Abstract
The yeasts transmitted from seeds to sprouts might be used as probiotics for host plants. To investigate the inheritable yeasts of rice plants for probiotics, the fungal internal transcribed spacer (ITS) regions (ITS1 and ITS2) in rice sprouts were analyzed by Illumina-based sequencing. The fungal genera Candida, Mortierella, Alternaria, Penicillium, and Tomentella were revealed by both ITS1 and ITS2 sequence analysis. The endophytic yeasts were isolated from rice sprouts by yeast selective medium. Compared with the negative controls, inoculation of isolate Y3 released 2.2 folds higher concentration of free phosphate in soybean meal broth. Most of the phytase activities were located in the yeast cell interiors. The shoot lengths, shoot fresh weights, and root fresh weights of inoculated seedlings increased by 35%, 80%, and 60% compared with the control seedlings, respectively. The results suggested that the rice sprouts contained diverse phytase-producing yeasts transmitted from seeds. These yeasts might be adopted as prospective probiotics to improve rice growth by increasing phosphate utilization efficacy.
Keywords: Endophyte, Rice, Mycobiome, Ascomycota, Plant growth-promoting
Introduction
Phosphorus is a limited resource playing a critical role in global agricultural production. The increasing mined phosphate prices demand alternative phosphate sources and the efficient phosphate utilization technology around the world (Brinch-Pedersen et al. 2014). Phytin is a mixed salt (K+, Ca2+, Mg2+ or Zn2+) with phytate, account for up to 85% of total organic phosphorus in seeds and straws (Higgins and Crittenden 2015). High levels of unutilized phytate lead to phosphorus runoff from oversupplied agricultural soils to aquatic ecosystems. The resulting eutrophication is a severe environmental risk (Brinch-Pedersen et al. 2014).
Phytase was viewed as a tool for determining the phytin degradation and phosphorus bioavailability (Brinch-Pedersen et al. 2014). It can be produced most notably in germinating seeds and microbes (Nasri et al. 2011; Higgins and Crittenden 2015). However, the mature plant and monogastric animals have only limited phytase activity in their nutrition absorption organs. Dietary phytase supplementation, transgenic plants, transgenic animals, and low-phytate crops have met with limited applications owing to poor consumer acceptance, industry hesitation, and technological constraint (Lei et al. 2013). Inoculation of phytase-producing probiotic microbes was proposed due to colonization stability in hosts.
Plants were colonized by some microbiota that can reach cell densities greater than the number of plant cells (Mendes et al. 2013). Each plant species hosts a genotype-specific core microbiome, dynamically responding to the environmental variables (Hirsch and Mauchline 2012). Compared with bacteria and mycorrhizal fungi, the potential to use yeasts as plant probiotics has been under-exploited (Amprayn et al. 2012). Unlike mycelial fungi, unicellular yeasts can not distribute within the intercellular space due to apical growth (Isaeva et al. 2010). However, some yeast species showed plant cell wall degradation enzyme which enables yeast cells to penetrate internal plant tissues from the surface through local cuticle damage. Plant starch-containing storage tissues of seeds nearly always contain yeasts that are able to actively reproduce in these tissues (Isaeva et al. 2010). During seed germination, the initially dormant seed undergoes passive and active changes in its physiological state (Ofek et al. 2011). Marked increase in phytase activity was reported in germinating seeds and was associated with a concomitant decrease in phytate content and an increase in that of phosphate (Nasri et al. 2011; Hui et al. 2016). Thus, the phytase activity of yeast might be involved in seed germination and plant growth promoting (Puppala et al. 2018). Phytase-producing ability is also one trait of animal probiotics (Tsang 2011; Andrabi et al. 2016). Some plant probiotic yeasts might be isolated from germinating seeds.
Rice is the staple food for the largest population in Asia. However, the yeast diversity of germinating rice seeds has not been reported. Until recently, most studies on probiotic strains focused on isolating microbes into pure culture, so the species that do not grow or grow very slowly in media were neglected (Porras-Alfaro and Bayman 2011). Compared to conventional cultivation methods, the sequencing approach for the analysis of fungal communities should provide much more information for probiotic yeast isolation (Suryanarayanan 2013; Sapkota et al. 2015).
The objective of this study was to rationally isolate the plant yeast probiotics from germinating rice seeds based on the analysis of fungal communities. In mycology, the internal transcribed spacer (ITS) region is the most commonly sequenced region for fungal systematics and taxonomy at and below the genus level (Nilsson et al. 2009). Due to restrictions in sequence read lengths, only ITS1 or ITS2 is normally analyzed in amplicon pyrosequencing. Therefore, in this study, the fungal communities based on different DNA barcodes (ITS1 and ITS2) were compared and the yeast probiotics were purposely isolated and characterized.
