Advertisement
Research ArticleImmunologyOncology Free access | 10.1172/JCI140837
MAL2 drives immune evasion in breast cancer by suppressing tumor antigen presentation
Yuanzhang Fang,1 Lifei Wang,1 Changlin Wan,1 Yifan Sun,1 Kevin Van der Jeught,1 Zhuolong Zhou,1 Tianhan Dong,2 Ka Man So,1 Tao Yu,1 Yujing Li,1 Haniyeh Eyvani,1 Austyn B. Colter,3 Edward Dong,1 Sha Cao,4 Jin Wang,5 Bryan P. Schneider,1,6,7 George E. Sandusky,3 Yunlong Liu,1,8 Chi Zhang,1,8 Xiongbin Lu,1,6,8 and Xinna Zhang1,6
1Department of Medical and Molecular Genetics,
2Department of Pharmacology and Toxicology,
3Department of Pathology and Laboratory Medicine, and
4Department of Biostatistics, Indiana University, School of Medicine, Indianapolis, Indiana, USA.
5Department of Pharmacology and Chemical Biology, Baylor College of Medicine, Houston, Texas, USA.
6Melvin and Bren Simon Cancer Center,
7Division of Hematology/Oncology, Department of Medicine, and
8Center for Computational Biology and Bioinformatics, Indiana University School of Medicine, Indianapolis, Indiana, USA.
Address correspondence to: Chi Zhang, Indiana University School of Medicine, 410 W. 10th Street, HS 5000, Indianapolis, Indiana 46202, USA. Phone: 317.278.9625; Email: [email protected]. Or to: Xiongbin Lu, Indiana University School of Medicine, 980 W. Walnut Street, R3.C218D, Indianapolis, Indiana 46202, USA. Phone: 317.274.4398; Email: [email protected]. Or to: Xinna Zhang, Indiana University School of Medicine, 980 W. Walnut Street, R3.C216B, Indianapolis, Indiana 46202, USA. Phone: 317.274.7931; Email: [email protected].
Authorship note: YF, LW, and CW contributed equally to this work.
Find articles by
Fang, Y.
in:
JCI
|
PubMed
|
Google Scholar
|

1Department of Medical and Molecular Genetics,
2Department of Pharmacology and Toxicology,
3Department of Pathology and Laboratory Medicine, and
4Department of Biostatistics, Indiana University, School of Medicine, Indianapolis, Indiana, USA.
5Department of Pharmacology and Chemical Biology, Baylor College of Medicine, Houston, Texas, USA.
6Melvin and Bren Simon Cancer Center,
7Division of Hematology/Oncology, Department of Medicine, and
8Center for Computational Biology and Bioinformatics, Indiana University School of Medicine, Indianapolis, Indiana, USA.
Address correspondence to: Chi Zhang, Indiana University School of Medicine, 410 W. 10th Street, HS 5000, Indianapolis, Indiana 46202, USA. Phone: 317.278.9625; Email: [email protected]. Or to: Xiongbin Lu, Indiana University School of Medicine, 980 W. Walnut Street, R3.C218D, Indianapolis, Indiana 46202, USA. Phone: 317.274.4398; Email: [email protected]. Or to: Xinna Zhang, Indiana University School of Medicine, 980 W. Walnut Street, R3.C216B, Indianapolis, Indiana 46202, USA. Phone: 317.274.7931; Email: [email protected].
Authorship note: YF, LW, and CW contributed equally to this work.
Find articles by Wang, L. in: JCI | PubMed | Google Scholar
1Department of Medical and Molecular Genetics,
2Department of Pharmacology and Toxicology,
3Department of Pathology and Laboratory Medicine, and
4Department of Biostatistics, Indiana University, School of Medicine, Indianapolis, Indiana, USA.
5Department of Pharmacology and Chemical Biology, Baylor College of Medicine, Houston, Texas, USA.
6Melvin and Bren Simon Cancer Center,
7Division of Hematology/Oncology, Department of Medicine, and
8Center for Computational Biology and Bioinformatics, Indiana University School of Medicine, Indianapolis, Indiana, USA.
Address correspondence to: Chi Zhang, Indiana University School of Medicine, 410 W. 10th Street, HS 5000, Indianapolis, Indiana 46202, USA. Phone: 317.278.9625; Email: [email protected]. Or to: Xiongbin Lu, Indiana University School of Medicine, 980 W. Walnut Street, R3.C218D, Indianapolis, Indiana 46202, USA. Phone: 317.274.4398; Email: [email protected]. Or to: Xinna Zhang, Indiana University School of Medicine, 980 W. Walnut Street, R3.C216B, Indianapolis, Indiana 46202, USA. Phone: 317.274.7931; Email: [email protected].
Authorship note: YF, LW, and CW contributed equally to this work.
Find articles by Wan, C. in: JCI | PubMed | Google Scholar
1Department of Medical and Molecular Genetics,
2Department of Pharmacology and Toxicology,
3Department of Pathology and Laboratory Medicine, and
4Department of Biostatistics, Indiana University, School of Medicine, Indianapolis, Indiana, USA.
5Department of Pharmacology and Chemical Biology, Baylor College of Medicine, Houston, Texas, USA.
6Melvin and Bren Simon Cancer Center,
7Division of Hematology/Oncology, Department of Medicine, and
8Center for Computational Biology and Bioinformatics, Indiana University School of Medicine, Indianapolis, Indiana, USA.
Address correspondence to: Chi Zhang, Indiana University School of Medicine, 410 W. 10th Street, HS 5000, Indianapolis, Indiana 46202, USA. Phone: 317.278.9625; Email: [email protected]. Or to: Xiongbin Lu, Indiana University School of Medicine, 980 W. Walnut Street, R3.C218D, Indianapolis, Indiana 46202, USA. Phone: 317.274.4398; Email: [email protected]. Or to: Xinna Zhang, Indiana University School of Medicine, 980 W. Walnut Street, R3.C216B, Indianapolis, Indiana 46202, USA. Phone: 317.274.7931; Email: [email protected].
Authorship note: YF, LW, and CW contributed equally to this work.
Find articles by
Sun, Y.
in:
JCI
|
PubMed
|
Google Scholar
|

1Department of Medical and Molecular Genetics,
2Department of Pharmacology and Toxicology,
3Department of Pathology and Laboratory Medicine, and
4Department of Biostatistics, Indiana University, School of Medicine, Indianapolis, Indiana, USA.
5Department of Pharmacology and Chemical Biology, Baylor College of Medicine, Houston, Texas, USA.
6Melvin and Bren Simon Cancer Center,
7Division of Hematology/Oncology, Department of Medicine, and
8Center for Computational Biology and Bioinformatics, Indiana University School of Medicine, Indianapolis, Indiana, USA.
Address correspondence to: Chi Zhang, Indiana University School of Medicine, 410 W. 10th Street, HS 5000, Indianapolis, Indiana 46202, USA. Phone: 317.278.9625; Email: [email protected]. Or to: Xiongbin Lu, Indiana University School of Medicine, 980 W. Walnut Street, R3.C218D, Indianapolis, Indiana 46202, USA. Phone: 317.274.4398; Email: [email protected]. Or to: Xinna Zhang, Indiana University School of Medicine, 980 W. Walnut Street, R3.C216B, Indianapolis, Indiana 46202, USA. Phone: 317.274.7931; Email: [email protected].
Authorship note: YF, LW, and CW contributed equally to this work.
Find articles by
Van der Jeught, K.
in:
JCI
|
PubMed
|
Google Scholar
|

1Department of Medical and Molecular Genetics,
2Department of Pharmacology and Toxicology,
3Department of Pathology and Laboratory Medicine, and
4Department of Biostatistics, Indiana University, School of Medicine, Indianapolis, Indiana, USA.
5Department of Pharmacology and Chemical Biology, Baylor College of Medicine, Houston, Texas, USA.
6Melvin and Bren Simon Cancer Center,
7Division of Hematology/Oncology, Department of Medicine, and
8Center for Computational Biology and Bioinformatics, Indiana University School of Medicine, Indianapolis, Indiana, USA.
Address correspondence to: Chi Zhang, Indiana University School of Medicine, 410 W. 10th Street, HS 5000, Indianapolis, Indiana 46202, USA. Phone: 317.278.9625; Email: [email protected]. Or to: Xiongbin Lu, Indiana University School of Medicine, 980 W. Walnut Street, R3.C218D, Indianapolis, Indiana 46202, USA. Phone: 317.274.4398; Email: [email protected]. Or to: Xinna Zhang, Indiana University School of Medicine, 980 W. Walnut Street, R3.C216B, Indianapolis, Indiana 46202, USA. Phone: 317.274.7931; Email: [email protected].
Authorship note: YF, LW, and CW contributed equally to this work.
Find articles by Zhou, Z. in: JCI | PubMed | Google Scholar
1Department of Medical and Molecular Genetics,
2Department of Pharmacology and Toxicology,
3Department of Pathology and Laboratory Medicine, and
4Department of Biostatistics, Indiana University, School of Medicine, Indianapolis, Indiana, USA.
5Department of Pharmacology and Chemical Biology, Baylor College of Medicine, Houston, Texas, USA.
6Melvin and Bren Simon Cancer Center,
7Division of Hematology/Oncology, Department of Medicine, and
8Center for Computational Biology and Bioinformatics, Indiana University School of Medicine, Indianapolis, Indiana, USA.
Address correspondence to: Chi Zhang, Indiana University School of Medicine, 410 W. 10th Street, HS 5000, Indianapolis, Indiana 46202, USA. Phone: 317.278.9625; Email: [email protected]. Or to: Xiongbin Lu, Indiana University School of Medicine, 980 W. Walnut Street, R3.C218D, Indianapolis, Indiana 46202, USA. Phone: 317.274.4398; Email: [email protected]. Or to: Xinna Zhang, Indiana University School of Medicine, 980 W. Walnut Street, R3.C216B, Indianapolis, Indiana 46202, USA. Phone: 317.274.7931; Email: [email protected].
Authorship note: YF, LW, and CW contributed equally to this work.
Find articles by Dong, T. in: JCI | PubMed | Google Scholar
1Department of Medical and Molecular Genetics,
2Department of Pharmacology and Toxicology,
3Department of Pathology and Laboratory Medicine, and
4Department of Biostatistics, Indiana University, School of Medicine, Indianapolis, Indiana, USA.
5Department of Pharmacology and Chemical Biology, Baylor College of Medicine, Houston, Texas, USA.
6Melvin and Bren Simon Cancer Center,
7Division of Hematology/Oncology, Department of Medicine, and
8Center for Computational Biology and Bioinformatics, Indiana University School of Medicine, Indianapolis, Indiana, USA.
Address correspondence to: Chi Zhang, Indiana University School of Medicine, 410 W. 10th Street, HS 5000, Indianapolis, Indiana 46202, USA. Phone: 317.278.9625; Email: [email protected]. Or to: Xiongbin Lu, Indiana University School of Medicine, 980 W. Walnut Street, R3.C218D, Indianapolis, Indiana 46202, USA. Phone: 317.274.4398; Email: [email protected]. Or to: Xinna Zhang, Indiana University School of Medicine, 980 W. Walnut Street, R3.C216B, Indianapolis, Indiana 46202, USA. Phone: 317.274.7931; Email: [email protected].
Authorship note: YF, LW, and CW contributed equally to this work.
Find articles by So, K. in: JCI | PubMed | Google Scholar
1Department of Medical and Molecular Genetics,
2Department of Pharmacology and Toxicology,
3Department of Pathology and Laboratory Medicine, and
4Department of Biostatistics, Indiana University, School of Medicine, Indianapolis, Indiana, USA.
5Department of Pharmacology and Chemical Biology, Baylor College of Medicine, Houston, Texas, USA.
6Melvin and Bren Simon Cancer Center,
7Division of Hematology/Oncology, Department of Medicine, and
8Center for Computational Biology and Bioinformatics, Indiana University School of Medicine, Indianapolis, Indiana, USA.
Address correspondence to: Chi Zhang, Indiana University School of Medicine, 410 W. 10th Street, HS 5000, Indianapolis, Indiana 46202, USA. Phone: 317.278.9625; Email: [email protected]. Or to: Xiongbin Lu, Indiana University School of Medicine, 980 W. Walnut Street, R3.C218D, Indianapolis, Indiana 46202, USA. Phone: 317.274.4398; Email: [email protected]. Or to: Xinna Zhang, Indiana University School of Medicine, 980 W. Walnut Street, R3.C216B, Indianapolis, Indiana 46202, USA. Phone: 317.274.7931; Email: [email protected].
Authorship note: YF, LW, and CW contributed equally to this work.
Find articles by Yu, T. in: JCI | PubMed | Google Scholar
1Department of Medical and Molecular Genetics,
2Department of Pharmacology and Toxicology,
3Department of Pathology and Laboratory Medicine, and
4Department of Biostatistics, Indiana University, School of Medicine, Indianapolis, Indiana, USA.
