuals) showing that BRAF is exclusively expressed in the tumors and always associated with activation of MAPK signaling. Total ERK1/2 protein was used as a loading control (ERK1/2, lower panel). nb, normal brain; PA, pilocytic astrocytoma.
FISH analysis of the BRAF locus in adult pilocytic and diffuse astrocytomas. To investigate the impact of copy number aberrations at the BRAF locus in adult pilocytic and diffuse astrocytomas, we investigated 28 tumors (3 pilocytic astrocytomas, 25 diffuse astrocytomas) by FISH on a tissue microarray (TMA). Based on the quality criteria outlined in Methods, 26 of the 28 cases could be evaluated. We found copy number gains of the BRAF locus in 16 of 26 tumors (62%). Six of these tumors (23%), all diffuse astrocytomas, showed a gain of BRAF without concomitant gain of the chromosome 7 centromeric probe, while 10 tumors (38%, all 3 pilocytic astrocytomas and 7 diffuse astrocytomas) displayed concomitant gains of the BRAF and the chromosome centromeric probes, indicating large genomic gains of chromosome 7 (data not shown).
Inhibition of the MAPK pathway in astrocytoma cell lines by MEK1/2 inhibitors. MAPK signaling component MEK1/2 constitutes the immediate downstream phosphorylation target of BRAF. MEK1/2 phosphorylation by BRAF can effectively be blocked in vitro by small molecule inhibitors, such as U0126. We treated 4 cell lines established from primary low-grade astrocytomas or oligoastrocytomas with U0126 and determined the impact of treatment on MAPK pathway activity and cell proliferation. One of these cell lines was derived from a pediatric patient with a pilocytic astrocytoma WHO grade I, and 3 were established from adult patients with WHO grade II tumors corresponding to diffuse astrocytoma (1 case) or oligoastrocytoma (2 cases), respectively. Apart from the cell line derived from the diffuse astrocytoma, all 3 other cell lines carried large copy number gains of chromosome arm 7q spanning the BRAF locus, as determined by array-CGH (data not shown). None of the tumor cell lines demonstrated an activating BRAF mutation, whereas all cell lines showed similar levels of BRAF mRNA and protein expression (data not shown). Using a broad range of different concentrations of the MEK1/2 inhibitor (range 1–20 μM), we found profound reduction of proliferation in all 4 cell lines (Figure 3, A and B). Biochemically, we could demonstrate that complete dephosphorylation of ERK1/2 is detectable as early as 30 minutes after incubation with 1 μM of the inhibitor and is maintained for at least 24 hours after a single dose of the drug (Figure 3C). Messenger RNA downregulation of the MAPK downstream target CCND1 varied between 2.4- and 7.5-fold for the different cell lines, reaching its maximum after 24 hours (Figure 3D). Reduced proliferation after treatment with U0126 was further confirmed by cell-cycle analysis, revealing arrest in G2/M phase (Figure 4, A, C, and E). Induction of apoptosis upon drug treatment was not observed. This was demonstrated both by the lack of a sub-G1 peak in the cell-cycle analysis (representing the apoptotic fraction of cells) and only a slight increase in annexin V–positive cells upon drug treatment (Figure 4, B, D, and F).
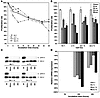 Figure 3
Figure 3Pharmacological inhibition of MAPK signaling in cell lines derived from low-grade gliomas. (A) Proliferation of glioma cells in vitro after treatment of 4 different cell lines derived from primary low-grade gliomas with the MEK1/2 inhibitor U0126 at a concentration of 20 μM as assessed by MTT assay over a time course of 48 hours. A °II, diffuse astrocytoma; OA, oligoastrocytoma. (B) Effective growth inhibition in all 4 cell lines can be achieved over a broad spectrum of concentrations ranging from 1 μM up to 20 μM. Medians and SDs of triplicate measurements at 48 hours are shown. (C) Dephosphorylation of ERK1/2 is readily detectable after 30 minutes and is maintained for 48 hours after a single dose of inhibitor at a concentration of 1 μM. (D) Maximal downregulation of CCND1 mRNA expression after treatment with MEK1/2 inhibitor U0126 at a concentration of μM was observed after 24 hours.
 Figure 4
Figure 4Functional analyses of low-grade glioma cell lines after pharmacological inhibition of MAPK signaling. Cell-cycle analysis by measurement of DNA content in NCH492 cells prior to (A) and 48 hours after treatment with 10 μM U0126 (C). Cells were arrested in G2/M upon treatment with U0126 and lacked a subG1-peak, indicating no increase in apoptosis. Early apoptosis rate was additionally determined by annexin V staining and was only slightly increased 24 hours after drug treatment (B and D). Puromycin was used as a positive control for apoptosis induction (E and F).