Materials and methods
Sample collection
Rice (Oryza sativa L. cv. Wusimi) seeds were collected from a rice field located in the suburb of Guangzhou, China (22°55′239″N, 113°31′074″E). The seeds were sterilized by serial immersion in 75% (v/v) ethanol for 5 min and sodium hypochlorite solution (5% available chlorine) for 5 min. Total 80 seeds were germinated on sterile filter papers soaked with water in Petri dishes without light at 25 °C. After 4 days, about 50 selected sprouts without retained seeds were collected for DNA extraction in each treatment.
DNA extraction, amplicon generation, and Illumina MiSeq sequencing
The total DNA of sprouts was extracted using E.Z.N.A. HP Plant DNA Kit (Omega, Norcross, GA, USA) according to the manufacturer’s instruction. The purity and concentration of DNA were monitored on 1% agarose gels.
The primers ITS1-1737F (GGAAGTAAAAGTCGTAACAAGG) and ITS2-2043R (GCTGCGTTCTTCATCGATGC) targeting the ITS1 hypervariable region, the primers ITS3_KYO2 (5′-GATGAAGAACGYAGYRAA-3′) and ITS4 (5′-TCCTCCGCTTATTGATATGC-3′) targeting the ITS2 region of fungal rRNA genes were adopted to analyze fungal taxa simultaneously (Schmidt et al. 2013). Both forward and reverse primers were tagged with adapter, pad, and linker sequencing (Schmidt et al. 2013). Each barcode sequence from multiple samples was pooled for one run of sequencing. All PCR reactions were performed by Applied Biosystems 2720 Thermal cycler in a total volume of 30 μL, sequencing on an Illumina MiSeq platform (Wang et al. 2016).
Combination and data preprocessing
Forward and reverse sequences were merged using FLASH, the qualities of clean tags were detected by QIIME (Magoc and Salzberg 2011; Caporaso et al. 2012; Bokulich et al. 2013). Tags with chimera were detected and removed using UCHIME Algorithm (Edgar et al. 2011; Haas et al. 2011). The retained sequences were clustered into operational taxonomic units (OTU) at 97% sequence similarity using the UPARSE-OTU and UPARSE-OTUref algorithms of UPARSE software package (Edgar et al. 2011). The representative sequence was assigned to the fungal taxa by previous methods (Nilsson et al. 2010; Kõljalg et al. 2013).
Isolation of yeasts from sprouts
The surface sterilized rice seeds germinated on wet filter paper at 25 °C under aseptic conditions. After 4 days, the sprouts were cut into small pieces (about 0.4 × 0.2 cm) and put into malt extract medium with 50 μg/mL chloramphenicol (Huankai, Guangzhou, China) to inhibit bacterial growth. Plates were incubated at 25 °C for 2 days for yeast growth.
Determination of phytase activity
The 0.5 mL yeast broth was inoculated into Erlenmeyer flasks (250 mL) containing 50 mL malt extract broth on a rotary shaker at 25 °C for 24 h. The cells were harvested by centrifugation at 8000g for 4 min at 4 °C. The supernatants were collected for extracellular phytase activity. Cell pellets were washed and disrupted by ultrasonication and centrifuged at 15,000g for 20 min to remove all cell debris. The phytase activity measurements were performed by previous methods (Olstorpe et al. 2009). The optical density at 750 nm was measured by UV–Vis spectrometer (Unico UV-2000, Shanghai, China). One unit of phytase was defined as the amount of enzyme required to liberate 1 µmol P from phytate per min per mL yeast culture.
Soybean meal fermentation
The feed diet composed of soybean meal (10 g/L) and yeast extract (5 g/L) was inoculated with 1% inoculum of yeast and incubated at 30 °C for 5 days. The soybean meal media without yeast inoculation were used as negative controls. The culture broth (5 mL) was collected and centrifuged (4000 rpm) to separate the insoluble mass and supernatants. The free P accumulation in the supernatant was measured by the method described previously (Olstorpe et al. 2009).
Pot experiments
Soil samples collected from the suburbs of Guangzhou was processed by previous methods (Wang et al. 2017). Rice seeds sterilized in bleaching solution (3% available chlorine) were inoculated by soaking in yeast cell suspension (105 cfu/mL) for 1 h. The seeds soaked in sterile water were adopted as the control treatment. The inoculated seeds were further sown in the soils and ten seeds were sown in each pot as one treatment. The soil moisture content was kept at the saturated condition. The plants were grown in a glasshouse at 25 °C and a 16/8 h day/night regime (Wang et al. 2017). After 2 weeks, the shoot length (cm), shoot and root fresh weight (g) were measured and compared.
Statistical analysis
Cluster analysis was preceded with the principal component analysis (PCA) using the QIIME software package. QIIME calculates both weighted and unweighted unifrac distances, which are phylogenetic measures of beta diversity (Kõljalg et al. 2013). Phylogenetic relations among different microbial taxa were further displayed by KRONA (Ondov et al. 2011). Alpha diversity indices of Chao1, ACE, Shannon, Simpson, and coverage were calculated to reflect the diversity and richness of the endophytic community in different samples (Kemp and Aller 2004).
