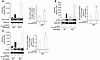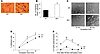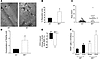Advertisement
Research ArticleBone biology Free access | 10.1172/JCI29092
Hyperactivation of p21ras and PI3K cooperate to alter murine and human neurofibromatosis type 1–haploinsufficient osteoclast functions
Feng-Chun Yang,1,2 Shi Chen,1,2 Alexander G. Robling,3 Xijie Yu,3 Todd D. Nebesio,1,2 Jincheng Yan,1,2 Trent Morgan,1,2 Xiaohong Li,1,2 Jin Yuan,1,2 Janet Hock,3 David A. Ingram,1,2,4 and D. Wade Clapp1,2,5
1Department of Pediatrics, 2Herman B. Wells Center for Pediatric Research, 3Department of Anatomy and Cell Biology, 4Department of Biochemistry and Molecular Biology, and 5Department of Microbiology and Immunology, Indiana University School of Medicine, Indianapolis, Indiana, USA.
Address correspondence to: D. Wade Clapp or Feng-Chun Yang, Indiana University School of Medicine, Herman B. Wells Center for Pediatric Research, Cancer Research Institute, 1044 West Walnut Street, R4 402, Indianapolis, Indiana 46202, USA. Phone: (317) 278-9290; Fax: (317) 274-8679; E-mail: [email protected] (D.W. Clapp) or [email protected] (F.-C. Yang).
Find articles by Yang, F. in: JCI | PubMed | Google Scholar
1Department of Pediatrics, 2Herman B. Wells Center for Pediatric Research, 3Department of Anatomy and Cell Biology, 4Department of Biochemistry and Molecular Biology, and 5Department of Microbiology and Immunology, Indiana University School of Medicine, Indianapolis, Indiana, USA.
Address correspondence to: D. Wade Clapp or Feng-Chun Yang, Indiana University School of Medicine, Herman B. Wells Center for Pediatric Research, Cancer Research Institute, 1044 West Walnut Street, R4 402, Indianapolis, Indiana 46202, USA. Phone: (317) 278-9290; Fax: (317) 274-8679; E-mail: [email protected] (D.W. Clapp) or [email protected] (F.-C. Yang).
Find articles by Chen, S. in: JCI | PubMed | Google Scholar
1Department of Pediatrics, 2Herman B. Wells Center for Pediatric Research, 3Department of Anatomy and Cell Biology, 4Department of Biochemistry and Molecular Biology, and 5Department of Microbiology and Immunology, Indiana University School of Medicine, Indianapolis, Indiana, USA.
Address correspondence to: D. Wade Clapp or Feng-Chun Yang, Indiana University School of Medicine, Herman B. Wells Center for Pediatric Research, Cancer Research Institute, 1044 West Walnut Street, R4 402, Indianapolis, Indiana 46202, USA. Phone: (317) 278-9290; Fax: (317) 274-8679; E-mail: [email protected] (D.W. Clapp) or [email protected] (F.-C. Yang).
Find articles by Robling, A. in: JCI | PubMed | Google Scholar
1Department of Pediatrics, 2Herman B. Wells Center for Pediatric Research, 3Department of Anatomy and Cell Biology, 4Department of Biochemistry and Molecular Biology, and 5Department of Microbiology and Immunology, Indiana University School of Medicine, Indianapolis, Indiana, USA.
Address correspondence to: D. Wade Clapp or Feng-Chun Yang, Indiana University School of Medicine, Herman B. Wells Center for Pediatric Research, Cancer Research Institute, 1044 West Walnut Street, R4 402, Indianapolis, Indiana 46202, USA. Phone: (317) 278-9290; Fax: (317) 274-8679; E-mail: [email protected] (D.W. Clapp) or [email protected] (F.-C. Yang).
Find articles by Yu, X. in: JCI | PubMed | Google Scholar
1Department of Pediatrics, 2Herman B. Wells Center for Pediatric Research, 3Department of Anatomy and Cell Biology, 4Department of Biochemistry and Molecular Biology, and 5Department of Microbiology and Immunology, Indiana University School of Medicine, Indianapolis, Indiana, USA.
Address correspondence to: D. Wade Clapp or Feng-Chun Yang, Indiana University School of Medicine, Herman B. Wells Center for Pediatric Research, Cancer Research Institute, 1044 West Walnut Street, R4 402, Indianapolis, Indiana 46202, USA. Phone: (317) 278-9290; Fax: (317) 274-8679; E-mail: [email protected] (D.W. Clapp) or [email protected] (F.-C. Yang).
Find articles by Nebesio, T. in: JCI | PubMed | Google Scholar
1Department of Pediatrics, 2Herman B. Wells Center for Pediatric Research, 3Department of Anatomy and Cell Biology, 4Department of Biochemistry and Molecular Biology, and 5Department of Microbiology and Immunology, Indiana University School of Medicine, Indianapolis, Indiana, USA.
Address correspondence to: D. Wade Clapp or Feng-Chun Yang, Indiana University School of Medicine, Herman B. Wells Center for Pediatric Research, Cancer Research Institute, 1044 West Walnut Street, R4 402, Indianapolis, Indiana 46202, USA. Phone: (317) 278-9290; Fax: (317) 274-8679; E-mail: [email protected] (D.W. Clapp) or [email protected] (F.-C. Yang).
Find articles by Yan, J. in: JCI | PubMed | Google Scholar
1Department of Pediatrics, 2Herman B. Wells Center for Pediatric Research, 3Department of Anatomy and Cell Biology, 4Department of Biochemistry and Molecular Biology, and 5Department of Microbiology and Immunology, Indiana University School of Medicine, Indianapolis, Indiana, USA.
Address correspondence to: D. Wade Clapp or Feng-Chun Yang, Indiana University School of Medicine, Herman B. Wells Center for Pediatric Research, Cancer Research Institute, 1044 West Walnut Street, R4 402, Indianapolis, Indiana 46202, USA. Phone: (317) 278-9290; Fax: (317) 274-8679; E-mail: [email protected] (D.W. Clapp) or [email protected] (F.-C. Yang).
Find articles by Morgan, T. in: JCI | PubMed | Google Scholar
1Department of Pediatrics, 2Herman B. Wells Center for Pediatric Research, 3Department of Anatomy and Cell Biology, 4Department of Biochemistry and Molecular Biology, and 5Department of Microbiology and Immunology, Indiana University School of Medicine, Indianapolis, Indiana, USA.
Address correspondence to: D. Wade Clapp or Feng-Chun Yang, Indiana University School of Medicine, Herman B. Wells Center for Pediatric Research, Cancer Research Institute, 1044 West Walnut Street, R4 402, Indianapolis, Indiana 46202, USA. Phone: (317) 278-9290; Fax: (317) 274-8679; E-mail: [email protected] (D.W. Clapp) or [email protected] (F.-C. Yang).
Find articles by Li, X. in: JCI | PubMed | Google Scholar
1Department of Pediatrics, 2Herman B. Wells Center for Pediatric Research, 3Department of Anatomy and Cell Biology, 4Department of Biochemistry and Molecular Biology, and 5Department of Microbiology and Immunology, Indiana University School of Medicine, Indianapolis, Indiana, USA.
Address correspondence to: D. Wade Clapp or Feng-Chun Yang, Indiana University School of Medicine, Herman B. Wells Center for Pediatric Research, Cancer Research Institute, 1044 West Walnut Street, R4 402, Indianapolis, Indiana 46202, USA. Phone: (317) 278-9290; Fax: (317) 274-8679; E-mail: [email protected] (D.W. Clapp) or [email protected] (F.-C. Yang).
Find articles by Yuan, J. in: JCI | PubMed | Google Scholar
1Department of Pediatrics, 2Herman B. Wells Center for Pediatric Research, 3Department of Anatomy and Cell Biology, 4Department of Biochemistry and Molecular Biology, and 5Department of Microbiology and Immunology, Indiana University School of Medicine, Indianapolis, Indiana, USA.
Address correspondence to: D. Wade Clapp or Feng-Chun Yang, Indiana University School of Medicine, Herman B. Wells Center for Pediatric Research, Cancer Research Institute, 1044 West Walnut Street, R4 402, Indianapolis, Indiana 46202, USA. Phone: (317) 278-9290; Fax: (317) 274-8679; E-mail: [email protected] (D.W. Clapp) or [email protected] (F.-C. Yang).
Find articles by Hock, J. in: JCI | PubMed | Google Scholar
1Department of Pediatrics, 2Herman B. Wells Center for Pediatric Research, 3Department of Anatomy and Cell Biology, 4Department of Biochemistry and Molecular Biology, and 5Department of Microbiology and Immunology, Indiana University School of Medicine, Indianapolis, Indiana, USA.