5Department of Pharmacology and Chemical Biology, Baylor College of Medicine, Houston, Texas, USA.
6Melvin and Bren Simon Cancer Center,
7Division of Hematology/Oncology, Department of Medicine, and
8Center for Computational Biology and Bioinformatics, Indiana University School of Medicine, Indianapolis, Indiana, USA.
Address correspondence to: Chi Zhang, Indiana University School of Medicine, 410 W. 10th Street, HS 5000, Indianapolis, Indiana 46202, USA. Phone: 317.278.9625; Email: [email protected]. Or to: Xiongbin Lu, Indiana University School of Medicine, 980 W. Walnut Street, R3.C218D, Indianapolis, Indiana 46202, USA. Phone: 317.274.4398; Email: [email protected]. Or to: Xinna Zhang, Indiana University School of Medicine, 980 W. Walnut Street, R3.C216B, Indianapolis, Indiana 46202, USA. Phone: 317.274.7931; Email: [email protected].
Authorship note: YF, LW, and CW contributed equally to this work.
Find articles by Li, Y. in: JCI | PubMed | Google Scholar
1Department of Medical and Molecular Genetics,
2Department of Pharmacology and Toxicology,
3Department of Pathology and Laboratory Medicine, and
4Department of Biostatistics, Indiana University, School of Medicine, Indianapolis, Indiana, USA.
5Department of Pharmacology and Chemical Biology, Baylor College of Medicine, Houston, Texas, USA.
6Melvin and Bren Simon Cancer Center,
7Division of Hematology/Oncology, Department of Medicine, and
8Center for Computational Biology and Bioinformatics, Indiana University School of Medicine, Indianapolis, Indiana, USA.
Address correspondence to: Chi Zhang, Indiana University School of Medicine, 410 W. 10th Street, HS 5000, Indianapolis, Indiana 46202, USA. Phone: 317.278.9625; Email: [email protected]. Or to: Xiongbin Lu, Indiana University School of Medicine, 980 W. Walnut Street, R3.C218D, Indianapolis, Indiana 46202, USA. Phone: 317.274.4398; Email: [email protected]. Or to: Xinna Zhang, Indiana University School of Medicine, 980 W. Walnut Street, R3.C216B, Indianapolis, Indiana 46202, USA. Phone: 317.274.7931; Email: [email protected].
Authorship note: YF, LW, and CW contributed equally to this work.
Find articles by Eyvani, H. in: JCI | PubMed | Google Scholar
1Department of Medical and Molecular Genetics,
2Department of Pharmacology and Toxicology,
3Department of Pathology and Laboratory Medicine, and
4Department of Biostatistics, Indiana University, School of Medicine, Indianapolis, Indiana, USA.
5Department of Pharmacology and Chemical Biology, Baylor College of Medicine, Houston, Texas, USA.
6Melvin and Bren Simon Cancer Center,
7Division of Hematology/Oncology, Department of Medicine, and
8Center for Computational Biology and Bioinformatics, Indiana University School of Medicine, Indianapolis, Indiana, USA.
Address correspondence to: Chi Zhang, Indiana University School of Medicine, 410 W. 10th Street, HS 5000, Indianapolis, Indiana 46202, USA. Phone: 317.278.9625; Email: [email protected]. Or to: Xiongbin Lu, Indiana University School of Medicine, 980 W. Walnut Street, R3.C218D, Indianapolis, Indiana 46202, USA. Phone: 317.274.4398; Email: [email protected]. Or to: Xinna Zhang, Indiana University School of Medicine, 980 W. Walnut Street, R3.C216B, Indianapolis, Indiana 46202, USA. Phone: 317.274.7931; Email: [email protected].
Authorship note: YF, LW, and CW contributed equally to this work.
Find articles by Colter, A. in: JCI | PubMed | Google Scholar
1Department of Medical and Molecular Genetics,
2Department of Pharmacology and Toxicology,
3Department of Pathology and Laboratory Medicine, and
4Department of Biostatistics, Indiana University, School of Medicine, Indianapolis, Indiana, USA.
5Department of Pharmacology and Chemical Biology, Baylor College of Medicine, Houston, Texas, USA.
6Melvin and Bren Simon Cancer Center,
7Division of Hematology/Oncology, Department of Medicine, and
8Center for Computational Biology and Bioinformatics, Indiana University School of Medicine, Indianapolis, Indiana, USA.
Address correspondence to: Chi Zhang, Indiana University School of Medicine, 410 W. 10th Street, HS 5000, Indianapolis, Indiana 46202, USA. Phone: 317.278.9625; Email: [email protected]. Or to: Xiongbin Lu, Indiana University School of Medicine, 980 W. Walnut Street, R3.C218D, Indianapolis, Indiana 46202, USA. Phone: 317.274.4398; Email: [email protected]. Or to: Xinna Zhang, Indiana University School of Medicine, 980 W. Walnut Street, R3.C216B, Indianapolis, Indiana 46202, USA. Phone: 317.274.7931; Email: [email protected].
Authorship note: YF, LW, and CW contributed equally to this work.
Find articles by Dong, E. in: JCI | PubMed | Google Scholar
1Department of Medical and Molecular Genetics,
2Department of Pharmacology and Toxicology,
3Department of Pathology and Laboratory Medicine, and
4Department of Biostatistics, Indiana University, School of Medicine, Indianapolis, Indiana, USA.
5Department of Pharmacology and Chemical Biology, Baylor College of Medicine, Houston, Texas, USA.
6Melvin and Bren Simon Cancer Center,
7Division of Hematology/Oncology, Department of Medicine, and
8Center for Computational Biology and Bioinformatics, Indiana University School of Medicine, Indianapolis, Indiana, USA.
Address correspondence to: Chi Zhang, Indiana University School of Medicine, 410 W. 10th Street, HS 5000, Indianapolis, Indiana 46202, USA. Phone: 317.278.9625; Email: [email protected]. Or to: Xiongbin Lu, Indiana University School of Medicine, 980 W. Walnut Street, R3.C218D, Indianapolis, Indiana 46202, USA. Phone: 317.274.4398; Email: [email protected]. Or to: Xinna Zhang, Indiana University School of Medicine, 980 W. Walnut Street, R3.C216B, Indianapolis, Indiana 46202, USA. Phone: 317.274.7931; Email: [email protected].
Authorship note: YF, LW, and CW contributed equally to this work.
Find articles by
Cao, S.
in:
JCI
|
PubMed
|
Google Scholar
|

1Department of Medical and Molecular Genetics,
2Department of Pharmacology and Toxicology,
3Department of Pathology and Laboratory Medicine, and
4Department of Biostatistics, Indiana University, School of Medicine, Indianapolis, Indiana, USA.
5Department of Pharmacology and Chemical Biology, Baylor College of Medicine, Houston, Texas, USA.
6Melvin and Bren Simon Cancer Center,
7Division of Hematology/Oncology, Department of Medicine, and
8Center for Computational Biology and Bioinformatics, Indiana University School of Medicine, Indianapolis, Indiana, USA.
Address correspondence to: Chi Zhang, Indiana University School of Medicine, 410 W. 10th Street, HS 5000, Indianapolis, Indiana 46202, USA. Phone: 317.278.9625; Email: [email protected]. Or to: Xiongbin Lu, Indiana University School of Medicine, 980 W. Walnut Street, R3.C218D, Indianapolis, Indiana 46202, USA. Phone: 317.274.4398; Email: [email protected]. Or to: Xinna Zhang, Indiana University School of Medicine, 980 W. Walnut Street, R3.C216B, Indianapolis, Indiana 46202, USA. Phone: 317.274.7931; Email: [email protected].
Authorship note: YF, LW, and CW contributed equally to this work.
Find articles by
Wang, J.
in:
JCI
|
PubMed
|
Google Scholar
|

1Department of Medical and Molecular Genetics,
2Department of Pharmacology and Toxicology,
3Department of Pathology and Laboratory Medicine, and
4Department of Biostatistics, Indiana University, School of Medicine, Indianapolis, Indiana, USA.
5Department of Pharmacology and Chemical Biology, Baylor College of Medicine, Houston, Texas, USA.
6Melvin and Bren Simon Cancer Center,
7Division of Hematology/Oncology, Department of Medicine, and
8Center for Computational Biology and Bioinformatics, Indiana University School of Medicine, Indianapolis, Indiana, USA.
Address correspondence to: Chi Zhang, Indiana University School of Medicine, 410 W. 10th Street, HS 5000, Indianapolis, Indiana 46202, USA. Phone: 317.278.9625; Email: [email protected]. Or to: Xiongbin Lu, Indiana University School of Medicine, 980 W. Walnut Street, R3.C218D, Indianapolis, Indiana 46202, USA. Phone: 317.274.4398; Email: [email protected]. Or to: Xinna Zhang, Indiana University School of Medicine, 980 W. Walnut Street, R3.C216B, Indianapolis, Indiana 46202, USA. Phone: 317.274.7931; Email: [email protected].
Authorship note: YF, LW, and CW contributed equally to this work.
Find articles by Schneider, B. in: JCI | PubMed | Google Scholar
1Department of Medical and Molecular Genetics,
2Department of Pharmacology and Toxicology,
3Department of Pathology and Laboratory Medicine, and
4Department of Biostatistics, Indiana University, School of Medicine, Indianapolis, Indiana, USA.
5Department of Pharmacology and Chemical Biology, Baylor College of Medicine, Houston, Texas, USA.
6Melvin and Bren Simon Cancer Center,
7Division of Hematology/Oncology, Department of Medicine, and
8Center for Computational Biology and Bioinformatics, Indiana University School of Medicine, Indianapolis, Indiana, USA.
Address correspondence to: Chi Zhang, Indiana University School of Medicine, 410 W. 10th Street, HS 5000, Indianapolis, Indiana 46202, USA. Phone: 317.278.9625; Email: [email protected]. Or to: Xiongbin Lu, Indiana University School of Medicine, 980 W. Walnut Street, R3.C218D, Indianapolis, Indiana 46202, USA. Phone: 317.274.4398; Email: [email protected]. Or to: Xinna Zhang, Indiana University School of Medicine, 980 W. Walnut Street, R3.C216B, Indianapolis, Indiana 46202, USA. Phone: 317.274.7931; Email: [email protected].
Authorship note: YF, LW, and CW contributed equally to this work.
Find articles by Sandusky, G. in: JCI | PubMed | Google Scholar
1Department of Medical and Molecular Genetics,
2Department of Pharmacology and Toxicology,
3Department of Pathology and Laboratory Medicine, and
4Department of Biostatistics, Indiana University, School of Medicine, Indianapolis, Indiana, USA.
5Department of Pharmacology and Chemical Biology, Baylor College of Medicine, Houston, Texas, USA.
6Melvin and Bren Simon Cancer Center,
7Division of Hematology/Oncology, Department of Medicine, and
8Center for Computational Biology and Bioinformatics, Indiana University School of Medicine, Indianapolis, Indiana, USA.
Address correspondence to: Chi Zhang, Indiana University School of Medicine, 410 W. 10th Street, HS 5000, Indianapolis, Indiana 46202, USA. Phone: 317.278.9625; Email: [email protected]. Or to: Xiongbin Lu, Indiana University School of Medicine, 980 W. Walnut Street, R3.C218D, Indianapolis, Indiana 46202, USA. Phone: 317.274.4398; Email: [email protected]. Or to: Xinna Zhang, Indiana University School of Medicine, 980 W. Walnut Street, R3.C216B, Indianapolis, Indiana 46202, USA. Phone: 317.274.7931; Email: [email protected].
Authorship note: YF, LW, and CW contributed equally to this work.
Find articles by
Liu, Y.
in:
JCI
|
PubMed
|
Google Scholar
|

1Department of Medical and Molecular Genetics,
2Department of Pharmacology and Toxicology,
3Department of Pathology and Laboratory Medicine, and
4Department of Biostatistics, Indiana University, School of Medicine, Indianapolis, Indiana, USA.
5Department of Pharmacology and Chemical Biology, Baylor College of Medicine, Houston, Texas, USA.
6Melvin and Bren Simon Cancer Center,
7Division of Hematology/Oncology, Department of Medicine, and
8Center for Computational Biology and Bioinformatics, Indiana University School of Medicine, Indianapolis, Indiana, USA.
Address correspondence to: Chi Zhang, Indiana University School of Medicine, 410 W. 10th Street, HS 5000, Indianapolis, Indiana 46202, USA. Phone: 317.278.9625; Email: [email protected]. Or to: Xiongbin Lu, Indiana University School of Medicine, 980 W. Walnut Street, R3.C218D, Indianapolis, Indiana 46202, USA. Phone: 317.274.4398; Email: [email protected]. Or to: Xinna Zhang, Indiana University School of Medicine, 980 W. Walnut Street, R3.C216B, Indianapolis, Indiana 46202, USA. Phone: 317.274.7931; Email: [email protected].