Statistical analysis of rice seedling growth data was carried out using the SPSS statistical package (version 16.0 for Windows, SPSS Inc.). For the pot experiments, data were represented as mean ± standard deviation (SD) of the three replicates. The data were subjected to analysis of variance (ANOVA) and treatment mean values were compared by Duncan’s multiple-range test. All analyses were performed at P ≤ 0.05.
Results
Fungal diversity and richness
Total 133,341 valid fungal sequences remained with an average ITS1 length of 260 bp and 31200 sequences remained with an average ITS2 length of 373 bp (Table 1). With ITS1 as the DNA barcode, the number of different fungal OTUs at the 97% similarity level was 407. Nevertheless, the number of fungal OTUs at the 97% similarity level was 772 in the same samples with ITS2 as the barcode. The coverage index calculated from sequences detected by the ITS1 and ITS2 barcodes were 0.99 and 0.97, respectively. Therefore, the amount of sequences for analysis was sufficient, and the sequences well represented the full proportion of fungal phylotypes in rice sprouts. The alpha diversity indices (ACE, Shannon, and Simpson) calculated from the fungal OTUs indicated that ITS2 revealed more diverse fungi than ITS1 when the same DNA was used to analyze fungal diversity in rice sprouts (Table 2).
Table 1.
The characteristics of effective tags in rice sprouts identified by the internal transcribed spacer (ITS) regions (i.e., barcodes ITS1 and ITS2) of fungal rRNA genes
| Barcode | Tags’ information | |||
|---|---|---|---|---|
| Number | Total length (bp) | Max length (bp) | Min length (bp) | |
| ITS1 | 133341 | 34,628,940 | 387 | 196 |
| ITS2 | 31200 | 11,649,244 | 410 | 214 |
Table 2.
The alpha diversity indices of fungal operational taxonomic units (OTUs) in rice sprouts identified by the internal transcribed spacer (ITS) regions (i.e., barcodes ITS1 and ITS2) of fungal rRNA genes
| Barcode | Alpha diversity indices | ||||
|---|---|---|---|---|---|
| Chao1a | ACE | Shannon | Simpson | Coverage | |
| ITS1 | 1531 ± 28.7 | 1452 ± 25.9 | 0.20 ± 0.02 | 0.03 ± 0.002 | 0.99 ± 0.02 |
| ITS2 | 1334 ± 23.1 | 1789 ± 27.4 | 2.28 ± 0.15 | 0.32 ± 0.03 | 0.97 ± 0.04 |
aBoth Chao1 and ACE describe an estimate of the total number of phylotypes in a source environment, and Chao1 is particularly appropriate for datasets in which most phylotypes are relatively rare in the community, ACE is appropriate for datasets in which some phylotypes occur more frequently. Both Shannon and Simpson indices comprehensively reflect the richness and evenness of community. Shannon index is more sensitive to the richness of the community and Simpson index is more sensitive to the evenness of the community. Coverage is a non-parametric estimator of the proportion of phylotypes in a library of infinite size that should be represented in a smaller library
Although the sprouts did not contact with fungi from the exterior environment during seed germination, diverse fungi, including 3 fungal phyla, 6 fungal classes, 10 fungal orders, 9 fungal families, 14 fungal genera, and unidentified fungi, were detected by ITS1 in rice sprouts. The ITS2 barcode detected 5 fungal phyla, 14 classes, 45 orders, 81 families, 133 genera, and unidentified fungi. Some novel fungal OTUs affiliated with unidentified fungi have not been reported as endophytes of rice.