Address correspondence to: D. Wade Clapp or Feng-Chun Yang, Indiana University School of Medicine, Herman B. Wells Center for Pediatric Research, Cancer Research Institute, 1044 West Walnut Street, R4 402, Indianapolis, Indiana 46202, USA. Phone: (317) 278-9290; Fax: (317) 274-8679; E-mail: [email protected] (D.W. Clapp) or [email protected] (F.-C. Yang).
Find articles by Ingram, D. in: JCI | PubMed | Google Scholar
1Department of Pediatrics, 2Herman B. Wells Center for Pediatric Research, 3Department of Anatomy and Cell Biology, 4Department of Biochemistry and Molecular Biology, and 5Department of Microbiology and Immunology, Indiana University School of Medicine, Indianapolis, Indiana, USA.
Address correspondence to: D. Wade Clapp or Feng-Chun Yang, Indiana University School of Medicine, Herman B. Wells Center for Pediatric Research, Cancer Research Institute, 1044 West Walnut Street, R4 402, Indianapolis, Indiana 46202, USA. Phone: (317) 278-9290; Fax: (317) 274-8679; E-mail: [email protected] (D.W. Clapp) or [email protected] (F.-C. Yang).
Find articles by Clapp, D. in: JCI | PubMed | Google Scholar
Published November 1, 2006 - More info
J Clin Invest. 2006;116(11):2880–2891. https://doi.org/10.1172/JCI29092.
© 2006 The American Society for Clinical Investigation
Received: May 15, 2006; Accepted: September 12, 2006
-
Abstract
Individuals with neurofibromatosis type 1 (NF1) have a high incidence of osteoporosis and osteopenia. However, understanding of the cellular and molecular basis of these sequelae is incomplete. Osteoclasts are specialized myeloid cells that are the principal bone-resorbing cells of the skeleton. We found that Nf1+/– mice contain elevated numbers of multinucleated osteoclasts. Both osteoclasts and osteoclast progenitors from Nf1+/– mice were hyperresponsive to limiting concentrations of M-CSF and receptor activator of NF-κB ligand (RANKL) levels. M-CSF–stimulated p21ras-GTP and Akt phosphorylation was elevated in Nf1+/– osteoclasts associated with gains of function in survival, proliferation, migration, adhesion, and lytic activity. These gains of function are associated with more severe bone loss following ovariectomy as compared with that in syngeneic WT mice. Intercrossing Nf1+/– mice and mice deficient in class 1A PI3K (p85α) restored elevated PI3K activity and Nf1+/– osteoclast functions to WT levels. Furthermore, in vitro–differentiated osteoclasts from NF1 patients also displayed elevated Ras/PI3K activity and increased lytic activity analogous to those in murine Nf1+/– osteoclasts. Collectively, our results identify a what we believe to be a novel cellular and biochemical NF1-haploinsufficient phenotype in osteoclasts that has potential implications for the pathogenesis of NF1 bone disease.
-
Introduction
Neurofibromatosis type 1 (NF1) is a common, pandemic genetic disorder that is characterized by a range of both malignant and nonmalignant manifestations and is caused by mutations in the NF1 gene. Neurofibromin, the protein encoded by NF1, functions as a GTPase-activating protein (GAP) for Ras. Though loss of both alleles of NF1 in tumorigenic cells is consistent with NF1 being a tumor suppressor gene, recent genetic evidence in murine models has indicated that nullizygous loss of Nf1 in the tumorigenic cells of plexiform neurofibromas (1) and optic gliomas (2) is necessary but not sufficient for tumor progression, though tumors occur with high penetrance when lineages of the tumor microenvironment are haploinsufficient at Nf1. These in vivo data provide evidence that haploinsufficiency of Nf1 in at least a subset of lineages has a role in the malignant manifestations of NF1 and also suggest a potential role for haploinsufficiency of Nf1 (NF1) in the nonmalignant pathogenesis of NF1.
Three recent reports, including a controlled trial using WHO criteria for osteoporosis and osteopenia, have provided evidence that NF1 patients have a significantly higher incidence of osteoporosis and osteopenia (3–5). Bone homeostasis is maintained by balancing skeletal matrix formation and resorption. Osteoclasts are specialized cells derived from the myeloid monocyte/macrophage lineage that successively adhere to bone matrix and resorb bone, while osteoblasts generate new skeletal matrix. Imbalances in the morphogenesis and remodeling of bone are known to lead to pathological perturbations of skeletal structure and function (6). The role of neurofibromin in modulating these processes is incompletely understood.
Genetically engineered mice harboring mutations at the Nf1 locus have both haploinsufficient (Nf1+/–) (7, 8) and nullizygous loss of Nf1 (Nf1–/–) and exhibit myeloid cell abnormalities (9, 10). Specifically, NF1 patients develop neurofibromas, which are infiltrated with mast cells, and recent genetic studies in mice suggest that Nf1+/– mast cells play a role in initiating tumorigenesis in neurofibromas (1). Further, mice transplanted with Nf1–/– fetal liver hematopoietic stem cells uniformly develop a myeloid leukemia (10, 11) that is highly reminiscent of the juvenile myelomonocytic leukemia observed in children with NF1 (12). Recent studies in Nf1+/– mast cells (7, 8, 13) and in Nf1–/– myeloid progenitors (14) link increased activation of the p21ras/PI3K signaling pathway to multiple neurofibromin-deficient mast cell and myeloid progenitor phenotypes.
The hematopoietic restricted protein Src homology 2–containing inositol-5-phosphatase (SHIP) decreases PI3K signaling by dephosphorylating its major substrate phosphatidylinositol 3,4,5-triphosphate (PIP3). Strikingly, SHIP–/– mice are severely osteoporotic due to increased numbers of hyperresorptive osteoclasts (15). Similar to mice transplanted with Nf1–/– stem cells (10, 11), SHIP–/– mice acquire a profound myeloproliferative disease secondary to increased PI3K activity and hypersensitivity to multiple hematopoietic cell growth factors. Given previous observations in myeloid mast cells that loss of a single allele of Nf1 increases signaling through the p21ras/PI3K pathway and the similarities between the myeloid cell phenotypes in Nf1–/– and SHIP–/– mice, it is intriguing that haploinsufficiency of Nf1 in osteoclasts may have a role in the osteoporosis that has recently been observed at an increased frequency in patients with NF1 (3–5). Therefore, we tested whether murine Nf1+/– osteoclasts and in vitro–differentiated osteoclasts from NF1 patients have increased p21ras/PI3K activity and an associated alteration in osteoclast fates in vitro and in vivo.
Here we show that haploinsufficiency of Nf1 (NF1) leads to the increased survival, proliferation, migration, adhesion, and lytic activity of both murine and human osteoclasts. We also provide pharmacologic and genetic evidence that the pathologic increase in multiple osteoclast functions is mediated by hyperactivation of a Ras/PI3K signaling axis. Collectively, these data provide what we believe to be the first cellular and biochemical evidence for an NF1-haploinsufficient phenotype in osteoclasts and could have important implications for the osteoporosis and osteopenia that occur in NF1 patients.
-
Results
Nf1+/– mice have increased numbers of osteoclast progenitors and multinucleated osteoclasts in vivo. Since osteoclasts are tissue-specific monocyte/macrophage progeny, we quantitated the number of macrophage and osteoclast progenitors per femur in Nf1+/– and WT mice in vivo by establishing clonogenic assays that promote the growth of monocyte/macrophage (CFU-macrophage [CFU-M]) and osteoclast (tartrate-resistant acid phosphatase–positive [TRAP+] CFU-M) progenitors. Following culture in semisolid media, the number of osteoclast and macrophage progenitor colonies was determined. Osteoclast progenitors were identified with TRAP, which stains them dark red, while macrophage progenitors lacking TRAP activity appear yellow (Figure 1A). In 5 independent experiments, Nf1+/– mice had significantly higher numbers of macrophage progenitors (Figure 1B) and osteoclast progenitors (Figure 1C) per femur as compared with WT mice. Thus, haploinsufficiency of Nf1 results in an increase in progenitors that have the potential to mature into fully differentiated osteoclasts.
