Advertisement
Research ArticleVirology Free access | 10.1172/JCI33402
Bim-mediated deletion of antigen-specific CD8+ T cells in patients unable to control HBV infection
A. Ross Lopes,1 Paul Kellam,1 Abhishek Das,1 Claire Dunn,1 Antonia Kwan,1 Joanna Turner,2,3 Dimitra Peppa,1,2,3 Richard J. Gilson,2,3 Adam Gehring,4 Antonio Bertoletti,4 and Mala K. Maini1,3
1Division of Infection and Immunity and 2Centre for Sexual Health and HIV Research, University College London, London, United Kingdom. 3Camden Primary Care Trust, London, United Kingdom. 4Singapore Institute for Clinical Sciences, Agency for Science Technology and Research, Centros, Singapore.
Address correspondence to: Mala K. Maini, Division of Infection and Immunity, Windeyer Institute of Medical Science, 46 Cleveland Street, University College London, London W1T 4JF, United Kingdom. Phone: 44-0-20-7679-9212; Fax: 44-0-20-7679-9652; E-mail: [email protected].
Find articles by Lopes, A. in: JCI | PubMed | Google Scholar
1Division of Infection and Immunity and 2Centre for Sexual Health and HIV Research, University College London, London, United Kingdom. 3Camden Primary Care Trust, London, United Kingdom. 4Singapore Institute for Clinical Sciences, Agency for Science Technology and Research, Centros, Singapore.
Address correspondence to: Mala K. Maini, Division of Infection and Immunity, Windeyer Institute of Medical Science, 46 Cleveland Street, University College London, London W1T 4JF, United Kingdom. Phone: 44-0-20-7679-9212; Fax: 44-0-20-7679-9652; E-mail: [email protected].
Find articles by Kellam, P. in: JCI | PubMed | Google Scholar
1Division of Infection and Immunity and 2Centre for Sexual Health and HIV Research, University College London, London, United Kingdom. 3Camden Primary Care Trust, London, United Kingdom. 4Singapore Institute for Clinical Sciences, Agency for Science Technology and Research, Centros, Singapore.
Address correspondence to: Mala K. Maini, Division of Infection and Immunity, Windeyer Institute of Medical Science, 46 Cleveland Street, University College London, London W1T 4JF, United Kingdom. Phone: 44-0-20-7679-9212; Fax: 44-0-20-7679-9652; E-mail: [email protected].
Find articles by Das, A. in: JCI | PubMed | Google Scholar
1Division of Infection and Immunity and 2Centre for Sexual Health and HIV Research, University College London, London, United Kingdom. 3Camden Primary Care Trust, London, United Kingdom. 4Singapore Institute for Clinical Sciences, Agency for Science Technology and Research, Centros, Singapore.
Address correspondence to: Mala K. Maini, Division of Infection and Immunity, Windeyer Institute of Medical Science, 46 Cleveland Street, University College London, London W1T 4JF, United Kingdom. Phone: 44-0-20-7679-9212; Fax: 44-0-20-7679-9652; E-mail: [email protected].
Find articles by Dunn, C. in: JCI | PubMed | Google Scholar
1Division of Infection and Immunity and 2Centre for Sexual Health and HIV Research, University College London, London, United Kingdom. 3Camden Primary Care Trust, London, United Kingdom. 4Singapore Institute for Clinical Sciences, Agency for Science Technology and Research, Centros, Singapore.
Address correspondence to: Mala K. Maini, Division of Infection and Immunity, Windeyer Institute of Medical Science, 46 Cleveland Street, University College London, London W1T 4JF, United Kingdom. Phone: 44-0-20-7679-9212; Fax: 44-0-20-7679-9652; E-mail: [email protected].
Find articles by Kwan, A. in: JCI | PubMed | Google Scholar
1Division of Infection and Immunity and 2Centre for Sexual Health and HIV Research, University College London, London, United Kingdom. 3Camden Primary Care Trust, London, United Kingdom. 4Singapore Institute for Clinical Sciences, Agency for Science Technology and Research, Centros, Singapore.
Address correspondence to: Mala K. Maini, Division of Infection and Immunity, Windeyer Institute of Medical Science, 46 Cleveland Street, University College London, London W1T 4JF, United Kingdom. Phone: 44-0-20-7679-9212; Fax: 44-0-20-7679-9652; E-mail: [email protected].
Find articles by Turner, J. in: JCI | PubMed | Google Scholar
1Division of Infection and Immunity and 2Centre for Sexual Health and HIV Research, University College London, London, United Kingdom. 3Camden Primary Care Trust, London, United Kingdom. 4Singapore Institute for Clinical Sciences, Agency for Science Technology and Research, Centros, Singapore.
Address correspondence to: Mala K. Maini, Division of Infection and Immunity, Windeyer Institute of Medical Science, 46 Cleveland Street, University College London, London W1T 4JF, United Kingdom. Phone: 44-0-20-7679-9212; Fax: 44-0-20-7679-9652; E-mail: [email protected].
Find articles by Peppa, D. in: JCI | PubMed | Google Scholar
1Division of Infection and Immunity and 2Centre for Sexual Health and HIV Research, University College London, London, United Kingdom. 3Camden Primary Care Trust, London, United Kingdom. 4Singapore Institute for Clinical Sciences, Agency for Science Technology and Research, Centros, Singapore.
Address correspondence to: Mala K. Maini, Division of Infection and Immunity, Windeyer Institute of Medical Science, 46 Cleveland Street, University College London, London W1T 4JF, United Kingdom. Phone: 44-0-20-7679-9212; Fax: 44-0-20-7679-9652; E-mail: [email protected].
Find articles by Gilson, R. in: JCI | PubMed | Google Scholar
1Division of Infection and Immunity and 2Centre for Sexual Health and HIV Research, University College London, London, United Kingdom. 3Camden Primary Care Trust, London, United Kingdom. 4Singapore Institute for Clinical Sciences, Agency for Science Technology and Research, Centros, Singapore.
Address correspondence to: Mala K. Maini, Division of Infection and Immunity, Windeyer Institute of Medical Science, 46 Cleveland Street, University College London, London W1T 4JF, United Kingdom. Phone: 44-0-20-7679-9212; Fax: 44-0-20-7679-9652; E-mail: [email protected].
Find articles by Gehring, A. in: JCI | PubMed | Google Scholar
1Division of Infection and Immunity and 2Centre for Sexual Health and HIV Research, University College London, London, United Kingdom. 3Camden Primary Care Trust, London, United Kingdom. 4Singapore Institute for Clinical Sciences, Agency for Science Technology and Research, Centros, Singapore.
Address correspondence to: Mala K. Maini, Division of Infection and Immunity, Windeyer Institute of Medical Science, 46 Cleveland Street, University College London, London W1T 4JF, United Kingdom. Phone: 44-0-20-7679-9212; Fax: 44-0-20-7679-9652; E-mail: [email protected].
Find articles by Bertoletti, A. in: JCI | PubMed | Google Scholar
1Division of Infection and Immunity and 2Centre for Sexual Health and HIV Research, University College London, London, United Kingdom. 3Camden Primary Care Trust, London, United Kingdom. 4Singapore Institute for Clinical Sciences, Agency for Science Technology and Research, Centros, Singapore.