Fungal composition and community structure
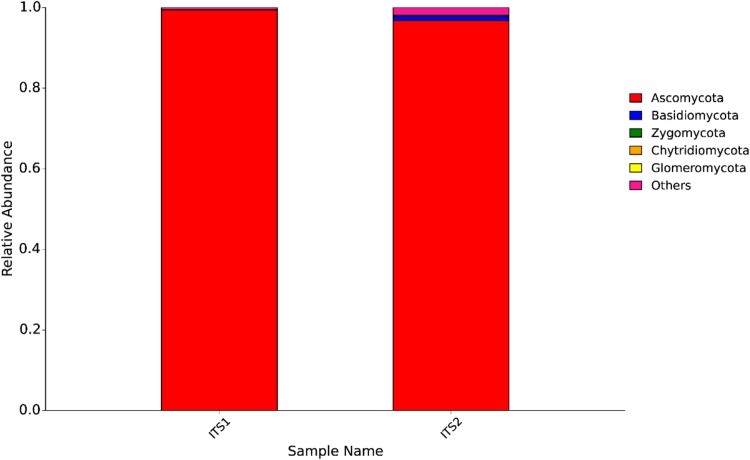
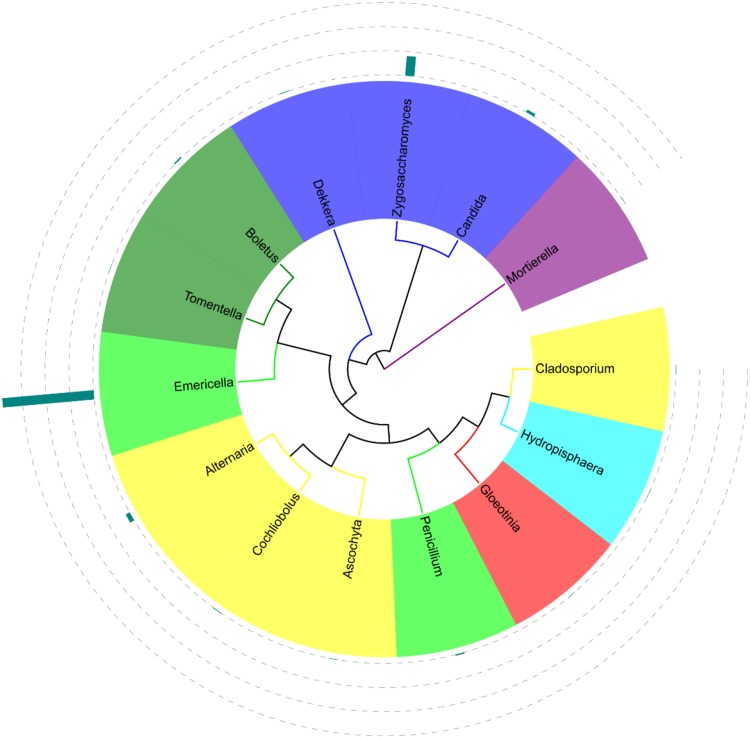
All the representative sequences of each OTU were classified into the domains fungi (100% of the total data set). Although the fungal proportions were different, Ascomycota and Basidiomycota were the dominant fungal taxa in sprouts based on both the ITS1 and ITS2 DNA barcodes (Fig. 1). The ITS1 barcode illustrated that 99.5% of ITS1 sequences could be assigned to the phylum level, Ascomycota was the dominant fungal phylum (99.1%), and Basidiomycota was the second most abundant (0.2%). Only 0.32% of the ITS1 sequences were affiliated with the class level, Eurotiomycetes was the most detected class (0.23%), and Saccharomycetes was the second most frequently detected (0.06%). Dominant orders detected by ITS1 included Eurotiales (0.23%), Saccharomycetales (0.06%), and Pleosporales (0.01%). Dominant families were Trichocomaceae (0.23%), Saccharomycetaceae (0.05%), Pleosporaceae (0.01%). Only 0.3% of ITS1 sequences could be assigned to the genus level. Emericells was the most detected genus (0.23%) and other dominant genera were Zygosaccharomyces (0.05%) and Alternaria (0.01%) (Fig. 2).
Fig. 1.
Abundances of different phyla in fungi in rice sprouts identified by the internal transcribed spacer (ITS) regions (i.e., barcodes ITS1 and ITS2) of fungal rRNA genes
Fig. 2.
Phylogenetic analysis of fungal taxa in rice sprouts identified by the internal transcribed spacer (ITS) region 1 (i.e., barcode ITS1) of fungal rRNA genes
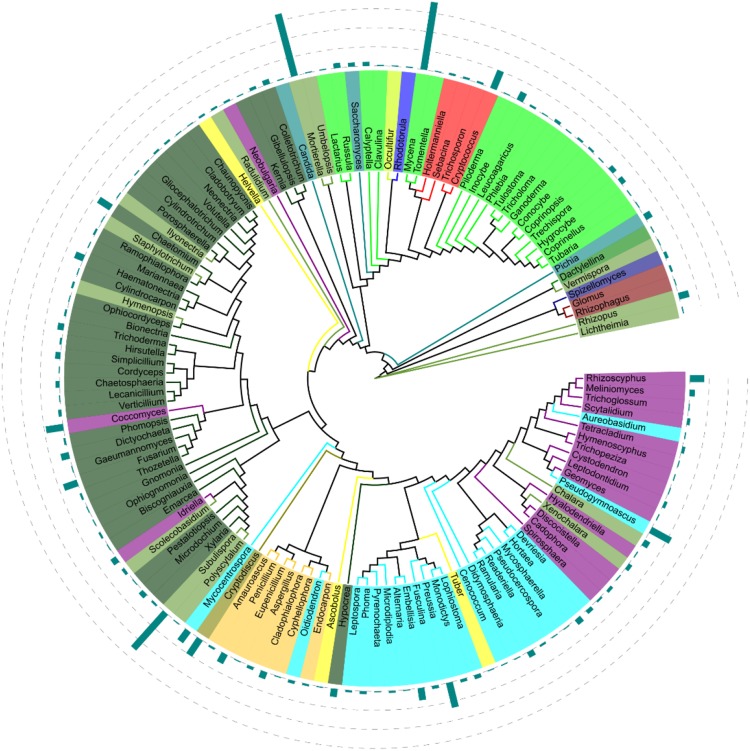
Total 99.8% of ITS2 sequences were assigned to the phylum level. The dominant fungal phyla included Ascomycota (96.8%), Basidiomycota (1.1%), Zygomycota (0.2%), and Chytridiomycota (0.03%) (Fig. 1). The 4.1% of ITS2 sequences could be assigned to the fungal class level. Dothideomycetes was the most dominant classes (0.87%) and other classes were Agaricomycetes (0.83%), Sordariomycetes (0.75%), and Leotiomycetes (0.73%). Pleosporales was the most dominant order (0.52%) and other frequently orders were Agaricales (0.42%), Helotiale (0.31%), and Hypocreales (0.21%). The dominant fungal families were Mycenaceae (0.24%), Mortierellaceae (0.20%), Nectriaceae (0.11%), Atheliaceae (0.10%), and Mycosphaerellaceae (0.10%). Compared with the ITS1 sequences (0.3%), higher proportion (2.1%) of ITS2 sequences were assigned to the genus level. Mycena and Mortierella were the most abundant genera. Other abundant genera included Subulispora (0.15%), Preussia (0.08%), Pseudogymnoascus (0.07%), and Cryptodiscus (0.06%) (Fig. 3). Candida, Mortierella, Alternaria, Penicillium, Tomentella were revealed by both ITS1 and ITS2 barcodes. Fusarium, Aspergillus, Penicillium, Phoma, Chaetomium, Trichoderma, Glomus, Kernia, Cladophialophora were detected by ITS2 barcode. Cladosporium was detected by ITS1 but not by ITS2 barcode in rice sprouts.
