Abstract
Terminally differentiating epidermal keratinocytes express a large number of structural and antimicrobial proteins that are involved in the physical barrier function of the stratum corneum and provide innate cutaneous host defense. Late cornified envelope (LCE) genes, located in the epidermal differentiation complex on chromosome 1, encode a family of 18 proteins of unknown function, whose expression is largely restricted to epidermis. Deletion of two members, LCE3B and LCE3C (LCE3B/C-del), is a widely replicated psoriasis risk factor that interacts with the major psoriasis-psoriasis risk gene HLA-C*06. Here we performed quantitative trait locus analysis, utilizing RNA-seq data from human skin and found that LCE3B/C-del was associated with a markedly increased expression of LCE3A, a gene directly adjacent to LCE3B/C-del. We confirmed these findings in a 3D skin model using primary keratinocytes from LCE3B/C-del genotyped donors. Functional analysis revealed that LCE3 proteins, and LCE3A in particular, have defensin-like antimicrobial activity against a variety of bacterial taxa at low micromolar concentrations. No genotype dependent effect was observed for the inside-out or outside-in physical skin barrier function. Our findings identify an unknown biological function for LCE3 proteins and suggest a role in epidermal host defense and LCE3B/C-del mediated psoriasis risk.
Keywords: Psoriasis, Late Cornified Envelope, LCE3B/C-del, defensing, skin barrier
INTRODUCTION
Psoriasis vulgaris is a common inflammatory skin disease determined by both genetic and environmental factors (Nestle et al., 2009). Based on a number of genome wide association studies (GWAS) and meta-analyses thereof, more than 60 susceptibility loci have been identified that account for 20–25% of psoriasis heritability (Tsoi et al., 2015b; Tsoi et al., 2012; Zuo et al., 2015). Psoriasis is characterized by dysregulated cutaneous immune responses involving innate immunity (TNF-alpha and NFkB pathways) and exaggerated Th1 and Th17 lymphocyte activation (Johnston et al., 2013; Jordan et al., 2012; Lowes et al., 2014). In addition, psoriasis-associated genes expressed by keratinocytes and with a presumed role in epidermal homeostasis have also been genetically implicated in psoriasis, including the late cornified envelope (LCE) genes (de Cid et al., 2009; Huffmeier et al., 2010) and the beta-defensins (Hollox et al., 2008). Despite the identification of many candidate genes, functional studies that explain the contribution of genetic polymorphism to psoriasis risk are largely lacking. A number of studies, however, have shown a plausible link between genes from susceptibility loci and immunobiological features of psoriasis such as an association between variation at TNFAIP3 and response to TNF-alpha blockade (Tejasvi et al., 2012), and the association between the IL12B risk allele and increased Th1-cytokine levels (Johnston et al., 2013). In addition, only very few risk loci involve coding variants that are amenable to experimental verification in animal models or in vitro cellular models of skin biology or inflammation (Jordan et al., 2012; Tsoi et al., 2012). Among the psoriasis susceptibility regions, the MHC class 1 gene HLA-C*06:02 (PSORS1) and the LCE region harboring a deletion of the LCE3B and LCE3C genes (originally designated PSORS4) (Capon et al., 2001), provide plausible candidates to investigate at the functional level. HLA-C*06:02 is by far the strongest psoriasis risk factor, with an odds ratio estimated to be between 2.6 to 5 in Caucasians (Genetic Analysis of Psoriasis et al., 2010; Nair et al., 2009), and the odds ratio for the LCE deletion (OR ~ 1.3) (Huffmeier et al., 2010) is one of the highest among the remaining psoriasis-associated loci (Tsoi et al., 2012). LCE genes are expressed only in epidermis and oral epithelia (Bergboer et al., 2011; Jackson et al., 2005) and are assumed to encode structural proteins with a role in epithelial barrier formation; however, no functional studies supporting this contention have been published so far. Remarkably, the LCE3 group, which encompasses the LCE3B and LCE3C genes, is under regulation of psoriasis-associated Th1 and Th17 cytokines (Bergboer et al., 2011; Niehues et al., 2015). LCE3B/C-del affects a 32 kb fragment in the epidermal differentiation complex (EDC) on chromosome 1, which is commonly deleted in the non-African population (allele frequency of LCE3B/C-del: 60–70%) (de Cid et al., 2009). In addition to the loss of the protein coding genes LCE3B and LCE3C, it also removes three intergenic fragments that harbor potential regulatory sequences (de Guzman Strong et al., 2010). In this study we show that LCE3B/C-del causes an upregulation of the flanking LCE3A gene. Our hypothesis-driven functional studies have revealed that these three proteins are unlikely to be involved in skin barrier function and rather represent antimicrobial proteins.
