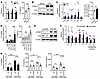Advertisement
Research ArticleImmunologyNeuroscience Free access | 10.1172/JCI133737
Type I interferon response drives neuroinflammation and synapse loss in Alzheimer disease
Ethan R. Roy,1,2 Baiping Wang,1 Ying-wooi Wan,3 Gabriel Chiu,1 Allysa Cole,1 Zhuoran Yin,4 Nicholas E. Propson,1,5 Yin Xu,1 Joanna L. Jankowsky,6 Zhandong Liu,7 Virginia M.-Y. Lee,8 John Q. Trojanowski,8 Stephen D. Ginsberg,9,10 Oleg Butovsky,4,11 Hui Zheng,1,3 and Wei Cao1,3
1Huffington Center on Aging,
2Translational Biology & Molecular Medicine Program, and
3Department of Molecular and Human Genetics, Baylor College of Medicine, Houston, Texas, USA.
4Ann Romney Center for Neurologic Diseases, Department of Neurology, Brigham and Women’s Hospital, Harvard Medical School, Boston, Massachusetts, USA.
5Molecular and Cellular Biology Program, Department of Molecular and Cellular Biology,
6Department of Neuroscience, and
7Department of Pediatrics-Neurology, Baylor College of Medicine, Houston, Texas, USA.
8Department of Pathology and Laboratory Medicine, University of Pennsylvania School of Medicine, Philadelphia, Pennsylvania, USA.
9Center for Dementia Research, Nathan Kline Institute, Orangeburg, New York, USA.
10Departments of Psychiatry, Neuroscience & Physiology and the NYU Neuroscience Institute, New York University Langone Medical Center, New York, New York, USA.
11Evergrande Center for Immunologic Diseases, Brigham and Women’s Hospital, Harvard Medical School, Boston, Massachusetts, USA.
Address correspondence to: Wei Cao or Hui Zheng, One Baylor Plaza, BCM 230, Baylor College of Medicine, Houston, Texas 77030, USA. Phone: 713.798.8523; Email: [email protected] (W. Cao). Phone: 713.798.5804; Email: [email protected] (H. Zheng).
Find articles by
Roy, E.
in:
JCI
|
PubMed
|
Google Scholar
|

1Huffington Center on Aging,
2Translational Biology & Molecular Medicine Program, and
3Department of Molecular and Human Genetics, Baylor College of Medicine, Houston, Texas, USA.
4Ann Romney Center for Neurologic Diseases, Department of Neurology, Brigham and Women’s Hospital, Harvard Medical School, Boston, Massachusetts, USA.
5Molecular and Cellular Biology Program, Department of Molecular and Cellular Biology,
6Department of Neuroscience, and
7Department of Pediatrics-Neurology, Baylor College of Medicine, Houston, Texas, USA.
8Department of Pathology and Laboratory Medicine, University of Pennsylvania School of Medicine, Philadelphia, Pennsylvania, USA.
9Center for Dementia Research, Nathan Kline Institute, Orangeburg, New York, USA.
10Departments of Psychiatry, Neuroscience & Physiology and the NYU Neuroscience Institute, New York University Langone Medical Center, New York, New York, USA.
11Evergrande Center for Immunologic Diseases, Brigham and Women’s Hospital, Harvard Medical School, Boston, Massachusetts, USA.
Address correspondence to: Wei Cao or Hui Zheng, One Baylor Plaza, BCM 230, Baylor College of Medicine, Houston, Texas 77030, USA. Phone: 713.798.8523; Email: [email protected] (W. Cao). Phone: 713.798.5804; Email: [email protected] (H. Zheng).
Find articles by
Wang, B.
in:
JCI
|
PubMed
|
Google Scholar
|

1Huffington Center on Aging,
2Translational Biology & Molecular Medicine Program, and
3Department of Molecular and Human Genetics, Baylor College of Medicine, Houston, Texas, USA.
4Ann Romney Center for Neurologic Diseases, Department of Neurology, Brigham and Women’s Hospital, Harvard Medical School, Boston, Massachusetts, USA.
5Molecular and Cellular Biology Program, Department of Molecular and Cellular Biology,
6Department of Neuroscience, and
7Department of Pediatrics-Neurology, Baylor College of Medicine, Houston, Texas, USA.
8Department of Pathology and Laboratory Medicine, University of Pennsylvania School of Medicine, Philadelphia, Pennsylvania, USA.
9Center for Dementia Research, Nathan Kline Institute, Orangeburg, New York, USA.
10Departments of Psychiatry, Neuroscience & Physiology and the NYU Neuroscience Institute, New York University Langone Medical Center, New York, New York, USA.
11Evergrande Center for Immunologic Diseases, Brigham and Women’s Hospital, Harvard Medical School, Boston, Massachusetts, USA.
Address correspondence to: Wei Cao or Hui Zheng, One Baylor Plaza, BCM 230, Baylor College of Medicine, Houston, Texas 77030, USA. Phone: 713.798.8523; Email: [email protected] (W. Cao). Phone: 713.798.5804; Email: [email protected] (H. Zheng).
Find articles by Wan, Y. in: JCI | PubMed | Google Scholar
1Huffington Center on Aging,
2Translational Biology & Molecular Medicine Program, and
3Department of Molecular and Human Genetics, Baylor College of Medicine, Houston, Texas, USA.
4Ann Romney Center for Neurologic Diseases, Department of Neurology, Brigham and Women’s Hospital, Harvard Medical School, Boston, Massachusetts, USA.
5Molecular and Cellular Biology Program, Department of Molecular and Cellular Biology,
6Department of Neuroscience, and
7Department of Pediatrics-Neurology, Baylor College of Medicine, Houston, Texas, USA.
8Department of Pathology and Laboratory Medicine, University of Pennsylvania School of Medicine, Philadelphia, Pennsylvania, USA.
9Center for Dementia Research, Nathan Kline Institute, Orangeburg, New York, USA.
10Departments of Psychiatry, Neuroscience & Physiology and the NYU Neuroscience Institute, New York University Langone Medical Center, New York, New York, USA.
11Evergrande Center for Immunologic Diseases, Brigham and Women’s Hospital, Harvard Medical School, Boston, Massachusetts, USA.
Address correspondence to: Wei Cao or Hui Zheng, One Baylor Plaza, BCM 230, Baylor College of Medicine, Houston, Texas 77030, USA. Phone: 713.798.8523; Email: [email protected] (W. Cao). Phone: 713.798.5804; Email: [email protected] (H. Zheng).
Find articles by Chiu, G. in: JCI | PubMed | Google Scholar
1Huffington Center on Aging,
2Translational Biology & Molecular Medicine Program, and
3Department of Molecular and Human Genetics, Baylor College of Medicine, Houston, Texas, USA.
4Ann Romney Center for Neurologic Diseases, Department of Neurology, Brigham and Women’s Hospital, Harvard Medical School, Boston, Massachusetts, USA.
5Molecular and Cellular Biology Program, Department of Molecular and Cellular Biology,
6Department of Neuroscience, and
7Department of Pediatrics-Neurology, Baylor College of Medicine, Houston, Texas, USA.
8Department of Pathology and Laboratory Medicine, University of Pennsylvania School of Medicine, Philadelphia, Pennsylvania, USA.
9Center for Dementia Research, Nathan Kline Institute, Orangeburg, New York, USA.
10Departments of Psychiatry, Neuroscience & Physiology and the NYU Neuroscience Institute, New York University Langone Medical Center, New York, New York, USA.
11Evergrande Center for Immunologic Diseases, Brigham and Women’s Hospital, Harvard Medical School, Boston, Massachusetts, USA.
Address correspondence to: Wei Cao or Hui Zheng, One Baylor Plaza, BCM 230, Baylor College of Medicine, Houston, Texas 77030, USA. Phone: 713.798.8523; Email: [email protected] (W. Cao). Phone: 713.798.5804; Email: [email protected] (H. Zheng).
Find articles by Cole, A. in: JCI | PubMed | Google Scholar
1Huffington Center on Aging,
2Translational Biology & Molecular Medicine Program, and
3Department of Molecular and Human Genetics, Baylor College of Medicine, Houston, Texas, USA.
4Ann Romney Center for Neurologic Diseases, Department of Neurology, Brigham and Women’s Hospital, Harvard Medical School, Boston, Massachusetts, USA.
5Molecular and Cellular Biology Program, Department of Molecular and Cellular Biology,
6Department of Neuroscience, and
7Department of Pediatrics-Neurology, Baylor College of Medicine, Houston, Texas, USA.
8Department of Pathology and Laboratory Medicine, University of Pennsylvania School of Medicine, Philadelphia, Pennsylvania, USA.
9Center for Dementia Research, Nathan Kline Institute, Orangeburg, New York, USA.
10Departments of Psychiatry, Neuroscience & Physiology and the NYU Neuroscience Institute, New York University Langone Medical Center, New York, New York, USA.
11Evergrande Center for Immunologic Diseases, Brigham and Women’s Hospital, Harvard Medical School, Boston, Massachusetts, USA.
Address correspondence to: Wei Cao or Hui Zheng, One Baylor Plaza, BCM 230, Baylor College of Medicine, Houston, Texas 77030, USA. Phone: 713.798.8523; Email: [email protected] (W. Cao). Phone: 713.798.5804; Email: [email protected] (H. Zheng).
Find articles by Yin, Z. in: JCI | PubMed | Google Scholar
1Huffington Center on Aging,
2Translational Biology & Molecular Medicine Program, and
3Department of Molecular and Human Genetics, Baylor College of Medicine, Houston, Texas, USA.
4Ann Romney Center for Neurologic Diseases, Department of Neurology, Brigham and Women’s Hospital, Harvard Medical School, Boston, Massachusetts, USA.
5Molecular and Cellular Biology Program, Department of Molecular and Cellular Biology,
6Department of Neuroscience, and
7Department of Pediatrics-Neurology, Baylor College of Medicine, Houston, Texas, USA.
8Department of Pathology and Laboratory Medicine, University of Pennsylvania School of Medicine, Philadelphia, Pennsylvania, USA.
9Center for Dementia Research, Nathan Kline Institute, Orangeburg, New York, USA.
10Departments of Psychiatry, Neuroscience & Physiology and the NYU Neuroscience Institute, New York University Langone Medical Center, New York, New York, USA.
11Evergrande Center for Immunologic Diseases, Brigham and Women’s Hospital, Harvard Medical School, Boston, Massachusetts, USA.
Address correspondence to: Wei Cao or Hui Zheng, One Baylor Plaza, BCM 230, Baylor College of Medicine, Houston, Texas 77030, USA. Phone: 713.798.8523; Email: [email protected] (W. Cao). Phone: 713.798.5804; Email: [email protected] (H. Zheng).
Find articles by Propson, N. in: JCI | PubMed | Google Scholar
1Huffington Center on Aging,
2Translational Biology & Molecular Medicine Program, and
3Department of Molecular and Human Genetics, Baylor College of Medicine, Houston, Texas, USA.
4Ann Romney Center for Neurologic Diseases, Department of Neurology, Brigham and Women’s Hospital, Harvard Medical School, Boston, Massachusetts, USA.
5Molecular and Cellular Biology Program, Department of Molecular and Cellular Biology,
6Department of Neuroscience, and
7Department of Pediatrics-Neurology, Baylor College of Medicine, Houston, Texas, USA.
8Department of Pathology and Laboratory Medicine, University of Pennsylvania School of Medicine, Philadelphia, Pennsylvania, USA.
9Center for Dementia Research, Nathan Kline Institute, Orangeburg, New York, USA.
10Departments of Psychiatry, Neuroscience & Physiology and the NYU Neuroscience Institute, New York University Langone Medical Center, New York, New York, USA.
11Evergrande Center for Immunologic Diseases, Brigham and Women’s Hospital, Harvard Medical School, Boston, Massachusetts, USA.
Address correspondence to: Wei Cao or Hui Zheng, One Baylor Plaza, BCM 230, Baylor College of Medicine, Houston, Texas 77030, USA. Phone: 713.798.8523; Email: [email protected] (W. Cao). Phone: 713.798.5804; Email: [email protected] (H. Zheng).
Find articles by Xu, Y. in: JCI | PubMed | Google Scholar
1Huffington Center on Aging,
2Translational Biology & Molecular Medicine Program, and
3Department of Molecular and Human Genetics, Baylor College of Medicine, Houston, Texas, USA.
4Ann Romney Center for Neurologic Diseases, Department of Neurology, Brigham and Women’s Hospital, Harvard Medical School, Boston, Massachusetts, USA.
5Molecular and Cellular Biology Program, Department of Molecular and Cellular Biology,
6Department of Neuroscience, and
7Department of Pediatrics-Neurology, Baylor College of Medicine, Houston, Texas, USA.