Fig. 3.
Phylogenetic analysis of fungal taxa in rice sprouts identified by the internal transcribed spacer (ITS) region 2 (i.e., barcode ITS2) of fungal rRNA genes
Isolation of probiotic yeasts and pot experiments
Endophytic yeast Candida was detected in rice sprouts by both ITS1 and ITS2 barcodes and further isolated from rice sprouts. Most of the sprout samples were overgrown by fungal growth on plates. Typical smooth yeast colonies were picked and examined for yeast cells by microscope. The white yeast Y3 with higher phytase activity was further identified as Candida sp. according to the ITS1-5.8S-ITS2 gene sequence analysis.
The phytase activity was not detected in the supernatant of yeast Y3 culture broth. The yeast Y3 cells could degrade sodium phytate after incubated for 24 h, however, the extracellular phytase was not detected. Most of the phytase activities were detected in the yeast cell debris after Y3 cells were disrupted by ultrasonication. The phytase might locate in cell walls.
The media contained soybean meal was inoculated with yeast Y3 and incubated at 30 °C for 5 days, the free phosphorus concentrations in broth increased. Compared with the negative controls, the inoculation of yeast Y3 released more free phosphate in broth (4.23 mmol/L) (P < 0.05). The yeast Y3 could release free P from the soybean meals and released 2.2 folds higher concentration of free phosphate than control treatment (P < 0.05).
The pot experiments illustrated the stimulation effects of Candida sp. Y3 in rice seedlings. The growth of rice seedlings inoculated with strain Y3 were promoted compared with the control seedlings. Shoot length, shoot fresh weight, and roots fresh weight were promoted by 35%, 80%, and 60%, respectively (Table 3).
Table 3.
The growth promotion effects of Candida sp. Y3 on rice seedlings inoculated
| Growth parameters | Seedlings without inoculation (controls) | Seedlings inoculated with Y3a |
|---|---|---|
| Shoot length (cm) | 7.5 ± 0.8a | 10. 2 ± 0.5b |
| Shoot fresh weight (g) | 4.5 ± 0.5a | 8.2 ± 0.4b |
| Root fresh weight (g) | 0.8 ± 0.2a | 1.3 ± 0.2b |
aDifferent letters (a and b) indicate the difference between seedlings was significant (P <0.05)
Discussion
Approximately, 60–90% of the phosphorus in grains is present in the form of phytic acid or phytate, the addition of phytase increases the availability of phosphorus for animal digestion and plant utilization (Marlida et al. 2010). The high price of commercial phytase restricts the application of phytase as feed supplement and soil fertilizer, a more economical alternative for phytase addition would be phytase-producing microbiota (Olstorpe et al. 2009). In the study, the phytase-producing yeasts were purposely isolated from rice sprouts according to the fungal community analysis. Phytase activity might be a contributing factor to yeast survival and proliferation within hosts (Tsang 2011). The free phosphate in the media completely suppressed the extracellular phytase activity and also reduced intracellular phytase activity (Olstorpe et al. 2009). Phytase-producing probiotic yeast must be further selected for application. In the study, the phytase-producing yeasts were isolated directly from rice sprouts as plant probiotics. Thus, the selection for plant probiotic yeasts based on mycobiome information is feasible. Furthermore, probiotic yeasts can prevent mold spoilage during postharvest storage of fruits and vegetables and promote plant growth (Fredlund et al. 2002; Nutaratat et al. 2014). Yeasts were useful for probiotics because that most yeasts are non-pathogen and do not produce mycotoxins or allergenic spores (Fredlund et al. 2002).