 Figure 1
Figure 1Nf1+/– mice have increased numbers of osteoclast progenitors and osteoclasts in vivo. Data in B, C, and E represent the mean ± SEM of 5 independent experiments. (A) Clonogenic assays were established to determine the number of CFU-Ms per femur. Representative TRAP– CFU-M (left panel) and TRAP+ CFU-M (right panel). (B) Total CFU-Ms per femur from Nf1+/– or WT mice. *P < 0.01, Nf1+/– CFU-Ms versus WT CFU-Ms by Student’s t test. (C) TRAP+ CFU-Ms per femur from mice of the indicated genotypes. *P < 0.01, Nf1+/– versus WT. (D) Representative photomicrographs (magnification, ×10) of WT and Nf1+/– distal femoral metaphyses following TRAP staining. Arrows indicate selected osteoclasts. (E) Average size of osteoclasts from Nf1+/– and WT mice. Ten high-power fields per experimental mouse were scored. **P < 0.05, Nf1+/– versus WT osteoclasts.
Given the increase in the number of osteoclast progenitors in Nf1+/– mice, we next determined the number of mature osteoclasts in Nf1+/– mice in vivo. The femurs of 7- to 8-week-old syngeneic Nf1+/– and WT mice were decalcified, and histological sections from the distal metaphysis were stained for the osteoclast enzyme TRAP. Strikingly, there was a marked increase in the number of multinucleated osteoclasts per unit of trabecular surface (Figure 1D) and in the size of individual osteoclasts among Nf1+/– mice (Figure 1E). These observations are reminiscent of the large multinucleated osteoclasts found in the SHIP–/– mice (15).
Nf1+/– osteoclasts have hyperactive Ras and Akt. The binding of M-CSF to its receptor (c-fms) causes a rapid increase in p21ras activity (16). Though heterozygosity of Nf1 alters p21ras activity in mast cells, it remains unclear whether heterozygous loss of Nf1 is sufficient to induce an increase in p21ras activity in other myeloid lineages. To investigate whether heterozygous loss of Nf1 alters p21ras activity in osteoclasts, Nf1+/– and WT BM mononuclear cells (BMMNCs) were cultured in M-CSF and receptor activator of NF-κB ligand (RANKL) for 7 days to enrich for a purified population of osteoclasts. Cells were then further purified by treatment with collagenase and dispase as previously described (17). Consistent with previous studies (17), 90% of the Nf1+/– and WT cultured cells were TRAP+ (data not shown). Following culture in the absence of growth factors for 4 hours, Nf1+/– and WT osteoclasts were stimulated with M-CSF and assayed for changes in p21ras-GTP levels. Levels of GTP-bound p21ras in cellular lysates were determined by precipitating the active GTPase with a GST-fusion of the p21ras binding domain of Raf-1 kinase in an effector pull-down assay. In 4 independent experiments, Nf1+/– osteoclasts had higher M-CSF–mediated p21ras-GTP levels compared with WT cells. Results of representative experiment are shown in Figure 2A (left panel). Overall, there was an approximately 2-fold increase in M-CSF–mediated Ras activation in Nf1+/– cells as compared with WT cells (Figure 2A, right panel).
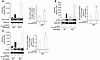 Figure 2
Figure 2Nf1+/– osteoclasts have elevated Ras and92 PI3K activity. (A) Ras activity in WT and Nf1+/– osteoclasts was measured at the indicated times following stimulation with M-SCF. A representative blot is shown (left panel). The relative increase in Ras activity in Nf1+/– osteoclasts compared with WT osteoclasts in 4 independent experiments is indicated (right panel). Data represent mean ± SEM. *P < 0.01, Ras activity of Nf1+/– osteoclasts versus WT osteoclasts. (B) Akt phosphorylation was measured at 0 and 2 minutes following stimulation with M-CSF. Results from 1 of 4 independent experiments are shown (left panel), and the relative increase (mean ± SEM; n = 4) in Akt phosphorylation in Nf1+/– osteoclasts compared with WT osteoclasts is indicated (right panel). **P < 0.01, Akt phosphorylation of Nf1+/– osteoclasts versus WT osteoclasts. (C) Erk phosphorylation was measured at the indicated levels following stimulation with M-CSF. A representative Western blot (left panel) is shown, and the relative mean increase (mean ± SEM; n = 4) in Erk phosphorylation in Nf1+/– osteoclasts is indicated (right panel). †P < 0.01, Nf1+/– osteoclasts versus WT osteoclasts.
Ras is at the apex of a series of signal transduction cascades that control a range of cellular functions in myeloid cells. Given the genetic evidence that activation of PI3K is important for bone resorption by osteoclasts (15), we compared Akt activity, a sensitive measure of PI3K activity, in Nf1+/– and WT cells. A 2- to 3-fold increase in M-CSF–stimulated Akt activity was observed in Nf1+/– osteoclasts (Figure 2B) compared with WT controls in 4 independent experiments. Erk activation is also required for osteoclastogenesis and is a known effector of p21ras (18). Erk phosphorylation was also elevated in Nf1+/– osteoclasts as compared with WT osteoclasts following M-CSF stimulation (Figure 2C). Collectively, these experiments demonstrate that haploinsufficiency of Nf1 alters Ras and 2 effector pathways that would be anticipated to modulate osteoclast formation and function.
Nf1+/– osteoclasts are hypersensitive to M-CSF and RANKL and have increased survival. Previous studies established that loss of neurofibromin increases the growth of Nf1–/– myeloid progenitors in response to low concentrations of multiple different growth factors (9). A possible mechanism for the increased number of macrophage and osteoclast progenitors in Nf1+/– mice is an increased sensitivity of these populations to low concentrations of either M-CSF or RANKL, 2 growth factors that are necessary and sufficient for osteoclast differentiation (6). We initially tested this hypothesis by culturing BMMNCs from Nf1+/– and WT mice in different concentrations of M-CSF and enumerated the number of CFU-Ms. Over a range of concentrations of M-CSF, increased numbers of macrophage progenitors were observed in Nf1+/– mice (Figure 3A). We next established liquid cultures of low-density BM cells from Nf1+/– and WT mice in varying concentrations of M-CSF (Figure 3B) and RANKL (Figure 3C). Over a wide range of concentrations of either M-CSF or RANKL, an increased number of osteoclasts was observed in Nf1+/– mice. Furthermore, fluorescence cytometric analysis of propidium iodide–stained (PI-stained) osteoclasts demonstrated that Nf1+/–osteoclasts have higher populations of multinucleated (≥4N) cells as compared with WT controls. A representative FACS image is shown in Figure 3D, and a representative photograph of the osteoclast culture is shown in Figure 3E.
 Figure 3
Figure 3Increase in the number of myeloid progenitors and TRAP+ cells in response to varying concentrations of M-CSF and RANKL. (A) Increase in the number of CFU-Ms per femur in response to M-CSF. Data represent the mean ± SEM of 5 experiments. *P < 0.01, Nf1+/– versus WT CFU-Ms by Student’s t test. (B and C) Evaluation of the ability of Nf1+/– and WT BMMNCs to form osteoclasts in response to M-CSF (B) and RANKL (C). Data represent the mean ± SEM of TRAP+ cells/5 × 104 LDMNCs from 5 experiments. #P < 0.01, Nf1+/– versus WT TRAP+ cells. (D) Osteoclast DNA content of cultured WT osteoclast (left panel) and Nf1+/– osteoclasts (right panel) was determined using fluorescence cytometry following PI staining. (E) Representative photomicrographs (magnification, ×20) of Nf1+/– and WT osteoclasts in liquid culture. Arrows indicate selected osteoclasts. (F) Osteoclast survival was determined by calculating annexin V– and PI-negative cell populations. ##P < 0.05 and **P < 0.01, Nf1+/– versus WT osteoclasts.
In addition to promoting growth at limiting concentrations of growth factors that may be particularly relevant at physiologic concentrations, Nf1–/– myeloid progenitors (14) and Nf1+/– mast cells (13) have increased survival associated with an increase in PI3K activity. To examine whether this phenotype is present in Nf1+/– osteoclasts, purified osteoclasts were cultured in M-CSF in the absence of serum, and apoptosis was determined sequentially over 24 hours using annexin V staining. Nf1+/– osteoclasts had decreased apoptosis as compared with WT osteoclasts, consistent with the increase in Akt phosphorylation in Nf1+/– osteoclasts (Figure 3F).
Nf1+/– osteoclasts have increased M-CSF–mediated migration, adhesion to αVβ3, and F-actin polymerization. Bone resorption by osteoclasts is linked to the migration and adherence of these cells to a local bone surface. To examine whether haploinsufficiency of Nf1 alters these cytoskeletal functions in osteoclasts, purified populations of TRAP+ cells were generated. Ten thousand TRAP+ cells from each genotype were placed in the upper chamber of a Transwell coated with vitronectin, and haptotaxis to M-CSF was determined. A representative photomicrograph of the migrating TRAP+ cells is shown in Figure 4A. In 5 independent experiments, a 2-fold increase in migration of the Nf1+/– osteoclasts was observed as compared with WT cells (Figure 4B).