Authorship note: YF, LW, and CW contributed equally to this work.
Find articles by
Zhang, C.
in:
JCI
|
PubMed
|
Google Scholar
|

1Department of Medical and Molecular Genetics,
2Department of Pharmacology and Toxicology,
3Department of Pathology and Laboratory Medicine, and
4Department of Biostatistics, Indiana University, School of Medicine, Indianapolis, Indiana, USA.
5Department of Pharmacology and Chemical Biology, Baylor College of Medicine, Houston, Texas, USA.
6Melvin and Bren Simon Cancer Center,
7Division of Hematology/Oncology, Department of Medicine, and
8Center for Computational Biology and Bioinformatics, Indiana University School of Medicine, Indianapolis, Indiana, USA.
Address correspondence to: Chi Zhang, Indiana University School of Medicine, 410 W. 10th Street, HS 5000, Indianapolis, Indiana 46202, USA. Phone: 317.278.9625; Email: [email protected]. Or to: Xiongbin Lu, Indiana University School of Medicine, 980 W. Walnut Street, R3.C218D, Indianapolis, Indiana 46202, USA. Phone: 317.274.4398; Email: [email protected]. Or to: Xinna Zhang, Indiana University School of Medicine, 980 W. Walnut Street, R3.C216B, Indianapolis, Indiana 46202, USA. Phone: 317.274.7931; Email: [email protected].
Authorship note: YF, LW, and CW contributed equally to this work.
Find articles by Lu, X. in: JCI | PubMed | Google Scholar
1Department of Medical and Molecular Genetics,
2Department of Pharmacology and Toxicology,
3Department of Pathology and Laboratory Medicine, and
4Department of Biostatistics, Indiana University, School of Medicine, Indianapolis, Indiana, USA.
5Department of Pharmacology and Chemical Biology, Baylor College of Medicine, Houston, Texas, USA.
6Melvin and Bren Simon Cancer Center,
7Division of Hematology/Oncology, Department of Medicine, and
8Center for Computational Biology and Bioinformatics, Indiana University School of Medicine, Indianapolis, Indiana, USA.
Address correspondence to: Chi Zhang, Indiana University School of Medicine, 410 W. 10th Street, HS 5000, Indianapolis, Indiana 46202, USA. Phone: 317.278.9625; Email: [email protected]. Or to: Xiongbin Lu, Indiana University School of Medicine, 980 W. Walnut Street, R3.C218D, Indianapolis, Indiana 46202, USA. Phone: 317.274.4398; Email: [email protected]. Or to: Xinna Zhang, Indiana University School of Medicine, 980 W. Walnut Street, R3.C216B, Indianapolis, Indiana 46202, USA. Phone: 317.274.7931; Email: [email protected].
Authorship note: YF, LW, and CW contributed equally to this work.
Find articles by Zhang, X. in: JCI | PubMed | Google Scholar
Authorship note: YF, LW, and CW contributed equally to this work.
Published September 29, 2020 - More info
J Clin Invest. 2021;131(1):e140837. https://doi.org/10.1172/JCI140837.
© 2021 American Society for Clinical Investigation
Received: June 1, 2020; Accepted: September 24, 2020
Related article:
Abstract
The success of tumor immunotherapy, while partial, confirms the existence and importance of tumor immunosurveillance. CD8+ T cell recognition of tumor-specific peptides bound to MHC class I (MHC-I) molecules is central to this process. In this issue of the JCI, Fang, Wang, et al. describe a unique tumor immunoevasion strategy based on endocytosis and degradation of MHC-I complexes mediated by the trafficking factor MAL2. Notably, MAL2 expression was associated with poor prognosis of breast cancer, and its downregulation enhanced CD8+ T cell recognition of breast cancer in various experimental models. This work demonstrates that a deeper understanding of tumor interference with MHC-I stability and trafficking has considerable potential for enhancing immunotherapies.
Authors
Devin Dersh, Jonathan W. Yewdell
-
Abstract
Immune evasion is a pivotal event in tumor progression. To eliminate human cancer cells, current immune checkpoint therapy is set to boost CD8+ T cell–mediated cytotoxicity. However, this action is eventually dependent on the efficient recognition of tumor-specific antigens via T cell receptors. One primary mechanism by which tumor cells evade immune surveillance is to downregulate their antigen presentation. Little progress has been made toward harnessing potential therapeutic targets for enhancing antigen presentation on the tumor cell. Here, we identified MAL2 as a key player that determines the turnover of the antigen-loaded MHC-I complex and reduces the antigen presentation on tumor cells. MAL2 promotes the endocytosis of tumor antigens via direct interaction with the MHC-I complex and endosome-associated RAB proteins. In preclinical models, depletion of MAL2 in breast tumor cells profoundly enhanced the cytotoxicity of tumor-infiltrating CD8+ T cells and suppressed breast tumor growth, suggesting that MAL2 is a potential therapeutic target for breast cancer immunotherapy.
-
Introduction
The immune system identifies and acts against tumor cells by adaptive cell reactions, which play a critical role in restricting tumor initiation and development. Cancer immunotherapy has become a promising therapeutic approach for cancers with no or limited specific, targeted therapies (1–3). However, various forms of immunotherapy, including checkpoint blockade immunotherapies, have been shown to boost T cell–mediated immune responses that lead to marked and sustained clinical responses in only a limited number of patients and cancer types (4–6). Antitumor immune responses require functional presentation of tumor antigens and a microenvironment that favors competent immune effectors (7–9). To execute cytotoxicity in cancer cells, CD8+ cytotoxic T lymphocytes (CTLs) recognize tumor antigens presented on the MHC-I of the cancer cell and trigger the cancer cell to undergo programmed cell death (10–12). Increased expression of MHC-I molecules can be of therapeutic significance since it makes tumor cells more susceptible to lysis induced by CTLs (13, 14). Actually, a primary mechanism by which tumors evade immune surveillance is to downregulate antigen processing and presentation activity, including deficient expression of the MHC-I complex components, inactive immunoproteasome machinery, and dysfunctional antigen transport in the cell (15).
The MHC-I complex displays peptide epitopes at the surface of cancer cells for recognition by CD8+ T cells (16). Presentation of peptides derived from tumor-associated antigens occurs by docking on the antigen-binding domain of the MHC-I, facilitating consequent identification and killing by CD8+ T cells. Like other proteins on the plasma membrane, the antigen-loaded MHC-I complex, if not recognized, is continuously removed from the cell surface for intracellular degradation or recycling to the cell surface (17, 18). To evade immune surveillance, cancer cells employ various mechanisms to downregulate the expression of MHC-I molecules or other proteins involved in antigen processing and presentation (18, 19). Downregulation appears to be more common than complete elimination of MHC-I expression because the latter renders cancer cells susceptible to the action of NK cells. Reduced MHC-I levels on the cell surface may not only avoid the attack from NK cells, but also diminish antigen presentation to evade CD8+ T cells (19).
On the plasma membrane, proteins that are destined to be recycled or degraded first need to be internalized from the cell surface. Internalization is a constitutive and indispensable event in all cell types for nutrient uptake, signal transduction, and recycling of plasma membrane components (20, 21). MHC-I molecules are internalized via non-clathrin–associated endocytosis, a type of endocytosis distinct from clathrin-dependent endocytosis (22). Regardless of the mode of internalization, internalized proteins arrive in early-stage endosomes in a short time (23). Typical markers of the sorting endosomes, with a luminal pH of approximately 6.3 to 6.8, are early endosome antigen-1 (EEA-1) and the GTPases (RAB5) (24). From these vesicles, the internalized cargo is sorted to late endosomes and lysosomes for degradation or redirection to the cell surface using a number of routes.
Among the main subtypes of breast cancer, triple-negative breast cancer (TNBC) often has poor clinical outcomes due to the lack of effective treatments and targeted therapies (25, 26). Immunotherapy, notably immune checkpoint blockade, has shown promise in treating patients with TNBC, but the response rates are up to 10% in unselected TNBC patients, which improve slightly even when patients are preselected on PD-L1 levels (27, 28). There is an unmet need to understand the potential mechanisms involved in immune evasion of breast cancer. Among the genes with highest significance associated with unfavorable prognosis in breast cancer, we identified MAL2, which encodes a transmembrane protein associated with protein endocytosis (29–33). Previous studies showed that MAL2 facilitates the delivery of membrane-bound proteins and exogenous cargos from the basolateral to the apical surface (29, 30, 32, 33). Here, we demonstrated that MAL2 was involved in endocytosis of the antigen-loaded MHC-I complex in breast tumor cells, by which MAL2 suppressed tumor antigen presentation and diminished the cytotoxicity of CD8+ T cells.
-
Results
MAL2 drives breast tumor progression in immunocompetent models. In a search for the genes significantly associated with unfavorable prognosis in breast cancer, we identified a list of 20 top-ranked genes with highest significance by using The Cancer Genome Atlas (TCGA) breast cancer data sets (Figure 1A). Distinct from the other genes in the list, MAL2 does not regulate the proliferation of breast cancer cells. We overexpressed or knocked down this gene in 2 murine TNBC cell lines (EO771 and 4T1) as well as in 1 human TNBC cell line (MDA-MB-468). Consistent with unbiased large-scale genome-wide studies showing that MAL2 is not a gene essential for breast cancer cell proliferation (34–37), we found that altered MAL2 levels had no or minimal effects on the proliferation of murine or human TNBC cells (Figure 1B and
 Figure 1
Figure 1MAL2 expression correlates with poor survival of breast cancer patients without affecting breast cancer cell proliferation. (A) Forest plot with HR is shown for overall survival analysis (best separation) of 20 top-ranked human genes that are negatively correlated with clinical outcomes in breast cancer. A total of 1,075 breast cancer patients from TCGA are included. (B) Cell proliferation was measured for mouse and human TNBC cells with different MAL2 levels (6 replicates under each condition). Data are presented as mean ± SD. Two-way ANOVA test was used for statistical analysis. (C–E) EO771-derived breast tumor growth in immunocompromised mice. EO771 cells with different Mal2 expression levels were orthotopically injected into female NU/J mice (5 × 104 cells per mouse). Tumor volume = 0.5 × length × width × width. Tumor images (C), tumor weights (D), and tumor growth curves (E) are shown here. Data are presented as mean ± SD. One-way and 2-way ANOVA tests were used for data analysis in tumor weight and tumor growth, respectively. *P < 0.05; **P < 0.01; ***P < 0.001.
Supplemental Figure 1A; supplemental material available online with this article; https://doi.org/10.1172/JCI140837DS1), and expression of the other 4 top-ranked genes (LRP11, PGK1, PCMT1, and FAM173B) had modest to significant effects on cancer cell proliferation (Supplemental Figure 1B). In line with this, orthotopic tumor growth of the EO771- and 4T1-derived tumors in immunocompromised nude (Nu/J) mice had no notable difference among control (WT), Mal2-overexpressing (OE), and Mal2-knockdown (KD) tumors (Figure 1, C–E, and Supplemental Figure 1C). MAL2 was highly expressed in each subtype of human breast cancer (estrogen receptor–positive/ progesterone receptor–positive, human epidermal growth factor receptor 2–positive, TNBC) (Supplemental Figure 1D). Our analysis of breast cancer in TCGA and Molecular Taxonomy of Breast Cancer International Consortium (METABRIC) cohorts showed that a higher expression level of MAL2 in breast cancer, especially in TNBC, was correlated with poorer patient survival (Figure 2A and Supplemental Figure 1E). In the immunocompetent mice, both EO771-derived (in C57BL/6 mice) and 4T1-derived (in BALB/c mice) orthotopic tumors with stable Mal2 overexpression came out with significantly higher volume and weight compared with the control tumors, whereas the tumors with Mal2-KD had strikingly lower volume and weight (Figure 2, B and C, and Supplemental Figure 1F). Tumor growth curves confirmed that the Mal2-OE tumors grew much faster than the ones in the Mal2-WT group, whereas Mal2-KD tumors barely grew over time (Figure 2D and Supplemental Figure 1F). The differences in tumor growth observed here suggest that Mal2 may modulate breast cancer growth by affecting the interaction of tumor cells with nontumor cells in the tumor microenvironment (TME), including immune cells.
 Figure 2
Figure 2MAL2 promotes breast tumor growth in immunocompetent models. (A) Kaplan-Meier survival analysis of patients with breast cancer (BRCA) or TNBC. Patients with high versus low expression of MAL2 were compared with respect to overall survival. The overall survival data of patients in TCGA and METABRIC cohorts are combined. Log rank test was used for statistical analysis. (B–D) EO771-derived breast tumor growth in immunocompetent mice (Mal2-WT: n = 9; Mal2-OE: n = 7; and Mal2-KD: n = 7). EO771 cells with different Mal2 expression levels were orthotopically injected into female C57BL/6 mice (5 × 104 cells per mouse). Tumor images (B), tumor weights (C), and tumor growth curves (D) are shown here. Data are presented as mean ± SD. One-way and 2-way ANOVA tests were used for data analysis in tumor weight and tumor growth, respectively. *P < 0.05; **P < 0.01; ***P < 0.001.