RESULTS
eQTL analysis of genes in the epidermal differentiation complex (EDC)
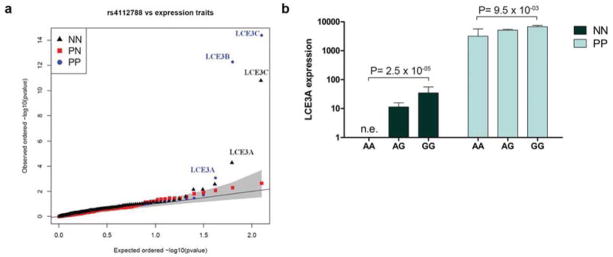
The association results between the deletion surrogate rs4112788 and expression traits are shown in Figure 1a. Within the EDC region, only the expression levels of LCE3A, LCE3B, and LCE3C are significantly (False Discovery Rate < 0.1) associated with the marker in both normal (NN) and psoriatic (PP) skin (Table 1). As expected, expression of both LCE3B and LCE3C are decreased in PP skin in the presence of the G allele of rs4112788, which is in linkage disequilibrium with LCE3B/C del. We observed the same result for LCE3C expression in NN, while LCE3B could not be assessed as it was not expressed in more than 20% NN skin samples. In fact, out of the 80 NN samples, we detected expression of LCE3A and LCE3C in 43 and 40 samples, respectively, but LCE3B was expressed only in one sample. In contrast, out of the 92 PP samples, we detected expression of LCE3A, LCE3B, and LCE3C in 92, 48, and 55 samples, respectively. Figure 1b shows that LCE3A expression levels were elevated in individuals with the GG genotype (surrogate for del/del) compared to individuals with the AA genotype, in both NN and PP samples. LCE3A is strongly and significantly (p=7.7×10−30) up-regulated in PP (mean expression level = 5.8 × 103) compared to NN skin (mean expression level = 20). The fold change difference in expression between GG/AA genotypes is 2.1 in PP samples, and the contrast is even higher in NN samples (mean expression level is 35 for GG and 0 for AA). We did not observe a significant genotype difference for LCE3A expression in PN skin samples, likely due to the smaller sample size. RNA-seq data showed that expression of LCE3A behaves similarly between NN and PN skin (NN = 4.6 vs. 4.5 in PN skin). See Table S4 for the normalized RNAseq expression data of all individuals.
Figure 1. In vivo eQTL analysis.
(a) Quantile-quantile plot of rs4112788 versus expression traits in the EDC region. Shaded area indicates 95% confidence interval of statistical significance. (b) Estimated effect size of LCE3B/C-del on in vivo LCE3A expression in the e-QTL data set. NN, normal skin; PN; non-lesional psoriatic skin; PP, lesional psoriatic skin. Error bars = SEM. n.e.=no expression.
Table 1. Top significant e-QTL results in NN and PP skin between rs4112788 versus EDC genes.