8Department of Pathology and Laboratory Medicine, University of Pennsylvania School of Medicine, Philadelphia, Pennsylvania, USA.
9Center for Dementia Research, Nathan Kline Institute, Orangeburg, New York, USA.
10Departments of Psychiatry, Neuroscience & Physiology and the NYU Neuroscience Institute, New York University Langone Medical Center, New York, New York, USA.
11Evergrande Center for Immunologic Diseases, Brigham and Women’s Hospital, Harvard Medical School, Boston, Massachusetts, USA.
Address correspondence to: Wei Cao or Hui Zheng, One Baylor Plaza, BCM 230, Baylor College of Medicine, Houston, Texas 77030, USA. Phone: 713.798.8523; Email: [email protected] (W. Cao). Phone: 713.798.5804; Email: [email protected] (H. Zheng).
Find articles by Jankowsky, J. in: JCI | PubMed | Google Scholar
1Huffington Center on Aging,
2Translational Biology & Molecular Medicine Program, and
3Department of Molecular and Human Genetics, Baylor College of Medicine, Houston, Texas, USA.
4Ann Romney Center for Neurologic Diseases, Department of Neurology, Brigham and Women’s Hospital, Harvard Medical School, Boston, Massachusetts, USA.
5Molecular and Cellular Biology Program, Department of Molecular and Cellular Biology,
6Department of Neuroscience, and
7Department of Pediatrics-Neurology, Baylor College of Medicine, Houston, Texas, USA.
8Department of Pathology and Laboratory Medicine, University of Pennsylvania School of Medicine, Philadelphia, Pennsylvania, USA.
9Center for Dementia Research, Nathan Kline Institute, Orangeburg, New York, USA.
10Departments of Psychiatry, Neuroscience & Physiology and the NYU Neuroscience Institute, New York University Langone Medical Center, New York, New York, USA.
11Evergrande Center for Immunologic Diseases, Brigham and Women’s Hospital, Harvard Medical School, Boston, Massachusetts, USA.
Address correspondence to: Wei Cao or Hui Zheng, One Baylor Plaza, BCM 230, Baylor College of Medicine, Houston, Texas 77030, USA. Phone: 713.798.8523; Email: [email protected] (W. Cao). Phone: 713.798.5804; Email: [email protected] (H. Zheng).
Find articles by Liu, Z. in: JCI | PubMed | Google Scholar
1Huffington Center on Aging,
2Translational Biology & Molecular Medicine Program, and
3Department of Molecular and Human Genetics, Baylor College of Medicine, Houston, Texas, USA.
4Ann Romney Center for Neurologic Diseases, Department of Neurology, Brigham and Women’s Hospital, Harvard Medical School, Boston, Massachusetts, USA.
5Molecular and Cellular Biology Program, Department of Molecular and Cellular Biology,
6Department of Neuroscience, and
7Department of Pediatrics-Neurology, Baylor College of Medicine, Houston, Texas, USA.
8Department of Pathology and Laboratory Medicine, University of Pennsylvania School of Medicine, Philadelphia, Pennsylvania, USA.
9Center for Dementia Research, Nathan Kline Institute, Orangeburg, New York, USA.
10Departments of Psychiatry, Neuroscience & Physiology and the NYU Neuroscience Institute, New York University Langone Medical Center, New York, New York, USA.
11Evergrande Center for Immunologic Diseases, Brigham and Women’s Hospital, Harvard Medical School, Boston, Massachusetts, USA.
Address correspondence to: Wei Cao or Hui Zheng, One Baylor Plaza, BCM 230, Baylor College of Medicine, Houston, Texas 77030, USA. Phone: 713.798.8523; Email: [email protected] (W. Cao). Phone: 713.798.5804; Email: [email protected] (H. Zheng).
Find articles by Lee, V. in: JCI | PubMed | Google Scholar
1Huffington Center on Aging,
2Translational Biology & Molecular Medicine Program, and
3Department of Molecular and Human Genetics, Baylor College of Medicine, Houston, Texas, USA.
4Ann Romney Center for Neurologic Diseases, Department of Neurology, Brigham and Women’s Hospital, Harvard Medical School, Boston, Massachusetts, USA.
5Molecular and Cellular Biology Program, Department of Molecular and Cellular Biology,
6Department of Neuroscience, and
7Department of Pediatrics-Neurology, Baylor College of Medicine, Houston, Texas, USA.
8Department of Pathology and Laboratory Medicine, University of Pennsylvania School of Medicine, Philadelphia, Pennsylvania, USA.
9Center for Dementia Research, Nathan Kline Institute, Orangeburg, New York, USA.
10Departments of Psychiatry, Neuroscience & Physiology and the NYU Neuroscience Institute, New York University Langone Medical Center, New York, New York, USA.
11Evergrande Center for Immunologic Diseases, Brigham and Women’s Hospital, Harvard Medical School, Boston, Massachusetts, USA.
Address correspondence to: Wei Cao or Hui Zheng, One Baylor Plaza, BCM 230, Baylor College of Medicine, Houston, Texas 77030, USA. Phone: 713.798.8523; Email: [email protected] (W. Cao). Phone: 713.798.5804; Email: [email protected] (H. Zheng).
Find articles by
Trojanowski, J.
in:
JCI
|
PubMed
|
Google Scholar
|

1Huffington Center on Aging,
2Translational Biology & Molecular Medicine Program, and
3Department of Molecular and Human Genetics, Baylor College of Medicine, Houston, Texas, USA.
4Ann Romney Center for Neurologic Diseases, Department of Neurology, Brigham and Women’s Hospital, Harvard Medical School, Boston, Massachusetts, USA.
5Molecular and Cellular Biology Program, Department of Molecular and Cellular Biology,
6Department of Neuroscience, and
7Department of Pediatrics-Neurology, Baylor College of Medicine, Houston, Texas, USA.
8Department of Pathology and Laboratory Medicine, University of Pennsylvania School of Medicine, Philadelphia, Pennsylvania, USA.
9Center for Dementia Research, Nathan Kline Institute, Orangeburg, New York, USA.
10Departments of Psychiatry, Neuroscience & Physiology and the NYU Neuroscience Institute, New York University Langone Medical Center, New York, New York, USA.
11Evergrande Center for Immunologic Diseases, Brigham and Women’s Hospital, Harvard Medical School, Boston, Massachusetts, USA.
Address correspondence to: Wei Cao or Hui Zheng, One Baylor Plaza, BCM 230, Baylor College of Medicine, Houston, Texas 77030, USA. Phone: 713.798.8523; Email: [email protected] (W. Cao). Phone: 713.798.5804; Email: [email protected] (H. Zheng).
Find articles by
Ginsberg, S.
in:
JCI
|
PubMed
|
Google Scholar
|

1Huffington Center on Aging,
2Translational Biology & Molecular Medicine Program, and
3Department of Molecular and Human Genetics, Baylor College of Medicine, Houston, Texas, USA.
4Ann Romney Center for Neurologic Diseases, Department of Neurology, Brigham and Women’s Hospital, Harvard Medical School, Boston, Massachusetts, USA.
5Molecular and Cellular Biology Program, Department of Molecular and Cellular Biology,
6Department of Neuroscience, and
7Department of Pediatrics-Neurology, Baylor College of Medicine, Houston, Texas, USA.
8Department of Pathology and Laboratory Medicine, University of Pennsylvania School of Medicine, Philadelphia, Pennsylvania, USA.
9Center for Dementia Research, Nathan Kline Institute, Orangeburg, New York, USA.
10Departments of Psychiatry, Neuroscience & Physiology and the NYU Neuroscience Institute, New York University Langone Medical Center, New York, New York, USA.
11Evergrande Center for Immunologic Diseases, Brigham and Women’s Hospital, Harvard Medical School, Boston, Massachusetts, USA.
Address correspondence to: Wei Cao or Hui Zheng, One Baylor Plaza, BCM 230, Baylor College of Medicine, Houston, Texas 77030, USA. Phone: 713.798.8523; Email: [email protected] (W. Cao). Phone: 713.798.5804; Email: [email protected] (H. Zheng).
Find articles by
Butovsky, O.
in:
JCI
|
PubMed
|
Google Scholar
|

1Huffington Center on Aging,
2Translational Biology & Molecular Medicine Program, and
3Department of Molecular and Human Genetics, Baylor College of Medicine, Houston, Texas, USA.
4Ann Romney Center for Neurologic Diseases, Department of Neurology, Brigham and Women’s Hospital, Harvard Medical School, Boston, Massachusetts, USA.
5Molecular and Cellular Biology Program, Department of Molecular and Cellular Biology,
6Department of Neuroscience, and
7Department of Pediatrics-Neurology, Baylor College of Medicine, Houston, Texas, USA.
8Department of Pathology and Laboratory Medicine, University of Pennsylvania School of Medicine, Philadelphia, Pennsylvania, USA.
9Center for Dementia Research, Nathan Kline Institute, Orangeburg, New York, USA.
10Departments of Psychiatry, Neuroscience & Physiology and the NYU Neuroscience Institute, New York University Langone Medical Center, New York, New York, USA.
11Evergrande Center for Immunologic Diseases, Brigham and Women’s Hospital, Harvard Medical School, Boston, Massachusetts, USA.
Address correspondence to: Wei Cao or Hui Zheng, One Baylor Plaza, BCM 230, Baylor College of Medicine, Houston, Texas 77030, USA. Phone: 713.798.8523; Email: [email protected] (W. Cao). Phone: 713.798.5804; Email: [email protected] (H. Zheng).
Find articles by Zheng, H. in: JCI | PubMed | Google Scholar
1Huffington Center on Aging,
2Translational Biology & Molecular Medicine Program, and
3Department of Molecular and Human Genetics, Baylor College of Medicine, Houston, Texas, USA.
4Ann Romney Center for Neurologic Diseases, Department of Neurology, Brigham and Women’s Hospital, Harvard Medical School, Boston, Massachusetts, USA.
5Molecular and Cellular Biology Program, Department of Molecular and Cellular Biology,
6Department of Neuroscience, and
7Department of Pediatrics-Neurology, Baylor College of Medicine, Houston, Texas, USA.
8Department of Pathology and Laboratory Medicine, University of Pennsylvania School of Medicine, Philadelphia, Pennsylvania, USA.
9Center for Dementia Research, Nathan Kline Institute, Orangeburg, New York, USA.
10Departments of Psychiatry, Neuroscience & Physiology and the NYU Neuroscience Institute, New York University Langone Medical Center, New York, New York, USA.
11Evergrande Center for Immunologic Diseases, Brigham and Women’s Hospital, Harvard Medical School, Boston, Massachusetts, USA.
Address correspondence to: Wei Cao or Hui Zheng, One Baylor Plaza, BCM 230, Baylor College of Medicine, Houston, Texas 77030, USA. Phone: 713.798.8523; Email: [email protected] (W. Cao). Phone: 713.798.5804; Email: [email protected] (H. Zheng).
Find articles by
Cao, W.
in:
JCI
|
PubMed
|
Google Scholar
|

Published January 9, 2020 - More info
J Clin Invest. 2020;130(4):1912–1930. https://doi.org/10.1172/JCI133737.
© 2020 American Society for Clinical Investigation
Received: September 23, 2019; Accepted: January 3, 2020
Related article:
Abstract
β-Amyloid aggregates found in brain plaques are viewed as triggers of cytotoxicity and neuroinflammation in Alzheimer disease (AD). However, the main β-amyloid (Aβ) species and what imbues the aggregates with such toxic potential are still not yet understood. In this issue of the JCI, Roy et al. show that Aβ complexed with nucleic acids triggers an antiviral type I interferon response in neuroglia, resulting in complement-mediated synapse elimination in AD models. These findings identify a putative endogenous immune signaling axis that drives neurodegeneration in AD and has strong implications for the development of precise therapeutic strategies.
Authors
Stefano Pluchino, Cory Willis
-
Abstract
Type I interferon (IFN) is a key cytokine that curbs viral infection and cell malignancy. Previously, we demonstrated a potent IFN immunogenicity of nucleic acid–containing (NA-containing) amyloid fibrils in the periphery. Here, we investigated whether IFN is associated with β-amyloidosis inside the brain and contributes to neuropathology. An IFN-stimulated gene (ISG) signature was detected in the brains of multiple murine Alzheimer disease (AD) models, a phenomenon also observed in WT mouse brain challenged with generic NA-containing amyloid fibrils. In vitro, microglia innately responded to NA-containing amyloid fibrils. In AD models, activated ISG-expressing microglia exclusively surrounded NA+ amyloid β plaques, which accumulated in an age-dependent manner. Brain administration of rIFN-β resulted in microglial activation and complement C3-dependent synapse elimination in vivo. Conversely, selective IFN receptor blockade effectively diminished the ongoing microgliosis and synapse loss in AD models. Moreover, we detected activated ISG-expressing microglia enveloping NA-containing neuritic plaques in postmortem brains of patients with AD. Gene expression interrogation revealed that IFN pathway was grossly upregulated in clinical AD and significantly correlated with disease severity and complement activation. Therefore, IFN constitutes a pivotal element within the neuroinflammatory network of AD and critically contributes to neuropathogenic processes.