Cell-cycle arrest in pilocytic astrocytoma cell lines upon stable silencing of BRAF. Since pilocytic astrocytomas were the main focus of this study, lentivirus-mediated silencing of BRAF by RNA interference was performed in the cell line derived from a pilocytic astrocytoma. This cell line was also selected because it carries a copy number gain of chromosome arm 7q including the BRAF locus (data not shown). Using 5 different shRNAs targeting BRAF, we achieved knockdown efficiencies between 80% and 95% as assessed by quantitative real-time PCR (data not shown). Nontargeting shRNA was used as a control. Stable transduction with 2 shRNAs (shRNA4 and shRNA5) resulted in a virtually complete inhibition of proliferation, thus precluding any subsequent analyses. In the cells with stable expression of 1 of the other 3 shRNAs against BRAF or a nontargeting shRNA, proliferation rate was determined by 3-[4,5-dimethylthiazol-2-yl]-2,5-diphenyltetrazolium bromide (MTT) assays. Equal amounts of cells were platted 24 hours prior to the analysis. Proliferation was significantly and reproducibly lower in all BRAF-silenced cell lines (Figure 5A). This effect was also evident upon microscopic investigation of the cells after 24 hours (Figure 5B). As demonstrated for the treatment with U0126, cells were cell-cycle arrested in G2/M phase upon silencing of BRAF, thus indicating a nonredundant role for BRAF in MAPK signaling in astrocytic tumors.
 Figure 5
Figure 5Stable silencing of BRAF expression in pilocytic astrocytoma cells. (A) Proliferation of NCH492 pilocytic astrocytoma cells after shRNA-mediated silencing of BRAF using 3 different shRNAs as assessed by MTT assay 24 hours after plating equal numbers of cells. Nontargeting shRNA was used as a reference. Severe growth inhibition was observed for all 3 shRNAs targeting BRAF. Error bars represent the SD between replicates. (B) Microscopic examination of the same samples at the time of MTT analysis showing growth arrest of cells upon BRAF knockdown. Original magnification, ×10. (C–F) Cell-cycle analysis 24 hours after plating equal numbers of cells either carrying nontargeting shRNA (C) or one of the shRNAs targeting BRAF (D–F). As observed for the treatment with MEK1/2 inhibitor, cells accumulated in the G2/M phase of the cell cycle.
-
Discussion
MAPK signaling in cancer. The RAS/RAF (MAPK) signaling cascade is one of the most prominent pathways for regulating cell growth, proliferation, differentiation, and apoptosis in malignant and nonmalignant cells. BRAF, an immediate downstream target of the Ras proteins, has been identified as a frequent target of activating mutations in precancerous lesions, e.g., hyperplastic polyps of the colon and large congenital nevi (29, 30). In addition, oncogenic activation by point mutations, translocations, or inversions has been observed in about 60% of malignant melanomas, 50% of thyroid carcinomas, and 15% of colorectal and ovarian cancers (21, 31–35). In adult gliomas, previous studies have detected BRAF mutations in a small fraction of glioblastomas (36, 37), and an individual case of oligodendroglioma has been reported (38). In addition, rare cases of gliomas were found to carry mutations in NRAS (36, 38), while large copy number gains involving genes of the MAPK pathway were detected by chromosomal CGH analysis in 38 of 87 gliomas (44%) investigated (38). In the present study, we systematically investigated pediatric pilocytic and low-grade diffuse astrocytomas for genomic imbalances by using high-resolution array-CGH and identified frequent BRAF gene duplication as what we believe is a novel mechanism leading to MAPK pathway activation in pediatric low-grade astrocytomas.
