Advertisement
Research ArticleHematology Free access | 10.1172/JCI30630
Disruption of erythroid K-Cl cotransporters alters erythrocyte volume and partially rescues erythrocyte dehydration in SAD mice
Marco B. Rust,1 Seth L. Alper,2 York Rudhard,1 Boris E. Shmukler,2 Rubén Vicente,1,3 Carlo Brugnara,4 Marie Trudel,5 Thomas J. Jentsch,1,3 and Christian A. Hübner1,6
1Zentrum für Molekulare Neurobiologie Hamburg, Universität Hamburg, Hamburg, Germany. 2Molecular and Vascular Medicine Unit and Renal Unit, Beth Israel Deaconess Medical Center and Department of Medicine, Harvard Medical School, Boston, Massachusetts, USA. 3Leibniz-Institut für Molekulare Pharmakologie and Max-Delbrück-Centrum für Molekulare Medizin, Berlin, Germany. 4Department of Laboratory Medicine, The Children’s Hospital, and Department of Pathology, Harvard Medical School, Boston, Massachusetts, USA. 5Institut de Recherches Cliniques de Montréal, Molecular Genetics and Development, Faculté de Médecine de l’Université de Montréal, Montreal, Quebec, Canada. 6Institut für Humangenetik, Universitätsklinikum Hamburg-Eppendorf, Hamburg, Germany.
Address correspondence to: Thomas J. Jentsch, FMP/MDC, Robert-Roessle-Strasse 10, D-13125 Berlin, Germany. Phone: 49-30-9406-2961; Fax: 49-30-9406-2960; E-mail: [email protected]. Or to: Christian A. Hübner, Institut für Klinische Chemie, Friedrich Schiller Universität Jena, Erlanger Allee 101, D-07747 Jena, Germany. Phone: 49-3641-9325001; E-mail: [email protected].
Find articles by Rust, M. in: JCI | PubMed | Google Scholar
1Zentrum für Molekulare Neurobiologie Hamburg, Universität Hamburg, Hamburg, Germany. 2Molecular and Vascular Medicine Unit and Renal Unit, Beth Israel Deaconess Medical Center and Department of Medicine, Harvard Medical School, Boston, Massachusetts, USA. 3Leibniz-Institut für Molekulare Pharmakologie and Max-Delbrück-Centrum für Molekulare Medizin, Berlin, Germany. 4Department of Laboratory Medicine, The Children’s Hospital, and Department of Pathology, Harvard Medical School, Boston, Massachusetts, USA. 5Institut de Recherches Cliniques de Montréal, Molecular Genetics and Development, Faculté de Médecine de l’Université de Montréal, Montreal, Quebec, Canada. 6Institut für Humangenetik, Universitätsklinikum Hamburg-Eppendorf, Hamburg, Germany.
Address correspondence to: Thomas J. Jentsch, FMP/MDC, Robert-Roessle-Strasse 10, D-13125 Berlin, Germany. Phone: 49-30-9406-2961; Fax: 49-30-9406-2960; E-mail: [email protected]. Or to: Christian A. Hübner, Institut für Klinische Chemie, Friedrich Schiller Universität Jena, Erlanger Allee 101, D-07747 Jena, Germany. Phone: 49-3641-9325001; E-mail: [email protected].
Find articles by Alper, S. in: JCI | PubMed | Google Scholar
1Zentrum für Molekulare Neurobiologie Hamburg, Universität Hamburg, Hamburg, Germany. 2Molecular and Vascular Medicine Unit and Renal Unit, Beth Israel Deaconess Medical Center and Department of Medicine, Harvard Medical School, Boston, Massachusetts, USA. 3Leibniz-Institut für Molekulare Pharmakologie and Max-Delbrück-Centrum für Molekulare Medizin, Berlin, Germany. 4Department of Laboratory Medicine, The Children’s Hospital, and Department of Pathology, Harvard Medical School, Boston, Massachusetts, USA. 5Institut de Recherches Cliniques de Montréal, Molecular Genetics and Development, Faculté de Médecine de l’Université de Montréal, Montreal, Quebec, Canada. 6Institut für Humangenetik, Universitätsklinikum Hamburg-Eppendorf, Hamburg, Germany.
Address correspondence to: Thomas J. Jentsch, FMP/MDC, Robert-Roessle-Strasse 10, D-13125 Berlin, Germany. Phone: 49-30-9406-2961; Fax: 49-30-9406-2960; E-mail: [email protected]. Or to: Christian A. Hübner, Institut für Klinische Chemie, Friedrich Schiller Universität Jena, Erlanger Allee 101, D-07747 Jena, Germany. Phone: 49-3641-9325001; E-mail: [email protected].
Find articles by Rudhard, Y. in: JCI | PubMed | Google Scholar
1Zentrum für Molekulare Neurobiologie Hamburg, Universität Hamburg, Hamburg, Germany. 2Molecular and Vascular Medicine Unit and Renal Unit, Beth Israel Deaconess Medical Center and Department of Medicine, Harvard Medical School, Boston, Massachusetts, USA. 3Leibniz-Institut für Molekulare Pharmakologie and Max-Delbrück-Centrum für Molekulare Medizin, Berlin, Germany. 4Department of Laboratory Medicine, The Children’s Hospital, and Department of Pathology, Harvard Medical School, Boston, Massachusetts, USA. 5Institut de Recherches Cliniques de Montréal, Molecular Genetics and Development, Faculté de Médecine de l’Université de Montréal, Montreal, Quebec, Canada. 6Institut für Humangenetik, Universitätsklinikum Hamburg-Eppendorf, Hamburg, Germany.
Address correspondence to: Thomas J. Jentsch, FMP/MDC, Robert-Roessle-Strasse 10, D-13125 Berlin, Germany. Phone: 49-30-9406-2961; Fax: 49-30-9406-2960; E-mail: [email protected]. Or to: Christian A. Hübner, Institut für Klinische Chemie, Friedrich Schiller Universität Jena, Erlanger Allee 101, D-07747 Jena, Germany. Phone: 49-3641-9325001; E-mail: [email protected].
Find articles by Shmukler, B. in: JCI | PubMed | Google Scholar
1Zentrum für Molekulare Neurobiologie Hamburg, Universität Hamburg, Hamburg, Germany. 2Molecular and Vascular Medicine Unit and Renal Unit, Beth Israel Deaconess Medical Center and Department of Medicine, Harvard Medical School, Boston, Massachusetts, USA. 3Leibniz-Institut für Molekulare Pharmakologie and Max-Delbrück-Centrum für Molekulare Medizin, Berlin, Germany. 4Department of Laboratory Medicine, The Children’s Hospital, and Department of Pathology, Harvard Medical School, Boston, Massachusetts, USA. 5Institut de Recherches Cliniques de Montréal, Molecular Genetics and Development, Faculté de Médecine de l’Université de Montréal, Montreal, Quebec, Canada. 6Institut für Humangenetik, Universitätsklinikum Hamburg-Eppendorf, Hamburg, Germany.
Address correspondence to: Thomas J. Jentsch, FMP/MDC, Robert-Roessle-Strasse 10, D-13125 Berlin, Germany. Phone: 49-30-9406-2961; Fax: 49-30-9406-2960; E-mail: [email protected]. Or to: Christian A. Hübner, Institut für Klinische Chemie, Friedrich Schiller Universität Jena, Erlanger Allee 101, D-07747 Jena, Germany. Phone: 49-3641-9325001; E-mail: [email protected].
Find articles by Vicente, R. in: JCI | PubMed | Google Scholar
1Zentrum für Molekulare Neurobiologie Hamburg, Universität Hamburg, Hamburg, Germany. 2Molecular and Vascular Medicine Unit and Renal Unit, Beth Israel Deaconess Medical Center and Department of Medicine, Harvard Medical School, Boston, Massachusetts, USA. 3Leibniz-Institut für Molekulare Pharmakologie and Max-Delbrück-Centrum für Molekulare Medizin, Berlin, Germany. 4Department of Laboratory Medicine, The Children’s Hospital, and Department of Pathology, Harvard Medical School, Boston, Massachusetts, USA. 5Institut de Recherches Cliniques de Montréal, Molecular Genetics and Development, Faculté de Médecine de l’Université de Montréal, Montreal, Quebec, Canada. 6Institut für Humangenetik, Universitätsklinikum Hamburg-Eppendorf, Hamburg, Germany.