Address correspondence to: Mala K. Maini, Division of Infection and Immunity, Windeyer Institute of Medical Science, 46 Cleveland Street, University College London, London W1T 4JF, United Kingdom. Phone: 44-0-20-7679-9212; Fax: 44-0-20-7679-9652; E-mail: [email protected].
Find articles by Maini, M. in: JCI | PubMed | Google Scholar
Published April 8, 2008 - More info
J Clin Invest. 2008;118(5):1835–1845. https://doi.org/10.1172/JCI33402.
© 2008 The American Society for Clinical Investigation
Received: July 26, 2007; Accepted: February 15, 2008
-
Abstract
HBV-specific CD8+ T cells are critical for a successful immune response to HBV infection. They are markedly diminished in number in patients who fail to control the virus, but the mechanisms resulting in their depletion remain ill defined. Here, we dissected the defective HBV-specific CD8+ T cell response associated with chronic HBV infection by gene expression profiling. We found that HBV-specific CD8+ T cells from patients with different clinical outcomes could be distinguished by their patterns of gene expression. Microarray analysis revealed that overlapping clusters of functionally related apoptotic genes were upregulated in HBV-specific CD8+ T cells from patients with chronic compared with resolved infection. Further analysis confirmed that levels of the proapoptotic protein Bcl2-interacting mediator (Bim) were upregulated in HBV-specific CD8+ T cells from patients with chronic HBV infection. Blocking Bim-mediated apoptosis enhanced recovery of HBV-specific CD8+ T cells both in culture and directly ex vivo. Consistent with evidence that Bim mediates apoptosis of CD8+ T cells expressing low levels of CD127 (IL-7R), the few surviving HBV-specific CD8+ T cells were CD127hi and had elevated levels of the antiapoptotic protein Mcl1, suggesting they were amenable to IL-7–mediated rescue from apoptosis. We therefore postulate that Bim-mediated attrition of HBV-specific CD8+ T cells contributes to the inability of these cell populations to persist and control viral replication.
-
Introduction
Chronic HBV (CHB) infection is characterized by decades of high-level viral replication, with circulating viremia in the order of hundreds of millions of copies per milliliter. In addition, subviral particles are produced at 104 - to 106-fold excess in comparison with complete virions, resulting in extremely high quantities of circulating surface antigen. This, along with the secreted form of core antigen, eAg, has been postulated to represent viral strategies to subvert the immune response (1, 2). One component of the antiviral response known to be critical to HBV control is the specific CD8+ T cell response (3). The HBV-specific CD8+ T cell response is clearly blunted in patients with chronic infection, with scanty responses of low frequency and limited specificity (4–6). This contrasts with the more robust responses found in patients naturally resolving the infection (5, 7).
The CD8+ T cell hyporesponsiveness of CHB infection has been attributed to high-dose antigenic deletion, analogous to that seen in the lymphocytic choriomeningitis virus (LCMV) model (8). However, responses are not completely deleted, since a few envelope-specific CD8+ T cells persist in some patients despite extremely high viral loads but are unable to exert effective antiviral function in vivo (1). Additional CD8+ T cell responses can be reconstituted upon reduction of viral load in chronic infection, occurring either spontaneously or with antiviral therapy (9, 10). However, these reconstituted responses have a limited lifespan (11) and are unable to mediate sustained viral suppression if antiviral agents are stopped. A better understanding of this defective antiviral response is required in order to develop immunotherapeutic strategies to enhance the durability of viral suppression for the hundreds of millions of patients chronically infected with HBV worldwide.
The paucity of HBV-specific CD8+ T cell responses persisting in patients unable to control viral replication has precluded a thorough investigation of mechanisms underlying their depletion. In order to obtain a more comprehensive and unbiased analysis of the CD8+ T cell defects associated with chronicity compared with resolution, we took advantage of advances in GeneChip technology. Gene arrays have mostly been used to characterize pathogen-induced changes in host cells, but we found they could also be applied to providing global screening of small populations of CD8+ T cells specifically recognizing virally infected cells.
In this study, we applied gene expression profiling to dissecting differences in the HBV-specific CD8+ T cell response associated with control versus chronicity. Of a cluster of apoptosis genes upregulated in the HBV-specific CD8+ T cells from patients with chronic infection, Bcl2-interacting mediator (Bim) was consistently and significantly induced at both mRNA and protein levels. The functional implication of these findings was explored by specific inhibition of apoptosis, demonstrating rescue of HBV-specific responses both in culture and directly ex vivo. The findings suggest a role for cross-tolerance to HBV antigens mediated through Bim-induced attrition.
-
Results
Dissecting the defective HBV-specific CD8+ T cell response by gene expression profiling revealed differentially expressed apoptosis-related genes. CD8+ T cells capable of recognizing HBV epitopes are barely detectable in most patients with high-level HBV replication; this paucity of HBV-specific CD8+ T cell responses has limited their characterization. In this study, we therefore applied gene expression profiling to allow simultaneous screening of a large number of potentially relevant pathways from small samples. Our strategy was to compare the gene expression profiles of HBV-specific CD8+ T cells from patients who had controlled HBV with those of the limited HBV-specific CD8+ T cells detectable in patients with CHB. Dual-color spotted glass microarrays were used, standardizing gene expression from the linearly amplified sample RNA against a universal reference.
HBV-specific CD8+ T cell responses were barely detectable in patients with high HBV load directly ex vivo. Instead, we took advantage of the few patients in this disease category from whom an adequate HBV-specific response (1%–17% of total CD8) could be detected after 10 days culture in vitro (Figure 1A and Supplemental Table 1; supplemental material available online with this article; doi:10.1172/JCI33402DS1). These frequencies were postulated to be sufficient to detect changes that could be ascribed to the virus-specific component based on the results of a previous gene-array study of unsorted CD8+ cells (12). HBV-specific CD8+ T cells were restimulated with cognate peptide for 6 hours in order to focus the transcriptional profile on those genes activated or repressed upon antigen encounter (data not shown and refs. 13, 14). Sufficient high-quality RNA was obtained from such samples to successfully hybridize microarrays, 7 from patients with CHB and 12 from patients with resolved HBV infection. To ensure that the transcriptional profile reliably represented HBV-specific genes and not those from nonspecific cells in the short-term cell lines, microarray data was also obtained from sorted (>95% pure) HBV-specific CD8+ T cells from both groups of patients (Figure 1A).
 Figure 1
Figure 1cDNA microarray data of HBV-specific CD8+ T cells from resolved and CHB patients. (A) Envelope-specific CD8+ T cells were purified by flow cytometric sorting of a short-term PBMC line from a chronic patient (middle and left plots, respectively). mRNA extracted from the purified cells was then profiled by dual-color cDNA microarray technology (right). (B) TreeView analysis of average linkage hierarchically clustered (with self-organized mapping) gene expression data. The top dendrogram represents the similarity between individual arrayed samples (vertical plane) based on the global gene expression profile; a yellow line segregates the main clusters: chronic (C) and resolved (R). The blue box highlights a section of the heat map where a group of genes exhibited marked upregulation in branch C compared with R (listed with original and current unigene references on the right; genes in red participate in apoptosis; genes overlapping with the SAM short list are indicated by asterisks). (C) SAM plot illustrating the most significant differentially regulated genes (false discovery rate, 1.3%) between the group with chronic and that with resolved HBV infection. (D) cDNA array data of 5 highly significant apoptosis-related genes. Error bars indicate mean ± SD.