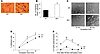 Figure 4
Figure 4Effect of heterozygosity of Nf1 on osteoclast haptotaxis and F-actin content in response to recombinant murine M-CSF. (A) Haptotaxis of WT and Nf1+/– osteoclasts in response to M-CSF. A representative photomicrograph (magnification, ×20) from 1 of 5 experiments. (B) Quantitative evaluation of migration in response to M-CSF. Results represent the mean ± SEM of 5 experiments. *P < 0.01, Nf1+/– versus WT by Student’s t test. (C) Photomicrographs (magnification, ×10) of M-CSF–stimulated osteoclast adhesion to αVβ3. Genotypes and length of adhesion are indicated. (D) Quantitative evaluation (mean ± SEM; n = 5) of osteoclast adhesion 10–60 minutes following incubation with M-CSF. Genotypes and length of adhesion are indicated. *P < 0.01, Nf1+/– versus WT osteoclasts by Student’s t test. (E) M-CSF–mediated F-actin polymerization. Osteoclasts were stimulated with 10 ng/ml M-CSF and fixed at the time points indicated. WT and Nf1+/– cells were examined in triplicate. Results are expressed as mean channel fluorescence (MCF). **P < 0.001, Nf1+/– osteoclasts versus WT osteoclasts by Student’s t test.
Since the initiation of bone resorption by osteoclasts is dependent on their ability to bind to the integrin αvβ3 on the bone surface (19–21), we next examined whether haploinsufficiency of Nf1 alters cell adhesion. Equivalent numbers of TRAP+ cells were cultured on vitronectin-coated plates, which express αvβ3, for 10–60 minutes in M-CSF, and the number of adherent TRAP+ cells was determined. In 4 independent experiments, we found that Nf1+/– osteoclasts had a significant increase in the number of cells adherent to vitronectin at all experimental time points following exposure to M-CSF (Figure 4, C and D). Furthermore, increased adherence to vitronectin was associated with an increase in F-actin content (Figure 4E).
Nf1+/– osteoclasts have increased capability to resorb bone in vitro and in vivo. Following adhesion, osteoclasts form a specialized cell–extracellular matrix to initiate degradation of bone matrix by secreting proteinases (6). This function is assessed in vitro by culturing osteoclasts on dentine slices and examining the number and area of “pits” that are resorbed. Ten thousand Nf1+/– and WT osteoclasts were cultured onto dentine slices for 24 hours in the presence of M-CSF and RANKL. Following the culture, the dentine slices were stained with toluidine blue, and the number of resorption pits was counted using light microscopy. We determined the area of all pits from 8–10 low-power fields per experiment from 4 independent studies. Representative fields showing bone resorption pits are shown in Figure 5A. The area of bone resorption on individual bone slices by Nf1+/– and WT osteoclasts is shown in Figure 5B, and the area of individual consecutively scored bone resorption pits of the 2 genotypes is shown in Figure 5C. Collectively, the data indicate that bone resorption by Nf1+/– osteoclasts is significantly greater than the resorption by WT osteoclasts.
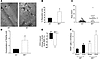 Figure 5
Figure 5Nf1+/– osteoclasts have increased bone resorption. (A) Osteoclasts were incubated on bone sections and stained with toluidine blue at the end of culture. The resorbed bone area stained dark blue. Representative photomicrographs (magnification, ×10) of the indicated genotypes are shown. (B) Quantitative evaluation of bone resorption. Results represent the mean area ± SEM of 5 independent experiments. *P < 0.01, WT versus Nf1+/– bone resorption. (C) The areas of individual bone resorption pits of the 2 genotypes are represented by individual symbols. The horizontal line represents the mean area of each genotype. *P < 0.01, WT versus Nf1+/–. (D) ELISA of serum TRAP5b activity. Results represent mean ± SEM (n = 6) using age- and sex-matched controls. Genotypes are indicated. #P < 0.05, Nf1+/– versus WT. (E) OVX-induced reduction (percent change compared with sham-operated animals) in BMD among adult female WT and Nf1+/– mice. Data represent the mean ± SEM (n = 7) of the indicated genotypes. *P < 0.01, WT versus Nf1+/– OVX mice by Student’s t test. (F) Osteoclast number per femur. Data represent the mean ± SEM (n = 7) of the indicated genotypes and treatment groups generated following ex vivo culture. ##P < 0.01, WT OVX versus WT sham; **P < 0.01, Nf1+/– sham versus WT sham; and †P < 0.01, Nf1+/– OVX versus Nf1+/– sham and Nf1+/– OVX versus WT OVX mice using ANOVA.
To examine whether Nf1+/–osteoclasts have increased activity in vivo, serum TRAP5b activity (22), an established measure of osteoclast lytic activity, was measured. The results for 6 Nf1+/– mice and age- and sex-matched controls indicate that serum from Nf1+/– mice has a significantly higher concentration of TRAP5b activity as compared with that from WT mice (Figure 5D). Similar data were obtained when urinary collagen crosslinks were measured (data not shown). Collectively, these functional data demonstrate that Nf1+/– mouse osteoclasts have an increase in lytic activity.
To assess whether Nf1 haploinsufficiency enhances osteoclast activity in response to a proresorptive challenge in vivo, 3-month-old Nf1+/– and WT mice underwent a bilateral ovariectomy (OVX) or sham surgery. OVX is an established experimental model of osteoporosis, a disease process that is becoming increasingly recognized in NF1 patients (3–5). Six weeks after the surgery, the mice were sacrificed, and both tibiae were dissected. Volumetric bone mineral density (BMD; mg/cm3) was measured in the proximal tibial metaphysis 1 mm distal to the proximal growth plate. As expected, the OVX procedure resulted in reduced BMD in both genotypes. However, the Nf1+/– OVX mice lost significantly more (by a factor of 2) bone mass than the WT OVX mice (Figure 5E), suggesting that a proresorptive challenge induces a greater osteoclastic response in Nf1-haploinsufficient mice. In an independent experiment, the number of osteoclasts generated following OVX or sham surgery was determined. As expected, Nf1+/– mice had increased numbers of osteoclast per femur following the sham surgery as compared with WT controls, and this increase was significantly elevated following OVX (Figure 5F), consistent with the more profound reduction in BMD found in Nf1+/– mice following OVX.
Introduction of the GTPase activating domains of NF1 restores normal Ras activity and in vitro functions. NF1 contains 60 exons and spans more than 250 kb in the genome. Though NF1 functions at least in part as a Ras-GAP, other functions have also been attributed to this complex gene (23, 24). To formally test the hypothesis that the gains of function observed in Nf1+/– osteoclasts are Ras dependent, we transduced Nf1+/– and WT osteoclast progenitors with either a retrovirus encoding the NF1 GAP-related domain (GRD) (MSCV-NF1 GRD-pac) or a control retrovirus (MSCV-pac) and performed functional and biochemical assays on the osteoclast progeny. We have shown previously that immunoprecipitated GRD peptides in transduced myeloid progenitors have GAP activity by stimulating phosphate release from GTP-loaded Ras (25). Interestingly, expressing the NF1 GRD in Nf1+/– osteoclasts restored migration to normal (Figure 6A). Given the specificity of the NF1 GRD in correcting the migration phenotype of Nf1+/– osteoclasts, we further tested whether this was due to a direct effect on Ras. We used a previously generated GAP-inactive mutant of NF1 GRD that harbors a mutation in the arginine finger loop (R1276P) (26). This mutation, which was identified in a family with a typical NF1 disease, greatly reduces neurofibromin GAP activity, and we have previously shown that the R1276P mutant recombinant protein has no catalytic activity in a GAP assay in myeloid progenitors (25). Expressing the R1276P mutant protein in Nf1+/– osteoclasts had no effect on migration (Figure 6A). Similar to the phenotype in migration, Nf1+/– osteoclasts transduced with the GRD sequences had a correction of bone resorption and adhesion comparable to that of WT osteoclasts, while Nf1+/– osteoclasts transduced with the R1276P mutation had no phenotypic correction comparable to that of WT osteoclasts (data not shown). To verify that the observed phenotypes were a consequence of differences in Ras activity, a Ras pull-down assay was performed. As expected, Nf1+/– osteoclasts transduced with the reporter construct had elevated levels of Ras-GTP compared with WT osteoclasts. Further, osteoclasts transduced with the GRD sequences corrected Ras activity to WT levels, while osteoclasts expressing the R1276P construct did not (Figure 6B). A similar pattern of correction was observed for Akt and Erk phosphorylation (data not shown). Collectively, these biochemical and functional data demonstrate that hyperactive Ras is necessary and sufficient for the gains of function observed in Nf1+/– osteoclasts.