-
Introduction
Accumulation of aggregated misfolded proteins is a pathologic hallmark of many neurodegenerative diseases, including Alzheimer disease (AD), the most predominant form of dementia. Accompanying the formation of β-amyloid (Aβ) plaques and neurofibrillary tangles, the brains of patients with AD are marked by conspicuous and chronic neuroinflammatory responses, manifested by reactive microgliosis, astrogliosis, and elevated levels of proinflammatory cytokines (1–3).
The type I interferon (IFN) cytokines are innately produced in response to viral infections and include multiple IFN-α subtypes, IFN-β, IFN-ε, IFN-κ, and IFN-ϖ. All type I IFN species signal through a common receptor complex, which initiates the JAK-STAT signaling pathway and drives the transcription of a large panel of IFN-stimulated genes (ISGs) (4–6). IFN is predominantly expressed by immune cells upon activation of intracellular innate immune sensors recognizing viral or self-derived nucleic acids (7–9). While best known for its ability to induce an antiviral state in cells via ISG induction, IFN is also involved in immune modulation and pathology under infectious, autoimmune, and various other conditions (4, 10, 11). Despite extensive examination in peripheral responses, IFN has been largely under-investigated in AD to date.
In AD brain, Aβ plaques are structurally and biochemically heterogeneous (12, 13). Besides Aβ peptides, nonprotein constituents, such as lipids, glycosaminoglycans, and nucleic acids, have been detected inside plaques (14–23). Previously, we have shown that soluble protein oligomers (i.e., amyloid precursors) have intrinsic affinity toward anionic cofactors, such as nucleic acids and glycosaminoglycans (GAGs), and that their interaction expedites the formation of amyloid fibrils (24). In particular, amyloid fibrils containing nucleic acids potently induced IFN production from peripheral immune cells by activating innate immune Toll-like receptors in vitro and in vivo, a response capable of initiating lupus-like systemic autoimmunity (25, 26). Moreover, patients receiving systemic IFN treatment and those with HIV-induced dementia display increased IFN activation in the central nervous system (CNS), which is associated with cognitive and psychiatric dysfunction (27–30).
Given its potential functional impact, we tested the hypothesis that CNS IFN response to immunogenic amyloid may participate neuroinflammation and neuropathology in AD. First, we assessed IFN pathway expression in multiple Aβ models and positively detected its upregulation in plaque-laden brains. Similar to periphery, NA+ amyloid elicited IFN response from CNS glial cells in vitro and in vivo. Importantly, we detected NA+ Aβ plaques accumulated in the brains of multiple Aβ models and activated microglia expressing ISGs surrounding them. Additional analysis demonstrated that IFN activated microglia and induced synapse elimination in a complement-dependent manner. Conversely, IFN blockade in mouse models of AD significantly dampened microglia activation and rescued synapse loss in vivo. Furthermore, we examined NA+ neuritic plaques and their relation to microglia activation in human AD brains and conducted transcriptome analysis to reveal profound IFN pathway activation in clinical AD. Together, our data identify an IFN-dependent program that is integral to neuroinflammation and neuropathology in AD.
-
Results
Prevalent IFN pathway activation in Aβ models and amyloid-induced IFN response. To study IFN pathway activation in AD-relevant brain amyloidosis, we analyzed transcriptional profiles of the hippocampus, a brain region crucial for learning and memory but vulnerable to pathology, in AD mouse models. Animal models with brain β amyloidosis differ in promoter usage, transgene products, genetic background, and kinetics of disease progression (31, 32). To conduct an unbiased survey, we collected hippocampal tissues from 4 distinct models: APPNL-G-F (33), 5XFAD, APP;tTa (34), and APP-PS1 (35) (Supplemental Table 1; supplemental material available online with this article; https://doi.org/10.1172/JCI133737DS1). The specific age of each model was chosen to ensure substantial accumulation of Aβ plaques in the parenchyma at the time of analysis. In all models, we detected significantly elevated expression of a panel of well-known genes specifically induced by IFN, including Ifi27I2a, Oas1, Irf7, and Cxcl10 (Figure 1, A–D), in conjunction with increased expression of glial markers and known AD-related proinflammatory factors, implying that the IFN pathway is activated concurrently with the ongoing neuroinflammation.
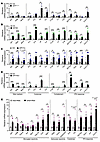 Figure 1
Figure 1Prevalent IFN pathway activation in mouse models of brain amyloidosis. (A–D) Transcriptional analysis of gliosis, inflammation, and IFN pathway markers in hippocampal tissues from 4 Aβ mouse models. (A) APPNL-G-F (6 months old, n = 4–5 mice/genotype), (B) 5XFAD (6 months old, n = 4–5 mice/genotype), (C) APP;tTA (18 months old, n = 10 mice/genotype), and (D) APP-PS1 (6.5 months old, n = 3 mice/genotype). (E) WT mice received RNA-containing amyloid (as depicted in Figure 2A) (n = 7 mice) or control preparation (n = 4 mice) via stereotaxic administration. Transcriptional analysis of hippocampi was performed and results from ipsilateral tissues are shown. Analysis of both contralateral and ipsilateral tissues is also presented in Supplemental Figure 6. For all panels, data are presented as mean ± SEM. *P < 0.05, **P < 0.01, ***P < 0.001 by 2-sided t tests.
To assess the brain’s cellular response to amyloid fibrils, we employed primary organotypic hippocampal slice culture, which retains most CNS cell types in both physiological and anatomical configurations in vitro (36, 37). These slices were exposed to generic amyloid prepared from human serum albumin (HSA) that contain either heparin (representing GAGs) or RNA (representing nucleic acids) (Figure 2A). Strikingly, RNA-containing amyloid stimulated the induction of CD68, a marker for general myeloid cell activation, in microglia, which was associated with the upregulated expression of Aif1, a gene encoding microglial marker Iba1, without increasing cell number (Figure 2A and Supplemental Figure 1). In addition, RNA-containing amyloid induced substantial secretion of TNF-α and CXCL10, a chemokine highly sensitive to IFN signaling (Figure 2A). In conjunction with these changes, RNA-containing amyloid upregulated the expression of ISGs, such as Ifi27I2a and Oas1 (Figure 2A). Despite harboring bona fide amyloid fibril structure (24), heparin-containing amyloid was immunologically inert. Primary mixed glial cells responded similarly to RNA-containing amyloid fibrils, but not to heparin-containing amyloid or nonfibril components (Supplemental Figure 2). In contrast, primary neurons did not react to RNA-containing amyloid (Supplemental Figure 3). These findings revealed a potent immunogenicity of NA-containing amyloid fibrils toward the glial compartment of the CNS milieu.
 Figure 2
Figure 2Nucleic acid–containing amyloid activates the IFN pathway by stimulating microglia in vitro. (A) Generation and treatment of generic amyloid composed of soluble protein oligomers (oHSA) and anionic cofactors (heparin or RNA) to organotypic hippocampal slice cultures. Quantification of CD68+ relative occupancy in Iba1+ microglia (n = 5–6 slices/treatment), transcript analysis (n = 5–16 slices/treatment), and secreted cytokine measurement (n = 2–11 supernatants/treatment) is presented. Slices derived from approximately 2–5 animals were used for each treatment group. Microglial CD68 staining is also shown in Supplemental Figure 1. (B) Quantification of Ifnb1 mRNA, secreted IFN-β, and CXCL10 in mixed glial cultures stimulated for 6 hours with RNA-containing amyloid or control mixture; n = 3 samples/treatment. Full results of additional interferon subtypes and with control treatments are shown in Supplemental Figure 4. (C) Levels of secreted CXCL10 and TNF-α from stimulated primary microglial cultures; n = 3 samples/treatment. For all panels, data are presented as mean ± SEM. *P < 0.05, **P < 0.01, ***P < 0.001 by 1-way ANOVA with Sidak’s correction (A), 2-sided t tests (B), or 2-way ANOVA with Tukey’s correction (C).
IFN response represents an immediate early event during viral infection. Accordingly, we detected both IFN-β transcript and protein secretion as early as 6 hours after exposure of cultured mixed glia to RNA-containing amyloid fibrils, in conjunction with significant CXCL10 release (Figure 2B). Heparin-containing amyloid was unable to induce IFN-β or CXCL10, and other IFN subtypes were not significantly altered in the cultures with any treatment (Supplemental Figure 4). To examine the contribution of microglia in the innate immune response to amyloids, we predepleted microglia from mixed glial cultures with liposome-encapsulated clodronate and found that the remaining cells significantly diminished their reactivity to RNA-containing amyloid (Supplemental Figure 5A). In agreement with this finding, purified primary microglia produced elevated levels of CXCL10 and TNF-α in response to RNA-containing amyloid in a dose-dependent manner (Figure 2C), suggesting NA-containing amyloid can directly activate microglia. To gain further insight into the immunogenicity of different Aβ amyloids, we stimulated primary mixed glial cells with Aβ42 peptides preincubated with either heparin or RNA, and detected increased levels of TNF-α and CXCL10 only from cultures treated with Aβ42 complexed with RNA (Supplemental Figure 5B). Together, NA-containing amyloid fibrils seemingly elicit innate immune response primarily from microglia in vitro.
To evaluate the brain’s response to NA+ amyloid in vivo, we injected generic RNA-containing amyloid or a control preparation containing equivalent amounts of HSA protein and RNA by unilateral stereotaxic injection into the hippocampus of WT mice. Transcriptional analysis of hippocampal tissue revealed that RNA-containing amyloid stimulated upregulation of a panel of ISGs (Ifi27I2a, Oas1, Irf7, Cxcl10), microglial markers (Aif1, Cd68, Trem2, Clec7a, Cst7), astrocytic reactivity markers (Gfap, Osmr, Serpina3n), and cytokines (Il1b, Tnf), highly analogous to the inflammatory profile from AD model brains as shown above (Figure 1E and Supplemental Figure 6). Therefore, the IFN pathway is prevalently activated as part of the neuroinflammatory response accompanying brain amyloidosis.
Nucleic acid–containing Aβ plaques engage microglia to confer IFN pathway activation. To examine the presence of NA+ β-amyloid in vivo, we stained brain tissues of the 5XFAD mouse model with fluorescent dyes specific to nucleic acids together with various antibodies against Aβ. Acridine orange (AO) labeled plaques and produced signals in green (525 nm) and red (650 nm) wavelengths, indicating the presence of both DNA and RNA, respectively (38) (Figure 3A). In young 5XFAD mice (3 months old), when plaques are relatively few and restricted primarily to the subiculum, a majority (~73%) of 6E10+ Aβ plaques were AO+ (Figure 3C). DAPI (4′,6-diamidino-2-phenylindole) and Hoechst 33342, which bind the minor groove of dsDNA, also labeled Aβ plaques, as did RNASelect, a probe that selectively labels RNA (Figure 3A). Moreover, we detected histone H3 signal associated with plaques, implying the presence of chromatin components within the plaques (Figure 3A).
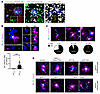 Figure 3
Figure 3Nucleic acid–containing Aβ plaques engage microglia in AD brain. (A) Representative confocal images of Aβ plaques in 5XFAD brain colabeled with a polyclonal antibody against human Aβ and fluorescent probes to detect nucleic acids. Insets show single- or dual-channel images of areas within dashed squares. Scale bar: 15 μm. (B) Representative confocal images of Aβ plaques in subicula of 3-month-old 5XFAD mice triple-labeled with monoclonal antibody 6E10 against Aβ, AO, and Clec7a, showing recruitment of Clec7a+ microglia to AO+ plaques (yellow arrowheads) versus AO– plaques (white arrowheads). White signal represents overlap of all 3 channels. Scale bars: 10 μm. Number of Clec7a+ cells surrounding plaque subtypes is quantified (n = 85 plaques from 3 animals). Mean ± SEM; ***P < 0.001 by 2-sided t test. (C and D) Representative confocal images of Aβ (6E10), AO, and Clec7a in 5XFAD mice at 3 ages with quantification of AO+ plaque prevalence (3 months old, n = 208 plaques from 3 animals; 6.5 months old, n = 198 plaques from 3 animals; 10 months old, n = 324 plaques from 3 animals) (C), and in other Aβ mouse models (n = 2–4 mice per strain) (D). White signal represents overlap of all 3 channels. Scale bars: 10 μm.