An initial qualitative comparison of total noncompartmentalized microarray data from resolved and CHB-specific CD8+ T cells was carried out using self-organized mapping followed by hierarchical clustering (mode: average linkage) with Cluster (15). TreeView visualization demonstrated that the primary data set branched into 2 main groups (Figure 1B; groups C and R). The majority of samples in group C (6/8) and group R (10/11) derived from individuals with chronic and resolved infection, respectively. The chronic patient whose HBV-specific CD8+ T cell gene expression branched with the resolved patients was the only CHB patient included in this analysis who did not have high HBV DNA (greater than 106 IU/ml). Thus, differences in gene expression induced upon HBV-specific activation were sufficient to allow segregation of samples according to clinical outcome. Among all the genes analyzed (5088 genes per array, spotted in duplicate), a clearly distinguishable cluster of genes was upregulated in chronic samples and downregulated in resolved samples (compared with reference RNA; Figure 1B). Closer inspection identified a subset of functionally related genes that exhibited a consistent divergence of expression level between the 2 groups. Of 20 genes in this hierarchical cluster with known functions, 16 had described roles in apoptosis. Two of these that are well characterized are the closely related BH3-only proapoptotic proteins Bim and Bid, the latter of which is the BH3-interacting domain death agonist.
In order to test for statistically robust differences in gene expression, normalized (median centered) cDNA array data were also processed by significance analysis of microarrays (SAM). This well-established bioinformatics tool utilized an algorithm (16) to calculate the fold change and statistical significance of any transcriptional differences between the resolved and chronic groups. Setting the acceptable median false discovery rate at 1.3%, we identified 105 differentially regulated genes (Supplemental Table 2). Seventy-six genes had increased expression, whereas 29 genes had reduced expression, in chronic compared with resolved HBV-specific CD8+ T cell samples (Figure 1C). Upregulated genes in HBV-specific CD8+ T cells from patients with CHB exhibited large fold increases (ranging from 2- to 9-fold). The proapoptotic mediator Bim, found to be 6.6-fold upregulated in the HBV-specific responses from chronic versus resolved patients, was the gene short-listed by SAM as showing the most statistically robust differences. In corroboration of our findings from the hierarchical cluster analysis, a large number of the genes in the SAM short list also participated in apoptotic events (data not shown). Moreover, multiple apoptosis-related genes from the 2 independently generated short lists overlapped (Figure 1B), confirming that a group of functionally related genes were transcriptionally dysregulated in antigen-specific CD8+ T cells in CHB infection.
A sample of 5 of the genes identified by both methods is presented in Figure 1D, with mean and SD of expression levels relative to the reference RNA (Mann-Whitney test). For each of these genes, expression was consistently increased in HBV-specific CD8+ T cells from CHB patients, both in samples from highly purified populations and from enriched 10-day cultures. These genes showing highly significant increases in HBV-specific responses from chronic compared with resolved patients are all involved in lymphocyte apoptotic pathways. Thus, data from 2 independent analyses revealed that overlapping clusters of apoptosis-related transcripts were dysregulated in the HBV-specific CD8+ T cell response associated with chronicity.
The proapoptotic molecule Bim was upregulated at the protein level in HBV-specific CD8+ T cells from patients with chronic infection. Intrinsic apoptosis is determined by a carefully balanced and complex group of pro- and antiapoptotic proteins of the Bcl2 family. Of these proteins, Bim, short-listed by both methods of data analysis, had the highest statistical significance among the 76 genes selected and was therefore chosen for further study. Bim has been found to be critical for the elimination of CD8+ T cells in a mouse model of chronic viral infection (17) and is also required for deletion of CD8+ T cells following cross-presentation of soluble antigen in the periphery (18). The array data showed more Bim transcripts for samples from CHB patients than for the resolved samples; these data were validated by quantitative PCR on a subset of the same samples used for the arrays (data not shown).
Next, we sought to confirm these data at the protein level for a larger sample of patients who had HBV-specific CD8+ T cell populations after 10 days in vitro expansion that could be costained with a Bim mAb (19 responses from resolved patients, 25 from CHB patients). These patients were all HIV and HCV negative, had not received antiviral or immunosuppressive treatment, and had similar demographic characteristics (e.g., sex and age; see Table 1 and Supplemental Table 1). Peptide-specific IFN-γ production was used primarily to identify HBV-specific populations because we have previously noted them to have a tetramer-negative, IFN-γ–positive phenotype in high-level carriers (1). Changes detected upon peptide restimulation should mimic those seen when the CD8+ T cells encounter their cognate antigen in the HBV-infected liver. Envelope 183–91– or core 18–27–specific responses were costained with a Bim mAb and examined by flow cytometry. Intracellular levels of Bim were stable over time in HBV-specific responses from patients who were sampled at repeated intervals while clinically stable (data not shown). HBV-specific CD8+ T cells from individuals with chronic infection were found to contain a significantly higher quantity of Bim protein compared with their counterparts in resolved individuals, in whom Bim was barely above background levels with an isotype control (Figure 2A; mean MFI of 52.3 compared with 20.7; P < 0.0001). Furthermore, an analysis of the chronic samples alone demonstrated that Bim expression was significantly higher in the HBV-specific population (IFN-γ+) compared with the total CD8+ T cell population of unrelated specificities (IFN-γ–) within individual patients (mean MFI of 52.3 compared with 33.6; P = 0.008) (Figure 2B).
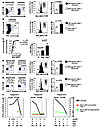 Figure 2
Figure 2Bim expression is increased at the protein level in HBV-specific CD8+ T cells from patients with CHB. (A) Representative example of Bim expression in HBV-specific CD8+ T cells from resolved and CHB patients (left and middle) and the cumulative data (right). Bim expression in resolved responses was similar to background levels with an isotype control. (B) Representative example of Bim expression in total CD8+ T cells (T) and HBV-specific CD8+ T cells (H) in a patient with CHB (left and middle) and cumulative data (right). (C) Correlation between viral load and Bim expression in HBV-specific CD8+ T responses in CHB patients (left) and relative levels of Bim expression in resolved (R) and CHB patients segregated according to eAg status (right). (D) Representative example of Bim expression directly ex vivo in HBV-specific CD8+ T cells in resolved and CHB individuals (left and middle) and cumulative data (right). (E) Bim expression directly ex vivo in tetramer-positive HBV-specific CD8+ T cell responses from resolved and CHB patients. Shown are examples of tetramer and Bim staining (left and middle) and summary data for all responses (right). Significance testing of all cumulative data by Mann-Whitney test. (F) Bim expression directly ex vivo in HBV-specific CD8+ T cells quantified over the course of acute HBV infection (using overlapping peptides in HLA-A2– patient R17 and using HLA-A2/HBV tetramers in 2 HLA-A2+ patients, R7 and R13). Bim levels in HBV-specific CD8+ T cells are plotted against HBV DNA, with serology and ALT in the acute and resolved (right of dotted line) phases indicated below. Error bars indicate mean ± SD.