MAL2 expression is correlated with low cytotoxicity of CD8+ T cells in tumors. For accurate assessment of cell population and activities in the TME, we developed a deconvolution method named Inference of Cell Types and Deconvolution (ICTD) (38). We analyzed bulk RNA-Seq databases of human breast tumors to evaluate the composition and activities of diverse cell types in the TME. Using TCGA (Figure 3A) and GSE32646 (Supplemental Figure 2A) breast cancer data sets, we found that low MAL2 expression levels were correlated with increased cytotoxicity of CD8+ T cells in TNBC tumors with high tumor-infiltrating T cell levels. Single-cell RNA-Seq (scRNA-Seq) analysis of TNBC tumor tissues revealed that MAL2 was primarily expressed in tumor cells rather than other cell types (Figure 3B and Supplemental Figure 2B). MAL2 expression did not appear to globally affect the cellular composition in the TME (Supplemental Figure 2C). Further investigation of T cell receptor (TCR) signaling from 115 TNBC cases in TCGA showed that tumors with high TCR signaling activity were enriched in the tumors with low MAL2 expression, whereas higher MAL2 expression was correlated with lower TCR signaling activity, suggesting an inhibitory role of MAL2 in T cell–mediated cytotoxicity (Figure 3C and Supplemental Table 1). Consistent with the bioinformatics analysis of the human TNBC database, we observed increased CD8+ T cell infiltration and tumor cell death (indicated by cleaved caspase-3 staining) in the Mal2-KD EO771-derived tumors in comparison with the Mal2-WT ones. However, the proliferation of tumor cells (indicated by Ki-67 staining) was not altered with Mal2 expression changes. By contrast, the Mal2-OE tumors exhibited reduced CD8+ T cell infiltration and cytotoxicity (Supplemental Figure 2D).
 Figure 3
Figure 3MAL2 expression is correlated with low cytotoxicity of tumor-infiltrating CD8+ T cells. (A) Violin plots representing relative cytotoxicity (RC) level of tumor-infiltrating CD8+ T cells and normalized expression levels of 3 cytotoxicity markers in TNBC samples with high or low MAL2 expression levels. The data of TNBC patients are from TCGA cohort. Unpaired 2-tailed t test was used for statistical analysis. (B) Expression of MAL2 was quantified by log(TPM+1) in immune cells, stromal cells, and TNBC tumor cells in the GSE75688 scRNA-Seq data set. One-way ANOVA test was used for statistical analysis. (C) MAL2 expression is negatively associated with the expression levels of TCR signaling genes in TNBC patient samples. The heatmap shows the expression levels of the TCR signaling genes in the samples. Each row is 1 TCR signaling gene and each column is 1 sample. The genes are ordered by their coexpression correlation with MAL2 (low to high from top to bottom). The histogram on the right illustrates the distribution of the coexpression correlation between MAL2 and the TCR signaling genes. x-axis, correlation values; y-axis: TCR gene counts. (D) CD8+ T cells isolated from EO771-derived tumors with different levels of Mal2 (WT, OE, and KD) in C57BL/6 mice were analyzed by flow cytometry for their activity indicated by IFN-γ, TNF-α, and GZMB levels in the cell. Flow cytometrical data are shown on the left, and the quantitative results are summarized on the right. ***P < 0.001, 1-way ANOVA test. Data are presented as mean ± SD and are representative of 3 independent experiments. (E) CD8+ T cells as above mentioned were analyzed for their secretion of cytokines (IFN-γ, TNF-α). Supernatants were collected and quantified by ELISA. ***P < 0.001, 2-way ANOVA test. Data are presented as mean ± SD and are representative of 3 independent experiments.
To determine whether Mal2 expression levels affect the activity of tumor-infiltrating T cells in vivo, we isolated the CD8+ T cells from the aforementioned EO771 tumors in C57BL/6 mice and evaluated their functionality. KD of Mal2 notably augmented the production of cytolytic granules (GZMB) and cytokines (IFN-γ, TNF-α) in the CD8+ T cells, indicating the active state of these cells. In comparison with the control tumors, increased levels of MAL2 in the tumor cells significantly suppressed granule and cytokine production of CD8+ T cells (Figure 3D). Using ELISA, we also confirmed that CD8+ T cells from the Mal2-OE tumors secreted less TNF-α and IFN-γ than their counterparts from the Mal2-WT tumors, in contrast to higher levels of TNF-α and IFN-γ secretion observed in the CD8+ T cells from the Mal2-KD tumors (Figure 3E). In addition to the EO771 tumor model, we also validated the negative correlation of Mal2 expression levels with CD8+ T cell activity in the 4T1-derived tumor models (Supplemental Figure 2E).
Next, we used a doxycycline-induced Mal2-KD model to eliminate the potential impact of Mal2 depletion on initial tumor establishment. EO771 cells expressing doxycycline-inducible Mal2 shRNA were injected orthotopically into C57BL/6 mice. When the tumors were established (~100 mm3), the mice were divided into 2 groups for treatment with vehicle or doxycycline. The mice treated with doxycycline had much smaller tumors in comparison with the vehicle-treated mice (Supplemental Figure 3, A and B), and the tumor growth rate was also decreased in the doxycycline-treated group (Supplemental Figure 3, C and D). CD8+ T cells isolated from the doxycycline-induced tumors accumulated more granules and cytokines in the cells (Supplemental Figure 3E) and secreted higher levels of TNF-α and IFN-γ (Supplemental Figure 3F). Taken together, the results suggested that Mal2 expression levels in tumor cells affected the immune response of breast tumors but not their proliferation.
MAL2 expressed in tumor cells directly inhibits CD8+ T cell cytotoxicity. To determine whether MAL2 expressed in tumor cells directly affects the cytotoxicity of CD8+ T cells, we generated a set of EO771 cell lines stably presenting a chicken ovalbumin peptide (OVA257-264) and expressing different levels of MAL2 (WT, OE, and KD). EO771-OVA cells were cocultured with mature CD8+ T cells isolated from splenocytes of an OT-I transgenic mouse that expressed TCR to recognize the OVA257-264 antigen in the context of H-2Kb, and T cell cytotoxicity was measured by lactate dehydrogenase (LDH) assay (39). Compared with the WT group, CD8+ T cell cytotoxicity on the Mal2-OE cells was decreased significantly, whereas the same T cells exhibited much higher cytotoxicity on the Mal2-KD cells (Figure 4A). In line with the cytotoxicity results, the OT-I T cells cocultured with the Mal2-KD cells secreted strikingly higher levels of TNF-α and IFN-γ than the ones cocultured with Mal2-WT tumor cells, but these cells were inactive when cocultured with Mal2-OE cells (Figure 4B). We also applied a mammary tumor organoid model to assess the effect of the tumor cell–expressing MAL2 on CD8+ T cell cytotoxicity ex vivo (40). When cocultured with tumor organoids generated from the EO771 tumors in C57BL/6 mice, the OT-I T cells exhibited significantly higher cytotoxicity on the Mal2-KD tumor organoids, but extremely lower cytotoxicity on the Mal2-OE tumor organoids, in comparison with their cytotoxicity on the control Mal2-WT tumor organoids (Figure 4, C and D). To determine whether the effect of MAL2 is dependent on the interaction between tumor cell antigen and the T cell receptor, we used an MHC-I–blocking antibody or control isotype antibody in the coculture of EO771-OVA and OT-I T cells. Blocking MHC-I dramatically reduced the cytotoxicity of OT-I T cells to similar levels against all the EO771-OVA cells expressing different levels of MAL2, suggesting that MHC-I–mediated antigen presentation was involved in the function of MAL2 (Figure 4E). Next, we tested whether MAL2 expression affected the CD8+ T cell cytotoxicity in human TNBC cells. NY-ESO-1–presenting human TNBC cells (MDA-MB-468) expressing different levels of MAL2 were cocultured with human T cells specific for the tumor-associated antigen NY-ESO-1 bound to HLA-A. Interestingly, when MAL2 was overexpressed, the cytotoxicity of NY-ESO-1–specific T cells was significantly decreased (Figure 4F). By contrast, the cytotoxicity of the NY-ESO-1–specific T cells was increased significantly when MAL2 was knocked down. This result was further supported by the T cell cytotoxicity assay using CD8+ T cells with TCR specific for the endogenous tumor-associated antigen MAGE-A10 in MDA-MB-468 cells (Figure 4G). Collectively, in both murine and human TNBCs, the MAL2 level in tumor cells negatively affected the cytotoxicity of CD8+ T cells.
 Figure 4
Figure 4Mal2 expression in tumor cells affects CD8+ T cell cytotoxicity. (A) EO771-OVA cells with different Mal2 expression levels (WT, OE, or KD) were cocultured with OT-I T cells, and T cell cytotoxicity was measured by LDH release assay. Two-way ANOVA test was used for statistical analysis. (B) EO771-OVA cells were cocultured with OT-I T cell with or without 50 ng/mL PMA for 5 hours. Supernatants were tested by ELISA for TNF-α and IFN-γ levels. Two-way ANOVA test was used for statistical analysis. (C) EO771 tumor organoids were cocultured with OT-I T cell for 30 hours. Spheroids with diameter greater than 100 μm were counted. Scale bar: 100 μm. One-way ANOVA test was used for statistical analysis. (D) EO771 tumor organoids from C were dissociated into single cells and stained with anti-EpCAM antibody and LIVE/DEAD cell stain. T cell cytotoxicity was assessed by percentages of dead tumor cells. Two-way ANOVA test was used for statistical analysis. (E) EO771-OVA cells were cocultured with OT-I T cells with control isotype or MHC-I–blocking antibody, and T cell cytotoxicity was measured. Two-way ANOVA test was used for statistical analysis. (F) NY-ESO-1–expressing human MDA-MB-468 cells with different MAL2 expression levels were cocultured with NY-ESO-1–specific T cells at the ratio of 1:10, and T cell cytotoxicity was measured. One-way ANOVA test was used for statistical analysis. (G) Human MDA-MB-468 cells with different MAL2 expression levels were cocultured with MAGE-A10 specific T cells at the ratio of 1:20 and T cell cytotoxicity was measured. One-way ANOVA test was used for statistical analysis. Data are presented as mean ± SD of 3 independent experiments in the figure. *P < 0.05; **P < 0.01; ***P < 0.001.
MAL2 downregulates tumor antigen presentation. Mass spectrometry analysis identified a number of MAL2-interacting proteins enriched in MHC-I–mediated antigen presentation, endocytosis/recycling, and RAB pathways (Figure 5A and Supplemental Table 2). Confocal 3D imaging analysis of the MHC-I complex on the TNBC cell (MDA-MB-468) membrane clearly showed that KD of MAL2 increased the percentage of the cells positive for MHC-I (human HLA), whereas overexpression of MAL2 suppressed MHC-I presence on the cell surface (Figure 5, B and C). In the membrane protein extraction analysis, MAL2 appeared to downregulate the total levels of human MHC-I components, HLA-A, B, and C, on the plasma membrane (Figure 5D). Flow cytometry assay further validated that Mal2 expression negatively affected OVA antigen presentation on EO771-OVA cells (Figure 6A). Consistently, the presentation of the MHC-I complex on the cell membrane was also inhibited by MAL2, although total mRNA levels (H-2Kb and HLA-A) remained unchanged (Figure 6B and Supplemental Figure 4A).
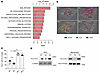 Figure 5
Figure 5MAL2 suppresses the presentation of MHC-I molecules on breast cancer cell membrane. (A) Mass spectrometry analysis showed that MAL2 was highly associated with MHC-I antigen presentation and endocytosis signaling. Gene Set Enrichment Analysis (GSEA) was performed to determine the signaling pathways. The curve shows enrichment ratios for the 10 top-ranked signaling pathways. Fisher’s exact test was used for statistical analysis. (B) Confocal 3D imaging of the MHC-I complex on the surface of MDA-MB-468 cells with different MAL2 levels. EGFR is a biomarker for cell surface proteins. (C) Quantitation results of confocal imaging analyses in B. One-way ANOVA test was used for statistical analysis. Data are presented as mean ± SD. *P < 0.05; **P < 0.01; ***P < 0.001. (D) Localization of human MHC-I proteins (HLA-A, B, C) in MDA-MB-468 cells was determined by immunoblotting analyses of cell fractions.