In AD brain, microglia surrounding Aβ plaques switch into amoeboid morphology and assume an altered cellular state (39, 40). Next, we examined plaques with and without nucleic acids in conjunction with the reactivity of adjacent microglia. Using AO to label NA+ plaques in 5XFAD brains, we detected a substantially increased microglial presence surrounding NA+ compared with NA– plaques, both by cell number counts and by volumetric measurements (Supplemental Figure 7). Cellular complexity analysis revealed a reactive morphology of these cells, in contrast to microglia associated with NA– plaques or that are not plaque associated (Supplemental Figure 7). Further, microglia associated with NA+ plaques (DAPI+) expressed more CD68 than those near NA– plaques (Supplemental Figure 7).
A subset of microglia in β-amyloid–laden brain display a unique molecular signature shared by the disease-associated microglia subtype (DAM) and the microglial neurodegenerative phenotype (MGnD) (41, 42). C-type lectin Clec7a selectively marks plaque-associated MGnD microglia. Strikingly, Clec7a was exclusively expressed by Iba1+ cells associated with NA+ plaques but not by those next to NA– plaques (Figure 3B and Supplemental Figure 8). Iba1+Clec7a+ cells also express Tmem119, a protein selectively marking brain-resident microglia but not CNS-associated macrophages (Supplemental Figure 9) (43, 44). Although NA+ plaques had a slightly larger diameter than NA– plaques, NA positivity, rather than plaque diameter, was better associated with the presence of Clec7a+ microglia (Supplemental Figure 7G). Of note, NA positivity did not seem to correlate with dystrophic neurites or axons present in the tissue (Supplemental Figure 10). Together, these results indicate that NA+ amyloid plaques induce microglial immune response in the AD brain.
Additionally, we examined NA+ plaques in 5XFAD brains at different ages and found that their occurrence increases with age (Figure 3C). To assess NA+ plaque deposition across models, we expanded the analysis to 6 other AD models, including APPswe/PSEN1ΔE9 (45), APP;tTa, APP-PS1, APPNL-G-F, APPNL-F (33), and APPDutch;PSEN1 (46) (Supplemental Table 1). Uniformly, NA+ plaques and accompanying Clec7a+ microglia were detected in all Aβ models analyzed (Figure 3D). Of note, NA+ Aβ deposition was also evident in cerebral amyloid angiopathy in APPDutch;PSEN1 brains.
STAT1 is a key signaling molecule for IFN pathway activation (4). In contrast to Clec7a– microglia, increased Stat1 was detected in Clec7a+ microglia surrounding Aβ plaques in the brains of 5XFAD mice (Figure 4A). Moreover, nuclear Stat1 signal was found enriched in PU.1+ nuclei (marking microglia) in young 5XFAD brain, which increased with age alongside the accumulation of NA+ plaques (Supplemental Figure 11). Last, we conducted an unbiased analysis of IFN pathway activation using the reported RNA-seq data set of CD11b+FCRLS+ microglia sorted into Clec7a+ and Clec7a– subsets from 9-month-old APP-PS1 mice or CD11b+FCRLS+Clec7a– microglia isolated from control mice (41). ISGs, in the form of a unique gene expression module, have been used to bioinformatically probe the activation of the IFN pathway in human diseases (47). Although all nucleated cells express IFN receptor and thus respond to IFN, some ISGs are cell type–specific. By employing a list of microglia-specific ISGs (48), we detected significantly upregulated expression of a large number of IFN pathway genes in Clec7a+ microglia over Clec7a– microglia from the same brains (Figure 4B).
 Figure 4
Figure 4IFN pathway activation is manifested in plaque-associated microglia in AD model. (A) Representative confocal image of an Iba1+Clec7a+ microglia and an Iba1+Clec7a– microglia (top) in relation to a methoxy-X04+ plaque in 5XFAD brain (3 months old) showing different levels of Stat1 expression (bottom). Cell body outlines (dashed lines) are superimposed on the Stat1/X04 merged image in the bottom panel for ease of visualization. Scale bar: 10 μm. (B) Heatmap of ISG expression determined by RNA-seq in sorted Clec7a+ and Clec7a– microglia from APP-PS1 versus control mice (n = 6, 9 months old). P less than 0.05 was considered significant (2-sided t tests). All individual ISGs with significant differential expression are listed on the right. GEO accession GSE101689.
Interestingly, microglia-specific ISGs include Axl, B2m, and Aif1, molecules previously identified among the MGnD gene signature (Figure 4B). One of the TAM receptor tyrosine kinases, AXL regulates microglial functions and promotes phagocytosis under inflammatory conditions (49, 50). In periphery, IFN stimulates the expression of MHC class I genes and promotes antigen processing and presentation during viral infection. In agreement with this, Clec7a+ microglia expressed elevated levels of H2-D1, H2-K1, Tap1, Tap2, Tapbp, and B2m (Figure 4B). In conjunction, Clec7a+ microglia upregulated ISGs functioning as costimulatory molecules, e.g., Cd72, Cd180, and Cd274 (i.e., Pd-l1) (Figure 4B). Immunoproteasome induction represents another hallmark of IFN activation, bridging the innate and adaptive immune responses (51). Hence, Clec7a+ microglia overexpressed multiple proteasome components, including Psmb8, Psmb9, Psmb10, Psme1, and Psme2 (Figure 4B). Among the ISGs selectively downregulated in Clec7a+ microglia was Irf1, a gene more sensitive to IFN-γ over IFN-α/β (52), consistent with our earlier observation that IFN-γ induction was absent in NA+ amyloid-stimulated glial cultures (Supplemental Figure 4). Physiological IFN response is tightly controlled by a panel of regulatory factors (53). We detected significantly downregulated expression of genes that are involved in restraining IFN production and/or receptor signaling, which include Socs3, Socs1, Irf2, Trim27, Pias1, Trim8, Smad3, and Ptpn1, in Clec7a+ microglia (Supplemental Figure 12). Consistent with the transcriptional data from APP-PS1 mice, we confirmed Axl protein expression in Stat1+ microglia surrounding Aβ plaques in 5XFAD brain (Supplemental Figure 13). Thus, the IFN pathway is activated exclusively in Clec7a+ microglia and concomitantly with the MGnD program. These data indicate that microglial activation is directly correlated with their close proximity to NA+ plaques and that the IFN pathway constitutes an integral element of neuroinflammation in AD models.
IFN activates microglia, initiates neuroinflammation, and leads to synapse loss. Given the pleiotropic functions of IFN, we examined its direct effects on CNS cells. In vitro, rIFN-β treatment not only induced ISGs and increased CXCL10 secretion, but also upregulated the expression of Aif1, Tnf, and Il1b in hippocampal slice cultures (Supplemental Figure 14). To assess its full effects in vivo, we injected rIFN-β intracerebroventricularly into WT mice (Figure 5A) and detected elevated expression of Cst7 and Spp1, 2 key genes associated with DAM/MGnD, and of cytokines Il1a and Il1b, in conjunction with ISGs (Figure 5B). Further, we found that microglia augmented the expression of CD68 and adopted a reactive morphology after rIFN-β exposure with reduced dendrite length, branch, and terminal points (Figure 5C and Supplemental Figure 15). In contrast to generic RNA-containing amyloid, rIFN-β did not stimulate the expression of Trem2 genes (Figure 5B) nor did it upregulate Clec7a in microglia, despite robustly enhanced nuclear Stat1 translocation (Figure 5D). This finding is consistent with the fact that neither Trem2 nor Clec7a was identified as a microglial ISG (48). Thus, IFN alone can directly activate microglia and induce a proinflammatory response, partially overlapping yet distinct from DAM/MGnD program.
 Figure 5
Figure 5IFN activates microglia, initiates neuroinflammation, and leads to synapse loss. (A) Schematic of rIFN-β administration into WT mice via bilateral intracerebroventricular (i.c.v.) stereotaxic injection. (B) Transcriptional analysis of ISGs, microglial markers, and cytokines in cortical tissue of mice 36 hours after vehicle (n = 6 mice) or rIFN-β (n = 5 mice) injection. (C) Representative confocal images of CD68 and 3D skeletonization of Iba1+ microglia from hippocampi (CA1) of vehicle- and rIFN-β–injected mice. Scale bar: 10 μm. Total dendrite length of microglia (n = 135–192 cells from 5–6 mice/treatment) and CD68+ occupancy (percentage of Iba1+ cell volume; n = 5–6 mice/treatment) is quantified. Additional analysis is shown in Supplemental Figure 15. (D) Left: Representative confocal images of Stat1, Iba1, and PU.1 on brain tissues from vehicle- and rIFN-β–injected mice. Insets show Stat1 single-channel images of areas within dashed squares, with PU.1+ nuclear areas outlined. Stat1 occupancy within PU.1+ nuclei is quantified (n = 3 mice/treatment). Right: Representative images of Iba1 and Clec7a in treated mice (n = 5–6 mice/treatment). Scale bars: 10 μm. (E) Representative high-magnification confocal images of CA1 dendritic spines and quantification of spine density in Thy1-eGFP mice that received bilateral i.c.v. administration of vehicle (n = 44 dendrites from 2 mice) or rIFN-β (n = 73 dendrites from 3 mice) for 36 hours. Scale bar: 1 μm. (F) Representative high-magnification confocal images of pre- and postsynaptic terminals labeled by synaptophysin (Syp) and PSD95, respectively, in hippocampi (CA1) of vehicle- and rIFN-β–injected mice. Scale bar: 2 μm. Quantification of puncta density for both synaptic compartments, and of degree of colocalization between the 2 markers; n = 5–6 mice/treatment. For all panels, data are presented as mean ± SEM, or median and quartiles (dendrite length). *P < 0.05, **P < 0.01, ***P < 0.001 by 2-sided t tests.
Previous reports suggested a negative effect of IFN on cognition and brain function (27–30). Healthy synapse is critical for memory and cognition. To facilitate the examination of synapse modification, we injected rIFN-β into the ventricles of a reporter mouse line expressing eGFP in a sparse subset of hippocampal neurons (54). Quantification of CA1 neuronal dendritic spines indicated a significant reduction in spine density by rIFN-β compared with vehicle treatment (Figure 5E). Additionally, high-resolution confocal imaging of pre- and postsynaptic puncta (marked by synaptophysin and PSD95, respectively) showed a specific loss in PSD95 puncta density, and a decrease in the colocalization (≤200 nm) of pre- and postsynaptic terminals with rIFN-β administration (Figure 5F). Conversely, we detected increased GFP+ puncta within microglia after rIFN-β treatment (Supplemental Figure 16), indicating enhanced uptake of neuronal material by activated microglia. These data together suggest that IFN is sufficient to activate microglia to cause synapse loss in the absence of amyloid pathology.
IFN drives microglial activation and synapse loss in AD models. To determine the contribution of IFN pathway activation in the context of AD pathology, we administered an antibody that specifically blocks the signaling of IFN receptor (55) into 3-month-old 5XFAD mice by intracerebroventricular injection. After 6 days of treatment, the brains were collected and the subiculum, where Aβ plaques first deposit, was subjected to detailed histological analysis. Suppression of IFN signaling in microglia was confirmed by reduced PU.1+ nuclear Stat1 (Figure 6A).
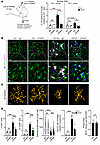 Figure 6
Figure 6IFN blockade dampens microglial activation in AD model. (A) Schematic of brain IFNAR blockade in control and 5XFAD mice (3 months old) by i.c.v. administration of mIgG1 (20 μg; n = 6 control, n = 6 5XFAD) or αIFNAR antibody (20 μg; n = 5 control, n = 6 5XFAD) for 6 days (left). Quantification of nuclear Stat1 levels in PU.1+ microglial nuclei and PU.1– nonmicroglial nuclei in subicula of treated mice; n = 3–5 mice/group (right). (B–D) Representative staining and 3D skeletonization of Clec7a+ microglia surrounding plaques in subicula of treated mice (B and C). White signal represents overlap of all 3 channels. Scale bars: 10 μm. Quantifications of total percentage of Iba1+ and CD68+ area, microglial dendrite length (n = 46–185 cells/group), Clec7a+ signal occupancy per microglia, and plaque load in subicula of treated mice (n = 5–6 mice/group) (D). Additional analysis is shown in Supplemental Figure 17. For all panels, data are presented as mean ± SEM, or median and quartiles (dendrite length). **P < 0.01, ***P < 0.001 by 1-way ANOVA with Sidak’s correction, or 2-sided t test (plaque load).