The level of Bim expressed in HBV-specific CD8+ T cells within the group of chronic patients tested correlated with viral load (r = 0.7, P < 0.0001; Figure 2C). Bim expression was increased in chronic compared with resolved responses regardless of eAg status, although there was a nonsignificant trend to further increases in eAg-positive patients (Figure 2C).
To exclude a bias from in vitro culture, we confirmed our findings for differences in Bim expression of HBV-specific CD8+ T cells directly ex vivo. In order to detect sufficient HBV-specific responses to costain with Bim directly ex vivo from the high-level HBV carriers of interest, we used pools of overlapping HBV peptides. We found that Bim levels were also significantly higher on HBV-specific CD8+ T cells responses sampled directly ex vivo from patients with chronic compared with resolved infection (Figure 2D). To circumvent any potential bias induced by the study of functional IFN-γ–producing cells, we also analyzed HBV-specific CD8+ T cells by tetramer staining. In the few patients with CHB in whom HBV-specific populations could be identified directly ex vivo with HLA-A2/HBV tetramers, we again found that Bim was significantly increased compared with such responses in resolved patients (Figure 2E). In contrast, HBV-specific CD8+ T cells sampled during acute symptomatic HBV infection at times of high viral load showed no induction of Bim expression compared with responses from the same patients after viremia had resolved (Figure 2F and Supplemental Figure 1). These data suggest that distortions in the interactions of the Bcl2 family mediated by an upregulation in Bim could be perturbing the fine equilibrium that ensures cell survival and instead favoring progression toward apoptosis during CHB.
Enhanced recovery of HBV-specific CD8+ T cells upon inhibition of apoptotic pathways in vitro. Bim is thought to act principally by activating Bax (19), permitting mitochondrial release of cytochrome c, which activates caspases, ultimately leading to cell death. We attempted to rescue HBV-specific CD8+ T cells that had upregulated Bim by interfering with caspase activity to block this intrinsic death cascade. Treatment of PBMCs from CHB patients with the irreversible pancaspase inhibitor zVAD-fmk prior to antigenic stimulation resulted in the expansion of a larger population of HBV-specific CD8+ T cells after 10 days of in vitro culture. Both core and envelope-specific responses could be reconstituted, with a mean 3-fold increase in virus-specific numbers compared with stimulation without the apoptosis inhibitor (Figure 3, A and B; P < 0.0001).
 Figure 3
Figure 3Rescue of in vitro–cultured HBV-specific CD8+ T cells derived from individuals with CHB. Representative flow cytometry plots and cumulative data (below) showing the effect of pancaspase inhibition on the detection of envelope and core-specific CD8+ T cells (A and B, respectively) in 10-day peptide-stimulated cultures of PBMCs from individuals with chronic infection. Differences in responses with and without caspase inhibition were calculated with the paired Student’s t test (P < 0.0001). (C) Representative plots of the detection of HBV-specific CD8+ T cells in short-term lines of PBMCs from an individual with chronic infection utilizing pools of overlapping peptides corresponding to the HBV precore/core (PreC/C), X, envelope (Ep1 and Ep2), and polymerase (Pp1–4) proteins with and without caspase inhibition. (D) Influenza A–specific CD8+ T cells detected in short-term lines from individuals with chronic infection ± caspase inhibition. (E) HBV-specific CD8+ T responses detected in short-term lines from resolved individuals with and without caspase inhibition. (F) HBV-specific CD8+ T cell rescue following specific inhibition of proapoptotic Bax in short-term lines from patients with chronic infection.
We then compared the rescue from Bim-mediated apoptosis that could be achieved for responses to each of the HBV proteins within individual patients. In order to do this, we applied 8 pools of peptides spanning the whole HBV genome, divided according to their protein specificity. In 4 patients with eAg+ high-level CHB, we were able to rescue additional responses to some or all of the pools of peptides spanning the different HBV proteins. An example shown in Figure 3C demonstrates that CD8+ T cell responses to only 3 pools were above background levels before rescue, whereas afterwards, responses were detectable in all pools. This suggested that responses were susceptible to Bim-mediated attrition regardless of their HBV specificity and that inhibition of this pathway held the potential to enhance the multispecificity of the HBV response.
The percentage of total CD8+ T cells did not increase in these experiments (data not shown; paired Student’s t test; P = 0.48), suggesting that this rescue was restricted to HBV-specific populations. This was supported by the fact that influenza-specific CD8+ T cell responses identified in the same patients with CHB infection were not increased by caspase inhibition (Figure 3D). In addition, HBV-specific CD8+ T cell responses expanded from patients who had resolved their infection were not prone to caspase-dependent apoptosis in vitro, as evidenced by their lack of rescue with the inhibitor (Figure 3E). Since Bim was already induced in vivo, our investigations were constrained by the need for downstream inhibition. However, we were able to block apoptosis directly downstream of Bim using a pentapeptide (VPLMK) that inhibits the proapoptotic mediator Bax by suppressing its mitochondrial translocation (20). This inhibitor was also capable of enhancing recovery of HBV-specific responses in culture (Figure 3F).
Rescue of HBV-specific CD8+ T cells from apoptosis directly ex vivo from patients with CHB. The experiments on short-term cell lines provided functional confirmation of the dysregulated apoptotic pathways identified by microarray profiling. They indicated that HBV-specific CD8+ T cells from patients with CHB infection are highly susceptible to apoptosis upon cognate-peptide restimulation in culture. To determine whether these populations were similarly apoptosis prone when circulating in patients with high viral load, we studied the effect of inhibition directly ex vivo. PBMCs from the same patients were stimulated with cognate peptide for 6 hours only in the presence of the caspase inhibitor and the responsive cells identified by intracellular IFN-γ production. Higher frequencies of both envelope and core-specific CD8+ T cells could be detected when caspase activity was blocked directly ex vivo (Figure 4). The fact that new functionally active responses became detectable after just 6 hours of culture indicated that these were generated by inhibition of apoptosis rather than an increase in proliferation. These data provided a direct ex vivo corroboration of our findings derived from HBV-specific CD8+ T cells cultured in vitro.
 Figure 4
Figure 4Direct ex vivo rescue of HBV-specific CD8+ T cells from patients with CHB. Representative flow cytometry plots and cumulative data (below) indicating direct ex vivo frequencies of HBV-specific CD8+ T cells in patients with chronic infection as detected following stimulation with viral peptide with and without treatment with the pancaspase inhibitor zVAD-fmk.