Oncogenic activation of BRAF by either gene duplications or activating mutations in pediatric low-grade astrocytomas. Our array-CGH analysis of 53 pilocytic and 13 diffuse astrocytomas from pediatric patients revealed that copy number imbalances are overall infrequent in these tumors. This finding is in line with results from a recently published array-CGH study of 44 pilocytic astrocytomas, which did not detect any copy number changes in 64% of the cases when using BAC-arrays with 1-Mb average resolution (10). Interestingly, this study reported that chromosomal gains were significantly more frequent and the number of affected chromosomes significantly greater in patients over 15 years old. In our cohort, only 3 of 66 patients (5%) were between 15 and 18 years old, with an average of 2 copy number aberrations per tumor. However, we identified circumscribed duplications at 7q34 as a prominent aberration in pediatric pilocytic and diffuse astrocytomas, with an overall frequency of 45% in our study population and a frequency of 53% in the pilocytic astrocytoma subgroup. BRAF was selected from 12 coding genes contained in this region, being the most plausible candidate oncogene. Further strengthening this assumption, we found activating mutations of BRAF at hot spot codon 600 in 6% of the tumors, with BRAF mutation and duplication appearing to be mutually exclusive. In addition, we found strong upregulation of BRAF mRNA and protein expression in all tumors carrying a duplication of the BRAF locus. These findings are remarkable since the molecular genetics of pediatric low-grade astrocytomas, in particular pilocytic astrocytomas, has been largely unknown. Besides the clinical observation that NF1 patients have an increased risk for these tumors, no other genetic aberration has consistently been detected in sporadic astrocytomas to date. While NF1-associated pilocytic astrocytomas show loss and inactivation of NF1 alleles, sporadic pilocytic astrocytomas lack NF1 mutations or reduced NF1 expression (39). Neurofibromin, the gene product of NF1, is involved in the repression of MAPK signaling by its GTPase domain (12). Interestingly, while not detecting BRAF gene duplications, Jones et al. (10) identified a small copy number gain of the RAF1 locus at 3p25 in 1 pilocytic astrocytoma (10). However, this aberration appears to be rare because it was not detected in any of the tumors investigated in our study. Two other studies found rare activating mutations of KRAS in low-grade astrocytomas (22, 23). Therefore, it is of particular interest that BRAF as a component of the same pathway is activated by duplications or activating mutations in about one-half of sporadic pilocytic and diffuse astrocytomas of WHO grade I or II, respectively. Although the fraction of patients showing tumor recurrence and/or malignant progression was too low to allow for a correlation between BRAF status and survival rates, we found that BRAF duplications are more frequently encountered in astrocytomas with noncerebellar localization and in tumors from patients with a residual tumor after surgery, with a substantial fraction of these patients requiring adjuvant treatment.
Activation of downstream targets of the RAS/RAF signaling pathway. After identifying BRAF duplication or mutation as a frequent event in low-grade astrocytomas, we were interested in studying downstream effectors of the MAPK pathway, such as ERK1/2 and CCND1. Phosphorylation of ERK1/2 was observed in all investigated pediatric astrocytomas to various levels, thus suggesting a constitutive MAPK pathway activation in these tumors, likely involving genetic or epigenetic aberrations yet to be uncovered in those tumors lacking BRAF alterations. CCND1 mRNA was not only highly overexpressed in most tumors in relation to nonneoplastic brain tissue but also showed significantly higher levels in tumors with BRAF duplication as compared with tumors retaining the ordinary 2 copies of this gene.
Role of BRAF in adult low-grade astrocytomas. Investigation of BRAF copy number status in low-grade astrocytomas from adult patients also revealed a high percentage of tumors with BRAF duplication (23%). In addition, large genomic gains of chromosome 7q involving the BRAF locus were present in 38% of cases. This is in line with other studies reporting trisomy of chromosome 7 as a common genomic aberration in adult low-grade astrocytomas (40, 41). The impact of BRAF duplications in the pathogenesis of astrocytomas in adults and the relationship between BRAF alteration and other genetic changes in diffuse astrocytomas, e.g., TP53 mutations, remains to be determined.
The MAPK pathway constitutes a potential drug target in low-grade astrocytomas. To provide first preclinical evidence for a potential role of specific pharmacological inhibitors of the MAPK pathway in the tailored treatment of low-grade astrocytomas, we made use of 4 cell lines established from primary low-grade gliomas. In all 4 cell lines, we found a significant reduction in tumor cell proliferation after treatment with a single dose of the MEK1/2 inhibitor U0126 at different concentrations. Furthermore, the observed rapid dephosphorylation of ERK1/2 and strong downregulation of CCND1 mRNA expression indicate an effective inhibition of the MAPK pathway in vitro. To investigate the level of apoptosis upon U0126 treatment, we performed cell-cycle analysis and annexin V staining. Cell-cycle analysis revealed an arrest in the G2/M phase and no increase in the apoptotic fraction upon drug treatment. This is in line with findings by other authors who demonstrated that MAPK activation is necessary for G2/M transition and that MEK1/2 inhibition leads to G2/M arrest (42–45). Annexin V staining, which is capable of detecting early apoptotic cells, showed a similar level of annexin V–positive cells upon MEK1/2 inhibition.
BRAF knockdown in pilocytic astrocytoma cells elicits growth arrest in G2/M phase. To study the specific role of BRAF in pilocytic astrocytomas, we silenced BRAF in a cell line derived from a pilocytic astrocytoma, which carries a copy number gain of chromosome arm 7q. To our knowledge, this is the first report on functional analyses performed in an established cell line derived from a pilocytic astrocytoma to date. Knockdown was performed by using lentiviral transduction of 5 different shRNAs targeting different regions within the BRAF transcript and compared with a control transduction using a nontargeting shRNA. Strikingly, 2 of the 5 shRNAs resulted in virtually complete inhibition of cell proliferation so that consecutive functional analyses could not be performed. The remaining 3 shRNAs caused profound inhibition of cell proliferation without obvious change in the morphologic appearance of the tumor cells. Functional analysis revealed accumulation of cells in the G2/M phase of the cell cycle and no evidence for a significant increase in apoptosis.