As expected, microglia in 5XFAD mice that received isotype control IgG displayed amoeboid morphology and expressed high levels of Clec7a as well as CD68 (Figure 6, B and C; Supplemental Figure 17). Remarkably, αIFNAR antibody treatment not only partially shifted the morphology of microglia toward homeostatic form, but also significantly decreased the overall Iba1+ and CD68+ signals, as well as the Clec7a+ signal per microglia, within plaque-laden areas of 5XFAD brains, despite unaltered Aβ plaque load (Figure 6, B–D; and Supplemental Figure 17). Reactive astrocytes in 5XFAD mice, quantified by GFAP staining, remained unchanged with IFNAR blockade (Supplemental Figure 18). We also observed that, despite the regional absence of Aβ plaques at this age, microglia in the hippocampal CA3 region displayed slightly reactive morphology in 5XFAD brain, which was completely restored to resting morphology with αIFNAR blockade (Supplemental Figure 19), suggesting a profound influence of IFN on microglia in different AD brain regions. To further validate the involvement of IFN in vivo, we administered αIFNAR blocking antibody to 10- to 12-month-old APPNL-G-F mice using the same experimental protocol and similarly detected significant reductions in microglia (both Iba1+ and CD68+ areas) compared with control IgG treatment, in the presence of comparable plaque load in the brain parenchyma (Supplemental Figure 20). Hence, IFN is likely required for sustaining microgliosis in different AD models.
Since IFN is potent in modulating synapses, we performed detailed synaptic puncta analyses to evaluate the effects of IFN blockade. While IgG-treated 5XFAD brains showed selective PSD95 density reduction and loss of colocalization between PSD95 and synaptophysin, αIFNAR blockade completely restored both PSD95 puncta density and pre- and postsynapse marker colocalization (Figure 7, A and C). We performed further imaging analysis to quantify engulfed synaptic puncta within the microglial cell bodies. Microglia from IgG-treated 5XFAD brain contained more internalized PSD95 puncta than microglia from control brains (Figure 7, B and D). In contrast, IFN blockade normalized the levels of internal PSD95 puncta relative to cell volume in 5XFAD, suggesting an essential role of IFN in promoting synapse loss in AD brains.
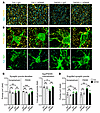 Figure 7
Figure 7IFN blockade rescues synapse loss in AD model. (A and C) Representative high-magnification images of pre- and postsynaptic terminals labeled against synaptophysin and PSD95, respectively, in subicula of control or 5XFAD mice treated with mIgG1 or αIFNAR as described in Figure 6A (A). Scale bar: 4 μm. Quantification of relative puncta density for both synaptic compartments, and of degree of colocalization between the 2 markers; n = 2–5 mice/group (C). (B and D) Representative high-magnification images of subicular microglia costained with synaptic markers in treated mice, showing engulfment of synaptic terminals (B). Scale bars: 4 μm. Quantification of engulfed puncta densities for both synaptic markers, normalized to total cell volumes; n = 2–5 mice/group (D). For all panels, data are presented as mean ± SEM. *P < 0.05, **P < 0.01, ***P < 0.001 by 1-way ANOVA with Sidak’s correction.
Although IFN-γ (i.e., type II interferon) was not induced by NA-containing amyloid in vitro (Supplemental Figure 4), several studies have previously implicated IFN-γ in the neuroinflammation and pathogenesis of AD (56–58). IFN-γ also signals through the JAK-STAT pathway and induces a panel of ISGs that largely overlap with ISGs triggered by type I interferon (59). To evaluate the relative contribution by IFN-γ, we administered αIFNGR blocking antibody to 3-month-old 5XFAD mice using a similar experimental protocol and examined the changes in microglial activation and synapse densities. In contrast to αIFNAR blockade, αIFNGR blockade did not alter microglial nuclear Stat1 levels, the activation of microglia, nor modify PSD95 puncta density reduction in 5XFAD brain (Supplemental Figure 21). Our results underline a selective and prominent role played by type I interferon in neuroinflammation and neuropathology in the presence of CNS β-amyloidosis.
IFN stimulates complement cascade activation and promotes C3-dependent synapse elimination. Although our data validated the notion that IFN-activated microglia are involved in synapse elimination, the underlying molecular mechanism was unclear. During development, microglia actively participate in the pruning of weak synapses (60), which is mediated via C1q and C3, 2 key members of the classical pathway of complement activation (61). Microglia have also been shown to excessively uptake synapses in a C3-dependent manner in AD models (62, 63) and during CNS viral infection (64). Given the prominent role of the complement cascade in microglia-mediated CNS synapse modulation, we questioned whether IFN directly affects complement expression. Previously, complement C4b was shown to be induced by brain IFN expression in microglia (65). Four additional complement cascade genes, which include C2, C3, Cd47, and Cfb, were also identified as genes induced by IFN (48), suggesting a potential direct crosstalk between IFN and complement cascade.
To obtain experimental evidence of the ability of IFN to promote complement activation, we treated hippocampal slice culture or mixed glial cultures with rIFN-β or vehicle. rIFN-β readily upregulated the levels of C1qa, C3, and C4b transcripts, and stimulated the production of C3 and C4 proteins in vitro (Figure 8, A and B, and Supplemental Figure 22). Stereotaxic rIFN-β injection in vivo similarly resulted in elevated expression of multiple complement genes and increased the levels of C3 protein selectively in astrocytes of WT brain (Figure 8C), suggesting complement activation downstream of IFN signaling.
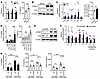 Figure 8
Figure 8IFN stimulates complement cascade activation. (A) Expression of complement transcripts and secreted C3 in slice cultures (n = 12 slices and n = 6–7 supernatants/treatment). Slices derived from approximately 2–5 animals were used for each treatment. (B) The levels of pStat1 and C3 proteins in mixed glial cultures treated with vehicle or 5 ng/mL rIFN-β in vitro (representative immunoblots from 2 independent experiments). (C) Expression of complement transcripts in brain tissue and quantification of C3 protein expression within GFAP+ astrocytes in mice injected with vehicle or rIFN-β in the experiment described in Figure 5A. (D) Levels of C3 mRNA and secreted C3 in hippocampal slices stimulated with generic amyloids and respective controls (n = 5–16 slices and n = 4–11 supernatants/treatment; experiment illustrated in Figure 2A). Slices derived from approximately 2–5 animals were used for each treatment group. (E) The levels of pStat1 and C3 proteins in mixed glial cultures treated with RNA-containing amyloid or control (representative immunoblots from 2 independent experiments). (F) Expression of complement transcripts in hippocampi of WT mice administered RNA-containing amyloid or control preparation in the experiment described in Figure 1E. (G) Expression of C3 mRNA (n = 5–6 slices/treatment) and secretion of C3 and CXCL10 (n = 8 supernatants/treatment) in hippocampal slices stimulated with RNA-containing amyloid or control preparation in the presence of either mIgG1 or αIFNAR antibody. Slices derived from approximately 2–5 animals were used for each treatment group. For all panels, data are presented as mean ± SEM. *P < 0.05, **P < 0.01, ***P < 0.001 by 2-sided t tests (A, C, F), or 1-way ANOVA with Sidak’s correction (D, G).
To study complement activation in response to amyloid, we exposed hippocampal slices or mixed glial cultures to different amyloid species or controls. In conjunction with IFN pathway activation (Figure 2A and Supplemental Figure 2), RNA-containing amyloid alone increased C3 transcript and protein expression as well as its secretion (Figure 8, D and E). Additionally, neuronal dendritic spine density was reduced by RNA-containing amyloid in cultured hippocampal slices (Supplemental Figure 23). Moreover, hippocampal injection of RNA-containing amyloid stimulated the expression of complement genes, and downregulated negative regulators of complement (Prelp and Cd55) (Figure 8F), concurrent with other neuroinflammatory changes in WT brain (Figure 1E). Therefore, NA+ amyloid potently triggers complement cascade activation.
Next, we examined the importance of IFN in complement activation by amyloid by selectively blocking IFNAR signaling. Both the transcription and secretion of C3, under influence of RNA-containing amyloid, were highly sensitive to IFNAR blockade, similar to CXCL10 (Figure 8G). Collectively, our data indicate that IFN is both necessary and required for complement activation in response to NA+ amyloid.
To validate these in vitro findings, we tested the levels of C3 in AD mice that received αIFNAR blocking antibody in experiments described earlier. In the hippocampi of 10- to 12-month-old APPNL-G-F mice that harbored Aβ plaque pathology, C3 mRNA was elevated in the group that received control IgG, as expected (Figure 9A). IFNAR blockade not only blunted ISG expression, as shown by Ifi27l2a mRNA reduction, but also effectively reduced C3 transcripts in APPNL-G-F mice to levels measured in control mice (Figure 9A).
 Figure 9
Figure 9IFN orchestrates C3-dependent synapse elimination. (A) Expression of C3 and Ifi27l2a mRNA in hippocampi of control or APPNL-G-F mice (10 to 12 months old) after i.c.v. injection with mIgG1 control (20 μg; n = 4 control, n = 7 APPNL-G-F) or αIFNAR antibody (20 μg; n = 3 control, n = 8 APPNL-G-F) for 6 days. Additional analysis is shown in Supplemental Figure 20. (B and C) Representative confocal images of GFAP, C3, and Aβ (6E10) in control or 5XFAD mice (3 months old) treated with IgG1 or αIFNAR antibody in the experiment described in Figure 6. Scale bar: 30 μm. Inset shows C3 signal within outlined cell body. Scale bar: 6 μm. See Supplemental Figure 18 for full images. C3 occupancy in astrocytes in treated mice is quantified (C). (D and E) WT and C3 null mice were treated with bilateral i.c.v. administration of vehicle (n = 3 WT, n = 4 C3–/–) or rIFN-β (n = 3 WT, n = 5 C3–/–) for 36 hours, similarly to the experiment in Figure 5A. (D) Quantification of relative puncta density for pre- and postsynaptic compartments (synaptophysin and PSD95, respectively), and of degree of colocalization between the 2 markers, in hippocampus (CA1). (E) Quantification of synaptic puncta densities engulfed by microglia for both synaptic markers, normalized to total cell volumes; n = 3–5 mice/group. Additional analysis of microglial reactivity is shown in Supplemental Figure 24. For all panels, data are presented as mean ± SEM. *P < 0.05, **P < 0.01, ***P < 0.001 by 1-way ANOVA with Sidak’s correction.
To further examine the C3 protein expression, we stained the brain sections of 5XFAD mice that received αIFNAR antibody or control IgG. In 5XFAD brains treated with IgG, we detected robust C3+ signal in GFAP+ astrocytes near amyloid plaques in the subiculum (Figure 9B and Supplemental Figure 18), consistent with what we have shown before (66). Although IFNAR blockade did not alter the amount of overall GFAP+ staining in the tissue (Supplemental Figure 18), the occupancy of C3 protein within the astrocytes was almost completely reduced to control levels (Figure 9C). Thus, these data show that C3 expression in AD mouse models is likely regulated by IFN.
To confirm that C3 is necessary for IFN-induced synapse elimination, we injected rIFN-β into the ventricles of WT or C3–/– mice and examined microglia activation and synaptic puncta (Figure 9, D and E, and Supplemental Figure 24). Microglia from mice lacking C3 reacted to rIFN-β by upregulating CD68 and adopting a reactive morphology, similarly to WT, as expected (Supplemental Figure 24). Despite the general activation of microglia, C3–/– brains were completely protected from rIFN-β–induced postsynaptic puncta loss (Figure 9D), which was correlated with the lack of microglial uptake of PSD95 (Figure 9E). Together, these results highlight a pivotal axis of amyloid-IFN-C3 in promoting synapse loss in AD models.
IFN pathway activation is manifested in human AD. Based on our cumulative findings in AD mouse models, we investigated the relevance of NA+ amyloids and the IFN pathway to human AD. We previously reported the detection of sequestered RNA species within neuritic plaques in human AD brains, as visualized by AO staining (17, 38). Here, this finding was confirmed in specimens of human AD brain tissues with the positive staining of neuritic plaques, but not diffuse plaques, with AO (Supplemental Figure 25). Similar patterns were observed using Hoechst dye recognizing DNA (Supplemental Figure 26). Of note, although all neuritic plaques were NA+, the degree of staining varied among plaques, which could be quantified by confocal microscopy and 3D image analysis (Supplemental Figure 25). In conjunction with labelling and volumetric measurements of IBA1+ microglia, we observed a strong correlation (r = 0.68, P < 0.0001) between the amount of AO signal and the volume of associated microglia among individual plaques (Figure 10A), suggesting that NA+ amyloids promote microglial reactivity in human AD.