HBV-specific CD8+ T cells persisting in the face of high antigen load are selectively enriched for high expression of CD127 and Mcl1. Bim mediates death of CD8+ T cells expressing low levels of the IL-7R α chain (CD127lo) (21, 22), a phenotype typically seen in situations of chronic antigenic stimulation (23–25). We therefore hypothesized that the bulk of HBV-specific CD8+ T cells have been subjected to Bim-mediated deletion; the few persisting in these patients may have escaped the effects of upregulated Bim because of high levels of CD127 expression. This is supported by the recent finding that persisting HBV-specific CD8+ T cells maintain high levels of CD127 expression in patients with chronic infection (6). IL-7–receptor–mediated rescue from Bim-induced apoptosis is regulated through the antiapoptotic molecule Mcl1, which binds specifically to Bim (26). To investigate whether persisting HBV-specific CD8+ T cells surviving after in vitro expansion without caspase inhibition had neutralized the proapoptotic drive of Bim, we examined their levels of Mcl1. Intracellular levels of Mcl1 were strikingly elevated in the core and envelope-specific CD8, with an MFI approximately double that seen in CD8+ T cells of the same specificity from patients who had resolved their HBV infections (Figure 5A). Mcl1 levels were also significantly higher (P = 0.0001) in HBV-specific CD8+ T cells than total CD8+ T cells in the same patients, whereas levels in influenza-specific CD8+ T cells were similar to those in total CD8+ T cells (Figure 5B). Mcl1 expression by HBV-specific CD8+ T cells correlated with viral load but was elevated in patients with chronic compared with resolved infection regardless of eAg status (Figure 5C). We confirmed that HBV-specific CD8+ T cells in patients with CHB infection were CD127hi, in line with published data (6). By costaining HBV-specific CD8+ T cells for CD127 and Mcl1, we found that all CD127hi CD8+ T cells of this specificity expressed high intracellular levels of Mcl1, consistent with their rescue through this mechanism (Figure 5D).
 Figure 5
Figure 5Mcl1 and CD127 expression of HBV-specific CD8+ T cells. (A) Intracellular staining for Mcl1 in core and envelope-specific CD8+ T cells expanded in vitro from individuals with chronic and resolved infection (left) with cumulative data (right). (B) Summary of Mcl1 levels in HBV-specific (H), total (T), and influenza-specific (I) CD8+ T cells from individuals with persistent HBV infection. (C) Correlation between viral load and the level of Mcl1 expression in HBV-specific CD8+ T responses from individuals with chronic infection (left) and relative levels of Mcl1 expression in resolved and persistently infected patients segregated according to eAg status (right). (D) Intracellular stain for Mcl1 in CD127hi populations of HBV-specific and total CD8+ T cells. Error bars indicate mean ± SD.
-
Discussion
Using microarray profiling, we found that HBV-specific CD8+ T cells from patients with different clinical outcomes could be distinguished by their global patterns of gene expression. A number of genes were highly upregulated in the HBV-specific CD8+ T cells from patients with uncontrolled HBV replication. Among these, a cluster of functionally related apoptotic genes was identified, sharing the intrinsic Bcl2 pathway. The most highly and consistently upregulated was Bim, which was confirmed by intracellular staining of HBV-specific CD8+ T cells from an extended patient cohort to be increased in the attenuated response associated with chronicity. Bim is one of the proapoptotic BH3-only group of proteins from the Bcl2 family that plays a central role in the initiation of apoptosis signaling in lymphocytes (27, 28).
Bim has been shown in murine models to be required for shutdown of the CD8+ T cell response in the setting of a superantigenic stimulus (29) or an acute viral infection (30). Conversely, downregulation of Bim is critical for CD8+ T cell memory survival in the absence of antigen (31). More relevant to the situation of persistent infection with HBV, Bim has also recently been found to regulate CD8+ T cell responses during chronic LCMV infection in mice (17). Bim mediated predominant loss of an immunodominant LCMV-specific CD8+ T cell response, which parallels the situation in CHB infection in which responses to an immunodominant core epitope become undetectable in patients with high viral loads (5); these responses could be recovered in our study following downstream inhibition of Bim-mediated apoptosis.
Bim mediates apoptosis of CD127lo CD8+ T cells (22) and is the major inducer of virus-specific CD8+ T cell apoptosis of this phenotype (21). In accordance with this, we found that the few HBV-specific CD8+ T cells surviving in this setting had maintained expression of CD127 (the IL-7 receptor α chain). This paradoxical maintenance of a CD127hi phenotype in the face of a chronic viral infection has been noted recently in other studies of both HBV (6) and HCV (32–34) infection but is at odds with the characteristic low levels seen in other human chronic viral infections (24, 25). We speculate that the bulk of HBV-specific CD8+ T cells with low levels of CD127 have already been subjected to Bim-mediated deletion and the scanty populations we are able to study are the exceptions. These CD127hi CD8+ T cells may be able to escape sensitization to apoptosis through Bim upregulation (23) by maximizing rescue signals from IL-7 through the antiapoptotic protein Mcl1 (26), which we found to be upregulated in the same populations. Mcl1, induced by IL-7, has been shown to play an essential role in mature lymphocyte survival by counteracting the proapoptotic effects of Bim (26). These CD8+ T cells, in which Bim is already induced, would be poised to die once cytokine signals became limiting (35). Thus, a small subpopulation of HBV-specific CD8-expressing CD127 may be selected by their ability to counteract Bim-mediated deletion. Alternatively, expression of CD127 may be a reflection of the recently primed status of the detectable HBV-specific CD8; such continuous recruitment of newly generated T cells to the ongoing response has recently been described in chronic LCMV infection (36). This would imply that the HBV-specific CD8+ T cell response in chronically infected individuals has a higher turnover than previously realized, in keeping with continuous attrition by apoptosis.
What drives the upregulation of Bim so that levels are specifically increased in HBV-specific CD8+ T cells from CHB when compared with either those in resolved patients or with total CD8+ T cell populations in CHB patients? One contributing factor may be the level of persistent antigenic drive and activation status of responding cells, since T cell receptor triggering has been shown to induce Bim in effector CD8+ T cells (37, 38). This is consistent with the previously noted preferential Bim-mediated deletion of immunodominant responses (17) and is in keeping with our data, which focused on 2 frequently recognized HBV epitopes (core 18–27 and envelope 183–191) from antigens that are produced at high concentrations in this infection. Once the hierarchy of HBV-specific CD8+ T cell responses restricted by diverse HLA alleles has been better defined, it will be important to investigate whether Bim levels are lower in any subdominant responses that are identified in chronic infection.
A more compelling explanation is that Bim is upregulated in HBV-specific responses associated with chronicity as a result of defective intrahepatic antigen presentation or cross-presentation of HBV antigens; this remains purely speculative at present. Investigation into the molecular basis of cross-tolerance has revealed that Bim is required for peripheral deletion of CD8+ T cells following cross-presentation of soluble antigen (18). Cross-presentation was shown to result in defective priming, such that CD8+ T cells underwent initial proliferation followed by deletion, which was abrogated in Bim-deficient mice (18). Large amounts of soluble surface antigen and eAg are produced in HBV infection, and surface antigen can access the class I processing pathway for cross-presentation (39). Liver sinusoidal endothelial cells, which are well positioned to efficiently take up exogenous soluble antigen from the circulation or released from infected hepatocytes, have been shown to induce cross-tolerance in CD8+ T cells (40). Cross-presentation of antigens released from apoptotic cells has also been associated with induction of tolerance in the liver (41, 42) and could affect responses to epitopes from all viral antigens; this may be pertinent to the inflamed liver in CHB, where we have recently demonstrated hepatocyte apoptosis mediated by TNF-related apoptosis-inducing ligand–expressing (TRAIL-expressing) NK cells (43). Antigen that is endogenously processed and presented by hepatocytes has also been shown to induce initial proliferation followed by deletion or anergy of responding CD8+ T cells (44, 45); whether this is mediated via Bim remains to be investigated. HBV-specific CD8+ T cells recognize antigen presented by HBV-infected human hepatocytes (46) and upon recognition become highly prone to apoptosis (A. Bertoletti et al., unpublished observations).