Address correspondence to: Thomas J. Jentsch, FMP/MDC, Robert-Roessle-Strasse 10, D-13125 Berlin, Germany. Phone: 49-30-9406-2961; Fax: 49-30-9406-2960; E-mail: [email protected]. Or to: Christian A. Hübner, Institut für Klinische Chemie, Friedrich Schiller Universität Jena, Erlanger Allee 101, D-07747 Jena, Germany. Phone: 49-3641-9325001; E-mail: [email protected].
Find articles by Brugnara, C. in: JCI | PubMed | Google Scholar
1Zentrum für Molekulare Neurobiologie Hamburg, Universität Hamburg, Hamburg, Germany. 2Molecular and Vascular Medicine Unit and Renal Unit, Beth Israel Deaconess Medical Center and Department of Medicine, Harvard Medical School, Boston, Massachusetts, USA. 3Leibniz-Institut für Molekulare Pharmakologie and Max-Delbrück-Centrum für Molekulare Medizin, Berlin, Germany. 4Department of Laboratory Medicine, The Children’s Hospital, and Department of Pathology, Harvard Medical School, Boston, Massachusetts, USA. 5Institut de Recherches Cliniques de Montréal, Molecular Genetics and Development, Faculté de Médecine de l’Université de Montréal, Montreal, Quebec, Canada. 6Institut für Humangenetik, Universitätsklinikum Hamburg-Eppendorf, Hamburg, Germany.
Address correspondence to: Thomas J. Jentsch, FMP/MDC, Robert-Roessle-Strasse 10, D-13125 Berlin, Germany. Phone: 49-30-9406-2961; Fax: 49-30-9406-2960; E-mail: [email protected]. Or to: Christian A. Hübner, Institut für Klinische Chemie, Friedrich Schiller Universität Jena, Erlanger Allee 101, D-07747 Jena, Germany. Phone: 49-3641-9325001; E-mail: [email protected].
Find articles by Trudel, M. in: JCI | PubMed | Google Scholar
1Zentrum für Molekulare Neurobiologie Hamburg, Universität Hamburg, Hamburg, Germany. 2Molecular and Vascular Medicine Unit and Renal Unit, Beth Israel Deaconess Medical Center and Department of Medicine, Harvard Medical School, Boston, Massachusetts, USA. 3Leibniz-Institut für Molekulare Pharmakologie and Max-Delbrück-Centrum für Molekulare Medizin, Berlin, Germany. 4Department of Laboratory Medicine, The Children’s Hospital, and Department of Pathology, Harvard Medical School, Boston, Massachusetts, USA. 5Institut de Recherches Cliniques de Montréal, Molecular Genetics and Development, Faculté de Médecine de l’Université de Montréal, Montreal, Quebec, Canada. 6Institut für Humangenetik, Universitätsklinikum Hamburg-Eppendorf, Hamburg, Germany.
Address correspondence to: Thomas J. Jentsch, FMP/MDC, Robert-Roessle-Strasse 10, D-13125 Berlin, Germany. Phone: 49-30-9406-2961; Fax: 49-30-9406-2960; E-mail: [email protected]. Or to: Christian A. Hübner, Institut für Klinische Chemie, Friedrich Schiller Universität Jena, Erlanger Allee 101, D-07747 Jena, Germany. Phone: 49-3641-9325001; E-mail: [email protected].
Find articles by Jentsch, T. in: JCI | PubMed | Google Scholar
1Zentrum für Molekulare Neurobiologie Hamburg, Universität Hamburg, Hamburg, Germany. 2Molecular and Vascular Medicine Unit and Renal Unit, Beth Israel Deaconess Medical Center and Department of Medicine, Harvard Medical School, Boston, Massachusetts, USA. 3Leibniz-Institut für Molekulare Pharmakologie and Max-Delbrück-Centrum für Molekulare Medizin, Berlin, Germany. 4Department of Laboratory Medicine, The Children’s Hospital, and Department of Pathology, Harvard Medical School, Boston, Massachusetts, USA. 5Institut de Recherches Cliniques de Montréal, Molecular Genetics and Development, Faculté de Médecine de l’Université de Montréal, Montreal, Quebec, Canada. 6Institut für Humangenetik, Universitätsklinikum Hamburg-Eppendorf, Hamburg, Germany.
Address correspondence to: Thomas J. Jentsch, FMP/MDC, Robert-Roessle-Strasse 10, D-13125 Berlin, Germany. Phone: 49-30-9406-2961; Fax: 49-30-9406-2960; E-mail: [email protected]. Or to: Christian A. Hübner, Institut für Klinische Chemie, Friedrich Schiller Universität Jena, Erlanger Allee 101, D-07747 Jena, Germany. Phone: 49-3641-9325001; E-mail: [email protected].
Find articles by Hübner, C. in: JCI | PubMed | Google Scholar
Published June 1, 2007 - More info
J Clin Invest. 2007;117(6):1708–1717. https://doi.org/10.1172/JCI30630.
© 2007 The American Society for Clinical Investigation
Received: October 12, 2006; Accepted: March 20, 2007
-
Abstract
K-Cl cotransport activity in rbc is a major determinant of rbc volume and density. Pathologic activation of erythroid K-Cl cotransport activity in sickle cell disease contributes to rbc dehydration and cell sickling. To address the roles of individual K-Cl cotransporter isoforms in rbc volume homeostasis, we disrupted the Kcc1 and Kcc3 genes in mice. As rbc K-Cl cotransport activity was undiminished in Kcc1–/– mice, decreased in Kcc3–/– mice, and almost completely abolished in mice lacking both isoforms, we conclude that K-Cl cotransport activity of mouse rbc is mediated largely by KCC3. Whereas rbc of either Kcc1–/– or Kcc3–/– mice were of normal density, rbc of Kcc1–/–Kcc3–/– mice exhibited defective volume regulation, including increased mean corpuscular volume, decreased density, and increased susceptibility to osmotic lysis. K-Cl cotransport activity was increased in rbc of SAD mice, which are transgenic for a hypersickling human hemoglobin S variant. Kcc1–/–Kcc3–/– SAD rbc lacked nearly all K-Cl cotransport activity and exhibited normalized values of mean corpuscular volume, corpuscular hemoglobin concentration mean, and K+ content. Although disruption of K-Cl cotransport rescued the dehydration phenotype of most SAD rbc, the proportion of the densest red blood cell population remained unaffected.
-
Introduction
K-Cl cotransporters (KCCs) mediate electroneutral symport of K+ and Cl– ions across the plasma membrane that is mostly outwardly directed and was originally described in rbc (1–3). K-Cl cotransport is mediated in a wide range of tissues by the polypeptide products of the 4 homologous SLC12 cation cotransporter superfamily genes SLC12A4/KCC1, SLC12A5/KCC2, SLC12A6/KCC3, and SLC12A7/KCC4 (4). These proteins contribute to regulatory volume decrease, transepithelial ion transport, and the control of the cytoplasmic Cl– concentration ([Cl–]i). Loss-of-function mutations in human KCC3 cause Andermann syndrome, which is characterized by severe peripheral neuropathy with variable agenesis of the corpus callosum (5). These clinical features are partially reproduced in Kcc3–/– mice (5, 6), which also display arterial hypertension and a slowly progressive deafness (6, 7). Two main 5′-splice variants of KCC3 have been identified, KCC3a and KCC3b, with distinct expression profiles (8). Specific roles of KCC1 have remained unknown in the absence of a Kcc1–/– mouse or a KCC1-associated human disease. The ubiquitous expression of KCC1 suggested a general role in cell volume regulation (9–11). In addition to KCC1, KCC3 has also been detected in rbc (11). rbc K-Cl cotransport has been implicated in the physiological reduction of cell volume accompanying maturation of reticulocytes and nascent erythrocytes (12, 13). K-Cl cotransport is substantially reduced in mature, circulating rbc but can be activated by urea, hypotonic challenge, thiol-reactive reagents such as N-ethylmaleimide, oxidants, and nonselective protein kinase inhibitors such as staurosporine (14, 15).
Erythroid K-Cl cotransport activity is increased in sickle cell disease (SCD) (16) and is thought to contribute to rbc dehydration and the formation of sickle cells (17, 18). SCD is generally caused by the hemoglobin (Hb) β-chain E6V mutation. This sickle Hb (HbS) causes rbc to change shape upon deoxygenation because of polymerization of the abnormal HbS. Sickled blood cells cause acute and chronic organ dysfunction due to vasoocclusive events in the microcirculation. The duration of the postoxygenation delay preceding the rapid phase of HbS polymerization is inversely proportional to the 30th-to-50th exponential power of HbS concentration. Thus, small increases in sickle cell volume, e.g., by inhibition of K-Cl cotransport, resulting in small decreases in intracellular HbS concentration (19), may greatly prolong the delay time of HbS polymerization. This prolongation, in turn, is hypothesized to delay deformation and sickling of sickle rbc until they escape the capillaries into larger venules, or even until they can be reoxygenated in the pulmonary circulation (20, 21). Hence, therapeutic inhibition of erythroid K-Cl cotransport has been proposed as an adjunct therapy for SCD. However, specific inhibitors of high potency are not yet available.