 Figure 10
Figure 10IFN pathway activation is manifested in human AD. (A) Examination of IBA1+ microglia surrounding neuritic plaques in human AD specimens (n = 3). Plaques were stained with Aβ (6E10) and nucleic acids with acridine orange (AO). Correlation analysis was performed with the total cell volume of recruited microglia and the level of AO+ signal inside plaques (n = 54 plaques from 3 AD cases; r = 0.68, P < 0.0001). Scale bar: 10 μm. Additional representative images are shown in Supplemental Figure 25. (B) Representative confocal images of neuritic plaque-associated microglia in human AD cases (n = 3) expressing ISGs: IFITM3 (left) and AXL (right). Insets show single-channel images of outlined areas. Scale bar: 10 μm. Signals in microglia from plaque-free areas are shown in Supplemental Figure 27. (C and D) Heatmaps showing group-averaged expression of human ISGs in postmortem brain tissue (parahippocampal gyrus) from 175 human subjects stratified by clinical dementia rating (CDR) (C) and Braak score (D). NCI indicates no cognitive impairment; MCI, mild cognitive impairment. Individual ISGs with differential expression are highlighted. See also Supplemental Figure 28 for other brain regions. (E and F) Correlations of IRF7 transcript level with CDR, Braak score (E), and mean neuritic plaque density (F). (G) Correlation of IRF7 transcript level with C3 transcript level. (H) Coexpression analysis between IFN pathway genes (yellow) and complement genes (green). Links inside the circle represent significant (P < 0.8 × 10–5) Spearman correlation coefficients between the genes (gray: r > 0.6; blue: r < 0.6). Coexpression analysis between IFN pathway genes and random gene sets is shown in Supplemental Figure 30 as a control. For A, E, F, and G, correlation coefficients (r) were computed using Pearson correlations (A, F, G) or Spearman correlations (E). All: P <0.0001. For A, F, and G, plots display one dot for each plaque (A) or subject (F, G), and linear regression line (solid) with 95% CI (dashed).
We then examined the protein expression of IFITM3 and AXL, 2 microglial ISGs (Figure 4B), to assess the activation of the IFN pathway. Both markers labeled a subset of IBA1+ microglia tightly associated with neuritic plaques in the human AD specimens (Figure 10B), whereas neither was evidently expressed by microglia in plaque-free regions of the same tissue (Supplemental Figure 27). Although αClec7a antibody failed to stain human brain tissue, our findings establish that TAM receptor AXL consistently marks plaque-associated microglia both in human and mouse brains (Figure 10B and Supplemental Figure 13). These observations indicate that, similar to AD models, microglia undergo IFN pathway activation upon recruitment to NA+ neuritic plaques in human AD.
In order to perform unbiased transcriptional assessment of IFN pathway activation in human brain samples, we first constructed a CNS-specific ISG panel of 306 genes, which included neuron-specific, astrocyte-specific, and microglia-specific ISGs deduced from mIFN treatment of respective primary murine cells (48, 67) (Supplemental Table 2). The human orthologs of these genes were used to probe the Mount Sinai Brain Bank, an RNA-Seq data set with accompanying neuropathological and clinical cognitive data from a large collection of Alzheimer dementia and healthy control brain tissues (68). This data set contains 4 brain regions, including parahippocampal gyrus (BM36-PHG), inferior frontal gyrus (BM44-IFG), superior temporal gyrus (BM22-STG), and frontal pole (BM10-FP), which displayed varying regional vulnerability to AD (69).
In PHG, the top-ranked brain region of relevance to AD pathology, overall ISG expression was grossly elevated in patients exhibiting clinical dementia (Figure 10C). The dementia group showed increased AXL, AIF1, and IFITM3, reflecting microglial activation by NA+ neuritic plaques (Figure 10B). In parallel with the transcriptome profile of Clec7a+ microglia in the Aβ mouse model (Figure 4B), PHG of patients with AD displayed increased expression of ISGs of several functional categories, such as MHC class I and antigen processing (HLA-A, HLA-B, HLA-C, HLA-F, TAP1, TAP2, TAPBP, TAPBPL, B2M), costimulatory molecules (CD40, CD86, CD74), and immunoproteasome components (PSMB8, PSMB9, PSMB10, PSME1, PSME2). In addition, the expression of ISG members from the tripartite motif (TRIM) family (TRIM5, TRIM14, TRIM21, TRIM22, TRIM25) and poly (ADP-ribose) polymerase (PARP) family (PARP9, PARP10, PARP12, PARP14) was escalated in the AD cohort (Figure 10C). On the other hand, CD47, an ISG that suppresses excessive microglia-mediated synaptic pruning (70), was downregulated in the mild cognitive impairment (MCI) group and even more so in AD (Figure 10C). Human ISG15 is a key negative regulator of IFN immunity (71). ISG15 levels increased moderately in MCI but declined profoundly in the AD cohort (Figure 10C). AD brains are also graded by Braak score based on postmortem histopathological evaluation (72). Interestingly, broad ISG expression in PHG was found positively associated with increasing Braak scores (Figure 10D). Similar patterns of ISG expression were detected in all 3 other brain regions included in the Mount Sinai Brain Bank (Supplemental Figure 28), suggesting a positive correlation between IFN pathway activation and cognitive decline as well as disease progression in clinical AD.
IRF7 is a master signaling mediator for IFN-dependent immune response (52, 73). In agreement with the overall ISG expression in AD, IRF7 expression in PHG correlated strongly with both clinical dementia rating (CDR, P < 0.0001) and Braak score (P < 0.0001) (Figure 10E). Consistent with our characterization of NA+ amyloid, IRF7 expression was also associated with neuritic plaque burden (Figure 10F; P < 0.0001).
Since we observed robust IFN-induced complement activation in mice, we examined the relationship between IFN and complement cascade in a humans. Most ISGs contain promoter elements that are responsive to JAK-STAT signaling downstream of IFN receptor activation (74). We surveyed the presence of interferon-stimulated responsive element (ISRE) and gamma interferon-activated sequence (GAS) motifs in the promoter regions of human complement genes. Among 45 complement cascade genes, 35 genes (78%) contain at least 1 putative (P < 10–5) binding site, implying potential widespread responsiveness to IFN activation (Supplemental Figure 29). In particular, the human C3 promoter harbors 4 ISRE and 3 GAS motifs, which may confer IFN responsiveness. Consistent with this, C3 expression was found to be highly correlated with IRF7 expression in the parahippocampal gyrus of patients with AD (Figure 10G).
To systematically evaluate the relationship between IFN and complement cascade in clinical AD, we computed the coexpressional relationship between human genes comprising both pathways and observed prominent positive correlations between the 2 pathways (Figure 10H). Such robust enrichment of positive coexpression correlation was absent between IFN pathway genes and 100 independent sets of random genes of the same size as the complement cascade, indicating a statistically meaningful connection between these 2 pathways (Supplemental Figure 30). In AD brain, the levels of ISGs encoding MHC class I, antigen processing, or immunoproteosome molecules were positively correlated with the levels of multiple complement components (Figure 10H). Similar strong correlation was evident with ISGs involved in immune response and signaling, such as DTX3L, GBP2, IFI16, and PIK3AP1. Conversely, multiple IFN pathway genes were negatively associated with complement member SERPING1, which is known to inhibit classical complement pathway activation via binding to C1R and C1S (75). Collectively, these data suggest that IFN pathway activation represents a prominent feature of neuroinflammation in human AD, where it may robustly interact with complement cascade.
-
Discussion
Although the heterogeneity of Aβ plaques and misfolded protein aggregates in general has long been recognized, the biological impacts of the cofactors in the complex amyloid are only being appreciated in recent years. In particular, heparan sulfate proteoglycans facilitate the spreading of proteopathic seeds and promote the deposition of Aβ plaques while inhibiting Aβ clearance (76, 77). Nucleic acids, when properly delivered intracellularly, can be sensed as pathogen- or danger-associated molecular patterns and trigger antiviral immunity (7, 9). However, the functional implication of NA+ plaques in AD has not been investigated. By direct comparison with GAG+ amyloid, we show that NA+ amyloid is highly immunogenic in the CNS milieu via activating microglia and stimulating a robust innate immune response, which is consistent with our previous findings on these amyloids with peripheral immune cells (25). We have also previously observed a comparable IFN induction by DNA+ amyloid and RNA+ amyloid and demonstrated a crucial role of nucleic acid sensing in IFN production (25).
Interestingly, we detected a widespread deposition of NA+ plaques in the brains of various β-amyloidosis models, accumulation of which is age dependent, exclusively correlated with microglia activation, yet independent of dystrophic neurites or axons. Previous studies have documented DAPI+ and Hoechst+ Aβ plaques in different models (22, 23). Our detailed examination using multiple NA-specific dyes indicates that both DNA and RNA are present within Aβ plaques in AD brain; however, the origin of plaque-sequestered nucleic acids is unclear. Interestingly, not only does Aβ naturally aggregate with DNA in vitro, Aβ species have also been located inside the nuclei of neurons in AD brains (20, 22). DAPI+ nuclear Aβ aggregates were shown to accumulate prior to the formation of extracellular DAPI+ Aβ plaques in 5XFAD brain, lending to a notion that neuritic plaques derive from intracellular amyloid and have a neuronal origin (22). Our detection of histone proteins together with DNA inside the plaque supports such a nuclear origin. In human AD brain, our data and other reports point to a similar codeposition of DNA and RNA within the plaques (17, 22). Interestingly, sequence analysis of RNA isolated from human AO+ neuritic plaques identified mRNA transcripts from cortical neurons (78), thus suggesting a neuronal origin of human plaque-associated nucleic acids. A minor fraction of NA– Aβ plaques exist in the subiculum of young 5XFAD brain, which intriguingly are devoid of activated microglia. Since a possible involvement of microglia in plaque formation was recently suggested (79), it is interesting and worthy to investigate the precise cellular process that prompts the formation of the NA+ plaques.
We have detected a universal activation of type I interferon pathway in mouse brains harboring Aβ plaques, regardless of differences among the AD models analyzed. Although genetic background can differentially influence plaque pathology, ISG expression was preserved nevertheless in outbred strains of 5XFAD mice, which harbor broad genetic diversity (Supplemental Figure 31) (80). These observations, in conjunction with the evident activation of the IFN pathway in human AD tissues, demonstrate a paramount presentation of IFN in AD pathology. We have also provided evidence to specifically implicate type I IFN, but not IFN-γ, in mediating neuroinflammation and neuropathology in AD models. This finding is distinct from the peripheral immune response stimulated by NA+ amyloid, in which we previously demonstrated that type I IFN led to IFN-γ production by NK cells, and that blocking either type I IFN or IFN-γ abolished the development of humoral autoimmunity (26).
Plaque-associated microglia are a central player driving neuroinflammation in AD models (81–83). Beyond the initial discovery and description, remaining to be answered are key questions regarding their generation and functional role in disease pathology. Here, our results postulate that: (a) NA+ amyloid represents an innate stimulus promoting the generation of plaque-associated MGnD microglia; (b) IFN is an essential element of microglial response and the neuroinflammatory network; and (c) microglia partake in pathogenic synapse elimination via an IFN-C3 axis in AD. Our findings on IFN’s involvement in synapse loss in AD are consistent with microglia-dependent learning deficits induced by brain IFN-β production (65). Moreover, chronic and dysregulated IFN expression in CNS in the absence of an infection is a major driver for type I interferonopathies, a group of hereditary CNS disorders (84–86). Interestingly, peripheral dysregulation of IFN production was also implicated in the pathogenesis of Parkinson’s disease, another CNS neurodegenerative condition (87).
Our analysis has revealed potent effects of IFN on activating CNS cells by inducing morphological changes and expression of proinflammatory factors, costimulatory and signaling molecules. However unlike NA+ amyloid, IFN alone is insufficient to instigate the full neuroinflammation program of DAM/MGnD (Supplemental Figure 32); yet, IFN signaling is required to sustain the ongoing microgliosis and overall neuroinflammation in the presence of β-amyloidosis. Myeloid receptor Trem2 is crucial for the generation of DAM/MGnD and plays a role in modifying Aβ pathology (41, 42, 88–90). Interestingly, Trem2 binds to ApoE and ApoJ, both of which are components of Aβ plaques (91–93), and the signaling pathway downstream of TREM2/DAP12 complex has been shown to limit IFN production in innate immune cells (8, 94). Therefore, it would be worthwhile to dissect these concurrent signaling events in microglia upon interacting with complex amyloid. In addition, microglia subset(s) containing an IFN signature in mouse brains have been detected from single-cell expression profiling studies (56, 65, 95). Whether plaque-associated microglia further segregate into IFN+ and IFN– subsets remains to be examined at the single-cell level.
Synapse loss is clinically associated with cognitive decline in AD (96–98). The profound activation of the IFN pathway in MGnD microglia supports a notion that this unique microglia subset participates in neuropathology via IFN-mediated synapse elimination. Complement has been implicated in synaptic pathologies in diverse neurological and neuropsychiatric diseases, in particular neurodegeneration (62, 63, 99, 100). Although neuronal activity can promote C3 expression during development (101), how CNS inflammation controls complement cascade activation remains elusive. Our study provides a major advance in the mechanistic understanding of this pathogenic process by identifying a crucial axis that connects IFN, complement, and synapse loss. We have shown that IFN controls the expression of multiple components of the complement cascade and mediates synapse elimination in a C3-depedent manner.