 Figure 6
Figure 6MAL2 downregulates antigen presentation on breast cancer cells. (A) MAL2 downregulates antigen presentation on EO771-OVA cells with different MAL2 levels. The antigen presentation efficiency was assessed by flow cytometry. MFI scores are presented for 3 independent experiments. One-way ANOVA test was used for statistical analysis. (B) The presentation of the MHC-I complex on the cell membrane is inhibited by MAL2. EO771 and MDA-MB-468 cells with different MAL2 levels were tested. One-way ANOVA test was used for statistical analysis. (C) MAL2 promotes the turnover rate of tumor antigen on the cell surface. EO771 cells with different Mal2 expression levels were preincubated with OVA257-264 peptides and OVA257-264 peptides were washed off, and then cells were collected at indicated times and stained for the MHC-I–presented OVA257-264. Two-way ANOVA test was used for statistical analysis. (D) Localization of the MAL2 protein in MDA-MB-468 cells. EGFR is biomarker for membrane proteins. EEA-1 and RAB7 are biomarkers for early and late stage endosomes, respectively, and LAMP-1 is a biomarker for lysosomes. Scale bar: 10 μm. (E) Quantification of (D). In this figure, data are presented as mean ± SD. *P < 0.05; **P < 0.01; ***P < 0.001.
We also assessed the turnover rate of the OVA antigen presented on the cell surface (Figure 6C). EO771 cells expressing varying levels of Mal2 were incubated with free OVA257-264 peptides to saturation, and then unbound OVA257-264 peptides were washed off. The OVA antigen presented on the cell surface was measured over time. It was evident that the OVA antigen bound to MHC-I on the cell surface had a markedly higher turnover rate when Mal2 was overexpressed. KD of Mal2 slowed down the turnover of the OVA antigen on the cell surface. Interestingly, in addition to its primary localization on the plasma membrane, MAL2 appeared to be associated with endosomes, indicated by its significant colocalization with EEA-1 and RAB7, 2 well-known markers for endosomes (Figure 6, D and E). The interaction of MAL2 with RAB7 was also shown in the results of mass spectrometry (Figure 5A and Supplemental Table 2). Like other cell surface receptors, MHC-I molecules are continuously removed from the surface, followed by intracellular degradation or recycling. We next asked whether MAL2 is involved in the endocytosis of the antigen-loaded MHC-I complex and protein degradation in tumor cells. Treatment of MDA-MB-468 cells with MG-132 (proteasome inhibitor) or filipin (endocytosis inhibitor) (41) increased HLA protein levels to a similar extent in the whole-cell lysates of MDA-MB-468 cells with varying levels of MAL2. However, only filipin treatment resulted in significantly increased levels of HLA on the plasma membrane of these cells, suggesting that MAL2 may regulate the endocytosis-mediated degradation of the MHC-I complex (Supplemental Figure 4B). Macropinocytosis is a special type of endocytosis, through which cancer cells take in extracellular macromolecules to support survival and growth in nutrient-deprived TMEs (42). We assessed the level of micropinocytosis by measuring the uptake of dextran, a large-molecule mass in MDA-MB-468 cells expressing different levels of MAL2. Ethylisopropylamiloride (EIPA) is a specific inhibitor for macropinocytosis (43). No significant difference was observed in the macropinocytosis of the cells with different levels of MAL2 in vitro (Supplemental Figure 4C) and in vivo (Supplemental Figure 4D). This result indicates that MAL2 is probably not involved in tumor cell metabolism by macropinocytosis.
Mal2 regulates tumor antigen presentation via endocytosis. Previous studies have shown that the MHC-I complex (or human HLA) potentially interacts with RAB7 for endocytosis (44, 45). Interestingly, mass spectrometric analysis identified both RAB7 and HLA proteins in the Mal2-containing protein complex (Figure 5A and Supplemental Table 2), suggesting that MAL2 may modulate the interaction between RAB7 and HLA. To test it, we first applied co-IP assays to detect whether MAL2 is within the same protein complex containing HLA and RAB7. We detected physical interaction of MAL2 with HLA-A (Figure 7A and Supplemental Figure 5A) and with RAB7 (Figure 7B and Supplemental Figure 5B) in human MDA-MB-468 cells. GFP was used as a negative control, which showed no interaction with either HLA or RAB7 (Figure 7, A and B). It has been noted that MAL2 is a heavily glycosylated protein, and its glycosylated form has a much higher molecular size (~40 kDa) compared with its unglycosylated form (19.1 kDa) (29, 46). Overexpression of exogenous MAL2 significantly increased the interaction between HLA-A and RAB7, whereas MAL2 KD decreased their interaction (Figure 7C). To examine how MAL2 regulates HLA-A and RAB7 interaction, we generated a series of MAL2 mutant forms on the consensus N-glycosylation site (N132) and potential glycosylation sites (N144, N147A, and N149A) (Figure 7D). Either MAL2 with a single glycosylation site mutated (N132A, N144A, N147A, N149A) or MAL2 with 3 other potential glycosylation sites (N144A/N147A/N149A, designated as 3N→3A) mutated retained partial glycosylation, whereas MAL2 with all 4 sites mutated (designated as 4N→4A) was completely unglycosylated (Figure 7E, left, and Supplemental Figure 5C, left). The co-IP results suggested that the functional interaction of MAL2 with HLA-A and RAB7 required the glycosylation of MAL2 because the MAL2 4N->4A mutant lost all binding activity with HLA-A and RAB7, whereas the N132A and 3N->3A mutants of MAL2 partially retained their binding activity (Figure 7E, right, and Supplemental Figure 5C, right). To examine the functional consequence of MAL2 glycosylation, we examined the effect of MAL2 on the level of HLA-A presented on the cell surface (Figure 7F). Overexpression of the MAL2 4N->4A mutant had no effect on the presentation of HLA-A on the cell surface, whereas WT or partially binding-deficient forms of MAL2 reduced HLA-A presentation. Other than the interaction with MHC-I molecules, the glycosylation mutations of MAL2 did not seem to affect its intracellular localization (Supplemental Figure 5E). Next, we applied an in situ proximity ligation assay (PLA) to assess the effect of MAL2 on the interaction of HLA-A and RAB7 in the cell. We found that MAL2 overexpression significantly enhanced the interaction between HLA and RAB7, shown by strong fluorescence signals (Figure 7G), whereas KD of MAL2 abolished this interaction. Flow cytometry assays also confirmed that MAL2 increased the coexistence of HLA-A and RAB7 in the endosomes isolated from MDA-MB-468 cells (Figure 7H). When treated with cycloheximide to block protein synthesis, MDA-MB-468 cells with exogenous MAL2 overexpression had a significantly higher degradation rate of MHC-I molecules (HLA) in comparison with the control cells, whereas MAL2 KD had the opposite effect (Supplemental Figure 5D), suggesting a critical role of MAL2 in the endocytosis-mediated protein degradation of the MHC-I complex.
 Figure 7
Figure 7MAL2 regulates tumor antigen presentation via endocytosis. (A) MAL2 is physically associated with HLA-A. MDA-MB-468 cells expressing flag-tagged MAL2 and HA-tagged HLA-A2 (HA-tagged GFP as negative control) were analyzed by co-IP and Western blotting using indicated antibodies. (B) MAL2 is physically associated with RAB7. MDA-MD-468 cells expressing flag-tagged MAL2 and HA-tagged RAB7 were analyzed as described in A. (C) MAL2 mediates the interaction of HLA-A and RAB7. MDA-MB-468 cells with different MAL2 expression levels were lysed and endogenous RAB7 was immunoprecipitated for Western blotting. (D) Illustration of MAL2 mutations on potential glycosylation sites. (E) Glycosylation of MAL2 is essential for the interaction of MAL2 with HLA-A. HA-tagged HLA-A2 and flag-tagged WT or mutant MAL2 were coexpressed in MDA-MD-468 cells. MAL2 was immunoprecipitated for Western blotting analysis. (F) WT but not unglycosylated MAL2 (4N->4A) regulates the presentation of human HLA on the cell membrane. MDA-MB-468 cells were transfected with control or MAL2 expression vectors and flow cytometry assay was applied to assess the presentation of HLA-A on the cell membrane. MFI scores are presented as mean ± SD of 3 independent experiments. (G) MAL2 regulates the interaction of HLA-A and RAB7 in MDA-MB-468 cells. Proximity ligation assay was applied, and fluorescence intensity scores are shown to indicate the extent of interaction. Red: colocalized HLA-A2 and RAB7 in situ; blue, DAPI for nucleus staining. (H) MAL2 regulates the endosome-mediated turnover of the MHC-I complex. Endosomes were isolated from MDA-MB-468 cells expressing different levels of MAL2, and then stained for HLA-A and RAB7 in flow cytometry assay. Quantitation data are presented as mean ± SD of 3 independent experiments. Statistical analyses in the figure were conducted using 1-way ANOVA test. *P < 0.05; **P < 0.01; ***P < 0.001.
MAL2 modulates the immune profile and response in tumors. To confirm whether Mal2 expression in tumor cells regulates T cell activities and cytotoxicity in vivo, we generated a scRNA-Seq data set of T cells (CD3+) isolated from EO771-derived tumors expressing different levels of Mal2 (WT, OE, and KD) by using the 10x Genomics Chromium system. Expression profiles of 11,884, 10,200, and 8,062 CD3+ T cells from the WT, OE, and KD tumors, respectively, were collected after exclusion of dead or low-quality cells. A total of 10 subtypes of the CD3+ T cells were identified by cell clustering analysis, including CD8+ effector T cells, CD8+ naive I T cells, CD8+ naive II T cells, NKT cells, CD4+ naive T cells, and T effector memory (Tem), Th1, Th2, Treg, and T stem cell memory (Tscm) cells (see details in Methods), which were illustrated in the t-SNE plot of the identified cell groups (Figure 8A and Supplemental Table 3). Compared with the T cells isolated from Mal2-WT tumors, which contained 35.4% of CD8+ effector T cells and 27.5% of CD8+ naive T cells, very few CD8+ effector T cells (0.1%) were observed in the Mal2-OE tumors, along with a much higher percentage of naive T cells (49.3%), including 18.5% of Tem cells, 14.7% of CD8+ naive T cells, and 16.1% of CD4+ naive T cells (Figure 8B). By contrast, Mal2-KD tumors were significantly more enriched (P < 1 × 10-10 by χ2 test) for CD8+ effector T cells (48.1%), and they contained fewer CD8+ naive T cells (8.3%) (Figure 8B). Expression levels of signature genes for cytotoxicity (Gzmb, Ifng, Cst7, and Ccl4), negative regulation of cytotoxicity (Tcf7, Ptpn22, Il7r, Lef1, and Peli1), and positive regulation of cytotoxicity (Il2ra, Hsph1, Il12rb1, Akt1, and Casp3) suggested that the activity of the CD8+ effector T cells was drastically suppressed in the Mal2-OE tumors in comparison with that of the Mal2-WT and Mal2-KD tumors (Figure 8C). We plotted the average expression levels of these marker genes in the CD8+ T cells under the 3 conditions (Mal2-WT, -OE, and -KD) (Figure 8D). The average expression levels of each function-associated gene (cytotoxicity, positive regulation, and negative regulation) in CD8+ T cells isolated from the tumors expressing varying levels of Mal2 are shown in Figure 8E. T cells from the Mal2-OE tumors displayed lower cytotoxicity and negative regulation of T cell cytotoxicity in comparison with the ones from the Mal2-WT tumors. However, T cells from the Mal2-KD tumors showed much higher cytotoxicity and positive regulation of T cell activity. Our data clearly suggested that suppression of Mal2 rescued the cytotoxicity of tumor-infiltrating CD8+ T cells, whereas high levels of Mal2 expression blocked the maturation and activation of T cells in the tumor. In addition to T cell analysis, we also analyzed other tumor-infiltrated immune cells by flow cytometry (Supplemental Figure 6). NK cells and DCs were decreased in the Mal2-OE tumors in comparison with the Mal2-WT tumors, whereas neutrophils were increased. By contrast, more NK cells and DCs were observed in the Mal2-KD tumors, suggesting activated innate immune response. However, no significant difference was seen for macrophage infiltration among these 3 subtypes of EO771 tumors.
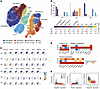 Figure 8
Figure 8Mal2 expression levels are negatively correlated with tumor infiltration and cytotoxicity of CD8+ T cells in vivo revealed by scRNA-Seq analysis. (A) A total of 27,827 CD3+ T cells were isolated from EO771 tumors with different Mal2 expression levels (WT, OE, and KD) and profiled with 10x Genomics. 10 groups are identified (by Louvain algorithm) and color marked over the t-SNE plot. (B) Distribution of T cell subtypes within EO771 tumors expressing different levels of Mal2 (WT, OE, and KD). (C) Functional marker gene expression of CD8+ T cells from the tumors expressing different levels of MAL2 over the low-dimensional t-SNE plot. (D) Relative gene expression heatmap of CD8+ T cells from the tumors expressing different levels of MAL2. Key genes responsible for cytotoxicity, negative regulation, and positive regulation of CD8+ T cells are shown. (E) The average expression levels of the T cell function genes in CD8+ T cells from the tumors expressing different levels of MAL2.