To study more definitively the role of erythroid K-Cl cotransport in health and SCD, we have created a Kcc1–/– mouse model. Through studies of this and the Kcc3–/– mouse, we show that KCC3 is the predominant KCC of mouse erythrocytes. Disruption of both rbc KCCs in mice resulted in impaired rbc volume regulation. To address the suggested role of K-Cl cotransport in SCD, we crossed KCC1- and KCC3-KO mice into the SAD mouse model of SCD, which reproduces the dehydrated red cell phenotype of human SCD (22, 23). Although disruption of K-Cl cotransport rescued the dehydration phenotype of most SAD rbc, the proportion of the densest red blood cell population remained unaffected.
-
Results
Kcc1–/– mice are viable and fertile. Deletion of the Slc12a4 gene exons 4 and 5, encoding the first 2 putative transmembrane spans of KCC1, results in a frameshift and premature termination (Figure 1A). Homologous recombination was verified by Southern blot analysis (Figure 1B). Northern blot analysis using a full-length KCC1 probe demonstrated that the transcript of the recombined allele was unstable, as shown for liver (Figure 1C) and brain (data not shown). Immunoblot analysis confirmed the broad expression pattern of the KCC1 polypeptide and its absence in Kcc1–/– mice (Figure 1D). Kcc1–/– mice were born at the expected Mendelian ratio (25.5% Kcc1+/+, 49.0% Kcc1+/–, and 25.5% Kcc1–/–; n = 168) and were viable and fertile. Their body weight and mortality were identical to those of WT (Kcc1+/+) littermates. They were indistinguishable in appearance and showed no motor deficits as assayed by rotarod or motor activity (data not shown). Auditory threshold, electroretinogram responses to light stimuli, and seizure threshold in response to the seizure-inducing agent flurothyl were unaltered (data not shown). Histological investigation of all major organ systems revealed no obvious abnormalities (data not shown). No signs of overt osteopetrosis were noted.
 Figure 1
Figure 1Generation of Kcc1–/– mice. (A) Partial genomic organization of Slc12a4 (top) and the targeting construct (middle). Exons are shown as vertical bars, loxP sites as arrowheads. Transient Cre expression resulted in excision of exons 4 and 5 (bottom), producing a frameshift and premature stop. DTA, diphtheria toxin A cassette. (B) An additional EcoRV site was exploited for Southern blot analysis and resulted in the approximately 6.5-kb KO compared with the approximately 10-kb Kcc1+/+ fragment with the probe shown in A. (C) Northern blot analysis of Kcc1+/+, Kcc1+/–, and Kcc1–/– liver tissue with a full-length KCC1 probe revealed some residual aberrant transcript. (D) A membrane protein immunoblot with a KCC1 antibody confirmed the absence of KCC1 in tissues of Kcc1–/– mice. (E) rbc membrane protein immunoblot demonstrated KCC3b expression in WT rbc and absence of KCC3a, KCC2, and KCC4 in erythrocytes of WT, Kcc1–/–Kcc3–/–, and SADKcc1–/–Kcc3–/– mice. Actin served as a loading control. Kidney served as a positive control for KCC1 and KCC3b, brain for KCC2 and KCC3a, and lung for KCC4. (F and G) Expression levels of KCC1 and KCC3 proteins in rbc ghosts of various genotypes. (F) KCC3b protein level was upregulated in ghosts lacking KCC1, but KCC1 levels were unchanged in KCC3-KO ghosts. (G) Levels of both KCC1 and KCC3b proteins were increased in ghosts from SAD mice. Protein levels were determined by Western blot analysis (see Supplemental Figure 1) and normalized to actin. Bars represent arithmetic means from 6 mice normalized to WT. Error bars represent SEM. *P < 0.05, **P < 0.005 compared with WT.
Kcc1–/–Kcc3–/– mice resemble Kcc3–/– mice in overall phenotype but have an increased mortality. The widespread expression pattern of KCC1 overlaps with that of KCC3, including in rbc (9–11). Immunoblot analysis of WT rbc ghost membranes demonstrated the presence of both KCC1 and KCC3b, but not of KCC3a, KCC2, and KCC4 (Figure 1E). Quantification of protein levels in Kcc1–/– ghosts revealed upregulation of KCC3b, whereas KCC1 protein levels were unchanged in Kcc3–/– ghosts (Figure 1F and Supplemental Figure 1; supplemental material available online with this article; doi:10.1172/JCI30630DS1). Coomassie staining of ghost membranes subjected to PAGE did not reveal any grossly evident alteration of polypeptide profile among different genotypes (Supplemental Figure 2). To generate mice deficient for both rbc KCCs, Kcc1–/–Kcc3+/– siblings were intercrossed. Kcc1–/–Kcc3–/– mice were born at the expected Mendelian ratio (25.0% Kcc1–/–, 50.6% Kcc1–/–Kcc3+/–, and 24.4% Kcc1–/–Kcc3–/–; n = 223), excluding a lethal developmental phenotype in the double-KO. The phenotype of Kcc1–/–Kcc3–/– mice closely resembled the extensively described phenotype of Kcc3–/– mice (6, 7). Although Kcc1–/–Kcc3–/– mice were fertile, litters from Kcc1–/–Kcc3–/– matings were very rare, and litter sizes were small. Kcc1–/–Kcc3–/– mice had an increased mortality (21.9%; n = 89) during the first 5 weeks of life compared with Kcc3–/– (11.1%; n = 121), Kcc1–/– (<1.0%), and WT mice (<1.0%). Throughout postnatal development, the body weight of Kcc1–/–Kcc3–/– mice was less than that of Kcc1–/– or Kcc3–/– littermates. P28 body weight of Kcc1–/–Kcc3–/– mice was 10.8 ± 0.5 g versus 12.8 ± 0.7 g for Kcc3–/– (P < 0.05), 14.7 ± 1.1 g for Kcc1–/–, and 14.5 ± 1.2 g for WT mice (n = 10 in each group). At age 8 weeks, the mean body weight of Kcc1–/–Kcc3–/– mice had increased to 18.2 ± 1.0 g compared with weights of 21.8 ± 1.1 g for Kcc3–/– (n = 10; P < 0.05), 23.3 ± 0.6 g for Kcc1–/– (n = 10), and 23.8 ± 0.5 g for WT mice (n = 12). The ratio of spleen weight to body weight (3.77% ± 0.41% in WT; n = 10) was reduced in Kcc1–/–Kcc3–/– mice (2.76% ± 0.18%; n = 10; P < 0.05) but not in Kcc1–/– (3.85% ± 0.49%; n = 21) or Kcc3–/– mice (3.54% ± 0.24% n = 11).
rbc K-Cl cotransport is mediated mainly by KCC3, and to a lesser extent by KCC1. rbc K-Cl cotransport, measured as Cl–-dependent K+ efflux in the presence of 1 mM ouabain (to inhibit the Na+/K+-ATPase) and 0.1 mM bumetanide (to inhibit NaK2Cl cotransport), was investigated under 3 conditions known to stimulate K-Cl cotransport, i.e., extracellular hypotonicity (~25% decrease in osmolarity), 0.5 M extracellular urea, and 1 μM staurosporine. Unexpectedly, K-Cl cotransport activity in Kcc1–/– rbc was moderately increased at extracellular hypotonicity (P < 0.05) (Figure 2, A and B), corresponding to the upregulation of KCC3b polypeptide abundance in Kcc1–/– rbc (Figure 1F). Under the other experimental conditions, K-Cl cotransport activity in Kcc1–/– rbc was similar to that of WT (Figure 2, A and B). However, K-Cl cotransport was moderately decreased in rbc of Kcc3+/– and Kcc1–/–Kcc3+/– mice (Figure 2, C and D) and decreased to a greater degree in rbc of Kcc3–/– mice under all conditions examined (Figure 2E). Hypotonicity-activated K-Cl cotransport appeared to be abolished in Kcc3–/– rbc, but staurosporine and urea stimulated a small remnant of K-Cl cotransport activity. However, all K-Cl cotransport activity stimulated by these 3 maneuvers was absent in rbc of Kcc1–/–Kcc3–/– mice (Figure 2F).