The crosstalk between microglia and astrocytes plays a critical role in neurodegenerative conditions, as shown by microglia-dependent generation of a neurotoxic subset of astrocytes (102, 103). These so-called A1 astrocytes abundantly express C3 protein (102). One of our interesting observations is that C3 protein, primarily produced by activated astrocytes in AD mouse brain, is highly sensitive to IFN signaling. In 5XFAD mice, astrogliosis was unaffected by acute IFNAR blockade despite C3 downregulation (Supplemental Figure 18). In APPNL-G-F mice, IFNAR blockade further reduced astrocyte reactivity (Supplemental Figure 20), which was correlated with the dampened expression of Il1a and Tnf, microglia-derived factors required for the generation of A1 astrocytes (Supplemental Figure 33). The reason behind these differing effects on astrocytes is unclear, although we suspect that the efficacy of acute IFN blockade may differ between the models, as 5XFAD mice manifest an overall higher magnitude of inflammation than APPNL-G-F mice. Additional studies with long-term IFN inhibition should be carried out to dissect how microglia and astrocytes cooperate to promote synapse elimination upon IFN pathway activation.
Amyloid burden in human brains is known to poorly correlate with the severity or duration of dementia (96, 104). Individuals without dementia can harbor significant amounts of both plaques and tangles, but remarkably they lack the robust glial activation present in their AD counterparts, a feature that is associated with preserved synapses and neurons (105). Our study has identified subsets of Aβ plaques in AD models that are immunogenic (i.e., NA+) or inert (i.e., NA–) with distinct consequential effects on synaptic health. It is plausible that plaques that accumulate in patients with AD and in high-pathology individuals without dementia differ in their fundamental immunogenicity, namely in terms of NA content. It is thus of great interest to examine more closely the molecular constituents of amyloid plaques in the brains of cognitively resilient individuals compared with AD cases.
In this study, we chose to demonstrate the functional involvement of the IFN pathway by employing a highly specific blocking antibody approach instead of a classical gene deletion approach, to minimize other confounding factors. It is well known that tonic IFN signaling is required for maintaining the homeostasis of the immune system and proper configuration of signaling networks (106–108). Importantly, neurons lacking germline IFN signaling were shown to undergo spontaneous neurodegeneration (109). Most notably, germline Ifnar1 deletion also alters the expression of APP and Aβ, which significantly obscured the interpretation of results from APPswe/PSEN1ΔE9;Ifnar1–/– mice (110).
Age represents the most influential risk factor in the development of AD. In aged mice and humans, IFN signaling in choroid plexus, an epithelial tissue located within the ventricles of the brain, has been implicated in hampering cognitive function and hippocampal neurogenesis (111). Moreover, microglia inside aged brain adopt an altered functional state with discernable IFN signature expression (65). Here, we have identified prominent microglial IFN response to NA+ Aβ plaques as a key factor promoting neuroinflammation and neuropathology in AD (Supplemental Figure 34). These findings collectively lend a rationale to explore whether modulating IFN pathway could be useful in alleviating AD.
-
Methods
Study approvals. All animal procedures were performed in accordance with NIH guidelines and with the approval of the Baylor College of Medicine Institutional Animal Care and Use Committee.
Parietal cortex samples from patients with AD were obtained from the University of Pennsylvania Center for Neurodegenerative Disease Research (CNDR). Written informed consent was obtained from all subjects. The cases used in this study are summarized in Supplemental Table 3.
RNA-seq data. All original RNA-seq data were previously deposited in the NCBI’s Gene Expression Omnibus database (GEO GSE101689).
-
Author contributions
WC conceptualized and supervised the study, and oversaw project administration. ERR, BW, YWW, SDG, HZ, and WC devised the study methodology. YWW and LZ contributed software. ERR, YWW, and WC performed formal study analysis. ERR, BW, YWW, GC, AC, and WC performed study investigation. ZY, NEP, YX, JLJ, VMYL, JQT, OB, and WC contributed resources. ERR, YWW, and WC wrote the original draft of the manuscript. ERR, JLJ, SDG, OB, HZ, and WC reviewed and edited the manuscript. JLJ, OB, HZ, and WC acquired funding for the study.
-
Acknowledgments
The study was funded by NIH grants AG057587 (WC and HZ), AG032051, AG020670, and AG054111 (HZ), NS092515 (JLJ), and AG051812 and AG054672 (OB); the Robert A. and Rene E. Belfer Family Foundation (JLJ); BrightFocus ADR A20183775 (WC); and the Cure Alzheimer’s Fund (OB). ERR was supported in part by TBMM T32 training grant (ST32GM088129). The authors would like to thank Markus Hofer for generously sharing the glial ISG lists. We also thank Takashi Saito and Takaomi Saido for generously providing their App knockin mouse lines (APPNL-G-F and APPNL-F). We acknowledge expert technical assistance from Nadia Aithmitti and Bianca Contreras.
Address correspondence to: Wei Cao or Hui Zheng, One Baylor Plaza, BCM 230, Baylor College of Medicine, Houston, Texas 77030, USA. Phone: 713.798.8523; Email: [email protected] (W. Cao). Phone: 713.798.5804; Email: [email protected] (H. Zheng).
-
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Copyright: © 2020, American Society for Clinical Investigation.
Reference information: J Clin Invest. 2020;130(4):1912–1930.https://doi.org/10.1172/JCI133737.
See the related Commentary at Intrinsic antiviral immunity drives neurodegeneration in Alzheimer disease.
-
References
- Perlmutter D. Preventing Alzheimer’s disease. J Am Coll Nutr. 2016;35(8):732–733.
- Hodson R. Alzheimer’s disease. Nature. 2018;559(7715):S1.
- Heppner FL, Ransohoff RM, Becher B. Immune attack: the role of inflammation in Alzheimer disease. Nat Rev Neurosci. 2015;16(6):358–372.
- McNab F, Mayer-Barber K, Sher A, Wack A, O’Garra A. Type I interferons in infectious disease. Nat Rev Immunol. 2015;15(2):87–103.
- Capobianchi MR, Uleri E, Caglioti C, Dolei A. Type I IFN family members: similarity, differences and interaction. Cytokine Growth Factor Rev. 2015;26(2):103–111.
- Schreiber G, Piehler J. The molecular basis for functional plasticity in type I interferon signaling. Trends Immunol. 2015;36(3):139–149.
- Tan X, Sun L, Chen J, Chen ZJ. Detection of microbial infections through innate immune sensing of nucleic acids. Annu Rev Microbiol. 2018;72:447–478.
- Gilliet M, Cao W, Liu YJ. Plasmacytoid dendritic cells: sensing nucleic acids in viral infection and autoimmune diseases. Nat Rev Immunol. 2008;8(8):594–606.
- Crowl JT, Gray EE, Pestal K, Volkman HE, Stetson DB. Intracellular nucleic acid detection in autoimmunity. Annu Rev Immunol. 2017;35:313–336.
- Zitvogel L, Galluzzi L, Kepp O, Smyth MJ, Kroemer G. Type I interferons in anticancer immunity. Nat Rev Immunol. 2015;15(7):405–414.
- Di Domizio J, Cao W. Fueling autoimmunity: type I interferon in autoimmune diseases. Expert Rev Clin Immunol. 2013;9(3):201–210.
- Qiang W, Yau WM, Lu JX, Collinge J, Tycko R. Structural variation in amyloid-β fibrils from Alzheimer’s disease clinical subtypes. Nature. 2017;541(7636):217–221.
- Liao L, et al. Proteomic characterization of postmortem amyloid plaques isolated by laser capture microdissection. J Biol Chem. 2004;279(35):37061–37068.
- Kiskis J, Fink H, Nyberg L, Thyr J, Li JY, Enejder A. Plaque-associated lipids in Alzheimer’s diseased brain tissue visualized by nonlinear microscopy. Sci Rep. 2015;5:13489. View this article via: PubMed Google Scholar
- van Horssen J, Wesseling P, van den Heuvel LP, de Waal RM, Verbeek MM. Heparan sulphate proteoglycans in Alzheimer’s disease and amyloid-related disorders. Lancet Neurol. 2003;2(8):482–492.
- McBride PA, Wilson MI, Eikelenboom P, Tunstall A, Bruce ME. Heparan sulfate proteoglycan is associated with amyloid plaques and neuroanatomically targeted PrP pathology throughout the incubation period of scrapie-infected mice. Exp Neurol. 1998;149(2):447–454.
- Ginsberg SD, Crino PB, Lee VM, Eberwine JH, Trojanowski JQ. Sequestration of RNA in Alzheimer’s disease neurofibrillary tangles and senile plaques. Ann Neurol. 1997;41(2):200–209.
- Gellermann GP, et al. Raft lipids as common components of human extracellular amyloid fibrils. Proc Natl Acad Sci USA. 2005;102(18):6297–6302.
- Bu G. Apolipoprotein E and its receptors in Alzheimer’s disease: pathways, pathogenesis and therapy. Nat Rev Neurosci. 2009;10(5):333–344.
- Jiménez JS. Protein-DNA interaction at the origin of neurological diseases: a hypothesis. J Alzheimers Dis. 2010;22(2):375–391.
- Ma J, Yee A, Brewer HB, Das S, Potter H. Amyloid-associated proteins alpha 1-antichymotrypsin and apolipoprotein E promote assembly of Alzheimer beta-protein into filaments. Nature. 1994;372(6501):92–94.
- Pensalfini A, et al. Intracellular amyloid and the neuronal origin of Alzheimer neuritic plaques. Neurobiol Dis. 2014;71:53–61.
- Uchida Y, Takahashi H. Rapid detection of Abeta deposits in APP transgenic mice by Hoechst 33342. Neurosci Lett. 2008;448(3):279–281.
- Di Domizio J, et al. Binding with nucleic acids or glycosaminoglycans converts soluble protein oligomers to amyloid. J Biol Chem. 2012;287(1):736–747.
- Di Domizio J, et al. Nucleic acid-containing amyloid fibrils potently induce type I interferon and stimulate systemic autoimmunity. Proc Natl Acad Sci USA. 2012;109(36):14550–14555.
- Huang X, et al. Neutrophils regulate humoral autoimmunity by restricting interferon-γ production via the generation of reactive oxygen species. Cell Rep. 2015;12(7):1120–1132.
- Gray F, et al. Neuropathology of early HIV-1 infection. Brain Pathol. 1996;6(1):1–15.
- Dipasquale O, et al. Interferon-α acutely impairs whole-brain functional connectivity network architecture - a preliminary study. Brain Behav Immun. 2016;58:31–39.
- Wachholz S, Eßlinger M, Plümper J, Manitz MP, Juckel G, Friebe A. Microglia activation is associated with IFN-α induced depressive-like behavior. Brain Behav Immun. 2016;55:105–113.
- Hayley S, Scharf J, Anisman H. Central administration of murine interferon-α induces depressive-like behavioral, brain cytokine and neurochemical alterations in mice: a mini-review and original experiments. Brain Behav Immun. 2013;31:115–127.
- Götz J, Bodea LG, Goedert M. Rodent models for Alzheimer disease. Nat Rev Neurosci. 2018;19(10):583–598.
- Jankowsky JL, Zheng H. Practical considerations for choosing a mouse model of Alzheimer’s disease. Mol Neurodegener. 2017;12(1):89.
- Saito T, et al. Single App knock-in mouse models of Alzheimer’s disease. Nat Neurosci. 2014;17(5):661–663.
- Jankowsky JL, et al. Persistent amyloidosis following suppression of Abeta production in a transgenic model of Alzheimer disease. PLoS Med. 2005;2(12):e355.
- Radde R, et al. Abeta42-driven cerebral amyloidosis in transgenic mice reveals early and robust pathology. EMBO Rep. 2006;7(9):940–946.
- Humpel C. Organotypic brain slice cultures: A review. Neuroscience. 2015;305:86–98.
- Opitz-Araya X, Barria A. Organotypic hippocampal slice cultures. J Vis Exp. 2011;(48):e2462.
- Ginsberg SD, Galvin JE, Chiu TS, Lee VM, Masliah E, Trojanowski JQ. RNA sequestration to pathological lesions of neurodegenerative diseases. Acta Neuropathol. 1998;96(5):487–494.
- Song WM, Colonna M. The identity and function of microglia in neurodegeneration. Nat Immunol. 2018;19(10):1048–1058.
- Hansen DV, Hanson JE, Sheng M. Microglia in Alzheimer’s disease. J Cell Biol. 2018;217(2):459–472.
- Krasemann S, et al. The TREM2-APOE pathway drives the transcriptional phenotype of dysfunctional microglia in neurodegenerative diseases. Immunity. 2017;47(3):566–581.e9.