By blocking Bim-mediated apoptosis, we were able to enhance recovery of HBV-specific CD8+ T cells in culture, providing functional confirmation of our microarray data and highlighting a potential strategy to enhance recovery of these populations. Blocking of PD-1/PD-L1 interactions has also recently been found to reverse some of the HBV-specific CD8+ T cell dysfunction after in vitro culture from patients with chronic infection (6). However, envelope-specific responses were not recovered by blocking the PD-1 pathway in vitro, suggesting they are subject to an alternative tolerizing mechanism, in line with the particularly large excess of surface antigen produced in these patients. In contrast, we were able to rescue functionally active responses of both core and envelope specificities upon blockade of Bim-mediated apoptosis. Data with individual and pooled overlapping peptides indicated the potential to reconstitute a response of enhanced multispecificity, as seen in patients resolving infection naturally (5, 6). Inhibition of this apoptosis pathway directly ex vivo also resulted in substantial rescue of HBV-specific CD8+ T cells, indicating that circulating responses are highly susceptible to apoptosis in patients with chronic infection. The positive correlation we found between viral load and Bim expression suggests that HBV-specific CD8+ T cells should become more susceptible to deletion as viral load increases; this is consistent with our own and published findings of a negative correlation between viral load and frequency of HBV-specific CD8+ T cells (5, 6).
We would only expect to be able to achieve a limited amount of reconstitution ex vivo, since most HBV-specific CD8+ T cells are likely to have already been tolerized by the persistent high antigen load in vivo. The short lifespan of HBV-specific responses reconstituted during antiviral therapy (11) suggests that a short-term reduction in viral load does not allow a full reversal of their propensity to apoptosis. A strategy that could block Bim induction (for example, with short-term use of cyclosporin A or FK506; ref. 37) rather than preventing apoptosis downstream once it is upregulated holds greater potential for reconstitution of effective HBV-specific responses. This raises the possibility of specifically reprogramming the HBV-specific CD8+ T cell susceptibility to Bim-mediated apoptosis in patients following the use of potent antivirals to first reduce viral load.
In conclusion, the profound HBV-specific CD8+ T cell hyporesponsiveness found in chronic infection is likely to represent the combined effect of multiple deletion and suppressor mechanisms related to the exceptionally high level of antigen load in these patients. In this study, a global, unbiased approach to dissecting these mechanisms highlighted a dysregulated apoptotic pathway. We postulate that cross-presentation of HBV antigens and subsequent Bim-mediated deletion contributes to the failure of CD8+ T cell responses in CHB infection. Interruption of this tolerizing mechanism may provide a new strategy to reconstitute more effective HBV responses in order to achieve a treatment strategy with sustained antiviral efficacy.
-
Methods
Patients. Forty-four patients were recruited with written informed consent; the Camden and Islington Primary Care Trust Local Research Ethics Committee approved this study (see Table 1 and Supplemental Table 1). HLA-A2 status was determined by flow cytometry (HLA-A2 surface staining; AbD Serotec). Nineteen patients had clinical, biochemical, and virological evidence of resolved HBV infection (recovery from acute hepatitis, normal alanine transaminase (ALT), anti-HBcAb+, HBsAg-, HBV DNA undetectable); of these, 3 (R7, R13, and R17; Supplemental Table 1) were also sampled during the acute symptomatic phase of primary HBV infection (anti-HBcIgM+, sAg+, HBV DNA high, ALT high). Twenty-five patients had clinical, biological, and virological evidence of CHB infection (HBsAg+, HBV DNA+, HBeAg+/–). These patients had no other causes of liver damage, were negative for HIV-1 and -2, HCV, and delta virus and had not received antiviral therapy or immunosuppressive drugs. HBsAg, anti-HBsAb, total and immunoglobulin M anti-HBcAb, HBeAg, anti-HBeAb, anti-HDV, anti-HCV, anti–HIV-1, and HIV-2 were determined by commercial enzyme immunoassays (Murex Diagnostics; Abbott; Ortho-Clinical Diagnostics; and Sanofi Diagnostics Pasteur). Serum HBV DNA load was determined by real-time PCR.
Antibodies and reagents used. Antibodies used were CD3-perCpCy5.5, CD8-Cy5.5, CD8-APC, CD127-PE, CD8PerCp-Cy5.5, IFN-γ–APC and Cytofix/Cytoperm, zVAD-fmk (BD Biosciences), IFN-γ–PE (R&D Systems), IFN-γ Secretion Assay Cell Enrichment and Detection kit (Miltenyi Biotech), Bim unconjugated (Alexis Biochemicals; Axxora), goat anti-rat IgG2a FITC (Bethyl Laboratories), Mcl1 unconjugated, goat anti-rabbit FITC (Insight Biotechnologies), brefeldin A, saponin, PBS (Sigma-Aldrich), and aMEM (Invitrogen). HBV c18–27, envelope 183–191, and polymerase 455–463 multimers were from Proimmune or were kindly provided by Alison Turner and Paul Klenerman (Nuffield Department of Medicine, University of Oxford, Oxford, United Kingdom).
Cell isolation, culture, and staining. PBMCs were separated from EDTA-treated venous blood on Ficoll. Virus-specific CD8-enriched lines were generated by culturing total PBMCs (0.3 × 106/200 μl/well) with 1 μM peptide (HBV core 18–27, HBV envelope 183–191 [genotype D/serotype ayw] or influenza A Mp58-66; Proimmune) in aMEM/10% FCS (Gibco; Invitrogen) with rIL-2 (Roche) supplemented on days 0 and 4 (20 U/ml) in 96-well round-bottom plates; antigen-specific CD8+ T cell frequencies were determined by intracellular cytokine staining and flow cytometry as described previously (5). Cells were surface stained for CD8 and CD127, followed by intracellular staining for IFN-γ, Bim, or Mcl1 after permeabilization, with appropriate negative controls for nonspecific staining.