In summary, we identified a mechanism of MAPK pathway activation in low-grade astrocytomas via duplication of the BRAF gene locus. This results in overexpression of BRAF at the mRNA and protein levels and consecutive activation of downstream signaling components. In addition, we found that a small percentage of pediatric low-grade astrocytomas carry BRAF point mutations as an alternative molecular mechanism for MAPK pathway activation. Other alterations in these tumors may activate the same pathway, such as overexpression of tyrosine kinase growth factor receptors, e.g., the PDGF receptor α (46), or in rare instances, NF1 inactivation or activating KRAS mutation. Thus, aberrant MAPK signaling is of paramount importance in the pathogenesis of pilocytic and diffuse astrocytomas. The fact that virtually all low-grade astrocytomas demonstrate an activation of downstream MAPK effectors suggests that targeted pharmacological inhibition of this pathway may constitute a promising approach in the treatment of these tumors.
-
Methods
Tumor material. The majority of astrocytoma samples from pediatric patients (n = 41) were collected between 1994 and 2002 at the Department of Pediatric Oncology, Würzburg University Hospital (Würzburg, Germany), and the Department of Pediatric Oncology, Mannheim University Hospital (Mannheim, Germany). Informed consent was obtained for all tumor samples either from the patient or the legal guardian. All diagnoses were confirmed by histological assessment of specimens obtained at neurosurgery by at least 2 neuropathologists according to the criteria of the WHO classification. Approval to link histological data to clinical data was obtained from the Institutional Review Board of the University of Würzburg. The collection and use of tumor samples was conducted according to the official statement of the National Ethics Council of the federal government of Germany (March 2004). Additional pediatric tumor tissue samples (n = 25) were obtained from the brain tumor tissue bank of the German Society for Pediatric Hematology and Oncology (GPOH). Approval to link histological data to clinical data was obtained for all samples collected in the specified tumor bank from the Ethical Committee of the University of Bonn, where the tumor bank is located.
Patient characteristics of all samples investigated by array-CGH are provided in Table 2. Tumor samples of low-grade astrocytomas (n = 28) from adult patients, including 3 pilocytic astrocytomas and 25 diffuse astrocytomas, were collected between 1996 and 2004 at the Department of Neurosurgery, University of Heidelberg. Approval to link histological data to clinical data was obtained by the Institutional Review Board of the University of Heidelberg. Informed consent was obtained from the patients directly or, in the case of minors, from their parents or legal guardians. Patient characteristics corresponding to these samples are provided in Supplemental Table 1. Permanent cell lines from primary cell cultures were established from a pediatric patient with pilocytic astrocytoma (WHO grade I; NCH492) and 3 adult patients with either a diffuse astrocytoma (WHO grade II; NCH134) or oligoastrocytomas (WHO grade II; NCH480b and NCH514) treated at the Department of Neurosurgery, University of Heidelberg. All primary cultures were propagated in vitro over 50 passages or more with a split ratio of 1:6 to 1:10. The in vitro experiments for this study were performed at passages 53, 10, 11, and 54, respectively. Total RNA and total protein extracted from nonneoplastic brain tissue (consisting of a pool of tissue samples from 5 donors; age range, 26–82 years) were purchased from BioChain.
Nucleic acid isolation. Extraction of high molecular weight DNA and RNA from frozen tumor samples was carried out by use of a salting-out method (47) or by ultracentrifugation over cesium chloride as previously described (48). Genomic DNA from peripheral blood mononuclear cells of healthy donors (pool of 10 donors; age range, 25–40 years for each sex) was isolated by use of the QIAGEN DNA Blood Midi-Kit. DNA quality was assessed on a 1% agarose gel, and total RNA quality and concentration was controlled with the Agilent 2100 bioanalyzer (Agilent Technologies).
Array-based comparative genomic hybridization. Array-CGH was carried out as previously described (26, 49, 50). Selection of genomic clones, isolation of BAC DNA, performance of degenerate oligonucleotide primed PCR, preparation of microarrays, labeling, hybridization, and washing procedures as well as data analysis were performed as outlined elsewhere (49, 50). Normalized log2 ratios as well as raw data sets of this study have been deposited in the NCBI Gene Expression Omnibus (accession numbers GSE8737 and GPL5713).
Statistics. To assess associations between BRAF copy number status and clinical subgroups, 2D contingency tables were calculated, and 2-sided χ2 tests were applied. A result was considered significant at P < 0.05. Statistical computations were performed with the statistical software environment R, version 2.6.1.