- Keren-Shaul H, et al. A unique microglia type associated with restricting development of Alzheimer’s disease. Cell. 2017;169(7):1276–1290.e17.
- Bennett ML, et al. New tools for studying microglia in the mouse and human CNS. Proc Natl Acad Sci USA. 2016;113(12):E1738–E1746.
- Jordão MJC, et al. Single-cell profiling identifies myeloid cell subsets with distinct fates during neuroinflammation. Science. 2019;363(6425):eaat7554.
- Jankowsky JL, et al. Mutant presenilins specifically elevate the levels of the 42 residue beta-amyloid peptide in vivo: evidence for augmentation of a 42-specific gamma secretase. Hum Mol Genet. 2004;13(2):159–170.
- Li H, et al. Vascular and parenchymal amyloid pathology in an Alzheimer disease knock-in mouse model: interplay with cerebral blood flow. Mol Neurodegener. 2014;9:28. View this article via: PubMed Google Scholar
- Chaussabel D, et al. A modular analysis framework for blood genomics studies: application to systemic lupus erythematosus. Immunity. 2008;29(1):150–164.
- Li W, Viengkhou B, Denyer G, West PK, Campbell IL, Hofer MJ. Microglia have a more extensive and divergent response to interferon-α compared with astrocytes. Glia. 2018;66(10):2058–2078.
- Zagórska A, Través PG, Lew ED, Dransfield I, Lemke G. Diversification of TAM receptor tyrosine kinase function. Nat Immunol. 2014;15(10):920–928.
- Fourgeaud L, et al. TAM receptors regulate multiple features of microglial physiology. Nature. 2016;532(7598):240–244.
- Murata S, Takahama Y, Kasahara M, Tanaka K. The immunoproteasome and thymoproteasome: functions, evolution and human disease. Nat Immunol. 2018;19(9):923–931.
- Ikushima H, Negishi H, Taniguchi T. The IRF family transcription factors at the interface of innate and adaptive immune responses. Cold Spring Harb Symp Quant Biol. 2013;78:105–116.
- Porritt RA, Hertzog PJ. Dynamic control of type I IFN signalling by an integrated network of negative regulators. Trends Immunol. 2015;36(3):150–160.
- Feng G, et al. Imaging neuronal subsets in transgenic mice expressing multiple spectral variants of GFP. Neuron. 2000;28(1):41–51.
- Dunn GP, et al. A critical function for type I interferons in cancer immunoediting. Nat Immunol. 2005;6(7):722–729.
- Mathys H, et al. Temporal tracking of microglia activation in neurodegeneration at single-cell resolution. Cell Rep. 2017;21(2):366–380.
- Park J, et al. A 3D human triculture system modeling neurodegeneration and neuroinflammation in Alzheimer’s disease. Nat Neurosci. 2018;21(7):941–951.
- Deczkowska A, Baruch K, Schwartz M. Type I/II interferon balance in the regulation of brain physiology and pathology. Trends Immunol. 2016;37(3):181–192.
- Liu SY, Sanchez DJ, Aliyari R, Lu S, Cheng G. Systematic identification of type I and type II interferon-induced antiviral factors. Proc Natl Acad Sci USA. 2012;109(11):4239–4244.
- Paolicelli RC, et al. Synaptic pruning by microglia is necessary for normal brain development. Science. 2011;333(6048):1456–1458.
- Stevens B, et al. The classical complement cascade mediates CNS synapse elimination. Cell. 2007;131(6):1164–1178.
- Wu T, et al. Complement C3 is activated in human AD Brain and is required for neurodegeneration in mouse models of amyloidosis and tauopathy. Cell Rep. 2019;28(8):2111–2123.e6.
- Hong S, et al. Complement and microglia mediate early synapse loss in Alzheimer mouse models. Science. 2016;352(6286):712–716.
- Vasek MJ, et al. A complement-microglial axis drives synapse loss during virus-induced memory impairment. Nature. 2016;534(7608):538–543.
- Deczkowska A, et al. Mef2C restrains microglial inflammatory response and is lost in brain ageing in an IFN-I-dependent manner. Nat Commun. 2017;8(1):717.
- Lian H, Litvinchuk A, Chiang AC, Aithmitti N, Jankowsky JL, Zheng H. Astrocyte-microglia cross talk through complement activation modulates amyloid pathology in mouse models of Alzheimer’s disease. J Neurosci. 2016;36(2):577–589.
- Wang J, Campbell IL. Innate STAT1-dependent genomic response of neurons to the antiviral cytokine alpha interferon. J Virol. 2005;79(13):8295–8302.
- Wang M, et al. The Mount Sinai cohort of large-scale genomic, transcriptomic and proteomic data in Alzheimer’s disease. Sci Data. 2018;5:180185. View this article via: PubMed Google Scholar
- Wang M, et al. Integrative network analysis of nineteen brain regions identifies molecular signatures and networks underlying selective regional vulnerability to Alzheimer’s disease. Genome Med. 2016;8(1):104.
- Lehrman EK, et al. CD47 protects synapses from excess microglia-mediated pruning during development. Neuron. 2018;100(1):120–134.e6.
- Zhang X, et al. Human intracellular ISG15 prevents interferon-α/β over-amplification and auto-inflammation. Nature. 2015;517(7532):89–93.
- Braak H, Braak E. Neuropathological stageing of Alzheimer-related changes. Acta Neuropathol. 1991;82(4):239–259.
- Honda K, et al. IRF-7 is the master regulator of type-I interferon-dependent immune responses. Nature. 2005;434(7034):772–777.
- Platanias LC. Mechanisms of type-I- and type-II-interferon-mediated signalling. Nat Rev Immunol. 2005;5(5):375–386.
- Merle NS, Church SE, Fremeaux-Bacchi V, Roumenina LT. Complement system part I - molecular mechanisms of activation and regulation. Front Immunol. 2015;6:262. View this article via: PubMed Google Scholar
- Holmes BB, et al. Heparan sulfate proteoglycans mediate internalization and propagation of specific proteopathic seeds. Proc Natl Acad Sci USA. 2013;110(33):E3138–E3147.
- Liu CC, et al. Neuronal heparan sulfates promote amyloid pathology by modulating brain amyloid-β clearance and aggregation in Alzheimer’s disease. Sci Transl Med. 2016;8(332):332ra44.
- Ginsberg SD, et al. Predominance of neuronal mRNAs in individual Alzheimer’s disease senile plaques. Ann Neurol. 1999;45(2):174–181.
- Spangenberg E, et al. Sustained microglial depletion with CSF1R inhibitor impairs parenchymal plaque development in an Alzheimer’s disease model. Nat Commun. 2019;10(1):3758.
- Neuner SM, Heuer SE, Huentelman MJ, O’Connell KMS, Kaczorowski CC. Harnessing genetic complexity to enhance translatability of Alzheimer’s disease mouse models: a path toward precision medicine. Neuron. 2019;101(3):399–411.e5.
- Wang S, Colonna M. Microglia in Alzheimer’s disease: A target for immunotherapy. J Leukoc Biol. 2019;106(1):219–227.View this article via: PubMed Google Scholar
- Hammond TR, Robinton D, Stevens B. Microglia and the brain: complementary partners in development and disease. Annu Rev Cell Dev Biol. 2018;34:523–544.
- Butovsky O, Weiner HL. Microglial signatures and their role in health and disease. Nat Rev Neurosci. 2018;19(10):622–635.
- Hofer MJ, Campbell IL. Type I interferon in neurological disease-the devil from within. Cytokine Growth Factor Rev. 2013;24(3):257–267.
- Rodero MP, Crow YJ. Type I interferon-mediated monogenic autoinflammation: The type I interferonopathies, a conceptual overview. J Exp Med. 2016;213(12):2527–2538.
- Goldmann T, et al. USP18 lack in microglia causes destructive interferonopathy of the mouse brain. EMBO J. 2015;34(12):1612–1629.
- Sliter DA, et al. Parkin and PINK1 mitigate STING-induced inflammation. Nature. 2018;561(7722):258–262.
- Wang Y, et al. TREM2 lipid sensing sustains the microglial response in an Alzheimer’s disease model. Cell. 2015;160(6):1061–1071.
- Jay TR, et al. TREM2 deficiency eliminates TREM2+ inflammatory macrophages and ameliorates pathology in Alzheimer’s disease mouse models. J Exp Med. 2015;212(3):287–295.
- Jay TR, et al. Disease progression-dependent effects of TREM2 deficiency in a mouse model of Alzheimer’s disease. J Neurosci. 2017;37(3):637–647.
- Yeh FL, Wang Y, Tom I, Gonzalez LC, Sheng M. TREM2 binds to apolipoproteins, including APOE and CLU/APOJ, and thereby facilitates uptake of amyloid-beta by microglia. Neuron. 2016;91(2):328–340.
- Bailey CC, DeVaux LB, Farzan M. The triggering receptor expressed on myeloid cells 2 binds apolipoprotein E. J Biol Chem. 2015;290(43):26033–26042.
- Atagi Y, et al. Apolipoprotein E is a ligand for triggering receptor expressed on myeloid cells 2 (TREM2). J Biol Chem. 2015;290(43):26043–26050.
- Hamerman JA, Jarjoura JR, Humphrey MB, Nakamura MC, Seaman WE, Lanier LL. Cutting edge: inhibition of TLR and FcR responses in macrophages by triggering receptor expressed on myeloid cells (TREM)-2 and DAP12. J Immunol. 2006;177(4):2051–2055.
- Hammond TR, et al. Single-cell RNA sequencing of microglia throughout the mouse lifespan and in the injured brain reveals complex cell-state changes. Immunity. 2019;50(1):253–271.e6.
- Terry RD, et al. Physical basis of cognitive alterations in Alzheimer’s disease: synapse loss is the major correlate of cognitive impairment. Ann Neurol. 1991;30(4):572–580.
- Scheff SW, DeKosky ST, Price DA. Quantitative assessment of cortical synaptic density in Alzheimer’s disease. Neurobiol Aging. 1990;11(1):29–37.
- DeKosky ST, Scheff SW. Synapse loss in frontal cortex biopsies in Alzheimer’s disease: correlation with cognitive severity. Ann Neurol. 1990;27(5):457–464.
- Tenner AJ, Stevens B, Woodruff TM. New tricks for an ancient system: Physiological and pathological roles of complement in the CNS. Mol Immunol. 2018;102:3–13.
- Morgan BP. Complement in the pathogenesis of Alzheimer’s disease. Semin Immunopathol. 2018;40(1):113–124.
- Schafer DP, et al. Microglia sculpt postnatal neural circuits in an activity and complement-dependent manner. Neuron. 2012;74(4):691–705.
- Liddelow SA, et al. Neurotoxic reactive astrocytes are induced by activated microglia. Nature. 2017;541(7638):481–487.
- Arranz AM, De Strooper B. The role of astroglia in Alzheimer’s disease: pathophysiology and clinical implications. Lancet Neurol. 2019;18(4):406–414.
- Serrano-Pozo A, Frosch MP, Masliah E, Hyman BT. Neuropathological alterations in Alzheimer disease. Cold Spring Harb Perspect Med. 2011;1(1):a006189. View this article via: PubMed Google Scholar
- Perez-Nievas BG, et al. Dissecting phenotypic traits linked to human resilience to Alzheimer’s pathology. Brain. 2013;136(Pt 8):2510–2526.View this article via: PubMed Google Scholar
- Abt MC, et al. Commensal bacteria calibrate the activation threshold of innate antiviral immunity. Immunity. 2012;37(1):158–170.
- Gough DJ, Messina NL, Clarke CJ, Johnstone RW, Levy DE. Constitutive type I interferon modulates homeostatic balance through tonic signaling. Immunity. 2012;36(2):166–174.
- Hwang SY, et al. A null mutation in the gene encoding a type I interferon receptor component eliminates antiproliferative and antiviral responses to interferons alpha and beta and alters macrophage responses. Proc Natl Acad Sci USA. 1995;92(24):11284–11288.
- Ejlerskov P, et al. Lack of neuronal IFN-β-IFNAR causes Lewy body- and Parkinson’s disease-like dementia. Cell. 2015;163(2):324–339.
- Minter MR, et al. Deletion of the type-1 interferon receptor in APPSWE/PS1ΔE9 mice preserves cognitive function and alters glial phenotype. Acta Neuropathol Commun. 2016;4(1):72.
- Baruch K, et al. Aging. Aging-induced type I interferon response at the choroid plexus negatively affects brain function. Science. 2014;346(6205):89–93.
-
Version history
- Version 1 (January 9, 2020): In-Press Preview
- Version 2 (March 9, 2020): Electronic publication
- Version 3 (April 1, 2020): Print issue publication



Copyright © 2024 American Society for Clinical Investigation
ISSN: 0021-9738 (print), 1558-8238 (online)