Note LCE3B is not expressed in more than 20% of the NN skin samples, therefore the eQTL analysis was not conducted.
| Skin type | Gene | Relative location to deletion | Effect | p | FDR |
|---|---|---|---|---|---|
|
| |||||
| Normal | LCE3C* | within | − | 1.60 × 10−11 | 1.96 × 10−9 |
| LCE3A* | downstream | + | 5.36 × 10−5 | 3.30 × 10−3 | |
| RP11-216N14.9 | downstream | + | 2.75 × 10−3 | 1.13 × 10−1 | |
| LCE1D | downstream | − | 7.1 × 10−3 | 0.18 | |
| LINGO4 | upstream | − | 7.29 × 10−3 | 0.18 | |
|
| |||||
| Psoriasis | LCE3C* | within | − | 4.09 × 10−15 | 5.11 ×10−13 |
| LCE3B* | within | − | 5.34 × 10−13 | 3.34 × 10−11 | |
|
| |||||
| LCE3A* | downstream | + | 8.73 × 10−4 | 3.64 × 10−2 | |
| LCE1D | downstream | − | 1.92 × 10−2 | 0.6 | |
| ILF2 | downstream | − | 3.49 × 10−2 | 0.67 | |
indicates significant at the false discovery rate (FDR<= 0.1) level. “Effect” refers to whether the gene is up (+) or down (−) regulated in the presence of the G allele for rs4112788 (surrogate for del/del).
Genotype dependent expression of LCE genes in 3D reconstructed epidermis
To substantiate these findings in vitro, and to determine LCE3B/C-del dependent epidermal morphology and differentiation, we generated 3D reconstructed epidermis from wt/wt (N=6) and del/del (N=6) keratinocytes. We have previously shown that in such a 3D reconstructed epidermis model, the spatio-temporal expression of LCE proteins is similar to in vivo epidermis (Niehues et al., 2015). Histopathological examination of the 3D reconstructed epidermis did not reveal obvious morphological differences between the two genotypes (H&E staining), nor did immunohistochemical staining reveal abnormalities in expression of differentiation proteins (Figure S1). Confirming what was observed in vivo (Figure 1a–b), qPCR analysis showed a significant effect of the LCE3B/C-del on LCE3A expression (Figure 2a–c): LCE3A mRNA expression was 6.6-fold higher in del/del keratinocytes compared to wt/wt keratinocytes (p<0.003). To mimic psoriasis in vitro and confirm the in vivo findings, we stimulated 3D reconstructed epidermis with psoriasis-associated cytokines. To verify the effect of cytokine stimulation, expression levels of known Th1 and Th1/Th17 responsive genes CXCL10 and DEFB4 were analyzed and found to be strongly induced irrespective of the LCE3B/C-del genotype. (Figure S2a–b). Following Th1-, but not Th17- cytokine stimulation, we found a significant genotype effect (4.7-fold, p<0.05, Figure 2b–c). Generally, Th1-cytokines induced expression of all tested LCE family genes (Figure 2d) whereas Th17-cytokines specifically increased LCE2 and LCE3 genes (Figure 2e). These in vitro findings corroborate the in vivo findings of the effect of LCE3B/C-del on LCE3A expression. The fold increase in 3D reconstructed epidermis of del/del keratinocytes is modest compared to the huge relative increase in vivo, which is caused by the relatively high basal expression of LCE3A in wt/wt cells in the skin equivalents, whereas LCE3A expression in vivo for the AA (wt/wt) genotype is zero.
Figure 2. In vitro mRNA analysis.
mRNA expression of (a) control, (b) Th1- and (c) Th17-cytokine stimulated cells (N=5 per group) in wt/wt and del/del 3D reconstructed epidermis. (d–e) mRNA expression upon Th1- and Th17-cytokine stimulation (N=10). Error bars = SEM. Red lines = control (wt/wt or unstimulated, set to 1), * P<0.05, ** P<0.005, ***P<0.0005.