 Figure 6
Figure 6Transduction of the GAP-related domains of NF1. (A) Osteoclasts were transduced with retroviral constructs expressing a selectable marker and the GRDs of NF1; the GRDs containing a point mutation that inactivates GAP activity (R1276P); or a construct encoding only the reporter transgene (Pac). M-CSF–mediated migration following antibiotic selection of transduced cells was measured. Results represent the mean ± SEM of 3 replicates from 1 of 4 independent experiments with similar results. *P < 0.01, Nf1+/– osteoclasts transduced with the Pac construct versus WT osteoclasts or Nf1+/– osteoclasts expressing the GRD or 1276 constructs. (B) Ras activity of osteoclasts at basal levels and 5 minutes following stimulation with M-CSF are shown. Genotypes and stimuli are indicated.
Genetic disruption of class 1A PI3K is sufficient to restore the gains of function in Nf1+/– osteoclasts. Given that the Nf1+/– phenotypes are consistent with hyperactivation of PI3K, we next tested whether genetic disruption of the p85α regulating subunit of class 1A PI3K would be sufficient to correct the Nf1-haploinsufficient phenotypes we observed. To conduct these studies, Nf1+/– mice were intercrossed with syngeneic mice that were heterozygous at the p85α allele, since p85α–/– mice die in utero. Fetal liver MNCs were isolated from F2 progeny that were mutant at the Nf1 locus (Nf1+/–p85α+/+), the p85α locus (Nf1+/+p85α–/–), or both loci (Nf1+/–p85α–/–) as described previously (8, 27), and their osteoclast formation upon stimulation with RANKL and serial concentrations of M-CSF was determined. Cells deficient only at the p85α locus formed significantly fewer numbers of osteoclasts as compared with WT cells (Figure 7, A and B). In addition, the increase in growth factor responsiveness of Nf1+/– MNCs to form osteoclasts was reduced to levels at or below those of WT osteoclasts in cells that were mutant at both loci (Figure 7A). The size of individual osteoclasts was also decreased in Nf1+/–p85α–/– osteoclasts as compared with osteoclasts mutant at the Nf1 locus only (Nf1+/–) (Figure 7B). Osteoclasts from F2 progeny were then isolated to evaluate the role of class 1A PI3K in modulating osteoclast migration, adhesion, and lytic activity. Osteoclasts mutant at both the Nf1 and p85α loci (Nf1+/–p85α–/–) had a significant reduction in migration (Figure 7C), adhesion (Figure 7D), pit formation (Figure 7E), and survival (Figure 7F) compared with Nf1+/– osteoclasts. The levels of migration, adhesion, and pit formation in p85α–/– osteoclasts were significantly lower than those in WT osteoclasts. Furthermore, osteoclasts that were mutant only at the p85α locus had profoundly reduced migration, adhesion, survival, and potential to form pits as compared with WT osteoclasts. To verify that genetic disruption of p85α reduces PI3K activity, Akt phosphorylation of purified osteoclast populations in response to M-CSF was examined (Figure 7G). As expected, Akt phosphorylation was significantly higher in the Nf1+/– osteoclasts as compared with WT osteoclasts. In addition, genetic disruption of p85α caused a reduction in Akt phosphorylation in cells that were WT or haploinsufficient at the Nf1 locus. Finally, osteoclasts that were mutant at both the Nf1+/– and the p85α locus had a significant reduction in Akt as compared with osteoclasts that were mutant at the p85α locus only. Collectively, these genetic data demonstrate a key role of PI3K in modulating elevated function in Nf1+/– osteoclasts with hyperactivated Ras activity and with normal RAS activity.
 Figure 7
Figure 7Genetic disruption of p85α restores M-CSF responsiveness of Nf1+/– osteoclasts. (A) MNCs from Nf1+/– and p85α–/– intercrossed fetal liver cells were stimulated with 0.1–50 ng/ml of M-CSF and 50 ng/ml RANKL. TRAP+ cells from 3 replicate wells/concentration were calculated. Results represent the mean ± SEM of 4 experiments. Statistical analyses were conducted using ANOVA. *P < 0.01, Nf1+/– osteoclasts versus WT osteoclasts; **P < 0.001, p85α–/– versus WT osteoclasts; #P < 0.01, Nf1+/– versus Nf1+/–p85α–/– osteoclasts. (B) Representative photographs (magnification, ×20) of osteoclasts from the 4 experimental groups. (C) M-CSF–mediated migration of Nf1+/– and p85α intercrossed osteoclasts. Results represent the mean ± SEM of 4 experiments. *P < 0.01, WT versus other experimental groups using ANOVA. **P < 0.001, p85α–/– versus WT osteoclasts; #P < 0.01, Nf1+/– versus Nf1+/– p85α–/– osteoclasts. (D) Evaluation of M-CSF–mediated adhesion. Results represent the mean ± SEM of 4 experiments. *P < 0.01, number of Nf1+/– osteoclasts versus WT osteoclasts; **P < 0.001, number of p85α–/– versus WT osteoclasts; #P < 0.01, number of Nf1+/– versus Nf1+/–p85α–/– osteoclasts. (E) Representative photomicrographs (magnification, ×10) of bone pits from the 4 F2 genotypes. (F) Osteoclast survival was evaluated by determining the annexin V–negative/PI-negative population. †P < 0.01, survival of Nf1+/– versus WT and Nf1+/– versus Nf1+/–p85α–/– cells; **P < 0.01 survival of p85α–/– versus WT osteoclasts using ANOVA. (G) Akt phosphorylation of the 4 F2 genotypes at basal levels and following M-CSF stimulation. Genotypes and length of stimulation are indicated.
The functional changes in Nf1+/– osteoclasts are conserved in human NF1 osteoclasts. To examine whether the alterations of osteoclast biological functions observed in murine Nf1+/– osteoclasts are conserved in NF1 patients, peripheral mononuclear cells (MNCs) were stimulated with M-CSF and RANKL to differentiate blood monocytes into osteoclasts as previously described (28). Neurofibromin levels in osteoclasts from NF1 patients were approximately 50% of the levels in control individuals. A representative Western blot from 2 patients and 2 unaffected controls is shown (Figure 8A). Though the total peripheral white blood cell counts of the 2 groups were similar (data not shown), the number and size of osteoclasts that were formed from MNCs of NF1 patients were significantly higher than those of unaffected control individuals (Figure 8B, left and right panels).
 Figure 8
Figure 8Osteoclasts generated from NF1 patient PBMCs and in response to M-CSF and have increased PI3K-dependent cytoskeletal functions. (A) Neurofibromin levels in unaffected controls and NF1 patients. (B) Number of osteoclasts formed from culture of 1 × 105 peripheral blood LDMNCs following M-SCF and RANKL stimulation for 14 days (left panel). Data represent the mean ± SEM for 5 individuals with NF1 and 5 unaffected sex- and age-matched controls. Representative photomicrographs (magnification, ×20) from the indicated genotypes (right panel) are shown. (C) Evaluation of M-CSF– and PI3K-mediated migration of osteoclasts from NF1 patients and controls. The genotypes and addition of M-CSF and the PI3K inhibitor LY294002 are indicated. Data represent the mean ± SEM of 4 experiments. *P < 0.01, osteoclasts from NF1 patients versus unaffected controls; **P < 0.01, number of osteoclasts that migrated following preincubation in 5 μM of LY294002 versus osteoclasts preincubated in vehicle. (D) Pit formation of osteoclasts. Osteoclasts from NF1 patients and unaffected controls were cultured on bone sections in the presence of M-CSF and LY294002 or the vehicle control for 14 days. (E) Akt phosphorylation in M-CSF–stimulated osteoclasts. The genotype, stimulus, inhibitor, and length of stimulation are indicated.
To determine whether osteoclasts from NF1 patients had increased M-CSF–directed migration as compared with control specimens and whether this cellular function was inhibited by PI3K inhibitors, osteoclasts were incubated in the presence of 5 μm LY294002, a concentration of the PI3K inhibitor that does not induce apoptosis, for 1 hour prior to initiation of the migration assay (Figure 8C). Osteoclasts from NF1 patients had a 2-fold increase in the migration of osteoclasts as compared with osteoclasts from unaffected controls. Preincubation of osteoclasts with a PI3K inhibitor inhibited migration in both cell types. Osteoclasts from NF1 patients and unaffected controls were then placed on dentine sections to evaluate lytic activity. The area of resorption of NF1 osteoclasts, similar to that of osteoclasts from Nf1+/– mice, was significantly larger than that of unaffected controls (Figure 8D).