FISH. Two-color interphase FISH was performed using FITC-labeled (RP4-726N20) and rhodamine-labeled (p7t1) probes localizing in the locus of interest (7q34; BRAF) and the centromere of chromosome 7, respectively (51). Pretreatment of slides, hybridization, posthybridization processing, and signal detection were performed as described previously (52). Samples showing sufficient FISH efficiency (>90% nuclei with signals) were evaluated by 2 independent investigators. Signals were scored in at least 50 nonoverlapping, intact nuclei. Metaphase FISH for verifying clone mapping position was performed using peripheral blood cell cultures of healthy donors as outlined previously (53).
Mutational analysis by DHPLC and genomic sequencing. Genomic DNA was amplified by using the GenomiPhi kit (GE Healthcare). Mutational analysis was focused on those genomic fragments of BRAF that were previously reported to carry activating mutations (15, 16, 21). Specific intronic primer pairs were used for PCR amplification of exons 6, 11, 12, 14, and 15. Primer sequences are available in Supplemental Table 4. DHPLC was used for mutation screening. PCR products showing an abnormal DHPLC profile were further investigated by direct DNA sequencing. Each mutation detected in an amplified DNA sample was confirmed by independent sequencing of the corresponding nonamplified tumor DNA.
Preparation of TMAs and immunohistochemistry. A custom TMA for the investigation of low-grade astrocytomas from adults was constructed. H&E-stained sections from all paraffin blocks were prepared to identify representative tumor regions as previously described (54). All sections were arrayed in triplicate when sufficient material was available.
Quantitative real-time PCR. To measure BRAF and CCND1 mRNA abundance in tumors, a reference of total RNA obtained from nonneoplastic human brain tissue samples of 5 individuals (age range, 26–82; BioChain) was used for tissue-specific normalization. Each cDNA sample was analyzed in triplicate with the ABI PRISM 7700 (Applied Biosystems) using Absolute SYBR Green ROX Mix (ABgene) according to the manufacturer’s instructions. Two endogenous housekeeping genes (PGK1, DCTN2) were used as internal standards. These genes were not regulated in expression profiling experiments comparing subsets of low-grade astrocytomas. Furthermore, they were tested in normal tissues from whole brain and cerebellum where they displayed very little variability. All primers were tested to exclude amplification from genomic DNA. The relative quantification of the RNA of interest in comparison with the housekeeping genes was calculated according to a previously published algorithm, with the modification that 2 housekeeping genes were used in the present study (55). Oligonucleotide sequences are available in Supplemental Table 5.
Western blotting. Protein fractions derived from cesium chloride ultracentrifugation were first subjected to dialysis against PBST overnight using a Slide-a-Lyzer MINI Dialysis cup (Pierce Biotechnology) to deplete residual cesium chloride. Polyclonal rabbit antibody against p44/42 (used as a loading control; Cell Signaling Technology), monoclonal rabbit antibody against phospho-p44/42 (Cell Signaling Technology), and monoclonal rabbit antibody against BRAF (Abcam) were all used at 1:1000 dilutions. Horseradish peroxidase–conjugated secondary antibodies (Abcam) were used in a 1:10000 dilution prior to the chemiluminescent detection of protein (ECL Plus kit; GE Healthcare).
Proliferation assay. To assess the proliferation of glioma cells in vitro before and after treatment with the MEK1/2 inhibitor U0126, the MTT-based CellTiter96 AQ Non-Radioactive Cell Proliferation Assay (Promega) was used following the manufacturer’s recommendations. All samples were assayed in triplicate, and experiments were performed in 2 biological replicas. U0126 was used at concentrations ranging from 1 μM to 20 μM. DMSO was used as a negative control. Serum starvation was performed for 4 hours prior to each experiment.
Cell-cycle analysis by flow cytometry. Cell-cycle analysis was performed 24 hours after drug treatment with U0126. Cells were fixed with ethanol, and DNA content was investigated by staining for 30 minutes at room temperature with a solution containing 0.1% Triton X-100 (Sigma-Aldrich), 20 μg/ml of propidium iodide (Invitrogen), and 0.2 mg/ml of DNase-free RNase A (QIAGEN). Cells were analyzed by flow cytometry using FACSCanto II (BD). FACSDiva software (BD) was used to quantify the distribution of cells in each cell-cycle phase: sub-G1 (apoptotic cells), G1, S, and G2/M.
Detection of early apoptosis by annexin V staining. Phosphatidylserine externalization as a measurement for early apoptosis was analyzed by flow cytometry. Cells were incubated with annexin V FITC (BD) and propidium iodide (Invitrogen) for 15 minutes in the dark, immediately followed by flow cytometry using a FACSCanto II (BD). Analysis was performed using the FACSDiva software.