LCE3B/C del is not associated with skin barrier alterations
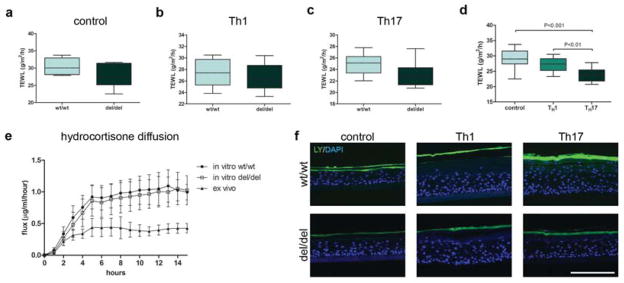
We asked whether genotype dependent alterations of LCE3A expression, i.e. the absence of LCE3B and LCE3C and the upregulation of LCE3A, had functional consequences that would provide a clue to its genetic association with psoriasis. Many epidermis-specific genes encode either structural proteins involved in physical barrier function (Candi et al., 2005) or small proteins that provide an antimicrobial barrier (Gallo and Hooper, 2012; Schroder and Harder, 2006). Limited proteomic data suggest that LCE proteins are part of the stratum corneum (SC), hence participate in physical skin barrier function, although direct proof is lacking. We therefore assessed trans-epidermal water loss (TEWL) from 3D reconstructed epidermis of wt/wt and del/del genotypes to unveil a possible effect on inside-out barrier function. No difference in TEWL was observed between the genotypes (Figure 3a–c). We noticed that cytokine-stimulated 3D reconstructed epidermis had a lower TEWL compared to control 3D constructs (control vs. Th17, p<0.001), but again no differences between the two genotypes were observed (Figure 3d). The outside-in barrier function was investigated by analysis of the penetration of hydrophilic and hydrophobic low molecular weight tracers. Quantitative determination of hydrocortisone diffusion by UPLC analysis through the SC did not reveal a difference between the two genotypes (Figure 3e). Furthermore, we did not observe LY penetration in constructs of any of the genotypes, under unstimulated and Th1- or Th17-cytokine conditions (Figure 3f). Collectively, these data indicate that LCE3B/C-del is not associated with altered skin barrier function.
Figure 3. Barrier function of LCE3B/C wt/wt and del/del 3D reconstructed epidermis.
(a–d) TEWL of unstimulated, Th1- or Th17-stimulated epidermis. Repeated measures ANOVA plus Bonferroni post hoc with P < 0.05 significance. (e) SC hydrocortisone-diffusion assay. (f) Lucifer yellow (LY) penetration in unstimulated, Th1 or Th17-cytokine stimulated epidermis. Scale bar = 100 μm. N=5 per genotype group. Error bars = SEM.
LCE3 proteins have antibacterial activity
Keratinocytes are a known source of antimicrobial peptides (AMPs) such as defensins, SLPI, SKALP/Elafin, psoriasin and LL37, many of which are small, cationic and/or cysteine-rich proteins (Harder et al., 2013). As LCE3 proteins are also small (9–11 kD), cationic (isoelectric points > 8.6) having an extremely high cysteine content (18–23%) we tested the hypothesis that LCE proteins are antimicrobial. Based on the finding that LCE3A expression is firmly increased in LCE3B/C-del carriers, we decided to study the antimicrobial activity for all three LCE3 proteins (LCE3A/B/C). We used chemically synthesized LCE3A, LCE3B and LCE3C which demonstrated clear antimicrobial activity against 10 commensal and pathogenic bacteria (Figure 4 and Table S3). Remarkably, even at submicromolar concentrations, activity was observed against a variety of taxa, including Gram-negative (Figure 4a–d) and Gram-positive (Figure 4e–j) microorganisms, including both aerobic and aerotolerant species. Notably, antimicrobial activity was present under the same experimental (low salt) conditions that reveal antibacterial activity of human beta-defensin 2 (hBD2), a prototypic cationic, epidermal cysteine-rich AMP (Harder et al., 1997). High salt concentrations inhibited antimicrobial activity of LCE3 proteins (Figure S3a). LCE3A appeared to be the strongest antimicrobial protein of the tested LCE’s, and its potency towards gram-negative bacteria is similar to the reported potency of hBD2 for E. coli and P. aeruginosa (Harder et al., 1997) (Figure S3b).