Preincubation of osteoclasts generated from NF1 patients with LY294002-inhibited pit formation. To verify that PI3K activity was increased in osteoclasts from NF1 patients and that the concentration of LY294002 utilized in the cell biology assays was sufficient to reduce PI3K activity in these primary cells, osteoclasts from NF1 patients and unaffected controls were quiesced for 4 hours in 1% bovine serum albumin. Cells were then pretreated with 5 μm LY294002 or the vehicle for 1 hour and stimulated with M-CSF. Akt phosphorylation was detected at basal levels and 5 minutes following stimulation. A representative Western blot from 3 independent experiments is shown in Figure 8E. Akt phosphorylation was 3-fold higher in osteoclasts from NF1 patients as compared with unaffected control patients. Furthermore, Akt activity was inhibited by the concentration of LY294002 used in the cell biology assays. Collectively, these data indicate that osteoclasts from NF1 patients have an increase in multiple osteoclast functions, including migration, osteoclast formation, and bone resorption, that ultimately promote lytic activity. Furthermore, as we found in the genetic studies of mice intercrossed to contain disruptions in both Nf1 and p85α, these cellular functions are dependent on PI3K activation.
-
Discussion
NF1 is a common, pandemic, autosomal-dominant genetic disorder that affects 1 in 3,000 individuals. The detection of somatic mutations in the residual normal NF1 allele within the cancers of individuals with NF1 is consistent with NF1 functioning as a tumor-suppressor gene. However evidence in selected lineages (2, 29–31) now indicates that analogous to recent discoveries in p53 (32) and p27 (33), gene dosage effects of NF1 alter cell fates and functions. The most recent studies using Nf1+/– cells have focused on the role of NF1 haploinsufficiency in lineages of the tumor microenvironment of plexiform neurofibromas and optic gliomas (2). However, a phenotype in long-term learning in Nf1+/– mice, similar to a spatial-visual discoordination observed in NF1 patients, has been established (34). The high frequency of nonmalignant manifestations in NF1 patients, including cerebrovascular disease (35), learning deficits, and osseous abnormalities, such as osteoporosis, suggest the importance of NF1 haploinsufficiency in multiple lineages. Recognition of the cellular and biochemical underpinnings of these physical findings is important in identifying specific molecular therapies and in disease treatment and prevention.
Many skeletal diseases occur as a consequence of a skeletal imbalance that favors bone resorption (36, 37). In response to extracellular signals, macrophage progenitors differentiate into monocytes that evaginate from the marrow and circulate in the peripheral blood. Recent discoveries have demonstrated that M-CSF together with signals from a family of biologically related tumor necrosis factor receptor–like proteins (particularly RANKL and its decoy ligand osteoprotegerin) control the induction of osteoclast differentiation and are key to the physiologic and pathophysiologic responses of osteoclasts (38, 39). Common chronic diseases such as arthritis or menopause functionally lead to an increase in production of M-CSF and RANKL, leading to increased osteoclast activation and consequent bone loss (36, 37). Our results demonstrating that, at limiting concentrations of M-CSF and RANKL, murine (Nf1+/–) and human NF1 monocytes have an increased potential to mature into multinucleated osteoclasts and an increased ability to adhere to bone and form polykaryons that lead to increased bone lysis are consistent with previous findings relating to other pathological causes of osteoporosis and osteopenia. Data demonstrating an increased number of multinucleated osteoclasts in Nf1+/– mice, an increase in TRAP5b in the serum of Nf1+/– mice, and an increase in bone resorption following OVX provide in vivo support of this concept.
The increases in osteoclast numbers and lytic activity reported here in murine and human NF1 heterozygous cells is consistent with recent initial studies showing that NF1 patients have decreased bone density (3–5). Previous work by Yu et al. (40) found that Nf1+/– mice also had a trend toward reduced total bone as compared with WT controls, though the difference was not statistically significant. There are established species-specific differences in skeletal development and in the weight forces between quadripedal mice and bipedal humans that influence skeletal density (41). These species differences could be important when considering bone remodeling disease manifestations in preclinical models. Additionally, even in chronic diseases, osteoporosis frequently develops over the course of decades as opposed to 2–6 months, a typical duration of experiments using murine models. Future studies to evaluate measurements of bone remodeling in NF1 patients and epidemiologic studies to evaluate the risks of bone fractures in young and elderly adults with NF1 are also indicated.
Recent studies have emphasized the role of phospholipids in cytoskeletal reorganization, as well as cell survival and proliferation (42). PIP3 in particular is a phospholipid that is a known modulator of these cellular functions. Myeloid progenitors, macrophages, and osteoclasts from SHIP–/– mice are exquisitely hypersensitive to multiple growth factors, including M-CSF. Strains of mice with complementary mutations influencing osteoclast differentiation and function include op/op mice (43), which have an inactivating mutation in the M-CSF receptor, and Gab2–/– mice (44), which contain a disruption of the Grb-2–associated binder adaptor protein that renders them unable to mediate RANKL-induced activation of Akt, JNK, and NF-κB. Consequently, both op/op mice and Grb2–/– mice have profound osteopetrosis. Data in Nf1+/– mice provide evidence for the first time to our knowledge that support the concept that subtle regulations of Ras activity in osteoclasts lead to osteoclasts and osteoclast precursors being hypersensitive to growth factor signals that modulate osteoclast functions in vitro and in vivo.
The paradigm observed in Nf1+/– osteoclasts is similar to that in studies using Nf1+/– mast cells, where limiting concentrations of kit ligand led to activation of a receptor in the same growth factor family as M-CSF (c-kit receptor tyrosine kinase). Previous studies in mast cells that were WT or haploinsufficient at the Nf1 locus have positioned the small RhoGTPase Rac as a key downstream effector of class I A PI3K (8, 45). Preferential inhibition of Nf1+/– osteoclast migration by a low concentration of the PI3K inhibitor (5 μM, LY294002) as compared with a Mek inhibitor (50 μM, PD98059) (85% vs. 20% mean reduction) is consistent with these results. Further, in preliminary studies in osteoclasts, we have also identified Rac2 as a class 1A PI3K effector that modulates the migration, adhesion, and lytic activity of osteoclasts that are heterozygous or WT at the Nf1 locus (our unpublished observations). These data suggest that alterations of the Ras/PI3K/Rac signaling pathway may ultimately be critical in osteoclast development and function. The availability of conditional knockout mice will also allow an understanding of the role of Nf1+/– osteoclast function in other skeletal manifestations in the context of nullizygous loss of Nf1 in selected mesenchymal cell populations.
Though bisphosphonates have been the cornerstone of nonspecific agents utilized to treat osteoporosis since the 1960s, recent work has begun to focus on more specific targeted therapies (6). Currently available molecular targets to Ras itself, such as farnesyltransferase inhibitors, have been disappointing, since farnesylation is not sufficient to inhibit posttranslational modification of K-ras and N-ras, the 2 most prevalent isoforms in myeloid lineages. Data in osteoclasts in this genetic model provide genetic, cellular, and biochemical support for further evaluation of experimental compounds to inhibit class1A PI3K activity that are currently in development.
-
Methods
Animals. Nf1+/– mice were obtained from Tyler Jacks of the Massachusetts Institute of Technology (Cambridge, Massachusetts, USA) (46). p85α+/– mice were provided by Lewis Cantley (Harvard University, Boston, Massachusetts, USA). All studies were approved by the Indiana University Laboratory Animal Research Center.
Generation of murine osteoclasts. BMMNCs from Nf1+/– and WT mice were cultured in α-MEM supplemented with 10% FBS (Biomeda) in the presence of human recombinant RANKL (50 ng/ml; PeproTech) and murine recombinant M-CSF (30 ng/ml; PeproTech) for 7 days. The medium was changed every 3 days. Osteoclasts were placed on plastic dishes, washed with α-MEM, and treated with 0.1% collagenase and 0.2% dispase to remove stromal cells. To identify osteoclasts at the end of culture, adherent cells were fixed with 10% formaldehyde in PBS, treated with ethanol-acetone (50:50), and stained for TRAP as described previously (47) using a Nikon TE2000-S microscope (Nikon Inc.). Images were taken by a QImaging camera and QCapture Pro software. Multinucleated, TRAP+ cells containing more than 3 nuclei were scored as mature osteoclasts. For a subset of experiments, MNCs from fetal liver of WT, Nf1+/–, p85α–/–, and Nf1+/–p85α–/– embryos were cultured as described above.