BRAF silencing by lentiviral transduction of shRNA. Lentiviral vectors were produced by cotransfection of HEK293T cells with the psPAX2, pMD2.G, and pLKO.1 constructs (MISSION TRC-Hs 1.0; ref. 56). Transfections were carried out using FuGENE 6 (Roche). Virus was harvested at 48 and 72 hours after transfection, and infections of NCH492 cells were carried out in the presence of 8 μg/ml of polybrene. Virus-containing supernatant was removed after 24 hours. Following transduction, cells were selected with 2.5 μg/ml puromycin. Five independent shRNA constructs targeting different regions of the BRAF mRNA transcript were used: MISSION shRNA NM_004333.2-1106s1c1 (shRNA1); NM_004333.2-838s1c1 (shRNA2); NM_004333.2-2267s1c1 (shRNA3); NM_004333.2-1538s1c1 (shRNA4); and NM_004333.2-304s1c10 (shRNA5). Nontargeting shRNA was used as a control.
-
Acknowledgments
We gratefully thank Leonore Senf for the collection and maintenance of the brain tumor bank in the department of W. Scheurlen. The brain tumor tissue bank of GPOH has kindly provided tumor samples and clinical data for this study. Stefanie Hofmann is acknowledged for excellent technical assistance.
Address correspondence to: Peter Lichter, Division Molecular Genetics, German Cancer Research Center (DKFZ), In Neuenheimer Feld 280, 69120 Heidelberg, Germany. Phone: 49-6221-424619; Fax: 49-6221-424639; E-mail: [email protected].
-
Footnotes
Nonstandard abbreviations used: array-CGH, array-based CGH; CGH, comparative genomic hybridization; DHPLC, denaturing HPLC; MTT, 3-[4,5-dimethylthiazol-2-yl]-2,5-diphenyltetrazolium bromide; NF1, neurofibromatosis type 1; TMA, tissue microarray.
Conflict of interest: The authors have declared that no conflict of interest exists.
Reference information: J. Clin. Invest.118:1739–1749 (2008). doi:10.1172/JCI33656
Stefan Pfister, Wibke G. Janzarik, and Marc Remke contributed equally to this work.
-
References
- Ohgaki, H., Kleihues, P. 2005. Population-based studies on incidence, survival rates, and genetic alterations in astrocytic and oligodendroglial gliomas. J. Neuropathol. Exp. Neurol. 64:479-489. View this article via: PubMed Google Scholar
- Broniscer, A., et al. 2007. Clinical and molecular characteristics of malignant transformation of low-grade glioma in children. J. Clin. Oncol. 25:682-689.
- Watanabe, K., et al. 1997. Incidence and timing of p53 mutations during astrocytoma progression in patients with multiple biopsies. Clin. Cancer Res. 3:523-530. View this article via: PubMed Google Scholar
- Orr, L., et al. 2002. Cytogenetics in pediatric low-grade astrocytomas. Med. Pediatr. Oncol. 38:173-177.
- Cheng, Y., et al. 2000. Pilocytic astrocytomas do not show most of the genetic changes commonly seen in diffuse astrocytomas. Histopathology. 37:437-444.
- Sanoudou, D., Tingby, O., Ferguson-Smith, M., Collins, V., Coleman, N. 2000. Analysis of pilocytic astrocytoma by comparative genomic hybridization. Br. J. Cancer. 82:1218-1222.
- Jenkins, R.B., et al. 1989. A cytogenetic study of 53 human gliomas. Cancer Genet. Cytogenet. 39:253-279.
- Bigner, S.H., McLendon, R.E., Fuchs, M., McKeever, P.E., Friedman, H.S. 1997. Chromosomal characteristics of childhood brain tumors. Cancer Genet. Cytogenet. 97:125-134.
- Zattara-Cannoni, H., et al. 1998. Are juvenile pilocytic astrocytomas benign tumors? A cytogenetic study in 24 cases. Cancer Genet. Cytogenet. 104:157-160.
- Jones, D., et al. 2006. Genomic analysis of pilocytic astrocytomas at 0.97 Mb resolution shows an increasing tendency toward chromosomal copy number change with age. J. Neuropathol. Exp. Neurol. 65:1049-1058. View this article via: PubMed Google Scholar
- Listernick, R., Ferner, R., Liu, G., Gutmann, D. 2007. Optic pathway gliomas in neurofibromatosis-1: Controversies and recommendations. Ann. Neurol. 61:189-198.
- Yunoue, S., et al. 2003. Neurofibromatosis type I tumor suppressor neurofibromin regulates neuronal differentiation via its GTPase–activating protein function toward Ras. J. Biol. Chem. 278:26958-26969.