Figure 4. Antimicrobial activity of LCE3 proteins.
LCE3A, B, C were incubated with gram negative (a–d) and gram positive commensal and pathogenic bacteria (e–j) between 0.1 to 10 μM protein. For specifications see Supplemental Table 4. 2×102 CFU/ml is the detection limit, therefore values < 2×102 CFU/ml are not depicted. Error bars = SEM. CFU = colony forming units.
As defensins can exert toxic effect on mammalian cells as well (Lichtenstein et al., 1986) we tested LCE proteins effects up to 10 μM on submerged keratinocytes cultures for 4 or 20 hours and did not encounter any effect on cell viability or morphology (Figure S4a–b).
DISCUSSION
LCE expression is largely restricted to skin and oral epithelial tissue, and is absent in cells of the immune system. This indicates that the biological mechanism conferring an increased psoriasis risk is linked to specific epithelial functions of these tissues. qPCR studies have shown that LCE3 gene expression is low to undetectable in normal skin, but is readily detectable in oral epithelia (Jackson et al., 2005). Upon skin injury and inflammation, however, LCE3 genes are induced (Bergboer et al., 2011; Jackson et al., 2005) suggesting that LCE3 proteins might be involved in skin barrier function or repair. The reported epistasis between the major risk factor HLA-C*06:02 and LCE3B/C-del (Chandra et al., 2016; de Cid et al., 2009; Huffmeier et al., 2010) supports the idea that an impaired barrier in LCE3B/C-deficient skin would facilitate the penetration of environmental antigens, which in turn could trigger an HLA-C*06:02 restricted immune response (Bergboer et al., 2012). Based on this assumption, we performed detailed analysis of barrier properties of the del/del and wt/wt genotypes. However, for the molecules studied here, our in vitro models did not reveal an impairment of outside-in and inside-out barrier function in LCE3B/C-del epidermis. As the LCE3 baseline expression levels are higher in stratified oral epithelia (tonsil, gingiva, pharynx) than in normal skin (Niehues et al., 2015), we cannot exclude the possibility that the impact of LCE3B/C-loss could lead to leaky oral epithelia that are prone to penetration of external antigens. The observation that many psoriasis patients have their first flare after streptococcal tonsillitis (McFadden et al., 2009; Valdimarsson et al., 2009) would be compatible with the oral cavity as site of entry of bacterial antigens. This potential mechanism clearly warrants further investigation.
An alternative explanation for increased penetration of external antigens in LCEB/C -deficient epidermis would point to a more direct antimicrobial role of these LCE proteins. Mammalian epidermis is known for its antimicrobial shield of keratinocyte-expressed AMPs (Harder et al., 2013). We found a rather broad defensin-like spectrum of antibacterial activity that includes Gram-positive and Gram-negative species, and anaerobic as well as aerotolerant organisms. In view of their high homology, other LCE members may have antimicrobial properties as well. It will be interesting to determine whether this activity extends to other organisms such as yeasts and fungi.