Rescue of HBV-specific CD8+ T cells. PBMCs were stimulated with individual viral peptides (HBV core 18–27 or HBV envelope 183–191; Proimmune) or pools of 15 mer peptides overlapping by 10 residues spanning the major proteins of HBV genotype B (Mimotopes); 8 pools comprised precore and core (peptides 1–6 and 1–35), X (peptides 1–29), envelope pool 1 (peptides 1–38), envelope pool 2 (peptides 39–76), polymerase pool 1 (peptides 1–42), polymerase pool 2 (peptides 43–84), polymerase pool 3 (peptides 85–126), and polymerase pool 4 (peptides 127–167). Stimulated cells were simultaneously treated with and without the pancaspase inhibitor zVAD-fmk (50 μM) or Bax inhibitor peptide VPMLK (20) (50 μM); culture medium was replenished with the inhibitor every 3 days and with IL-2 on day 4. IFN-γ+ virus-specific CD8+ T cells were determined by flow cytometry after 10 days as described above. Direct ex vivo analysis of IFN-γ+ virus-specific CD8+ T cells was performed following stimulation with viral peptide (10 μM) with and without inhibitor as above for 6–12 hours, with the addition of brefeldin A (10 μg/ml) after 1 hour.
Microarray analysis. For microarray applications, PBMCs (~0.5 × 106 cells) were lysed following peptide restimulation (5 hours) and mRNA extracted (Dynal mRNA Direct microkit; Invitrogen) according to the manufacturer’s instructions. Highly purified virus-specific CD8+ T cells were obtained by FACS sorting with a MoFlo Sorter (Dako) or by magnetic bead purification after labeling cells with multimers (Proimmune) or IFN-γ catch reagent (Miltenyi Biotech) according to the manufacturers’ instructions. Dual-color microarray analysis was conducted as previously described (47). In brief, mRNA was purified and amplified twice (AmpliScribe T7-Flash transcription kit; EPICENTRE Biotechnologies), and 5 μg (quantified with an Agilent Bioanalyzer) was Cy5 labeled (Amersham Biosciences). This was cohybridized with 5 μg of Cy3-labeled Human Universal Reference aRNA (Stratagene) to Human Genome Mapping Project cDNA arrays. The hybridization mix consisted of 12 μl × 20 SSPE (Sigma-Aldrich), 1.1 μl 0.5M EDTA (Sigma-Aldrich), 2 μl poly d(A) (Amersham Biosciences), and 2 μl tRNA (Sigma-Aldrich) in a final volume of 45 μl corrected with Tris-EDTA buffer. 1 μl of 10% SDS (Sigma-Aldrich) was added, and the sample was incubated at 98°C for 2 minutes, followed by 37°C for 20 minutes. 1 μl of ×100 Denhardt’s solution (Sigma-Aldrich) was added, spun for 15 minutes, dispensed onto the array, and incubated in a humidified chamber (Ambion) at 65°C overnight. Arrays were washed (×2 SSPE at 50°C, ×2 SSPE at room temperature, ×1 SSPE at room temperature, ×0.1 SSPE at room temperature), spun dry (3 minutes), and scanned (Axon; GenePix software).
Cy5 and Cy3 fluorescence intensities for each gene spot were exported as an Excel-compatible file. This was followed by subtraction of specific local background/spot, exclusion of data below an assigned negative signal threshold, and calculation of log2 median and mean ratios for Cy5 and Cy3 signals/spot. Data were normalized (median centering of arrays and genes), and self-organizing maps (SOMs) were produced with Cluster and visualized in TreeView (15). For SAM, normalized data were processed and a short list was selected at an appropriate false discovery rate.
Statistics. All data were tested using the nonparametric Mann-Whitney test except when specifically stated that the paired Student’s t test (2 tailed) was used. P < 0.05 was considered significant.
-
Acknowledgments
We are grateful to the staff and patients at the Mortimer Market Centre for blood samples. A.R. Lopes, A. Das, and C. Dunn were funded by the Medical Research Council. M.K. Maini was funded by an MRC Clinician Scientist Fellowship.
Address correspondence to: Mala K. Maini, Division of Infection and Immunity, Windeyer Institute of Medical Science, 46 Cleveland Street, University College London, London W1T 4JF, United Kingdom. Phone: 44-0-20-7679-9212; Fax: 44-0-20-7679-9652; E-mail: [email protected].
-
Footnotes
Nonstandard abbreviations used: ALT, alanine transaminase; Bim, Bcl2-interacting mediator; CHB, chronic HBV; LCMV, lymphocytic choriomeningitis virus; SAM, significance analysis of microarrays.
Conflict of interest: The authors have declared that no conflict of interest exists.
Reference information: J. Clin. Invest.118:1835–1845 (2008). doi:10.1172/JCI33402
-
References
- Reignat, S., et al. 2002. Escaping high viral load exhaustion: CD8 cells with altered tetramer binding in chronic hepatitis B virus infection. J. Exp. Med. 195:1089-1101.
- Chen, M.T., et al. 2004. A function of the hepatitis B virus precore protein is to regulate the immune response to the core antigen. Proc. Natl. Acad. Sci. U. S. A. 101:14913-14918.
- Thimme, R., et al. 2003. CD8(+) T cells mediate viral clearance and disease pathogenesis during acute hepatitis B virus infection. J. Virol. 77:68-76.
- Maini, M.K., et al. 2000. The role of virus-specific CD8(+) cells in liver damage and viral control during persistent hepatitis B virus infection. J. Exp. Med. 191:1269-1280.
- Webster, G.J., et al. 2004. Longitudinal analysis of CD8+ T cells specific for structural and nonstructural hepatitis B virus proteins in patients with chronic hepatitis B: implications for immunotherapy. J. Virol. 78:5707-5719.
- Boni, C., et al. 2007. Characterization of hepatitis B virus (HBV)-specific T-cell dysfunction in chronic HBV infection. J. Virol. 81:4215-4225.
- Maini, M.K., et al. 1999. Direct ex vivo analysis of hepatitis B virus-specific CD8(+) T cells associated with the control of infection. Gastroenterology. 117:1386-1396.
- Moskophidis, D., Lechner, F., Pircher, H., Zinkernagel, R.M. 1993. Virus persistence in acutely infected immunocompetent mice by exhaustion of antiviral cytotoxic effector T cells. Nature. 362:758-761.
- Rehermann, B., Lau, D., Hoofnagle, J.H., Chisari, F.V. 1996. Cytotoxic T lymphocyte responsiveness after resolution of chronic hepatitis B virus infection. J. Clin. Invest. 97:1655-1665.
- Boni, C., et al. 2001. Lamivudine treatment can overcome cytotoxic T-cell hyporesponsiveness in chronic hepatitis B: new perspectives for immune therapy. Hepatology. 33:963-971.
- Boni, C., et al. 2003. Transient restoration of anti-viral T cell responses induced by lamivudine therapy in chronic hepatitis B. J. Hepatol. 39:595-605.
- Vine, A.M., et al. 2004. The role of CTLs in persistent viral infection: cytolytic gene expression in CD8+ lymphocytes distinguishes between individuals with a high or low proviral load of human T cell lymphotropic virus type 1. J. Immunol. 173:5121-5129. View this article via: PubMed Google Scholar
- Teague, T.K., et al. 1999. Activation changes the spectrum but not the diversity of genes expressed by T cells. Proc. Natl. Acad. Sci. U. S. A. 96:12691-12696.
- Abdalla, A.O., et al. 2003. Kinetics of cytokine gene expression in human CD4+ and CD8+ T-lymphocyte subsets using quantitative real-time PCR. Scand. J. Immunol. 58:601-606.