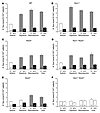 Figure 2
Figure 2K-Cl cotransport activity in rbc of WT and KO mice. K-Cl cotransport was determined by measuring K+ efflux in the presence or absence of Cl– (replaced with SFA–) in isotonic or hypotonic conditions and in the presence of staurosporine (1 μM) or urea (500 mM) added to the isotonic solution. Gray bars indicate a significant difference between stimulated and isotonic K+ fluxes in the presence of Cl–. Black bars indicate a significant difference between K+ fluxes in a single condition in the presence and absence of Cl–. WT K-Cl cotransport activity (A) was unaltered in rbc of Kcc1–/– mice (B). (C, D, and E) Disruption of Kcc3 reduced K-Cl cotransport activity in all conditions examined, with an apparent gene-dosage effect. (F) In rbc of Kcc1–/–Kcc3–/–mice, equivalent K+ fluxes in the presence and absence of Cl– indicated complete loss of K-Cl cotransport activity. Numbers in parentheses refer to the number of independent samples in each category.
Kcc1–/–Kcc3–/– mice have reduced rbc numbers but are not anemic. rbc counts of Kcc1–/–Kcc3–/– mice, but not of single KO littermates, were mildly decreased compared with those of WT mice (P < 0.05; Table 1). However, these lower values did not reflect anemia, since Hb values in all strains were within the normal ranges for the parental C57BL6/J and SV129 strains (Mouse Genome Informatics Database, The Jackson Laboratory; http://www.informatics.jax.org/). The lack of a hemolytic anemia in Kcc1–/–Kcc3–/– mice was corroborated by WT values of serum bilirubin and lactate dehydrogenase (data not shown) and by normal reticulocyte counts as determined by brilliant cresyl violet staining of blood smears and by ADVIA analysis. Histological analysis of spleen, liver, and bone marrow by Prussian blue staining did not reveal increased iron deposition as a consequence of increased rbc turnover (ref. 24 and data not shown).
Disruption of K-Cl cotransport affects rbc volume. Although rbc mean corpuscular volume (MCV) of Kcc1–/– mice and Kcc3–/– mice was not altered, MCV was significantly increased in Kcc1–/–Kcc3–/– mice (53.3 ± 0.4 fl; n = 10) compared with WT values (50.4 ± 0.6 fl; n = 10; P < 0.005) (Table 1). The reticulocyte MCV as determined by ADVIA analysis was similarly unchanged in Kcc1–/– (61.8 ± 0.6 fl; n = 31) compared with WT mice (61.2 ± 0.6, n = 35) but elevated in Kcc1–/–Kcc3–/– mice (65.1 ± 0.7 fl; n = 23; P < 0.001), with a corresponding reduction in reticulocyte corpuscular Hb concentration mean (CHCM). The density distribution of rbc was determined for Kcc1–/–, Kcc3–/–, and Kcc1–/–Kcc3–/– mice using 2 methods. Whereas the phthalate density distribution of rbc was unaltered in Kcc1–/– and Kcc3–/– rbc, Kcc1–/–Kcc3–/– rbc distributions were shifted toward lower densities, suggesting that loss of both KCC1 and KCC3 is necessary to alter steady-state volume homeostasis of rbc (Figure 3A). The shift of K-Cl cotransport–deficient rbc toward lower densities was confirmed by discontinuous Stractan gradient centrifugation (Figure 3B) and quantitative analysis of the resulting fractions (Figure 3C). To address the role of K-Cl cotransport in determination of rbc surface area/volume ratio, rbc osmotic fragility was tested by exposure to NaCl concentrations ranging from 154 mM (0.90%) to 60 mM (0.35%). Whereas the osmotic resistance of rbc from Kcc1–/–or Kcc3–/– mice resembled that of control rbc, the osmotic resistance of Kcc1–/–Kcc3–/– rbc was decreased (Figure 3D).
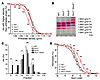 Figure 3
Figure 3Hematologic characterization of Kcc-KO mice. (A) rbc densities determined by centrifugation through phthalate mixtures of defined density. (B) rbc densities determined by centrifugation through discontinuous Stractan gradients. FI, fraction I. (C) Distinct rbc bands were collected and the rbc numbers quantified to evaluate relative number of rbc per fraction. Kcc1–/–and Kcc3–/–rbc densities were indistinguishable from WT, but Kcc1–/–Kcc3–/–rbc densities were reduced. (D) Osmotic lysis curve shows significantly increased osmotic sensitivity only for rbc lacking both KCCs. *P ≤ 0.05; **P ≤ 0.005.
The densest rbc of SAD mice are not rescued by disruption of rbc K-Cl cotransport. rbc dehydration produced by elevated K-Cl cotransport activity is thought to be a key factor in accelerated HbS polymerization in deoxygenated sickle cells (17). Indeed, overall K-Cl cotransport activity was markedly elevated in rbc of SAD mice at basal isotonic conditions (Figure 4A), as previously shown for rbc of both SAD mice (25) and SCD patients (26, 27). This may be due in part to upregulation of protein levels of both KCC1 and KCC3b in SAD rbc (Figure 1G). However, the increased levels of these proteins may be largely influenced by the roughly 2-fold increase in SAD mice of reticulocytes (Tables 1 and 2), cells that express more cotransport activity than mature rbc. Activators of K-Cl cotransport, hypotonicity or urea, did not increase K-Cl cotransport activity in SAD rbc to levels higher than those observed in WT rbc (compare Figure 4A and Figure 2A). As observed for rbc of WT and Kcc1–/–Kcc3–/– mice, rbc of SAD or Kcc1–/–Kcc3–/– SAD mice (designated SADKcc1–/–Kcc3–/– mice) did not express detectable amounts of KCC2 or KCC4 protein (Figure 1E). Disruption of KCC1 in SAD mice did not affect basal rbc K-Cl cotransport activity or the magnitude of stimulated activity (Figure 4B). Neither hypotonicity nor urea increased K-Cl cotransport activity in SADKcc3–/– rbc (Figure 4C). Residual basal K-Cl cotransport activity was evident in SADKcc3–/– rbc and was further reduced in SADKcc1–/–Kcc3–/– rbc (Figure 4D). K-Cl cotransport activity in SADKcc1–/–Kcc3–/– rbc was not sensitive to stimulation by hypotonicity or urea (Figure 4D). Consistent with previous studies on human sickle cells (16) and SAD rbc (23), the K+ content of SAD rbc was substantially reduced compared with that of controls (Figure 4E). Neither a deficiency of KCC1 or KCC3 alone (Supplemental Table 1) nor disruption of both KCCs affected K+ content of non-SAD rbc (Figure 4E). However, lack of KCC3 partially restored SAD rbc K+ content; and in SADKcc1–/–Kcc3–/– rbc, K+ content was comparable to that in WT (Figure 4E). Na+ content did not differ significantly among red cells from SAD mice and SAD mice lacking one or both KCC polypeptides (Supplemental Table 1).
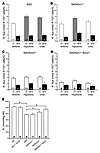 Figure 4
Figure 4rbc K-Cl cotransport activity and K+ content of SAD mice of various Kcc genotypes. (A–D) K+ efflux from rbc measured as described in Methods in the presence of Cl– or upon its substitution by SFA–, measured under isotonic or hypotonic conditions and in the presence of 500 mM urea added to the isotonic solutions. Gray bars indicate a significant difference between isotonic and stimulated K+ fluxes. Black bars indicate a significant difference between K+ fluxes in a single condition in the presence and absence of Cl–. (A) The elevated isotonic K-Cl cotransport activity of SAD rbc (compare with WT rbc in Figure 2A) was further stimulated by hypotonicity and by urea. Absence of KCC1 had minimal effect on K-Cl cotransport activity (B), whereas absence of KCC3 reduced K-Cl cotransport activity (C), and absence of both KCC1 and KCC3 nearly abolished cotransport activity (D). (E) Disruption of KCC1 and KCC3 did not alter rbc K+ content in the absence of the SAD transgene but rescued nearly completely the reduced K+ content of SAD rbc. *P ≤ 0.05.