Culture of myeloid progenitors. Osteoclast progenitors were examined in clonogenic assays as described previously (38). The colonies were fixed in a citrate/acetone/formaldehyde solution and histochemically stained for TRAP activity to detect osteoclast progenitors.
Osteoclast adhesion assay. Osteoclast precursors (1 × 105 cells/ml) were placed into 24-well plates coated with 20 μg/ml vitronectin (Takara Bio Inc.) supplemented with M-CSF (30 ng/ml) as described previously (48).
Osteoclast migration assay. Migration of osteoclasts was evaluated using a Transwell assay as described previously, with minor modifications (48). The cells were lifted from the plates by adding 0.05% trypsin and 0.2% EDTA 4Na in HBSS. MNCs previously cultured in M-CSF and RANKL for 6 days were examined to identify TRAP+ cells. Equivalent numbers of TRAP+ cells/well were loaded onto the upper chamber and allowed to migrate through an 8-μm polycarbonate filter coated with vitronectin for 15 hours in a humidified incubator at 37°C to M-CSF (30 ng/ml), 0.1% BSA in α-MEM placed in the bottom chamber. TRAP+ cells per field were then counted using a Nikon TE 2000-S microscope (Nikon Inc.).
F-actin polymerization. Actin polymerization was measured as described previously (48). Osteoclasts were stimulated with 30 ng/ml of M-CSF following deprivation of serum and growth factors for 3 hours. Cells were fixed and permeabilized prior to staining by phalloidin-FITC solution (Sigma-Aldrich), then resuspended in PBS/0.1% BSA for fluorescence cytometric analysis.
Bone resorption assay. Bone resorption was examined as described previously, with minor modifications (28). Single-cell suspensions of purified osteoclasts were seeded onto dentine slices (15) (ALPCO Diagnostic), incubated at 37°C, 5% CO2, in the presence of M-CSF and RANKL. Following culture, the slices were rinsed with PBS, left overnight in 1 M ammonium hydroxide, and then stained with 1% toluidine blue in 0.5% sodium tetraborate solution. For human pit assays, MNCs were cultured on dentine slices for 14 days and then processed with 1% toluidine blue in 0.5% sodium tetraborate solution. The number of resorptive areas, or “pits,” per low-power field on each bone slice was counted using reflective light microscopy. The area (mm2) of each pit was evaluated by measuring width and length using QCapture Pro (version 5.1) by an investigator who was blinded to the experimental groups.
TRAP assay. Bone resorptive activity was measured using a solid-phase immunofixed enzyme activity assay according to the manufacturer’s instructions (SBA Sciences).
OVX studies. Twelve-week-old WT and Nf1+/– female mice were subjected to OVX or sham surgery. Six weeks following surgery, mice were sacrificed, and both tibiae were dissected. Each tibia was placed in a plastic tube filled with 70% ethanol and centered in the gantry of a Norland Stratec XCT Research SA+ pQCT (Stratec Medizintechnik). Two cross-sectional slices were scanned on each proximal tibial metaphysis, at a point 1 mm distal to the proximal growth plate, using 0.46-mm collimation (4 × 105 counts/s) and 0.08-mm voxel size. For each slice, the x-ray source was rotated through 180° of projection (1 block). Total volumetric BMD was collected from each of the 2 slices and averaged over each bone.
Osteoclast ploidy and apoptosis analysis. BMMNCs were cultured in the presence of M-CSF and RANKL for 7 days. Osteoclasts were then lifted from the plates by adding trypsin-EDTA and resuspended in PBS containing 0.1% Triton-X 100 and 0.5% RNase (ribonuclease A). Cellular DNA was labeled with 500 ng/ml PI (Calbiochem). The ploidy in the stained cells was analyzed on a FACScan flow cytometer (BD). Ten thousand events were acquired and analyzed. For apoptosis analysis, serum- and cytokine-depleted osteoclasts were collected and resuspended in binding buffer (10 mM HEPES/NaOH [pH 7.4], 140 mM NaCl, 2.5 mM CaCl2), containing annexin V–FITC (Pharmingen) and PI and analyzed by FACS.
Evaluation of biochemical activities. Osteoclasts were deprived of serum and growth factors for 4 hours and stimulated with 30 ng/ml M-CSF. Ras activation was determined using a pull-down assay based on the Ras binding domain of Raf1 (Upstate). Akt and Erk phosphorylation were determined by Western blotting using phosphospecific antibodies for Akt and Erk as described previously (7, 29, 48).
Generation of osteoclasts from human NF1 patients. Monocytes from age- and sex-matched NF1 patients and healthy adults were isolated from the peripheral blood following Ficoll-Hypaque separation and differentiated into osteoclasts as described previously (49).
Statistics. Two-tailed Student’s t test was used to evaluate statistical difference. P values less than 0.05 were considered significant.
-
Acknowledgments
We thank Natascha Karlova for administrative support and Linda Di Migglio for reading the manuscript. The authors also thank M. Yu (Department of Medicine, Indiana University School of Medicine) for statistical support. This work was supported by NIH grant RO1 CA74177-06 (to D.W. Clapp) and Department of Defense grants NF043032 (to F.-C. Yang) and NF043019 (to D.A. Ingram).
Address correspondence to: D. Wade Clapp or Feng-Chun Yang, Indiana University School of Medicine, Herman B. Wells Center for Pediatric Research, Cancer Research Institute, 1044 West Walnut Street, R4 402, Indianapolis, Indiana 46202, USA. Phone: (317) 278-9290; Fax: (317) 274-8679; E-mail: [email protected] (D.W. Clapp) or [email protected] (F.-C. Yang).
-
Footnotes
Nonstandard abbreviations used: BMD, bone mineral density; BMMNC, BM mononuclear cell; CFU-M, CFU-macrophage; GAP, GTPase-activating protein; GRD, GAP-related domain; MNC, mononuclear cell; NF1, neurofibromatosis type 1; OVX, ovariectomy; PI, propidium iodide; RANKL, receptor activator of NF-κB ligand; SHIP, Src homology 2–containing inositol-5-phosphatase; TRAP, tartrate-resistant acid phosphatase.
Conflict of interest: The authors have declared that no conflict of interest exists.
Reference information: J. Clin. Invest.116:2880–2891 (2006). doi:10.1172/JCI29092
Feng-Chun Yang and Shi Chen contributed equally to this work.
-
References
- Zhu, Y., Ghosh, P., Charnay, P., Burns, D.K., Parada, L.F. 2002. Neurofibromas in NF1: Schwann cell origin and role of tumor environment. Science. 296:920-922.
- Bajenaru, M.L., et al. 2003. Optic nerve glioma in mice requires astrocyte Nf1 gene inactivation and Nf1 brain heterozygosity. Cancer Res. 63:8573-8577. View this article via: PubMed Google Scholar
- Kuorilehto, T., et al. 2005. Decreased bone mineral density and content in neurofibromatosis type 1: lowest local values are located in the load-carrying parts of the body. Osteoporos. Int. 16:928-936.
- Lammert, M., et al. 2005. Decreased bone mineral density in patients with neurofibromatosis 1. Osteoporos. Int. 16:1161-1166.
- Illes, T., Halmai, V., de Jonge, T., Dubousset, J. 2001. Decreased bone mineral density in neurofibromatosis-1 patients with spinal deformities. Osteoporos. Int. 12:823-827.
- Boyle, W.J., Simonet, W.S., Lacey, D.L. 2003. Osteoclast differentiation and activation. Nature. 423:337-342.
- Ingram, D.A., et al. 2001. Hyperactivation of p21(ras) and the hematopoietic-specific Rho GTPase, Rac2, cooperate to alter the proliferation of neurofibromin-deficient mast cells in vivo and in vitro. J. Exp. Med. 194:57-69.
- Yang, F.C., et al. 2003. Neurofibromin-deficient Schwann cells secrete a potent migratory stimulus for Nf1+/– mast cells. J. Clin. Invest. 112:1851-1861.
- Zhang, Y.Y., et al. 1998. Nf1 regulates hematopoietic progenitor cell growth and ras signaling in response to multiple cytokines. J. Exp. Med. 187:1893-1902.
- Largaespada, D.A., Brannan, C.I., Jenkins, N.A., Copeland, N.G. 1996. Nf1 deficiency causes Ras-mediated granulocyte/macrophage colony stimulating factor hypersensitivity and chronic myeloid leukaemia. Nat. Genet. 12:137-143.