Depletion of MAL2 enhances CD8+ T cell cytotoxicity in human TNBC tumors. We applied patient-derived tumor organoid models to determine whether MAL2 expression in tumor cells affects CD8+ T cell cytotoxicity ex vivo. MAL2 expression in fresh primary tumor cells was manipulated via lentiviral MAL2 or its shRNA expression (Supplemental Figure 7A). Tumor cells with different levels of MAL2 expression were then mixed with cancer-associated fibroblasts (CAFs) (tumor cell vs. CAF = 1:1) from the same tumor tissue to form patient-derived tumor organoids. When the spheroid diameter reached 100 μm, organoids were cocultured with preactivated autologous CD8+ T cells isolated from the same tumor tissue. Tumor organoid dissociation and T cell cytotoxicity were monitored (illustrated in Supplemental Figure 7B). As expected, the MAL2-OE organoids exhibited much more resistance to CD8+ T cell killing compared with the MAL2-WT organoids. By contrast, the MAL2-KD organoids were more vulnerable to the CD8+ T cells (Figure 9, A and B). Flow cytometry assay also showed that CD8+ T cells exhibited significantly higher cytotoxicity on the MAL2-KD organoids, but much lower cytotoxicity on the MAL2-OE organoids compared with their cytotoxicity on the control MAL2-WT organoids (Figure 9C). Consistently, KD of MAL2 notably augmented the production of cytolytic granules (GZMB) and cytokines (IFN-γ, TNF-α) of CD8+ T cells, indicating the active state of CD8+ T cells (Figure 9D and Supplemental Figure 7C). Increased levels of MAL2 in tumor cells significantly suppressed granule and cytokine production of CD8+ T cells, whereas CD8+T cells from the MAL2-KD spheroids had much higher cytotoxicity (Figure 9, C and D, and Supplemental Figure 7C).
 Figure 9
Figure 9Depletion of MAL2 enhances CD8+ T cell cytotoxicity in human TNBC tumors. (A) MAL2 levels in tumor cells affect the cytotoxicity of the tumor-infiltrating CD8+ T cells in human TNBC tumors. Tumor cells isolated from fresh TNBC tumor tissues (patient #1 and #2) were transduced with lentiviral MAL2 or its shRNA. The tumor cells formed spheroids with CAFs isolated from the same tumor tissue. The tumor spheroids were cocultured with preactivated CD8+ T cell isolated from the same tumor tissue to detect the T cell cytotoxicity (spheroid dissociation rates). Representative images of spheroids are shown. Scale bar: 100 μm. (B) Quantitative data are presented as mean ± SD of represented images from 3 parallel experiments in A. (C) Spheroids from A were digested into single cells, which were stained for EpCAM (tumor cells), anti-CD140a (CAFs), and LIVE/DEAD dead-cell stain. Flow cytometry data show the CD8+ T cell cytotoxicity in patient-derived CAF-tumor spheroids with different MAL2 expression levels. (D) The CD8+T cells collected from A were analyzed for their activity. The CD8+ T cells were incubated with 50 ng/mL PMA, 1 μg/mL ionomycin, and 5 μg/mL brefeldin A for 5 hours, and were stained with antibodies against IFN-γ, TNF-α, and GZMB. Quantitative data are presented as mean ± SD of 3 technical experiments. (E) Correlation of MAL2 levels (pathological scores) with tumor-infiltrating CD8+ T cell cytotoxicity (determined by GZMB levels) in human breast tumor tissue microarrays. Immunohistochemical analyses of MAL2, CD8, and GZMB in human TNBC TMA slides were conducted, and quantitative results were obtained as described in Supplemental Methods. Statistical analyses were conducted using 1-way ANOVA test. *P < 0.05; **P < 0.01; ***P < 0.001.
To confirm the findings above, we also expressed NY-ESO-1 in human TNBC cells isolated from patient-derived xenograft (PDX) tissues, manipulated MAL2 expression as mentioned above (by using the lentiviral system), and then formed PDX-derived tumor organoids with the matched mouse CAFs. The spheroids were cocultured with preactivated NY-ESO-1–specific CD8+ T cells to test the cytotoxicity ex vivo (Supplemental Figure 8, A–C). The NY-ESO-1–specific CD8+ T cells cocultured with the MAL2-OE organoids displayed much lower cytotoxicity than the ones cocultured with the MAL2-WT organoids. However, coculturing with the MAL2-KD organoids significantly boosted T cell activity ex vivo.
Additionally, we analyzed human breast tumor tissue microarray to assess the effect of MAL2 levels on the cytotoxicity of tumor-infiltrating CD8+ T cells. A total of 260 breast tumor tissue samples (260 cores from 185 cases) were processed by double-staining (MAL2 and CD8a or GZMB) IHC. Tumor tissue samples were stained with anti-CD8a antibody for tumor-infiltrating CD8+ T cells and with anti-GZMB for cytotoxicity. Two consecutive TMA slides were used for each staining. The results showed that breast tumor tissues with a relatively high expression level of MAL2 (pathological scores 2–3) had significantly lower cytotoxicity of CD8+ T cells than those with low MAL2 expression (pathological scores 0–1) (Figure 9E, Supplemental Figure 8D, and Supplemental Tables 4 and 5), confirming our results from animal tumor models.
-
Discussion
Immune checkpoint blockade therapy has yielded promising results in both early and advanced stages of human cancers and is expected to substantially improve overall cancer prognosis. However, durable response rates of immunotherapy remain relatively modest in a significant number of cancer patients (15, 28, 47). Unlike most breast cancer subtypes that are not inherently immunogenic and typically have low T cell infiltration, TNBC is characterized by greater tumor immune infiltration, a predictive marker for responses to immunotherapy and favorable clinical outcomes. The inefficacy of immunotherapy in TNBCs is potentially caused by a myriad of other factors, including suppressive TME, tumor heterogeneity, inactive T effectors, and immune checkpoints (48, 49). As a major immune response to eliminate tumor cells in the body, CD8+ T cell–mediated cytotoxicity is dependent on its recognition of tumor-specific antigens via TCR. In fact, immune evasion of cancer is often associated with suppression of tumor-specific antigens and the absence of antigen-loaded MHC complex on the surface of tumor cells (50, 51). A functional antigen presentation pathway in the tumor cell consists of immunoproteasome-mediated antigen processing, antigen binding and transport in the ER, and antigen transport to the cell surface through the Golgi apparatus (52). If not used or recognized, the antigen-loaded MHC-I complex is destined for degradation or recycling by endocytosis (53). In this study, we found that the transmembrane protein MAL2 promotes the turnover of the antigen-bound MHC-I complex, thereby providing a mechanism of resistance to the CD8+ T cell–mediated immune attack. Consistent with this mechanism, expression of MAL2 in tumor cells was correlated with poor survival of patients with TNBC. Bioinformatics analysis also revealed that TNBC tumors with lower CD8+ T cell activities had relatively higher expression of MAL2 in tumor cells (Figure 3, A and C).
Major breakthroughs in cancer immunotherapy, particularly the capacity of antibodies targeting the immune checkpoints such as PD-1 and CTLA-4, have elicited much interest in understanding of tumor antigen presentation. Given the central role of CD8+ cytotoxic T cells in immune checkpoint blockade therapies, it is not surprising that mutations or transcriptional silencing in the genes encoding components of the MHC-I antigen processing pathway or the immune response pathway have emerged as a frequent cause of therapeutic resistance in a number of cancers, including neuroblastoma, small-cell lung cancer, and Merkel cell carcinoma (54–57). However, even with normal MHC-I expression, there must be alternative or parallel routes for tumor cells to survive immune surveillance, one of which is to upregulate the degradation and recycling of the MHC-I complex. Our results showed that high expression of MAL2 decreased the stability and level of the antigen-loaded MHC-I on the cell membrane, leading to ineffective antigen presentation. With low presentation efficiency, these tumor cells have more chances to evade recognition by CD8+ T cells, thereby greatly increasing the survival opportunity of the MAL2-high malignant cells in the tumor. Our scRNA-Seq analysis revealed that Mal2 overexpression in mouse tumor cells resulted in dampened maturation of CD8+ T cells and suppression of their cytotoxicity. Moreover, downregulation of the MHC-I antigen presentation machinery was also observed in metastatic TNBC, suggesting that spreading cancer cells maintain the phenotype of the primary tumor and thus the ability to evade immune surveillance in metastasis (58).
By interacting with RAB7 and MHC-I molecules, MAL2 may lead the endocytic MHC-I molecules to the late-stage endosome for degradation. Depletion of MAL2 significantly increases the antigen presentation on tumor cells and activates the cytotoxicity of CD8+ T cells. This finding has potential clinical applications for stratifying patients based on their immune responsiveness and thereby identifying patients suited for immunotherapy. Currently, in clinics, the primary biomarker for PD1/PD-L1 antibody therapy is the expression level of PD-L1 on cancer cells (59–62). However, administration of PD1/PD-L1 antibodies to TNBC patients with high PD-L1 tumors is unlikely to benefit patients whose tumors lack MHC-I–mediated antigen presentation, suggesting that MAL2 levels could be included in the biomarker panel for immune checkpoint therapies.
In summary, this study identifies MAL2 as a key player that downregulates CD8+ T cell cytotoxicity via suppression of the MHC-I–associated antigen presentation, and thus promotes tumor immune evasion. We demonstrated that inhibiting MAL2 is potentially an effective strategy for cancer immunotherapy. Further translational studies are warranted to transform this finding to therapeutics.
-
Methods
For additional details, see Supplemental Methods.
Tissue culture. Human breast cancer cell lines (MDA-MB-468 and HCC1954 from ATCC), the human HER293 cell line (ATCC), and murine mammary tumor cell lines EO771 (CH3 Biosystems) and 4T1 (ATCC) were cultured in DMEM supplemented with 10% FBS and 1% penicillin/streptomycin. For primary breast cancer cell culture, tumor tissues were collected and digested with MACS tumor dissociation kit (mouse, 130-096-730; human, 130-095-929; Miltenyi Biotec). Tumor tissues were sliced into small pieces in diameter around 2–4 mm, then incubated in 37°C water bath for 40 minutes and dissociated using the gentleMACS Octo Dissociator (Miltenyi Biotec). After centrifugation at 300 g for 7 minutes, single cells were cultured in DMEM/F-12 medium supplemented with 5% FBS, 1% penicillin/streptomycin, 20 ng/mL EGF, 0.5 mg/mL hydrocortisone, 100 ng/mL cholera toxin, and 10 μg/mL insulin. After 1–2 days of culture, cells were trypsinized, washed, and ready for the next procedures.
Cell proliferation assay. For the WST-1 cell proliferation assay, cells were seeded at a density of 5,000 cells per well (96-well-plate) in growth media and incubated at 37°C with 5% CO2 for 48 hours. Cell growth was analyzed using Takara PreMix WST-1 Cell Proliferation Assay System. For the IncuCyte cell proliferation assay, cells were seeded at a density of 500 cells per well (96-well plate) in growth media and incubated at 37°C with 5% CO2. Growth medium was replaced every 48 hours for 6 days. Cell growth was monitored and analyzed using IncuCyte S3 Live-Cell Analysis System.
Animal studies. C57/BL6, BALB/c, and NU/J nude mice were purchased from The Jackson Laboratory. Breast cancer cells were harvested and resuspended in serum-free DMEM/F-12 medium (Gibco) and then injected into the mammary fat pad (50 μL per site) from the fourth pair of 5-week-old female mouse mammary glands (right side). Tumors were measured in 2 dimensions using a manual caliper. Tumor volume was calculated using the following formula: V = 0.5 × length × width × width. Tumor volume was measured every 3 days. When harvesting, tumors were sliced into 3 parts: a small piece was fixed in formalin overnight and then in 70% ethanol for histopathological analysis; the second small piece was snap-frozen in liquid nitrogen to prepare cell lysate for immunoblotting analysis, and the remaining tumors were digested with the MACS tumor dissociation kit for single-cell analyses.