The MCV of SAD rbc (44.8 ± 0.7 fl) was decreased compared with that of WT (50.4 ± 0.6 fl; P < 0.005) (Table 2). CHCM was correspondingly increased (32.4 ± 0.7 g/dl for SAD versus 27.8 ± 0.2 g/dl for WT; P < 0.005). Absence of KCC1 and KCC3 in SAD mice raised MCV to values comparable to those in WT mice and significantly decreased CHCM. In contrast, the absence of either KCC1 or KCC3 in SAD mice did not alter MCV, although CHCM of SADKcc3–/– mice (31.7 ± 0.2 g/dl) was slightly decreased compared with that of SAD mice (32.4 ± 0.7 g/dl; P < 0.005) (Table 2). As dense rbc are most likely to sickle upon deoxygenation, we determined the rbc density profiles for WT, SADKcc1–/–, SADKcc3–/–, and SADKcc1–/–Kcc3–/– mice. SAD rbc were of higher density (Figure 5, A–C) than WT rbc, as previously reported (22, 28). Compared with SAD rbc, the density profile of most SADKcc1–/–Kcc3–/– rbc mice was shifted toward lower-density values, but there was no significant effect of the double KO on the density of the densest 10%–15% rbc. The increased osmotic resistance of SAD rbc (Figure 5D), similar to that of human sickle cells, was partially normalized toward WT values in SAD cells lacking Kcc1 and Kcc3 (Figure 5C).
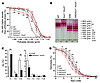 Figure 5
Figure 5Hematological characterization of SAD mice of various Kcc genotypes. (A) Phthalate density profile of SAD mice of various Kcc genotypes. (B) rbc density distribution revealed by discontinuous Stractan gradient centrifugation. (C) Disruption of erythroid K-Cl cotransport in SADKcc1–/–Kcc3–/–mice shifted the density distribution profile toward lower densities only in the low-density fractions, but not in high-density fractions. (D) Osmotic fragility curve. The osmotic resistance phenotype of SAD rbc was partially rescued by the absence of both KCC3 and KCC1. *P ≤ 0.05; **P ≤ 0.005.
-
Discussion
KCCs regulate cell volume and contribute to transepithelial ion transport. Early Northern blot analysis studies revealed that KCC1 transcripts are ubiquitously expressed, leading investigators to suggest a fundamental role in cell volume regulation (29), in particular in rbc (9). To address the physiological roles of Kcc1, we disrupted the Kcc1 gene in mice. Surprisingly, Kcc1–/– mice lacked any obvious clinical phenotype. In vitro analysis of Kcc1–/– rbc revealed no reduction in K-Cl cotransport activity under basal or stimulated conditions (Figure 2B).
Although KCC4 was detected in human erythroid precursor cells and reticulocytes by RT-PCR (30), our immunoblot analysis detected only KCC1 and KCC3b polypeptides in mouse rbc. K-Cl cotransport activity was partially decreased in Kcc3+/– rbc, more severely reduced in Kcc3–/– rbc (Figure 2, C and E), but undiminished in Kcc1–/– rbc. However, under hypotonic stimulation, K-Cl cotransport activity was surprisingly increased in Kcc1–/– compared with WT rbc, likely reflecting increased KCC3b protein levels in rbc lacking KCC1. In our experimental conditions, KCC3b was the dominant erythroid KCC, consistent with the biophysical characteristics of sheep rbc K-Cl cotransport, which better match the known properties of KCC3 than those of KCC1. For instance, the rank order of anion-dependent K+ transport in sheep rbc, [Br– > Cl– > I– > SCN–], coincides better with the rank order of anion-dependent K+ transport for human KCC3a [Br– > Cl– > (I– = SCN–)] (8) than that for rabbit KCC1 [Cl– > (SCN = Br–) > I–] (31, 32) expressed in Xenopus oocytes. However, the KCC gene product responsible for most erythroid K-Cl cotransport may be species specific, as the anion rank order measured in human rbc, [Cl– > Br– > I–] (2), suggests a significant contribution by KCC1. Possible species differences in regulation of erythroid K-Cl cotransport are highlighted by a recent study of human rbc from Andermann syndrome patients with truncating mutations in the KCC3 gene (33). This study documented decreased N-ethylmaleimide–stimulated K-Cl cotransport without reduction of staurosporine-stimulated K-Cl cotransport, in contrast to our results with mouse Kcc3–/– rbc. Kcc3–/– rbc displayed some K-Cl cotransport activity (Figure 2E), which was absent in rbc of Kcc1–/–Kcc3–/– mice and thus likely reflects KCC1 activity. Since K-Cl cotransport was measured in total rbc, different contributions of KCC1 and KCC3 to the K-Cl cotransport activity of rbc subpopulations of different developmental age or density cannot be excluded. Indeed, overall K-Cl cotransport activity was shown to be higher in reticulocytes and young erythrocytes but almost inactive in nonstimulated mature sheep rbc (14, 15). This observation led to the speculation that K-Cl cotransport reduces rbc volume during rbc maturation (12, 13). Erythrocyte MCV was elevated in Kcc1–/–Kcc3–/– mice (Table 1), but not in Kcc1–/– and Kcc3–/– mice, suggesting an abnormal regulation of rbc volume. Bone marrow histology did not indicate any morphological evidence for an rbc maturation defect (data not shown), although Kcc1–/–Kcc3–/– mice exhibited a small change in hematocrit. The density profile shift toward lower densities in Kcc1–/–Kcc3–/– rbc supports the idea that KCC1 and KCC3 are also active in nonstimulated mature rbc. Indeed, the rbc osmotic fragility of Kcc1–/–Kcc3–/– mice was increased in vitro.
The rate of sickling of deoxygenated rbc strongly depends on the rbc HbS concentration. Small reductions in the HbS concentration produced by therapeutic elevation of rbc volume have improved the pathological changes of sickle disease (34). As K-Cl cotransport activity is increased in rbc from SCD patients (35), K-Cl cotransport–mediated rbc dehydration is believed to be important in controlling the rate of deoxygenation-induced red cell sickling. The KCC may therefore be an attractive pharmacological target in the treatment of SCD. An increase in rbc Mg2+ content ([Mg2+]i) is known to inhibit K-Cl cotransport activity, and Mg2+ was shown to be decreased in rbc of some SCD patients (36). Oral Mg2+ supplementation elevated erythrocyte [Mg2+]i, decreased rbc K-Cl cotransport activity, and prevented rbc dehydration in SAD mice and in a small group of patients (25, 37). However, no small-molecule inhibitors of K-Cl cotransport with sufficient specificity and potency are available to allow clinical trial. To investigate the contribution of K-Cl cotransport to the pathogenesis of SCD, we disrupted rbc K-Cl cotransport in the SAD mouse model of sickle disease. SAD mice have circulating irreversibly sickled cells and develop chronic organ and tissue pathology resembling that of human sickle disease (22). Our data confirm that K-Cl cotransport activity is increased in SAD mice and demonstrate that this is at least in part due to an upregulation of KCC1 and KCC3b protein levels. Erythrocyte MCV was increased in SADKcc1–/–Kcc3–/– mice compared with SAD mice (Table 2). The increased rbc osmotic resistance of SAD rbc (an expression of cell dehydration) was modified in SADKcc1–/–Kcc3–/– rbc and approached that of WT rbc (Figure 5D). Mean corpuscular Hb concentration was strongly elevated in SAD mice, as observed previously by Trudel et al. (28) and De Franceschi et al. (23). Loss of KCC1 and KCC3 in SAD mice significantly decreased CHCM, but CHCM of SADKcc1–/–Kcc3–/– mice remained slightly higher than that of WT controls (Table 2). Density distribution analysis clearly demonstrated that loss of erythroid K-Cl cotransport in SAD mice increased the proportion of light, less-dense rbc subpopulations (Figure 5, A–C). However, the fraction containing the densest, dehydrated rbc remained unaltered. Lew and Bookchin have suggested that this dense fraction of human sickle cells originates from fast-track dehydration of reticulocytes mediated in substantial part by the elevated K-Cl cotransport activity of these young cells (17). The current data suggest that formation of the densest red cell fraction in SAD mice does not require K-Cl cotransport. Comparative investigation of the transport activity of specific density fractions of human sickle and mouse SAD red cells may further elucidate the relative contributions to dehydration of K-Cl cotransport and KCNN4 (Gardos channel) activity. Results of preliminary in vitro dithionite sickling experiments (38) trended toward a reduced proportion of abnormally shaped deoxygenated rbc from SADKcc1–/–Kcc3–/– mice, which, however, did not differ statistically from that of deoxygenated SAD rbc (data not shown). The severe neurodegeneration of Kcc1–/–Kcc3–/– and SADKcc1–/–Kcc3–/– mice, which resembles that described for Kcc3–/– mice, precluded an in vivo analysis of rbc sickling under conditions of acute or chronic hypoxia.