Analysis of fungal diversity is essential for selection for yeast probiotics (Berlec 2012). To our knowledge, this is the first report on fungal diversity in rice sprouts using the Illumina-based analysis. Our results suggested that Ascomycota was the dominant fungal phyla in rice sprouts. The results were consistent with previous reports that many endophytic fungi belonged to Ascomycota (Suryanarayanan 2013). Some endophytic Ascomycota in mature plants could transmit vertically from seeds and sprouts (Suryanarayanan 2013). The potential of endophytic Ascomycota for biocontrol and rice growth promotion has to be assessed in future research (Remlein-Starosta et al. 2016). In the study, Basidiomycota, Glomeromycota, and Zygomycota were all detected in sprouts, indicating that the sprouts contained diverse fungi derived from seeds.
Fungal endophytes have been isolated from surface-sterilized leaves, roots, and seeds of rice. They were considered as saprotrophic fungi or potential pathogens (Fisher and Petrini 1992). Alternaria, Cladosporium, Fusarium, and Phoma have been frequently isolated from seeds (Fisher and Petrini 1992). The fungal genera were also detected in sprouts germinated under sterile conditions in this study. Therefore, the fungal taxa were considered to be seed-borne and they were not detrimental to the growth of rice sprouts. The effects of these fungal taxa on rice sprouts would be further studied. Dark septate endophytes (DSEs) are rich endophytic fungal consortium in wild rice (Oryza granulate) roots, whereas the arbuscular mycorrhizal fungi were absent (Yuan et al. 2010a, b). DSEs and plants may establish a mutualistic interaction. Arbuscular mycorrhizal fungi (AMF) Glomus is frequently isolated from the roots of rice (Oryza sativa) (Vallino et al. 2009). In this study, the DSE group of Cladophialophora and the AMF group of Glomus were detected in rice sprouts. The beneficial fungi in rice roots might transmit from soils or sprouts. Some endophytic fungi, such as Chaetomium, are found in rice sprouts (Naik et al. 2009). Compared with previous results, most rice endophytic fungal taxa were detected in rice sprouts. Illumina-based analysis on endophytic fungal communities could provide more fungal information than previous methods. Phylogenetic analysis of endophytic mycobiota in roots of wild rice has shown that Basidiomycota is frequently detected (Yuan et al. 2010a). In the study, Ascomycota was the abundant fungal phylum in rice sprouts, whereas some Basidiomycota might be lost during the domestication process.
Conclusions
In conclusion, a diverse fungal community (5 fungal phyla, 14 classes, 45 orders, 81 families, and 133 genera) was detected in rice sprouts under aseptic conditions. Both ITS1 and ITS2 revealed that Ascomycota was the dominant fungal phylum in rice sprouts. The yeasts and other fungal taxa Glomus, Cladophialophora, and Chaetomium derived from sprouts might proliferate and transmit in plant interiors during the growth stages. These fungi might transmit into different organs of mature plants and should be isolated as probiotic fungi.
Accession number(s)
The sequence data have been deposited in the National Center for Biotechnology Information (NCBI) Sequence Read Archive (SRA) under the accession number SRP062152.
Acknowledgements
This study was supported by grants from Natural Science Foundation of Guangdong Province (No. 2018A0303130071), the Research Fund Program of Guangdong Province Key Laboratory for Climate Change and Natural Disaster Studies (2017CCND004) and Educational Commission of Guangdong Province of China (No. 2016KTSCX045).
Compliance with ethical standards
Conflict of interest
No potential conflict of interest was reported by the authors.
Contributor Information
Aiping Zhu, Email: [email protected].
Hongming Tan, Email: [email protected].
Lixiang Cao, Phone: 86- 20-84110238, Email: [email protected].