- Eisen, M.B., Spellman, P.T., Brown, P.O., Botstein, D. 1998. Cluster analysis and display of genome-wide expression patterns. Proc. Natl. Acad. Sci. U. S. A. 95:14863-14868.
- Tusher, V.G., Tibshirani, R., Chu, G. 2001. Significance analysis of microarrays applied to the ionizing radiation response. Proc. Natl. Acad. Sci. U. S. A. 98:5116-5121.
- Grayson, J.M., Weant, A.E., Holbrook, B.C., Hildeman, D. 2006. Role of Bim in regulating CD8+ T-cell responses during chronic viral infection. J. Virol. 80:8627-8638.
- Davey, G.M., et al. 2002. Peripheral deletion of autoreactive CD8 T cells by cross presentation of self-antigen occurs by a Bcl-2-inhibitable pathway mediated by Bim. J. Exp. Med. 196:947-955.
- Weber, A., et al. 2007. BimS-induced apoptosis requires mitochondrial localization but not interaction with anti-apoptotic Bcl-2 proteins. J. Cell Biol. 177:625-636.
- Sawada, M., Hayes, P., Matsuyama, S. 2003. Cytoprotective membrane-permeable peptides designed from the Bax-binding domain of Ku70. Nat. Cell Biol. 5:352-357.
- Pellegrini, M., et al. 2004. Loss of Bim increases T cell production and function in interleukin 7 receptor-deficient mice. J. Exp. Med. 200:1189-1195.
- Wojciechowski, S., et al. 2006. Bim mediates apoptosis of CD127(lo) effector T cells and limits T cell memory. Eur. J. Immunol. 36:1694-1706.
- Kaech, S.M., et al. 2003. Selective expression of the interleukin 7 receptor identifies effector CD8 T cells that give rise to long-lived memory cells. Nat. Immunol. 4:1191-1198.
- Paiardini, M., et al. 2005. Loss of CD127 expression defines an expansion of effector CD8+ T cells in HIV-infected individuals. J. Immunol. 174:2900-2909. View this article via: PubMed Google Scholar
- van Leeuwen, E.M., et al. 2005. IL-7 receptor alpha chain expression distinguishes functional subsets of virus-specific human CD8+ T cells. Blood. 106:2091-2098.
- Opferman, J.T., et al. 2003. Development and maintenance of B and T lymphocytes requires antiapoptotic MCL-1. Nature. 426:671-676.
- O’Connor, L., et al. 1998. Bim: a novel member of the Bcl-2 family that promotes apoptosis. EMBO J. 17:384-395.
- Bouillet, P., et al. 1999. Proapoptotic Bcl-2 relative Bim required for certain apoptotic responses, leukocyte homeostasis, and to preclude autoimmunity. Science. 286:1735-1738.
- Hildeman, D.A., et al. 2002. Activated T cell death in vivo mediated by proapoptotic bcl-2 family member bim. Immunity. 16:759-767.
- Pellegrini, M., Belz, G., Bouillet, P., Strasser, A. 2003. Shutdown of an acute T cell immune response to viral infection is mediated by the proapoptotic Bcl-2 homology 3-only protein Bim. Proc. Natl. Acad. Sci. U. S. A. 100:14175-14180.
- Sabbagh, L., et al. 2006. A critical role for TNF receptor-associated factor 1 and Bim down-regulation in CD8 memory T cell survival. Proc. Natl. Acad. Sci. U. S. A. 103:18703-18708.
- Bengsch, B., et al. 2007. Analysis of CD127 and KLRG1 expression on hepatitis C virus-specific CD8+ T cells reveals the existence of different memory T-cell subsets in the peripheral blood and liver. J. Virol. 81:945-953.
- Radziewicz, H., et al. 2007. Liver-infiltrating lymphocytes in chronic human hepatitis C virus infection display an exhausted phenotype with high levels of PD-1 and low levels of CD127 expression. J. Virol. 81:2545-2553.
- Penna, A., et al. 2007. Dysfunction and functional restoration of HCV-specific CD8 responses in chronic hepatitis C virus infection. Hepatology. 45:588-601.
- Bosque, A., et al. 2007. The induction of Bim expression in human T-cell blasts is dependent on nonapoptotic Fas/CD95 signaling. Blood. 109:1627-1635.
- Vezys, V., et al. 2006. Continuous recruitment of naive T cells contributes to heterogeneity of antiviral CD8 T cells during persistent infection. J. Exp. Med. 203:2263-2269.
- Sandalova, E., Wei, C.H., Masucci, M.G., Levitsky, V. 2004. Regulation of expression of Bcl-2 protein family member Bim by T cell receptor triggering. Proc. Natl. Acad. Sci. U. S. A. 101:3011-3016.
- Sandalova, E., Hislop, A.D., Levitsky, V. 2006. T-cell receptor triggering differentially regulates bim expression in human lymphocytes from healthy individuals and patients with infectious mononucleosis. Hum. Immunol. 67:958-965.
- Jin, Y., Shih, W.K., Berkower, I. 1988. Human T cell response to the surface antigen of hepatitis B virus (HBsAg). Endosomal and nonendosomal processing pathways are accessible to both endogenous and exogenous antigen. J. Exp. Med. 168:293-306.
- Limmer, A., et al. 2000. Efficient presentation of exogenous antigen by liver endothelial cells to CD8+ T cells results in antigen-specific T-cell tolerance. Nat. Med. 6:1348-1354.
- Albert, M.L., Sauter, B., Bhardwaj, N. 1998. Dendritic cells acquire antigen from apoptotic cells and induce class I-restricted CTLs. Nature. 392:86-89.
- Berg, M., et al. 2006. Cross-presentation of antigens from apoptotic tumor cells by liver sinusoidal endothelial cells leads to tumor-specific CD8+ T cell tolerance. Eur. J. Immunol. 36:2960-2970.
- Dunn, C., et al. 2007. Cytokines induced during chronic hepatitis B virus infection promote a pathway for NK cell-mediated liver damage. J. Exp. Med. 204:667-680.
- Bertolino, P., Trescol-Biemont, M.C., Rabourdin-Combe, C. 1998. Hepatocytes induce functional activation of naive CD8+ T lymphocytes but fail to promote survival. Eur. J. Immunol. 28:221-236.
- Morimoto, J., Tan, X., Teague, R.M., Ohlen, C., Greenberg, P.D. 2007. Induction of tolerance in CD8+ T cells to a transgenic autoantigen expressed in the liver does not require cross-presentation. J. Immunol. 178:6849-6860. View this article via: PubMed Google Scholar
- Gehring, A.J., et al. 2007. The level of viral antigen presented by hepatocytes influences CD8 T-cell function. J. Virol. 81:2940-2949.
- Baugh, L.R., Hill, A.A., Brown, E.L., Hunter, C.P. 2001. Quantitative analysis of mRNA amplification by in vitro transcription. Nucleic Acids Res. 29:E29.
-
Version history
- Version 1 (April 8, 2008): No description
- Version 2 (May 1, 2008): No description



Copyright © 2024 American Society for Clinical Investigation
ISSN: 0021-9738 (print), 1558-8238 (online)