T cell isolation and effector function analysis. CD8+ T cell negative selection kits (Miltenyi Biotec) were used to isolate peripheral T cells from mouse spleens (39). To analyze tumor-infiltrating T cells, the tumors were first digested with collagenase IV (Sigma-Aldrich), and then the tumor-infiltrating leukocytes were isolated by centrifugation at 40%–70% Percoll (GE) gradient. To measure the effector function of CD8+ T cells, T cells were first incubated in the presence of 5 μg/mL BFA with 1 μM ionomycin and 50 ng/mL PMA for 5 hours, then stained with APC/Cy7 conjugated anti-CD8a antibody (BioLegend, then stained with APC/Cy7 CD8a antibody (BioLegend, catalog 100714 for mouse, and catalog 300926 for human). Cells were then fixed and permeabilized with 4% paraformaldehyde (PFA) and stained with APC anti-IFN-γ antibody (Biolegend, catalog 505810 for mouse, and catalog 502512 for human), PE anti-TNF-α antibody (Biolegend, catalog 506306 for mouse, and catalog 502909 for human), and PerCP/Cyanine5.5 anti-human/mouse Granzyme B (BioLegend, catalog 372212. Generally, in order to gate cytokines or granule-producing cells, unstimulated T cells or T cells stained by an isotype control antibody were used as negative controls. This gating strategy was applied in all the flow cytometry analyses unless otherwise indicated.
Measurement of CD8+ T cell cytotoxicity. For mouse CD8+ T cell cytotoxicity assays, splenocytes were isolated from female OT-I mice and stimulated with OVA257-264 (Sigma-Aldrich, S7951) in the presence of 10 ng/mL IL-2 for 3 days (39). The T cells were then centrifuged and cultured in fresh medium containing 10 ng/mL IL-2. After 2 more days, most cells in the culture were CTLs. To measure the cytotoxicity of CD8+ T cells, CTLs were pulsed with 2 nM OVA257-264 for 30 minutes. After washing CTLs and EO771 cells 3 times with PBS, we mixed the CTLs and EO771 cells (1 × 105) in the coculture medium (phenol-free RPMI 1640, 2% FBS) at ratios of 5:1, 1:1, or 1:5. After 6 hours, the CytoTox 96 nonradioactive cytotoxicity kit (Promega) was used to measure the cytotoxic efficiency by quantifying the release of endogenous LDH from EO771 cells.
For human CD8+ T cell cytotoxicity assays, anti–NY-ESO-1 T cells (Cellero, 1093-4493OC19) or anti–MAGE-A10 T cells (Cellero, 1125-4530DE19) were stimulated with NY-ESO-1 peptide (Cellero, 1095) or MAGE-A10 peptide (Cellero, 1138), respectively, in the presence of 10 ng/mL IL-2 for 2 days. The T cells were then centrifuged and cultured in fresh medium containing 10 ng/mL IL-2 for 2 more days. Anti–NY-ESO-1 T cells and anti–MAGE-A10 T cells were pulsed with NY-ESO-1 or MAGE-A10 peptide, respectively, for 30 minutes. After washing T cells and MDA-MB-468 cells 3 times with PBS, we mixed the T cells and MDA-MB-468 cells (1 × 105) in the coculture medium (phenol-free RPMI 1640, 2% FBS) at the ratio of 10:1 (anti–NY-ESO-1) or 20:1 (anti–MAGE-A10). After 24 hours, CytoTox 96 nonradioactive cytotoxicity kit (Promega) was used to measure the cytotoxic efficiency by quantifying the release of endogenous LDH from MDA-MB-468 cells.
MAL2 protein expression and mass spectrometry. Flag-tagged human MAL2 protein was overexpressed in HEK293T cells. Whole-cell lysates were prepared by incubating cells in 0.5% NP-40 lysis buffer on ice for 30 minutes. After clearing by centrifugation, cell lysates were incubated with anti-FLAG M2 Affinity Gel (Sigma-Aldrich) for 12 hours at 4°C, washed 3 times with lysis buffer without NP-40, and the Flag-MAL2 protein-containing complex was eluted from the gel with FLAG peptides and subjected to digestion using a DigestPro automated digestion unit (CEM Corp.). The resulting peptides were separated and delivered using an Ultimate 3000 UHPLC (Thermo Fisher Scientific) at 300 nL/minute by C18 fritted microcapillary columns. Peptides were analyzed by positive ion mode using an Orbitrap Fusion Lumos mass spectrometer (Thermo Fisher Scientific).
Immunofluorescence. Immunofluorescence was performed as described previously (63). In brief, cells (5 × 103 per well) were seeded into Millicell EZ 8-well glass slides and cultured overnight. The cells were then rinsed with PBS and fixed with 4% PFA for 10 minutes at room temperature. Fixed cells were incubated with 0.5% Triton X-100/PBS and blocked with 0.2% BSA/PBS. Cells were then incubated with primary antibody overnight at 4 °C, followed by incubation with Alexa Fluor 488-conjugated goat anti-rabbit IgG antibody (Invitrogen, catalog A32731, dilution 1:300), Alexa Fluor 488-conjugated goat anti-mouse IgG antibody (Invitrogen, catalog A32723, dilution 1:300), Alexa Fluor 647-conjugated donkey anti-rat IgG antibody (Invitrogen, catalog A-21247, dilution 1:300), or 647-conjugated donkey anti-mouse IgG antibody (Invitrogen, catalog A32787, dilution 1:300) for 1 hour at room temperature. DAPI (Sigma-Aldrich) staining was performed after antibody staining. Samples were mounted with mounting medium (Sigma-Aldrich) and fluorescent images were taken using the Leica TCS SP8 (upright high-speed multiphoton) confocal imaging system. 3D images were built and analyzed with Imaris ×64 8.1.2 software.
Proximity ligation assay. Proximity ligation assay (PLA) was performed with Duolink In Situ Red Starter Kit Mouse/Rabbit (MilliporeSigma). Briefly, MDA-MB-468 cells (MAL2-WT, MAL2-OE, and MAL2-KD) were seeded on the 8-well Millicell EZ glass slide (2,000 cells per well). After 12 hours incubation, cells were fixed and permeabilized. Cells were blocked for 1 hour in a heated humidity chamber at 37°C and then incubated with anti–HLA-A2 (Abcam, catalog ab168405) and anti-RAB7 (Abcam, catalog ab50533) antibodies for another 2 hours. Duolink PLA Probes were added, and then the ligation and amplification were applied at 37°C following the instructions of the assay kit. After the final wash, the slides were mounted using Duolink In Situ Mounting Media with DAPI. Images were taken under a Leica confocal microscope and analyzed with ImageJ (NIH).
Endosome isolation. Endosomes were isolated using Trident Endosome Isolation Kit (GeneTex, GTX35192). Briefly, 3 × 107 cells from each sample were collected and washed once with cold PBS. The supernatant was removed completely, and the cell pellet was resuspended in 500 μL buffer A. The cell suspension was incubated on ice for 5–10 minutes and vortexed vigorously for 10–30 seconds, and then transferred to a filter cartridge. To remove the mitochondria, the cell suspension was centrifuged at 16,000 g for 30 seconds. The pellet was resuspended by vortexing briefly and centrifuged at 700 g for 2–3 minutes. The supernatant was carefully transferred to a fresh 1.5 mL tube and centrifuged at 16,000 g at 4°C for 30–60 minutes. The resulting supernatant was transferred to a fresh 1.5 mL tube, added with buffer B and vortexed briefly. The tube was incubated at 4°C overnight, and centrifuged at 10,000 g for 30 minutes at 4°C. After removing the supernatant, the pellet contained isolated endosomes.
Breast tumor organoid culture. Tumor cells and CAFs with a concentration of 2 × 105 cells per milliliter were suspended into a mixture of pre-cold breast cancer culture medium (40) and Matrigel (Corning, 354234) with a ratio of 1:1. The cells were split into each well of a Costar 6-well culture plate with an ultralow attachment surface (Corning) and incubated in a humidified 37°C, 5% CO2 incubator. The medium was replaced by fresh 10% Matrigel breast cancer culture medium every 2 days, and the organoids were collected by using cell strainers with 40 μm nylon mesh (Thermo Fisher Scientific). At day 7, the organoids with diameter between 70 and 100 μm were collected by the corresponding cell strainers (Thermo Fisher Scientific) and gently resuspended in Matrigel-free DMEM/F-12 medium containing 10% FBS, 1% penicillin/streptomycin, and 2 ng/mL IL2. Organoid cell numbers were counted by digesting the organoids with trypLe Express (Thermo Fisher Scientific) at 37°C with a shaking velocity of 500 rpm for 15 minutes. The organoids were mixed with OT-I–specific CD8+ T cells with a ratio of 1:5 (organoid cell vs. T cell) in the Costar 6-well culture plate. After coculturing for 24–30 hours, the killing efficiency was evaluated by optical imaging and flow cytometry.
Single-cell RNA-Seq data analysis of T cells with altered levels of MAL2. We generated in-house single-cell RNA-Seq data of T cells collected from mouse TNBC tumors of Mal2 overexpression, KD, and control conditions. Raw sequencing data were processed by Cell Ranger for creation of a FASTQ file, alignment to mm10 genome, and generation of a gene expression count matrix. Gene expression was normalized by TPM. Cell clustering analysis was conducted by Seurat v3 with default parameters. Subtypes of T cells were annotated by selected markers as illustrated in Figure 8A (Supplemental Table 3) (64–66). Differential gene expression states and relative expression level were computed by using a left truncated mixture Gaussian model. T cell cytotoxicity levels were estimated by the average gene expression level of selected gene markers Ccl4, Cst7, Gzmb, and Ifng. Genes involved in negative and positive regulation of T cell functions were collected from the Gene Ontology database. Standard laboratory practice random procedures were used for cell line groups and mice of the same age and sex. During the evaluation of experiments and results, researchers were not blindly assigned.
Immunoblotting analysis. Immunoblotting analysis was described in the Supplemental Methods. See complete unedited blots in the supplemental material.
Data availability. scRNA-Seq data that support the findings of this study have been deposited in NCBI’s Gene Expression Omnibus (GEO GSE144099). Other source data and statistics are provided in Supplemental Tables 1–5.
Code availability. Custom codes used to support the findings of this research are available from the corresponding author (Chi Zhang) upon reasonable request.
Statistics. The number of mice required was calculated by performing a power analysis using data from a small pilot experiment. Values represent mean ± SD unless otherwise indicated. Breast cancer data sets were obtained from TCGA database (https://gdac.broadinstitute.org/) or otherwise indicated. One-way or 2-way ANOVA tests or an unpaired 2-tailed t test were used to determine the P value, and the statistical significance was set at P less than 0.05. Technical copies were defined by the tests or assays performed multiple times on the same samples. Unless otherwise stated, the data were tested for normal distribution and applied to the Student’s t test (if normally distributed) or the nonparametric Mann-Whitney U test. Kaplan-Meier plots were generated using the GraphPad Prism 7.0 survival calculation tool, and significance was calculated at P less than 0.05 by a 2-tailed log rank test.
Study approval. All animal experiments were performed in accordance with the NIH animal use guidelines, and the protocols were approved by the Animal Care and Use Committee of Indiana University School of Medicine.
-
Author contributions
YF, LW, YS, KVJ, TD, TY, Y. Li, HE, ED, XL, and XZ designed and performed the experiments. As for the 3 co–first authors, YF designed, conducted, and coordinated cell-based and animal studies; LW was in charge of molecular and biochemical studies; and CW performed and coordinated scRNA-Seq analysis and bioinformatics analysis. While they equally contributed to the study, the authorship order was assigned based on the amount of results they provided. CW, KS, SC Y. Liu, and CZ conducted bioinformatics and statistical analyses. ZZ established human breast tumor organoid models. JW established and provided the PDX model. BPS provided guidance in analysis of clinical samples and databases. ABC and GES analyzed tumor tissue microarrays. YF, CZ, XL, and XZ wrote and revised the manuscript.
-
Acknowledgments
We thank MA Alonso for sharing the anti-Mal2 antibodies. We thank Indiana Center for Biological Microscopy, the Center for Medical Genomics, the Flow Cytometry Resource Facility of Indiana University Simon Cancer Center (IUSCC), and the Department of Pathology and Laboratory Medicine at Indiana University School of Medicine for providing us with instruments and technical support. We also thank Tissue Procurement & Distribution Core and the Tissue Bank, which are IUSCC facilities that provided us with service and technical support for human breast cancer tissue sample procurement and processing. This study is funded partly by the Vera Bradley Foundation for Breast Cancer Research (to XZ and XL), American Cancer Society Institutional Research Grant 16-192-31 (to XZ), NIH grants R01CA203737 and R01CA206366 (to XL), and Indiana University School of Medicine Strategic Research Initiative (to XL).
Address correspondence to: Chi Zhang, Indiana University School of Medicine, 410 W. 10th Street, HS 5000, Indianapolis, Indiana 46202, USA. Phone: 317.278.9625; Email: [email protected]. Or to: Xiongbin Lu, Indiana University School of Medicine, 980 W. Walnut Street, R3.C218D, Indianapolis, Indiana 46202, USA. Phone: 317.274.4398; Email: [email protected]. Or to: Xinna Zhang, Indiana University School of Medicine, 980 W. Walnut Street, R3.C216B, Indianapolis, Indiana 46202, USA. Phone: 317.274.7931; Email: [email protected].