LCE3 proteins are small (9–10 kDa), cationic proteins (isoelectric points about 9.0) and their antimicrobial activity is inhibited at high ionic strength. LCE3 proteins share these properties with many previously described AMPs such as human beta defensin -1, human beta defensin-2, human alpha-defensin-1, SLPI and LL37. All these proteins loose antimicrobial activity in aqueous solutions of physiological ionic strength, which raises the question whether the observed in vitro activity has any relevance under physiological conditions. It has been argued that in vitro assays poorly capture the physiological conditions in which these proteins work in vivo, such as stratum corneum of the skin, or mucous surfaces of the oral cavity or respiratory system. Epidermis-expressed LCE proteins or peptides derived thereof will end up in the stratum corneum, which is composed of lipids and crosslinked proteins, and devoid of free water. The (bound) water content of stratum corneum is extremely variable (Thakoersing et al., 2013), and to our knowledge the ionic composition and concentration of the stratum corneum is unknown. The antimicrobial activity of AMPs is obviously concentration dependent, and so is their salt sensitivity. Although, AMPs are typically tested in the low micromolar range, their actual concentration in vivo may be orders of magnitude higher. We have modeled the epidermal concentration of hBD-2 using an in vitro reconstructed skin model, which suggested that a concentration of 300 μM is reached (Jansen et al., 2009), which is far higher than required for antimicrobial activity, and which may offset te effect of high ionic strength. For all these reasons, the in vitro antimicrobial assay conditions are quite distinct from the environment in which AMPs like LCE and defensins operate, and in vitro models to test AMPs under these tissue-specific conditions are currently lacking. Indirect evidence that these molecules are genuine antimicrobial agents in vivo, is provided by the observation that mice in which these genes have been knocked out, have a compromised innate immune system (Moser et al., 2002).
A variety of AMP mechanisms that mediate bacterial killing or toxin inactivation have been described, including membrane permeabilization and depolarization (Selsted and Ouellette, 2005), peptide nanonets (Chu et al., 2012) and local protein unfolding (Kudryashova et al., 2014). LCE3 proteins cause a rapid killing which is reminiscent of the kinetics of killing by human neutrophil defensins where the outer and inner membrane of Escherichia coli are sequentially permeabilized within 30–60 minutes (Lehrer et al., 1989). Further analysis is needed to better understand the mechanism of bacterial killing and its relevance in the antimicrobial shield of the epidermis.
How can we integrate these findings into a comprehensive picture of the pathogenesis of psoriasis? From an evolutionary perspective, the LCE3B/C indel is likely a derived rather than an ancestral allele, as the deletion has not been reported in non-human primate genomes. In addition, the frequency of the indel is only 37% in African populations but it is between 62–64% in East-Asian, South-Asian, American and European populations (Bassaganyas et al., 2013; Genomes Project et al., 2010). This may possibly indicate a selective advantage in out-of-Africa populations. In view of the antimicrobial activity of LCE3 proteins, the deletion could cause a different antimicrobial shield in the epidermis. In wt/wt individuals (AA genotype), LCE3A is not expressed in normal skin, and also the expression levels of LCE3B/Care also very low or absent. LCE3B/C-del thus causes the LCE3A gene to be expressed at significant levels in normal skin of these individuals (Figure 1b). The loss of LCE3B/C would not have a noticeable effect in normal skin, as they were not expressed anyway. The net result of the indel would be a quantitatively (and possibly qualitatively) different epidermal host defense repertoire. Speculatively, this could provide an evolutionary benefit, and hence favor the spread of LCE3B/C-del. An increased risk to develop psoriasis could be regarded as the evolutionary cost of having this stronger antimicrobial shield in the epidermis and oral cavity.
As there is an epistatic interaction with HLA*C06:02 (Tsoi et al., 2012), LCE3B/C-del could alter the self-peptide profile of the skin by altering the balance of LCE3A self-peptides, compared to LCE3B and LCE3C. It could also alter the cutaneous microbiome in such a way as to promote the survival and colonization of taxa that elicit an HLA*C06:02 restricted response. A precedent comes from a recent microbiome study showing that filaggrin deficiency in ichthyosis vulgaris patients causes marked changes in the bacterial species composition (Zeeuwen et al., 2016). Microbiome studies on psoriasis have been performed, and shifts in microbiome composition have been reported, but unfortunately none of these studies have included the host HLA*C06:02 and LCE3B/C-del genotype. In addition to psoriasis, the LCE3B/C-del has been associated with different other diseases, including lupus (Lu et al., 2011) and rheumatoid arthritis (Docampo et al., 2010), our findings will provide further insights for the causal mechanism of this locus to the associated autoimmune disorders. Clearly, future studies should aim to establish the effects of LCE3B/C-del on the tissue microbiome, and their potential effects on HLA*C06:02 dependent immune processes that lead to disease.