- Razzaque, M.A., et al. 2007. Germline gain-of-function mutations in RAF1 cause Noonan syndrome. Nat. Genet. 39:1013-1017.
- Pandit, B., et al. 2007. Gain-of-function RAF1 mutations cause Noonan and LEOPARD syndromes with hypertrophic cardiomyopathy. Nat. Genet. 39:1007-1012.
- Niihori, T., et al. 2006. Germline KRAS and BRAF mutations in cardio-facio-cutaneous syndrome. Nat. Genet. 38:294-296.
- Rodriguez-Viciana, P., et al. 2006. Germline mutations in genes within the MAPK pathway cause cardio-facio-cutaneous syndrome. Science. 311:1287-1290.
- Bentires-Alj, M., Kontaridis, M.I., Neel, B.G. 2006. Stops along the RAS pathway in human genetic disease. Nat. Med. 12:283-285.
- Kratz, C., Niemeyer, C., Zenker, M. 2007. An unexpected new role of mutant Ras: perturbation of human embryonic development. J. Mol. Med. 85:227-235.
- Bos, J.L., et al. 1987. Prevalence of ras gene mutations in human colorectal cancers. Nature. 327:293-297.
- Bentires-Alj, M., et al. 2004. Activating mutations of the noonan syndrome-associated SHP2/PTPN11 gene in human solid tumors and adult acute myelogenous leukemia. Cancer Res. 64:8816-8820.
- Davies, H., et al. 2002. Mutations of the BRAF gene in human cancer. Nature. 417:949-954.
- Sharma, M., Zehnbauer, B., Watson, M., Gutmann, D. 2005. RAS pathway activation and an oncogenic RAS mutation in sporadic pilocytic astrocytoma. Neurology. 65:1335-1336.
- Janzarik, W., et al. 2007. Further evidence for a somatic KRAS Mutation in a low-grade Astrocytoma. Neuropediatrics. 38:1-3.
- Sharma, M.K., et al. 2007. Distinct genetic signatures among pilocytic astrocytomas relate to their brain region origin. Cancer Res. 67:890-900.
- Rorive, S., et al. 2006. Exploring the distinctive biological characteristics of pilocytic and low-grade diffuse astrocytomas using microarray gene expression profiles. J. Neuropathol. Exp. Neurol. 65:794-807. View this article via: PubMed Google Scholar
- Solinas-Toldo, S., et al. 1997. Matrix-based comparative genomic hybridization: Biochips to screen for genomic imbalances. Genes Chromosomes Cancer. 20:399-407.
- Weber, R.G., et al. 2006. Frequent loss of chromosome 9, homozygous CDKN2A//p14ARF//CDKN2B deletion and low TSC1 mRNA expression in pleomorphic xanthoastrocytomas. Oncogene. 26:1088-1097.
- Hoischen, A., et al. 2008. Comprehensive characterization of genomic aberrations in gangliogliomas by CGH, array-based CGH and interphase FISH. Brain Pathol.In press
- Rosenberg, D.W., et al. 2007. Mutations in BRAF and KRAS differentially distinguish serrated versus non-serrated hyperplastic aberrant crypt foci in humans. Cancer Res. 67:3551-3554.
- Dessars, B., et al. 2007. Chromosomal translocations as a mechanism of BRAF activation in two cases of large congenital melanocytic nevi. J. Invest. Dermatol. 127:1468-1470.
- Rajagopalan, H., et al. 2002. Tumorigenesis: RAF/RAS oncogenes and mismatch-repair status. Nature. 418:934.
- Brose, M.S., et al. 2002. BRAF and RAS mutations in human lung cancer and melanoma. Cancer Res. 62:6997-7000. View this article via: PubMed Google Scholar
- Xu, X., Quiros, R.M., Gattuso, P., Ain, K.B., Prinz, R.A. 2003. High prevalence of BRAF gene mutation in papillary thyroid carcinomas and thyroid tumor cell lines. Cancer Res. 63:4561-4567. View this article via: PubMed Google Scholar
- Kimura, E.T., et al. 2003. High prevalence of BRAF mutations in thyroid cancer: genetic evidence for constitutive activation of the RET/PTC-RAS-BRAF signaling pathway in papillary thyroid carcinoma. Cancer Res. 63:1454-1457. View this article via: PubMed Google Scholar
- Ciampi, R., et al. 2005. Oncogenic AKAP9–BRAF fusion is a novel mechanism of MAPK pathway activation in thyroid cancer. J. Clin. Invest. 115:94-101.
- Knobbe, C.B., Reifenberger, J., Reifenberger, G. 2004. Mutation analysis of the Ras pathway genes NRAS, HRAS, KRAS and BRAF in glioblastomas. Acta Neuropathologica. 108:467-470.