Our work demonstrates the important contribution of K-Cl cotransport to the regulation of mouse rbc volume, an effect that is enhanced in the presence of transgenic human sickling Hb SAD (HbSAD). This volume regulation is mediated principally by KCC3 and to a lesser degree by KCC1. The genetic disruption of K-Cl cotransport results in increased MCV, decreased CHCM and cell density, and increased susceptibility to osmotic lysis. Although these changes are also observed in K-Cl cotransport–deficient SAD red cells, the proportion of densest rbc in SAD mice remained undiminished. Hence, activation of K-Cl cotransport is a main determinant for the volume regulation of young rbc in SAD mice but may be not required for the generation of the densest fraction of rbc in SAD mice. This result, if extrapolated to human sickle disease, predicts that inhibitors of K-Cl cotransport used alone will not be maximally effective in reversing the red cell dehydration that predisposes to sickling. Inhibitors of another major volume regulatory pathway of red cells, the Ca2+-activated K+ channel KCNN4, attenuated the cell dehydration phenotype of sickle cells both in humans and in SAD mice (26, 39), but studies on sickle-cell mice lacking erythroid Kcnn4 expression are not yet available. Inhibition of both volume-regulatory pathways in sickle red cells may suffice to normalize the cell dehydration phenotype and maximize the delay time for deoxygenation-induced sickling.
-
Methods
All animal experiments were approved by Amt für Gesundheit und Verbraucherschutz, University of Hamburg, and by the Institutional Animal Care and Use Committee (IACUC) of Beth Israel Deaconess Medical Center.
Disruption of Slc12a4 in mice. A genomic clone of murine Slc12a4, the gene encoding KCC1, was isolated from a 129/Sv mouse genomic library (λFixII; Stratagene). The region encompassing exons 4 and 5 was flanked by loxP sites by insertion of a loxP site into the intron between exons 5 and 6 by PCR mutagenesis and by insertion of a floxed neomycin resistance (neo) cassette between exons 3 and 4 into a blunted genomic BamHI restriction site. A diphtheria toxin A (DTA) cassette was introduced at the 5ι end of the targeting construct as a negative selection marker. The linearized vector was electroporated into R1 ES cells (derived from the SVJ strain). G418-resistant clones that had undergone homologous recombination (Figure 1) were transfected with a plasmid expressing Cre-recombinase. Resulting clones were checked for the extent of DNA excision. Cells with a deletion of a fragment encompassing exons 4 and 5 and the neo cassette were used for the generation of the KO line. The recombinational excision resulted in a Kcc1–/– gene encoding a protein lacking aa 115–182, with a frameshift in codon 182 leading to termination after 36 residues of neosequence. These cells were injected into C57BL/6 blastocysts that were implanted into foster mothers. Resulting male chimeras were bred with C57BL/6 females. Heterozygous animals stemming from 2 different ES cell clones were inbred to yield mutant mouse lines. Routine genotyping was done by multiplex PCR using primers mKCC1-Seq12-s (5′-AGGCCAGAGCCAAGGAGTCTG-3′), mKCC1-r (5′-GGAGATTACAGGAGTGTACTG-3′), and mKCC1-Seq13-a (5′-ATCCCAGCAACCATATGGTGG-3′) to yield amplification products of 355 bp (WT) or 270 bp (KO).
Additional animal models used in this study. The generation of Kcc3–/– mice and SAD mice has been described previously (6, 28). Mating of Kcc1+/–Kcc3+/– mice resulted in Kcc1–/–, Kcc3–/–, and Kcc1–/–Kcc3–/– mice and the appropriate controls. SAD mice were mated with Kcc1+/–, Kcc3+/–, and Kcc1+/–Kcc3+/– mice to obtain SADKcc1–/–, SADKcc3–/–, and SADKcc1–/–Kcc3–/– mice. Studies were performed in a mixed 129SV/C57BL6 background using littermates (Figures 3–5) or age- and sex-matched animals of the same genetic backgrounds (Figure 2) as controls. SAD mouse genotyping was performed by 2 types of diagnostic multiplex genomic PCR. The human HbSAD transgene primer pair sad-s1 (5′-GACTCCTGAGGAGAAGTCTGCCGT-3′) and SAD-Seq6-a (5′-CACCGAGCACTTTCTTGCCATGAGC-3′) with annealing at 55°C yielded a 210-bp SAD transgene amplification product, while the control primer pair of mKA1-Seq37 (5′-AACAGCAGCGAGCCCGAGTAGTG-3′) and mKA1-Seq38-a (5′-TAAGAACTAGACCCAGGGTACAATG-3′) yielded a 388-bp control mouse genomic amplification product. Alternatively, the SAD primer pair SAD.A (5′-CCAACTCCTAAGCCAGTGCCAGAAG-3′) and SAD.B (5′-ACATCAAGGGTCCCATAGACTCACC-3′) with annealing at 60°C yielded a 685-bp SAD transgene amplification product, while the control primer pair of KCC1.E20F (5′-ACCTCCGCCTGGAAGCTGAAG-3′) and KCC1.CtM (5′-CTCAGGARTAGATGGTGATGACTTC-3′) yielded a 1,297-bp mouse KCC1 genomic amplification product. Human Hb α2 genotyping was also performed to verify cosegregation of both transgenes. Primers HBA2-Seq1-s (5′-CCGGTCAACTTCAAGGTGAGC-3′) and HBA2-Seq2-a (5′-TACCGAGGCTCCAGCTTAACG-3′) yielded a 301-bp band specific for Hb α2.
Northern blot analysis. Total RNA was prepared from WT, Kcc1+/–, and Kcc1–/– liver. Ten micrograms each was subjected to agarose electrophoresis and subsequently blotted onto a nylon membrane as described previously (6) and hybridized with a murine full-length Kcc1 cDNA probe.
Antibodies. Polyclonal antibodies were raised in rabbits against the N-terminal mouse KCC1 peptide sequence PVDGPRRGDYDNLEG that was coupled to BSA via a C-terminal cysteine. One rabbit serum specifically detected KCC1 in membrane-enriched protein lysates in immunoblot analysis as confirmed using Kcc1–/– tissues. Likewise, antibodies were raised against N-terminal peptide sequences of murine KCC3a (RVRFSSRESVPETSR) and KCC3b (KVEDPEEGAAGPLSP). Peptide affinity-purified antibodies were used in immunoblots: rb α-KCC1, rb α-KCC3a, rb α-KCC3b, rb α-KCC2, and gp α-KCC4 (all 1:500) (6, 40, 41); rb α-actin (1:4,000; Sigma-Aldrich).
Immunoblot. Membrane proteins were enriched from tissues as described previously (6), except that prior to tissue dissection, animals were deeply anesthetized followed by transcardial perfusion with PBS to clear tissues from blood. rbc ghosts were prepared from venous blood drawn from the vena cava into heparinized syringes. Blood cells were harvested by centrifugation, and ghosts were prepared by several cycles of washing with H2O followed by centrifugation until the red heme color cleared. Equal amounts of protein were separated by SDS–PAGE, blotted onto nitrocellulose, and labeled with primary antibodies. Secondary antibodies were conjugated to HRP (Chemicon) and detected by chemiluminescence (SuperSignal West; Pierce). Quantification was done with the Chemi-Smart 5000 image acquisition system (Vilber Lourmat) and ImageJ 1.36b software (http://rsb.info.nih.gov/ij/).
Density distribution profile. Venous blood was drawn from retro-orbital venous plexus with heparinized hematocrit capillaries (Brand) and collected into EDTA-coated tubes (SARSTEDT) to prevent coagulation. The density distribution was determined using mixtures of methyl phthalate (Fluka; Sigma-Aldrich) and di-η-butyl phthalate (Sigma-Aldrich) (42). rbc were spun through phthalate ester mixtures with densities ranging from 1.086 to 1.118 g/ml at 12,000 g for 15 minutes in a hematocrit centrifuge (Biofuge Haemo; Heraeus), and relative heights of rbc columns below and above each phthalate ester phase were quantified.