- Zhang, Y., Taylor, B.R., Shannon, K., Clapp, D.W. 2001. Quantitative effects of Nf1 inactivation on in vivo hematopoiesis. J. Clin. Invest. 108:709-715.
- Shannon, K.M., et al. 1994. Loss of the normal NF1 allele from the bone marrow of children with type 1 neurofibromatosis and malignant myeloid disorders. N. Engl. J. Med. 330:597-601.
- Hiatt, K., et al. 2004. Loss of the nf1 tumor suppressor gene decreases fas antigen expression in myeloid cells. Am. J. Pathol. 164:1471-1479. View this article via: PubMed Google Scholar
- Donovan, S., See, W., Bonifas, J., Stokoe, D., Shannon, K.M. 2002. Hyperactivation of protein kinase B and ERK have discrete effects on survival, proliferation, and cytokine expression in Nf1-deficient myeloid cells. Cancer Cell. 2:507-514.
- Takeshita, S., et al. 2002. SHIP-deficient mice are severely osteoporotic due to increased numbers of hyper-resorptive osteoclasts. Nat. Med. 8:943-949.
- Guidez, F., Li, A.C., Horvai, A., Welch, J.S., Glass, C.K. 1998. Differential utilization of Ras signaling pathways by macrophage colony-stimulating factor (CSF) and granulocyte-macrophage CSF receptors during macrophage differentiation. Mol. Cell. Biol. 18:3851-3861. View this article via: PubMed Google Scholar
- Miyazaki, T., et al. 2000. Reciprocal role of ERK and NF-kappaB pathways in survival and activation of osteoclasts. J. Cell Biol. 148:333-342.
- Lee, S.E., et al. 2002. The phosphatidylinositol 3-kinase, p38, and extracellular signal-regulated kinase pathways are involved in osteoclast differentiation. Bone. 30:71-77. View this article via: PubMed Google Scholar
- Aubin, J.E. 1992. Osteoclast adhesion and resorption: the role of podosomes. J. Bone Miner. Res. 7:365-368. View this article via: PubMed Google Scholar
- McHugh, K.P., et al. 2000. Mice lacking beta3 integrins are osteosclerotic because of dysfunctional osteoclasts. J. Clin. Invest. 105:433-440.
- Sanjay, A., et al. 2001. Cbl associates with Pyk2 and Src to regulate Src kinase activity, alpha(v)beta(3) integrin-mediated signaling, cell adhesion, and osteoclast motility. J. Cell Biol. 152:181-195.
- Janckila, A.J., Takahashi, K., Sun, S.Z., Yam, L.T. 2001. Naphthol-ASBI phosphate as a preferred substrate for tartrate-resistant acid phosphatase isoform 5b. J. Bone Miner. Res. 16:788-793.
- Woods, S.A., et al. 2002. Aberrant G protein signaling in nervous system tumors. J. Neurosurg. 97:627-642. View this article via: PubMed Google Scholar
- Kim, H.A., Ratner, N., Roberts, T.M., Stiles, C.D. 2001. Schwann cell proliferative responses to cAMP and Nf1 are mediated by cyclin D1. J. Neurosci. 21:1110-1116. View this article via: PubMed Google Scholar
- Hiatt, K.K., Ingram, D.A., Zhang, Y., Bollag, G., Clapp, D.W. 2001. Neurofibromin GTPase-activating protein-related domains restore normal growth in Nf1–/– cells. J. Biol. Chem. 276:7240-7245.
- Klose, A., et al. 1998. Selective disactivation of neurofibromin GAP activity in neurofibromatosis type 1. Hum. Mol. Genet. 7:1261-1268.
- Lu-Kuo, J.M., Fruman, D.A., Joyal, D.M., Cantley, L.C., Katz, H.R. 2000. Impaired kit- but not FcepsilonRI-initiated mast cell activation in the absence of phosphoinositide 3-kinase p85alpha gene products. J. Biol. Chem. 275:6022-6029.
- Shinoda, K., et al. 2003. Resting T cells negatively regulate osteoclast generation from peripheral blood monocytes. Bone. 33:711-720.
- Ingram, D.A., et al. 2000. Genetic and biochemical evidence that haploinsufficiency of the Nf1 tumor suppressor gene modulates melanocyte and mast cell fates in vivo. J. Exp. Med. 191:181-188.
- Johannessen, C.M., et al. 2005. The NF1 tumor suppressor critically regulates TSC2 and mTOR. Proc. Natl. Acad. Sci. U. S. A. 102:8573-8578.
- Atit, R.P., Mitchell, K., Nguyen, L., Warshawsky, D., Ratner, N. 2000. The neurofibromatosis type 1 (Nf1) tumor suppressor is a modifier of carcinogen-induced pigmentation and papilloma formation in C57BL/6 mice. J. Invest. Dermatol. 114:1093-1100.
- Venkatachalam, S., et al. 1998. Retention of wild-type p53 in tumors from p53 heterozygous mice: reduction of p53 dosage can promote cancer formation. EMBO J. 17:4657-4667.
- Fero, M.L., Randel, E., Gurley, K.E., Roberts, J.M., Kemp, C.J. 1998. The murine gene p27Kip1 is haplo-insufficient for tumour suppression. Nature. 396:177-180.
- Costa, R.M., et al. 2001. Learning deficits, but normal development and tumor predisposition, in mice lacking exon 23a of Nf1. Nat. Genet. 27:399-405.
- Rosser, T.L., Vezina, G., Packer, R.J. 2005. Cerebrovascular abnormalities in a population of children with neurofibromatosis type 1. Neurology. 64:553-555. View this article via: PubMed Google Scholar
- Rodan, G.A., Martin, T.J. 2000. Therapeutic approaches to bone diseases. Science. 289:1508-1514.
- Raisz, L.G. 2005. Pathogenesis of osteoporosis: concepts, conflicts, and prospects. J. Clin. Invest. 115:3318-3325.
- Lacey, D.L., et al. 1998. Osteoprotegerin ligand is a cytokine that regulates osteoclast differentiation and activation. Cell. 93:165-176.
- Yasuda, H., et al. 1998. Osteoclast differentiation factor is a ligand for osteoprotegerin/osteoclastogenesis-inhibitory factor and is identical to TRANCE/RANKL. Proc. Natl. Acad. Sci. U. S. A. 95:3597-3602.
- Yu, X., et al. 2005. Neurofibromin and its inactivation of Ras are prerequisites for osteoblast functioning. Bone. 36:793-802.
- Jiang, H., Moreau, M., Raso, V.J., Russell, G., Bagnall, K. 1995. A comparison of spinal ligaments — differences between bipeds and quadrupeds. J. Anat. 187:85-91. View this article via: PubMed Google Scholar
- Brazil, D.P., Yang, Z.Z., Hemmings, B.A. 2004. Advances in protein kinase B signalling: AKTion on multiple fronts. Trends Biochem. Sci. 29:233-242.
- Kodama, H., Nose, M., Niida, S., Yamasaki, A. 1991. Essential role of macrophage colony-stimulating factor in the osteoclast differentiation supported by stromal cells. J. Exp. Med. 173:1291-1294.
- Wada, T., et al. 2005. The molecular scaffold Gab2 is a crucial component of RANK signaling and osteoclastogenesis. Nat. Med. 11:394-399.
- Munugalavadla, V., Borneo, J., Ingram, D.A., Kapur, R. 2005. p85alpha subunit of class IA PI-3 kinase is crucial for macrophage growth and migration. Blood. 106:103-109.
- Jacks, T., et al. 1994. Tumour predisposition in mice heterozygous for a targeted mutation in Nf1. Nat. Genet. 7:353-361.
- Suda, T., Jimi, E., Nakamura, I., Takahashi, N. 1997. Role of 1 alpha,25-dihydroxyvitamin D3 in osteoclast differentiation and function. Methods Enzymol. 282:223-235.
- Yang, F.C., et al. 2000. Rac2 stimulates Akt activation affecting BAD/Bcl-XL expression while mediating survival and actin function in primary mast cells. Immunity. 12:557-568.
- Fujikawa, Y., et al. 2001. The effect of macrophage-colony stimulating factor and other humoral factors (interleukin-1, -3, -6, and -11, tumor necrosis factor-alpha, and granulocyte macrophage-colony stimulating factor) on human osteoclast formation from circulating cells. Bone. 28:261-267.
-
Version history
- Version 1 (November 1, 2006): No description



Copyright © 2024 American Society for Clinical Investigation
ISSN: 0021-9738 (print), 1558-8238 (online)