- Basto, D., et al. 2005. Mutation analysis of B-RAF gene in human gliomas. Acta Neuropathologica. 109:207-210.
- Jeuken, J., et al. 2007. RAS/RAF pathway activation in gliomas: the result of copy number gains rather than activating mutations. Acta Neuropathologica. 114:121-133.
- Wimmer, K., et al. 2002. Mutational and expression analysis of the NF1 gene argues against a role as tumor suppressor in sporadic pilocytic astrocytomas. J. Neuropathol. Exp. Neurol. 61:896-902. View this article via: PubMed Google Scholar
- Wessels, P., et al. 2002. Gain of chromosome 7, as detected by in situ hybridization, strongly correlates with shorter survival in astrocytoma grade 2. Genes Chromosomes Cancer. 33:279-284.
- Hirose, Y., et al. 2003. Grade II astrocytomas are subgrouped by chromosome aberrations. Cancer Genet. Cytogenet. 142:1-7.
- Wright, J.H., et al. 1999. Mitogen-activated protein kinase kinase activity is required for the G2/M transition of the cell cycle in mammalian fibroblasts. Proc. Natl. Acad. Sci. U. S. A. 96:11335-11340.
- Roberts, E.C., et al. 2002. Distinct cell cycle timing requirements for extracellular signal-regulated kinase and phosphoinositide 3-kinase signaling pathways in somatic cell mitosis. Mol. Cell. Biol. 22:7226-7241.
- Hayne, C., Tzivion, G., Luo, Z. 2000. Raf-1/MEK/MAPK pathway is necessary for the G2/M transition induced by nocodazole. J. Biol. Chem. 275:31876-31882.
- Knauf, J.A., et al. 2006. Oncogenic RAS induces accelerated transition through G2/M and promotes defects in the G2 DNA damage and mitotic spindle checkpoints. J. Biol. Chem. 281:3800-3809.
- Hermanson, M., et al. 1992. Platelet-derived growth factor and its receptors in human glioma tissue: expression of messenger RNA and protein suggests the presence of autocrine and paracrine loops. Cancer Res. 52:3213-3219. View this article via: PubMed Google Scholar
- Miller, S.A., Dykes, D.D., Polesky, H.F. 1988. A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res. 16:1215. View this article via: PubMed Google Scholar
- van den Boom, J., et al. 2003. Characterization of gene expression profiles associated with glioma progression using oligonucleotide-based microarray analysis and real-time reverse transcription-polymerase chain reaction. Am. J. Pathol. 163:1033-1043. View this article via: PubMed Google Scholar
- Mendrzyk, F., et al. 2005. Genomic and protein expression profiling identifies CDK6 as novel independent prognostic marker in medulloblastoma. J. Clin. Oncol. 23:8853-8862.
- Zielinski, B., et al. 2005. Detection of chromosomal imbalances in retinoblastoma by matrix-based comparative genomic hybridization. Genes Chromosomes Cancer. 43:294-301.
- Rocchi, M., Archidiacono, N., Ward, D., Baldini, A. 1991. A human chromosome 9-specific alphoid DNA repeat spatially resolvable from satellite 3 DNA by fluorescent in situ hybridization. Genomics. 9:517-523.
- Bubendorf, L., et al. 1999. Survey of gene amplifications during prostate cancer progression by high-throughput fluorescence in situ hybridization on tissue microarrays. Cancer Res. 59:803-806. View this article via: PubMed Google Scholar
- Lichter, P., Cremer, T., Borden, J., Manuelidis, L., Ward, D. 1988. Delineation of individual human chromosomes in metaphase and interphase cells by in situ suppression hybridization using recombinant DNA libraries. Hum. Genet. 80:224-234.
- Freier, K., et al. 2003. Tissue microarray analysis reveals site-specific prevalence of oncogene amplifications in head and neck squamous cell carcinoma. Cancer Res. 63:1179-1182. View this article via: PubMed Google Scholar
- Pfaffl, M.W. 2001. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29:e45. View this article via: PubMed Google Scholar
- Moffat, J., et al. 2006. A lentiviral RNAi library for human and mouse genes applied to an arrayed viral high-content screen. Cell. 124:1283-1298.
- Ohgaki, H., Kleihues, P. 2005. Population-based studies on incidence, survival rates, and genetic alterations in astrocytic and oligodendroglial gliomas. J. Neuropathol. Exp. Neurol. 64:479-489.
-
Version history
- Version 1 (April 8, 2008): No description
- Version 2 (May 1, 2008): No description



Copyright © 2024 American Society for Clinical Investigation
ISSN: 0021-9738 (print), 1558-8238 (online)