References
- Amprayn K, Rose MT, Kecskes M, Pereg L, Nguyen HT, Kennedy IR. Plant growth promoting characteristics of soil yeasts (Candida tropicalis HY) and its effectiveness for promoting rice growth. Appl Soil Ecol. 2012;61:295–299. doi: 10.1016/j.apsoil.2011.11.009. [DOI] [Google Scholar]
- Andrabi ST, Bhat B, Gupta M, Bajaj BK. Phytase-producing potential and other functional attributes of lactic acid bacteria isolates for prospective probiotic applications. Probiotics Antimicrob Proteins. 2016;8:121–129. doi: 10.1007/s12602-016-9220-3. [DOI] [PubMed] [Google Scholar]
- Berlec A. Novel techniques and findings in the study of plant microbiota: search for plant probiotics. Plant Sci. 2012;193–194:96–102. doi: 10.1016/j.plantsci.2012.05.010. [DOI] [PubMed] [Google Scholar]
- Bokulich N, Subramanian S, Faith J, Gevers D, Gordon J, Knight R, Mills D, Caporaso J. Quality-filtering vastly improves diversity estimates from illumine amplicon sequencing. Nat Methods. 2013;10:57–59. doi: 10.1038/nmeth.2276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brinch-Pedersen H, Madsen CK, Holme IB, Dionisio G. Increased understanding of the cereal phytase complement for better mineral bioavailability and resource management. J Cereal Sci. 2014;59:373–381. doi: 10.1016/j.jcs.2013.10.003. [DOI] [Google Scholar]
- Caporaso J, Lauber C, Walters W, et al. Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J. 2012;6:1621–1624. doi: 10.1038/ismej.2012.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R, Haas B, Clemente J, Quince C, Knight R. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics. 2011;27:2194–2200. doi: 10.1093/bioinformatics/btr381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher PJ, Petrini O. Fungal saprobes and pathogens as endophytes of rice (Oryza sativa L.) New Phytol. 1992;120:137–143. doi: 10.1111/j.1469-8137.1992.tb01066.x. [DOI] [Google Scholar]
- Fredlund E, Druvefors U, Boysen ME, Lingsten KJ, Schnurer J. Physiological characteristics of the biocontrol yeast Pichia anomala J121. FEMS Yeast Res. 2002;2:395–402. doi: 10.1016/S1567-1356(02)00098-3. [DOI] [PubMed] [Google Scholar]
- Haas B, Gevers D, Earl A, Feldgarden M, et al. Chimeric 16S rRNA sequence formation and detection in Sanger and 454-pyrosequenced PCR amplicons. Genome Res. 2011;21:494–504. doi: 10.1101/gr.112730.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins NF, Crittenden PD. Phytase activity in lichens. New Phytol. 2015;208:544–554. doi: 10.1111/nph.13454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirsch P, Mauchline T. Who’s who in the plant root microbiome? Nat Biotechnol. 2012;30:961–962. doi: 10.1038/nbt.2387. [DOI] [PubMed] [Google Scholar]
- Hui Q, Yang R, Shen C, Zhou Y, Gu Z. Mechanism of calcium lactate facilitating phytic acid degradation in soybean during germination. J Agric Food Chem. 2016;64:5564–5573. doi: 10.1021/acs.jafc.6b01598. [DOI] [PubMed] [Google Scholar]
- Isaeva OV, Glushakova AM, Garbuz SA, Kachalkin AV, Chernov IY. Endophytic yeast fungi in plant storage tissues. Biol Bull. 2010;37:26–34. doi: 10.1134/S1062359010010048. [DOI] [PubMed] [Google Scholar]
- Kemp P, Aller J. Bacterial diversity in aquatic and other environments: what 16S rDNA libraries can tell us. FEMS Microbiol Ecol. 2004;47:161–177. doi: 10.1016/S0168-6496(03)00257-5. [DOI] [PubMed] [Google Scholar]
- Kõljalg U, Nilsson R, Abarenkov K, Tedersoo L, Taylor A, Larsson K. Towards a unified paradigm for sequence-based identification of fungi. Mol Ecol. 2013;22:5271–5277. doi: 10.1111/mec.12481. [DOI] [PubMed] [Google Scholar]
- Lei XG, Weaver JD, Mullaney E, Ullah AH, Azain MJ. Phytase, a new life for an “old” enzyme. Annu Rev Anim Biosci. 2013;1:283–309. doi: 10.1146/annurev-animal-031412-103717. [DOI] [PubMed] [Google Scholar]
- Magoč T, Salzberg S. FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics. 2011;27:2957–2963. doi: 10.1093/bioinformatics/btr507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marlida Y, Delfita R, Adnadi P, Ciptaan G. Isolation, characterization and production of phjytase from endophytic fungus its application for feed. Pak J Nutr. 2010;9:471–474. doi: 10.3923/pjn.2010.471.474. [DOI] [Google Scholar]
- Mendes R, Garbeva P, Raaijmakers J. The rhizosphere microbiome: significance of plant beneficial, plant pathogenic, and human pathogenic microorganisms. FEMS Microbiol Rev. 2013;37:634–663. doi: 10.1111/1574-6976.12028. [DOI] [PubMed] [Google Scholar]
- Naik BS, Shashikala J, Krishnamurthy YL. Study on the diversity of endophytic communities from rice (Oryza sativa L.) and their antagonistic activities in vitro. Microbiol Res. 2009;164:290–296. doi: 10.1016/j.micres.2006.12.003. [DOI] [PubMed] [Google Scholar]