Discontinuous Stractan gradient. Stractan/polyarabinogalactan (Fluka; Sigma-Aldrich) solutions and buffered saline with potassium and glucose (BSKG; in mM: 134 NaCl, 8.6 Na2HPO4, 1.4 NaH2PO4, 5 KCl, 10 glucose, and 1 % BSA; pH 7.4; osmolarity 290–295 mOsm) were prepared as described previously (22). Nine different 400-μl fractions of Stractan/BSKG solutions with densities ranging from 1.0822 g/ml to 1.1102 g/ml were successively layered into 4.2 ml ultraclear centrifuge tubes (Beckman Coulter). Venous blood was drawn retro-orbitally with heparinized hematocrit capillaries and collected into EDTA-coated tubes. Twenty microliters blood diluted with 180 μl BSKG was layered on top of the Stractan gradient and centrifuged 30 minutes at 23,400 g in a TST 60.4 rotor (Kontron) at 20°C. For quantitative analysis, experiments were repeated with six 600-μl Stractan fractions with densities ranging from 1.082 to 1.100 g/ml for rbc from WT, Kcc1–/–, Kcc3–/–, and Kcc1–/–Kcc3–/– mice and with densities ranging from 1.093 to 1.110 for rbc from SAD, SADKcc1–/–, SADKcc3–/–, and SADKcc1–/–Kcc3–/– mice. Distinct rbc bands were collected and the rbc numbers quantified using cell counter Z1 (Beckman Coulter) to evaluate relative number of rbc per fraction.
Resistance to osmotic lysis. Venous blood drawn retro-orbitally was collected into reaction tubes containing approximately one-fourth of the final volume of 4.3% (wt/vol) Na citrate. Twenty microliters citrated blood was incubated for 2 hours at 37°C in solutions of NaCl in water, concentrations ranging from 0.0% to 0.7% (wt/vol). rbc were pelleted at 1,000 g for 5 minutes, and the supernatants were incubated in a 0.1% SDS solution containing (in mM) 0.8 KCN, 0.6 K3Fe(CN)6, 1 KH2PO4 to transform Hb into cyanohemoglobin, which was quantified photometrically at a wavelength of 546 nm. The degree of rbc hemolysis was normalized to complete hemolysis in H2O.
Hematological parameters. Venous blood was drawn retro-orbitally and collected into EDTA-coated tubes. Hemogram and reticulocyte counts were analyzed automatically using the ADVIA 120 Hematology System (Bayer). Reticulocyte counts were also determined by brilliant cresyl violet staining of blood smears.
Determination of rbc K-Cl cotransport activity. K-Cl cotransport activity, defined as net ouabain- and bumetanide-resistant K+ efflux, was examined as described previously (43). In brief, venous blood was drawn retro-orbitally into EDTA-coated tubes or via cardiac or caval puncture into heparinized tubes. Twenty-four microliters of whole blood was diluted with 276 μl normal mouse saline (in mM: 165 NaCl, 2 KCl, 0.15 MgCl, 10 Tris/Mops; pH 7.4 at 25°C) to evaluate hematological parameters using the ADVIA 120 Hematology System (Bayer). The remaining rbc were washed 4–6 times in choline wash solution (CWS; in mM: 172 choline chloride, 10 sucrose, 10 Tris-Mops; pH 7.4 at 4°C) and resuspended in CWS to approximately 50% hematocrit. rbc were suspended at 2.5% hematocrit in ice-cold isotonic (340 mOsm; 160 mM NaCl) or hypotonic (260 mOsm; 115 mM NaCl) NaCl solutions containing (in mM): 1 MgCl2, 10 glucose, 1 ouabain, 0.01 or 0.1 bumetanide, and 10 Tris-Mops (pH 7.4 at 37°C). KCC fluxes in the presence of 0.01 mM bumetanide (Figure 4) were consistently approximately 10% higher than in the presence of 0.1 mM bumetanide (Figure 2) for red cells of WT, SAD, and Kcc1–/– mice, whether stimulated by hypotonicity, staurosporine, or urea. These results were consistent with equivalent, low bumetanide sensitivities of KCC1 and KCC3 activities under our net efflux assay conditions of nominally K+ free flux medium. Some isotonic solutions contained 1 μM staurosporine or 500 mM urea as indicated. In Cl–-free solutions, sulfamate (SFA–) substituted for Cl–. rbc suspensions were transferred into a water bath and incubated at 37°C. Triplicate samples of cell suspension were taken after 5 and 25 minutes, centrifuged at 700 g for 4 minutes at 4°C, and the supernatants harvested for measurement of the K+ concentrations by atomic absorption spectroscopy (PerkinElmer). K+ efflux rates were calculated from the linear regression slopes of medium K+ concentrations at 5 and 25 minutes. In room air CO2-equilibrated sulfamate solutions at 37°C, intracellular chloride was effectively depleted throughout the efflux measurement period (43, 44).
Determination of rbc cation content. rbc were washed 4 times in CWS and thereafter resuspended at a hematocrit of 50% in CWS. rbc suspensions were diluted 50-fold in 0.02% Pluronic PE 6400 (BASF), a nonionic detergent, to lyse rbc. Supernatants were further diluted 10-fold in 0.02% Pluronic PE 6400 to determine K+ concentration by atomic absorption spectrometry (PerkinElmer).
Statistics. Values are expressed as mean ± SEM. For unpaired 2-sample 2-tailed Student’s t tests, P ≤ 0.05 was considered significant. Values of K+ efflux and of hematological indices were compared among all genotypes by least-squares ANOVA with proportionate weighting for constant coefficient of variation and with Bonferroni-Holm and Bonferroni-Hochberg step-down corrections for multiple samples (JMP [version 6; SAS Institute] and Excel [2003; Microsoft] yielded similar results). For ANOVA, P < 0.05 was considered significant. Density distribution profile, quantitative analyses of Stractan gradients, and osmotic resistance were compared by 2-way ANOVA with Bonferroni correction.
-
Acknowledgments
We acknowledge M. Kolster, S. Bauer, E.H. Kim, B. Leclair, A.K. Stuart-Tilley, and L.C. Pong for excellent technical assistance and L.L. Peters for helpful discussion. We thank Clive Ellory for generously hosting M.B. Rust while he received instruction on the performance of rubidium flux assays. This work was supported by grants from the Deutsche Forschungsgemeinschaft to T.J. Jentsch and C.A. Hübner, by NIH grants HL077765 and HL15157 (Boston Comprehensive Sickle Cell Center) to S.L. Alper and C. Brugnara, and from the Canadian Institutes of Health Research to M. Trudel.
Address correspondence to: Thomas J. Jentsch, FMP/MDC, Robert-Roessle-Strasse 10, D-13125 Berlin, Germany. Phone: 49-30-9406-2961; Fax: 49-30-9406-2960; E-mail: [email protected]. Or to: Christian A. Hübner, Institut für Klinische Chemie, Friedrich Schiller Universität Jena, Erlanger Allee 101, D-07747 Jena, Germany. Phone: 49-3641-9325001; E-mail: [email protected].
-
Footnotes
Nonstandard abbreviations used: BSKG, buffered saline with potassium and glucose; CHCM, corpuscular Hb concentration mean; CWS, choline wash solution; Hb, hemoglobin; HbS, sickle Hb; HbSAD, SAD Hb; KCC, K-Cl cotransporter; MCV, mean corpuscular volume; SCD, sickle cell disease; SFA–, sulfamate.
Conflict of interest: The authors have declared that no conflict of interest exists.
Reference information: J. Clin. Invest.117:1708–1717 (2007). doi:10.1172/JCI30630
Christian A. Hübner’s present address is: Institut für Klinische Chemie und Laboratoriumsmedizin, Friedrich Schiller Universität Jena, Jena, Germany.
Marco B. Rust’s present address is: European Molecular Biology Laboratory, Mouse Biology Programme, Monterotondo, Italy.
-
References
- Lauf, P.K., Theg, B.E. 1980. A chloride dependent K+ flux induced by N-ethylmaleimide in genetically low K+ sheep and goat erythrocytes. Biochem. Biophys. Res. Commun. 92:1422-1428.
- Dunham, P.B., Stewart, G.W., Ellory, J.C. 1980. Chloride-activated passive potassium transport in human erythrocytes. Proc. Natl. Acad. Sci. U. S. A. 77:1711-1715.
- Kregenow, F.M. 1971. The response of duck erythrocytes to nonhemolytic hypotonic media. Evidence for a volume-controlling mechanism. J. Gen. Physiol. 58:372-395.
- Gamba, G. 2005. Molecular physiology and pathophysiology of electroneutral cation-chloride cotransporters. Physiol. Rev. 85:423-493.
- Howard, H.C., et al. 2002. The K-Cl cotransporter KCC3 is mutant in a severe peripheral neuropathy associated with agenesis of the corpus callosum. Nat. Genet. 32:384-392.