-
Footnotes
LW’s present address is: Institute of Medicinal Biotechnology, Chinese Academy of Sciences & Peking Union Medical College, Beijing, China.
Conflict of interest: The authors have declared that no conflict of interest exists.
Copyright: © 2021, American Society for Clinical Investigation.
Reference information: J Clin Invest. 2021;131(1):e140837.https://doi.org/10.1172/JCI140837.
See the related Commentary at Immune MAL2-practice: breast cancer immunoevasion via MHC class I degradation.
-
References
- Ribas A, Wolchok JD. Cancer immunotherapy using checkpoint blockade. Science. 2018;359(6382):1350–1355.
- Kwa MJ, Adams S. Checkpoint inhibitors in triple-negative breast cancer (TNBC): where to go from here. Cancer. 2018;124(10):2086–2103.
- Keren L, et al. A structured tumor-immune microenvironment in triple negative breast cancer revealed by multiplexed ion beam imaging. Cell. 2018;174(6):1373–1387.e19.
- Rosenberg SA, Restifo NP. Adoptive cell transfer as personalized immunotherapy for human cancer. Science. 2015;348(6230):62–68.
- Simoni Y, et al. Bystander CD8+ T cells are abundant and phenotypically distinct in human tumour infiltrates. Nature. 2018;557(7706):575–579.
- Maleki Vareki S, Garrigós C, Duran I. Biomarkers of response to PD-1/PD-L1 inhibition. Crit Rev Oncol Hematol. 2017;116:116–124.
- Charoentong P, et al. Pan-cancer immunogenomic analyses reveal genotype-immunophenotype relationships and predictors of response to checkpoint blockade. Cell Rep. 2017;18(1):248–262.
- Rosenthal R, et al. Neoantigen-directed immune escape in lung cancer evolution. Nature. 2019;567(7749):479–485.
- Zhang AW, et al. Interfaces of malignant and immunologic clonal dynamics in ovarian cancer. Cell. 2018;173(7):1755–1769.e22.
- McGranahan N, et al. Clonal neoantigens elicit T cell immunoreactivity and sensitivity to immune checkpoint blockade. Science. 2016;351(6280):1463–1469.
- Łuksza M, et al. A neoantigen fitness model predicts tumour response to checkpoint blockade immunotherapy. Nature. 2017;551(7681):517–520.
- Chen L, Azuma T, Yu W, Zheng X, Luo L, Chen L. B7-H1 maintains the polyclonal T cell response by protecting dendritic cells from cytotoxic T lymphocyte destruction. Proc Natl Acad Sci U S A. 2018;115(12):3126–3131.
- Bukur J, Jasinski S, Seliger B. The role of classical and non-classical HLA class I antigens in human tumors. Semin Cancer Biol. 2012;22(4):350–358.
- Donia M, et al. Acquired immune resistance follows complete tumor regression without loss of target antigens or IFNgamma signaling. Cancer Res. 2017;77(17):4562–4566.
- Spranger S, Gajewski TF. Impact of oncogenic pathways on evasion of antitumour immune responses. Nat Rev Cancer. 2018;18(3):139–147.
- Neefjes J, Jongsma ML, Paul P, Bakke O. Towards a systems understanding of MHC class I and MHC class II antigen presentation. Nat Rev Immunol. 2011;11(12):823–836.
- Blum JS, Wearsch PA, Cresswell P. Pathways of antigen processing. Annu Rev Immunol. 2013;31:443–473.
- Leone P, Shin EC, Perosa F, Vacca A, Dammacco F, Racanelli V. MHC class I antigen processing and presenting machinery: organization, function, and defects in tumor cells. J Natl Cancer Inst. 2013;105(16):1172–1187.
- Beatty GL, Gladney WL. Immune escape mechanisms as a guide for cancer immunotherapy. Clin Cancer Res. 2015;21(4):687–692.
- Grant BD, Donaldson JG. Pathways and mechanisms of endocytic recycling. Nat Rev Mol Cell Biol. 2009;10(9):597–608.
- van Endert P. Intracellular recycling and cross-presentation by MHC class I molecules. Immunol Rev. 2016;272(1):80–96.
- Eyster CA, et al. Discovery of new cargo proteins that enter cells through clathrin-independent endocytosis. Traffic. 2009;10(5):590–599.
- Naslavsky N, Caplan S. The enigmatic endosome - sorting the ins and outs of endocytic trafficking. J Cell Sci. 2018;131(13):jcs216499.
- Jovic M, Sharma M, Rahajeng J, Caplan S. The early endosome: a busy sorting station for proteins at the crossroads. Histol Histopathol. 2010;25(1):99–112.View this article via: PubMed Google Scholar
- Foulkes WD, Smith IE, Reis-Filho JS. Triple-negative breast cancer. N Engl J Med. 2010;363(20):1938–1948.
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2017. CA Cancer J Clin. 2017;67(1):7–30.
- Adams S, et al. Pembrolizumab monotherapy for previously treated metastatic triple-negative breast cancer: cohort A of the phase II KEYNOTE-086 study. Ann Oncol. 2019;30(3):397–404.
- Adams S, et al. Pembrolizumab monotherapy for previously untreated, PD-L1-positive, metastatic triple-negative breast cancer: cohort B of the phase II KEYNOTE-086 study. Ann Oncol. 2019;30(3):405–411.
- Madrid R, et al. The formin INF2 regulates basolateral-to-apical transcytosis and lumen formation in association with Cdc42 and MAL2. Dev Cell. 2010;18(5):814–827.
- Byrne JA, et al. MAL2 and tumor protein D52 (TPD52) are frequently overexpressed in ovarian carcinoma, but differentially associated with histological subtype and patient outcome. BMC Cancer. 2010;10:497. View this article via: PubMed Google Scholar
- de Marco MC, Puertollano R, Martínez-Menárguez JA, Alonso MA. Dynamics of MAL2 during glycosylphosphatidylinositol-anchored protein transcytotic transport to the apical surface of hepatoma HepG2 cells. Traffic. 2006;7(1):61–73.
- Marazuela M, Acevedo A, García-López MA, Adrados M, de Marco MC, Alonso MA. Expression of MAL2, an integral protein component of the machinery for basolateral-to-apical transcytosis, in human epithelia. J Histochem Cytochem. 2004;52(2):243–252.
- de Marco MC, et al. MAL2, a novel raft protein of the MAL family, is an essential component of the machinery for transcytosis in hepatoma HepG2 cells. J Cell Biol. 2002;159(1):37–44.
- Marcotte R, et al. Essential gene profiles in breast, pancreatic, and ovarian cancer cells. Cancer Discov. 2012;2(2):172–189.
- Blomen VA, et al. Gene essentiality and synthetic lethality in haploid human cells. Science. 2015;350(6264):1092–1096.
- Wang T, et al. Identification and characterization of essential genes in the human genome. Science. 2015;350(6264):1096–1101.
- Silva JM, et al. Profiling essential genes in human mammary cells by multiplex RNAi screening. Science. 2008;319(5863):617–620.
- Chang W, et al. ICTD: Inference of cell types and deconvolution — a next generation deconvolution method for accurate assess cell population and activities in tumor microenvironment [preprint]. https://doi.org/10.1101/426593 Posted on bioRxiv September 27, 2018.
- Yang W, et al. Potentiating the antitumour response of CD8(+) T cells by modulating cholesterol metabolism. Nature. 2016;531(7596):651–655.
- Sachs N, et al. A living biobank of breast cancer organoids captures disease heterogeneity. Cell. 2018;172(1-2):373–386.e10.
- Rodal SK, Skretting G, Garred O, Vilhardt F, van Deurs B, Sandvig K. Extraction of cholesterol with methyl-beta-cyclodextrin perturbs formation of clathrin-coated endocytic vesicles. Mol Biol Cell. 1999;10(4):961–974.
- Koivusalo M, et al. Amiloride inhibits macropinocytosis by lowering submembranous pH and preventing Rac1 and Cdc42 signaling. J Cell Biol. 2010;188(4):547–563.
- Commisso C, et al. Macropinocytosis of protein is an amino acid supply route in Ras-transformed cells. Nature. 2013;497(7451):633–637.
- Wang Z, Zhang L, Qiao A, Watson K, Zhang J, Fan GH. Activation of CXCR4 triggers ubiquitination and down-regulation of major histocompatibility complex class I (MHC-I) on epithelioid carcinoma HeLa cells. J Biol Chem. 2008;283(7):3951–3959.
- Sapmaz A, et al. USP32 regulates late endosomal transport and recycling through deubiquitylation of Rab7. Nat Commun. 2019;10(1):1454.
- Marazuela M, Martín-Belmonte F, García-López MA, Aranda JF, de Marco MC, Alonso MA. Expression and distribution of MAL2, an essential element of the machinery for basolateral-to-apical transcytosis, in human thyroid epithelial cells. Endocrinology. 2004;145(2):1011–1016.
- Emens LA, et al. Long-term clinical outcomes and biomarker analyses of atezolizumab therapy for patients with metastatic triple-negative breast cancer: a phase 1 study. JAMA Oncol. 2019;5(1):74–82.
- Hida AI, et al. Prognostic and predictive impacts of tumor-infiltrating lymphocytes differ between triple-negative and HER2-positive breast cancers treated with standard systemic therapies. Breast Cancer Res Treat. 2016;158(1):1–9.
- Bergin ART, Loi S. Triple-negative breast cancer: recent treatment advances. F1000Res. 2019;8:F1000 Faculty Rev-1342.
- Borcherding N, Kolb R, Gullicksrud J, Vikas P, Zhu Y, Zhang W. Keeping tumors in check: a mechanistic review of clinical response and resistance to immune checkpoint blockade in cancer. J Mol Biol. 2018;430(14):2014–2029.
- Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144(5):646–674.
- Hewitt EW. The MHC class I antigen presentation pathway: strategies for viral immune evasion. Immunology. 2003;110(2):163–169.
- Montealegre S, van Endert PM. Endocytic recycling of MHC class I molecules in non-professional antigen presenting and dendritic cells. Front Immunol. 2018;9:3098. View this article via: PubMed Google Scholar
- Burr ML, et al. An evolutionarily conserved function of polycomb silences the MHC class I antigen presentation pathway and enables immune evasion in cancer. Cancer Cell. 2019;36(4):385–401.e8.
- Patel SJ, et al. Identification of essential genes for cancer immunotherapy. Nature. 2017;548(7669):537–542.
- Pan D, et al. A major chromatin regulator determines resistance of tumor cells to T cell-mediated killing. Science. 2018;359(6377):770–775.
- Paulson KG, et al. Acquired cancer resistance to combination immunotherapy from transcriptional loss of class I HLA. Nat Commun. 2018;9(1):3868.
- Pedersen MH, Hood BL, Beck HC, Conrads TP, Ditzel HJ, Leth-Larsen R. Downregulation of antigen presentation-associated pathway proteins is linked to poor outcome in triple-negative breast cancer patient tumors. Oncoimmunology. 2017;6(5):e1305531.
- Yi M, et al. Biomarkers for predicting efficacy of PD-1/PD-L1 inhibitors. Mol Cancer. 2018;17(1):129.
- Page DB, Postow MA, Callahan MK, Allison JP, Wolchok JD. Immune modulation in cancer with antibodies. Annu Rev Med. 2014;65:185–202.
- Goswami S, Aparicio A, Subudhi SK. Immune checkpoint therapies in prostate cancer. Cancer J. 2016;22(2):117–120.
- Sharma P, Hu-Lieskovan S, Wargo JA, Ribas A. Primary, adaptive, and acquired resistance to cancer immunotherapy. Cell. 2017;168(4):707–723.
- Liu Y, et al. Targeting 17q23 amplicon to overcome the resistance to anti-HER2 therapy in HER2+ breast cancer. Nat Commun. 2018;9(1):4718.
- Mahnke YD, Brodie TM, Sallusto F, Roederer M, Lugli E. The who’s who of T-cell differentiation: human memory T-cell subsets. European journal of immunology. 2013;43(11):2797–2809.
- Kanhere A, et al. T-bet and GATA3 orchestrate Th1 and Th2 differentiation through lineage-specific targeting of distal regulatory elements. Nat Commun. 2012;3:1268. View this article via: PubMed Google Scholar
- Burchill MA, Yang J, Vogtenhuber C, Blazar BR, Farrar MA. IL-2 receptor beta-dependent STAT5 activation is required for the development of Foxp3+ regulatory T cells. J Immunol. 2007;178(1):280–290.
-
Version history
- Version 1 (September 29, 2020): In-Press Preview
- Version 2 (January 4, 2021): Print issue publication



Copyright © 2024 American Society for Clinical Investigation
ISSN: 0021-9738 (print), 1558-8238 (online)