- Nasri N, Kaddour R, Rabhi M, Plassard C, Lachaal M. Effect of salinity on germination, phytase activity and phytate content in lettuce seedling. Acta Physiol Plant. 2011;33:935–942. doi: 10.1007/s11738-010-0625-4. [DOI] [Google Scholar]
- Nilsson R, Ryberg M, Abarenkov K, Sjökvist E, Kristiansson E. The ITS region as a target for characterization of fungal communities using emerging sequencing technologies. FEMS Microbiol Lett. 2009;296:97–101. doi: 10.1111/j.1574-6968.2009.01618.x. [DOI] [PubMed] [Google Scholar]
- Nilsson RH, Veldre V, Hartmann M, Unterseher M, et al. An open source software package for automated extraction of ITS1 and ITS2 from fungal ITS sequences for use in high-throughput community assays and molecular ecology. Fungal Ecol. 2010;3:284–287. doi: 10.1016/j.funeco.2010.05.002. [DOI] [Google Scholar]
- Nutaratat P, Srisuk N, Arunrattiyakorn P, Limtong S. Plant growth-promoting traits of epiphytic and endophytic yeasts isolated from rice and sugar cane leaves in Thailand. Fungal Biol. 2014;118:683–694. doi: 10.1016/j.funbio.2014.04.010. [DOI] [PubMed] [Google Scholar]
- Ofek M, Hadar Y, Minz D. Colonization of cucumber seeds by bacteria during germination. Environ Microbiol. 2011;13:2794–2807. doi: 10.1111/j.1462-2920.2011.02551.x. [DOI] [PubMed] [Google Scholar]
- Olstorpe M, Schnurer J, Passoth V. Screening of yeast strains for phytase activity. FEMS Yeast Res. 2009;9:478–488. doi: 10.1111/j.1567-1364.2009.00493.x. [DOI] [PubMed] [Google Scholar]
- Ondov B, Bergman N, Phillippy A. Interactive metagenomic visualization in a Web browser. BMC Bioinform. 2011;12:385. doi: 10.1186/1471-2105-12-385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porras-Alfaro A, Bayman P. Hidden fungi, emergent properties: endophytes and microbiomes. Annu Rev Phytopathol. 2011;49:291–315. doi: 10.1146/annurev-phyto-080508-081831. [DOI] [PubMed] [Google Scholar]
- Puppala KR, Naik T, Shaik A, Dastager S, Kumar R, Khire J, Dharne M. Evaluation of Candida tropicalis (NCIM 3321) extracellular phytase having plant growth promoting potential and process development. Biocatal Agric Biotechnol. 2018;13:225–235. doi: 10.1016/j.bcab.2017.12.013. [DOI] [Google Scholar]
- Remlein-Starosta D, Krzymińska J, Kowalska J, Bocianowski J. Evaluation of yeast-like fungi to protect Virginia mallow (Sida hermaphrodita) against Sclerotinia sclerotiorum. Can J Plant Sci. 2016;96:243–251. doi: 10.1139/cjps-2015-0230. [DOI] [Google Scholar]
- Sapkota R, Knorr K, Jørgensen LN, O’Hanlon KA, Nicolaisen M. Host genotype is an important determinant of the cereal phyllosphere mycobiome. New Phytol. 2015;207:1134–1144. doi: 10.1111/nph.13418. [DOI] [PubMed] [Google Scholar]
- Schmidt PA, Bálint M, Greshake B, Bandow C, Römbke J, Schmitt I. Illumina metabarcoding of a soil fungal community. Soil Biol Biochem. 2013;65:128–132. doi: 10.1016/j.soilbio.2013.05.014. [DOI] [Google Scholar]
- Suryanarayanan T. Endophyte research: going beyond isolation and metabolite documentation. Fungal Ecol. 2013;6:561–568. doi: 10.1016/j.funeco.2013.09.007. [DOI] [Google Scholar]
- Tsang PW. Differential phytate utilization in Candida species. Mycopathologia. 2011;172:473–479. doi: 10.1007/s11046-011-9453-3. [DOI] [PubMed] [Google Scholar]
- Vallino M, Greppi D, Novero M, Bonfante P, Lupotto E. Rice root colonisation by mycorrhizal and endophytic fungi in aerobic soil. Ann Appl Biol. 2009;154:195–204. doi: 10.1111/j.1744-7348.2008.00286.x. [DOI] [Google Scholar]
- Wang W, Zhai Y, Cao L, Tan H, Zhang R. Endophytic bacterial and fungal microbiota in sprouts, roots and stems of rice (Oryza sativa L.) Microbiol Res. 2016;188–189:1–8. doi: 10.1016/j.micres.2016.04.009. [DOI] [PubMed] [Google Scholar]
- Wang W, Li Y, Qin W, Sun C, Tan H, Cao L. Using Illumina-based sequence analysis to guide probiotic candidate selection and isolation. Probiotics Antimicrob Proteins. 2017 doi: 10.1007/s12602-017-9298-2. [DOI] [PubMed] [Google Scholar]
- Yuan Z, Zhang C, Lin F, Kubicek CP. Identity, diversity, and molecular phylogeny of endophytic mycobiota in the roots of rare wild rice (Oryza granulate) from a nature reserve in Yunnan, China. Appl Environ Microbiol. 2010;76:1642–1652. doi: 10.1128/AEM.01911-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan Z, Lin F, Zhang C, Kubicek CP. A new species of Harpophora (Magnaporthaceae) recovered from healthy wild rice (Oryza granulate) roots, representing a novel member of a beneficial dark septate endophyte. FEMS Microbiol Lett. 2010;307:94–101. doi: 10.1111/j.1574-6968.2010.01963.x. [DOI] [PubMed] [Google Scholar]