- Boettger, T., et al. 2003. Loss of K-Cl co-transporter KCC3 causes deafness, neurodegeneration and reduced seizure threshold. EMBO J. 22:5422-5434.
- Rust, M.B., et al. 2006. Neurogenic mechanisms contribute to hypertension in mice with disruption of the K-Cl cotransporter KCC3. Circ. Res. 98:549-556.
- Mercado, A., et al. 2005. NH2-terminal heterogeneity in the KCC3 K+-Cl– cotransporter. Am. J. Physiol. Renal Physiol. 289:F1246-F1261.
- Pellegrino, C.M., Rybicki, A.C., Musto, S., Nagel, R.L., Schwartz, R.S. 1998. Molecular identification and expression of erythroid K:Cl cotransporter in human and mouse erythroleukemic cells. Blood Cells Mol. Dis. 24:31-40.
- Bize, I., Guvenc, B., Buchbinder, G., Brugnara, C. 2000. Stimulation of human erythrocyte K-Cl cotransport and protein phosphatase type 2A by n-ethylmaleimide: role of intracellular Mg++. J. Membr. Biol. 177:159-168.
- Lauf, P.K., et al. 2001. K-Cl co-transport: immunocytochemical and functional evidence for more than one KCC isoform in high K and low K sheep erythrocytes. Comp. Biochem. Physiol. A Mol. Integr. Physiol. 130:499-509.
- Hall, A.C., Ellory, J.C. 1986. Evidence for the presence of volume-sensitive KCl transport in ‘young’ human red cells. Biochim. Biophys. Acta. 858:317-320.
- Brugnara, C., Tosteson, D.C. 1987. Cell volume, K transport, and cell density in human erythrocytes. Am. J. Physiol. 252:C269-C276. View this article via: PubMed Google Scholar
- Hall, A.C., Ellory, J.C. 1986. Effects of high hydrostatic pressure on ‘passive’ monovalent cation transport in human red cells. J. Membr. Biol. 94:1-17.
- Lauf, P.K., Perkins, C.M., Adragna, N.C. 1985. Cell volume and metabolic dependence of NEM-activated K+-Cl– flux in human red blood cells.
Am. J. Physiol. 249:C124-C128. View this article via: PubMed Google Scholar
- Brugnara, C., Bunn, H.F., Tosteson, D.C. 1986. Regulation of erythrocyte cation and water content in sickle cell anemia. Science. 232:388-390.
- Lew, V.L., Bookchin, R.M. 2005. Ion transport pathology in the mechanism of sickle cell dehydration. Physiol. Rev. 85:179-200.
- Joiner, C.H., Rettig, R.K., Jiang, M., Risinger, M., Franco, R.S. 2007. Urea stimulation of KCl cotransport induces abnormal volume reduction in sickle reticulocytes. Blood. 109:1728-1735.
- Eaton, W.A., Hofrichter, J. 1990. Sickle cell hemoglobin polymerization. Adv. Protein Chem. 40:63-279. View this article via: PubMed Google Scholar
- Christoph, G.W., Hofrichter, J., Eaton, W.A. 2005. Understanding the shape of sickled red cells. Biophys. J. 88:1371-1376. View this article via: PubMed Google Scholar
- Brugnara, C. 2001. Therapeutic strategies for prevention of sickle cell dehydration. Blood Cells Mol. Dis. 27:71-80.
- Trudel, M., et al. 1994. Sickle cell disease of transgenic SAD mice. Blood. 84:3189-3197. View this article via: PubMed Google Scholar
- De Franceschi, L., et al. 1994. Treatment with oral clotrimazole blocks Ca2+-activated K+ transport and reverses erythrocyte dehydration in transgenic SAD mice. A model for therapy of sickle cell disease. J. Clin. Invest. 93:1670-1676.
- Kong, Y., et al. 2004. Loss of α-hemoglobin–stabilizing protein impairs erythropoiesis and exacerbates β-thalassemia. J. Clin. Invest. 114:1457-1466.
- De Franceschi, L., Beuzard, Y., Jouault, H., Brugnara, C. 1996. Modulation of erythrocyte potassium chloride cotransport, potassium content, and density by dietary magnesium intake in transgenic SAD mouse. Blood. 88:2738-2744. View this article via: PubMed Google Scholar
- Brugnara, C., Kopin, A.S., Bunn, H.F., Tosteson, D.C. 1985. Regulation of cation content and cell volume in hemoglobin erythrocytes from patients with homozygous hemoglobin C disease. J. Clin. Invest. 75:1608-1617.
- Bize, I., Taher, S., Brugnara, C. 2003. Regulation of K-Cl cotransport during reticulocyte maturation and erythrocyte aging in normal and sickle erythrocytes. Am. J. Physiol. Cell Physiol. 285:C31-C38. View this article via: PubMed Google Scholar
- Trudel, M., et al. 1991. Towards a transgenic mouse model of sickle cell disease: hemoglobin SAD. EMBO J. 10:3157-3165. View this article via: PubMed Google Scholar
- Gillen, C.M., Brill, S., Payne, J.A., Forbush (3rd.), B. 1996. Molecular cloning and functional expression of the K-Cl cotransporter from rabbit, rat, and human. A new member of the cation-chloride cotransporter family. J. Biol. Chem. 271:16237-16244.
- Crable, S.C., et al. 2005. Multiple isoforms of the KCl cotransporter are expressed in sickle and normal erythroid cells. Exp. Hematol. 33:624-631.
- Mercado, A., Song, L., Vazquez, N., Mount, D.B., Gamba, G. 2000. Functional comparison of the K+-Cl– cotransporters KCC1 and KCC4. J. Biol. Chem. 275:30326-30334.
- Lauf, P.K., et al. 1992. Erythrocyte K-Cl cotransport: properties and regulation. Am. J. Physiol. 263:C917-C932. View this article via: PubMed Google Scholar
- Lauf, P.K., Adragna, N.C., Dupre, N., Bouchard, J.P., Rouleau, G.A. 2006. K-Cl cotransport in red blood cells from patients with KCC3 isoform mutants. Biochem. Cell Biol. 84:1034-1044. View this article via: PubMed Google Scholar
- Rosa, R.M., et al. 1980. A study of induced hyponatremia in the prevention and treatment of sickle-cell crisis. N. Engl. J. Med. 303:1138-1143. View this article via: PubMed Google Scholar
- Canessa, M., Spalvins, A., Nagel, R.L. 1986. Volume-dependent and NEM-stimulated K+,Cl– transport is elevated in oxygenated SS, SC and CC human red cells. FEBS Lett. 200:197-202.
- Olukoga, A.O., Erasmus, R.T., Adewoye, H.O. 1989. Erythrocyte and plasma magnesium status in Nigerians with diabetes mellitus. Ann. Clin. Biochem. 26:74-77. View this article via: PubMed Google Scholar
- De Franceschi, L., et al. 1997. Oral magnesium supplements reduce erythrocyte dehydration in patients with sickle cell disease. J. Clin. Invest. 100:1847-1852.
- Asakura, T., Mayberry, J. 1984. Relationship between morphologic characteristics of sickle cells and method of deoxygenation. J. Lab. Clin. Med. 104:987-994. View this article via: PubMed Google Scholar
- Stocker, J.W., et al. 2003. ICA-17043, a novel Gardos channel blocker, prevents sickled red blood cell dehydration in vitro and in vivo in SAD mice. Blood. 101:2412-2418.
- Hübner, C.A., et al. 2001. Disruption of KCC2 reveals an essential role of K-Cl cotransport already in early synaptic inhibition. Neuron. 30:515-524.
- Boettger, T., et al. 2002. Deafness and renal tubular acidosis in mice lacking the K-Cl co-transporter Kcc4. Nature. 416:874-878.
- Danon, D., Marikovsky, V. 1964. Determination of density distribution of red cell population. J. Lab. Clin. Med. 64:668-674. View this article via: PubMed Google Scholar
- Armsby, C.C., Brugnara, C., Alper, S.L. 1995. Cation transport in mouse erythrocytes: role of K+-Cl– cotransport in regulatory volume decrease.
Am. J. Physiol. 268:C894-C902. View this article via: PubMed Google Scholar
- Brahm, J. 1977. Temperature-dependent changes of chloride transport kinetics in human red cells. J. Gen. Physiol. 70:283-306.
-
Version history
- Version 1 (June 1, 2007): No description



Copyright © 2024 American Society for Clinical Investigation
ISSN: 0021-9738 (print), 1558-8238 (online)