In conclusion, our study establishes two findings regarding functional genomics of the psoriasis risk factor LCE3B/C-del. Firstly, we show that LCE3B/C-del, a risk allele for psoriasis, is significantly associated with the elevated expression of the upstream LCE3A gene in human skin. Secondly, we have uncovered a hitherto unknown antibacterial function of the proteins encoded by these three genes of the LCE3 family. Overall, these findings suggest a central role for LCE3A in epidermal host defense and LCE3B/C-del mediated psoriasis risk.
MATERIALS AND METHODS
QTL analysis of skin biopsies
All subjects involved in this study provided written informed consent according to the Helsinki Guidelines and approved by the Institutional Review Board of the University of Michigan Medical School. To evaluate the associations between the LCE3B/C-del and expression traits in the EDC region (between 151.5–154 Mb of chromosome 1), we utilized our RNA-seq cohort for psoriasis (Tsoi, 2014; Tsoi et al., 2015a) to measure the gene expression levels in normal (NN), uninvolved (PN), and psoriatic (PP) skin samples, respectively. Most of the samples in the RNA-seq cohort were genotyped in our Exomechip cohort. Altogether 80 non-psoriatic controls and 92 psoriatic patients with both genetic and expression data were included, with 27 of the patients also providing expression data for PN skin. The samples and data processing procedures for the RNA-seq data were described previously (Li et al., 2014; Tsoi et al., 2015a). Briefly, we employed Tophat and Cufflink to align reads and estimate expression levels, respectively. We included in the eQTL analysis only genes in the EDC region (chromosome 1: 151.5–154Mb) that were expressed in at least 20% of the samples in the condition being studied (i.e. NN, PN, or PP skin). For each expression trait we removed expression outliers (i.e. three times the interquartile range smaller than the lower quartile or three times the interquartile range larger than the upper quartile), and we further performed inverse normal transformation on each gene’s expression values. Genetic marker rs4112788 has been reported to be a close proxy of LCE3B/C-del (de Cid et al., 2009), and it was genotyped in the Exomechip cohort, therefore we used this SNP as a surrogate for the deletion and correlated its genotypes with expression traits using a linear model. The eQTL analysis was performed for NN, PN, and PP skin separately.
LCE3B/C-del genotyping of keratinocytes used for in vitro analyses
All subjects involved in this study were healthy (no skin diseases) and provided written informed consent according to the Helsinki Guidelines and approved by the local Medical Ethics Committee in The Netherlands (CMO Regio Arnhem-Nijmegen, The Netherlands). Keratinocytes were isolated and genotyped for their LCE3B/C-del status as previously described (de Cid et al., 2009). The non-deletion allele (i.e. containing LCE3B and 3C) is designated as wild type (wt). The deletion allele is designated del. Genotypes are referred to as wt/wt, del/wt and del/del.
3D reconstructed epidermis
3D reconstructed epidermis was generated as described previously (Niehues et al., 2016). To model the response of Th1- or Th 17 T-cells, combinations of cytokines were added into the medium during days 5 – 8 of the air-liquid interface culture (Th1 mix: 10 ng/ml IL-1α, 0.5 ng/ml TNF-1α, 250 U/ml IFN-γ; Th17 mix: 50 ng/ml IL-17, 50 ng/ml IL-22; Preprotech, Rocky Hill, NJ).
Quantitative real-time PCR
RNA isolation, cDNA synthesis, qPCR analysis and primer design was performed as described previously (Bergboer et al., 2011; Zeeuwen et al., 2008) (Table S1). Target gene expression was normalized to the expression of the housekeeping gene human acidic ribosomal phosphoprotein P0 (RPLP0). The ΔΔCt method was used to calculate relative mRNA expression levels (Livak and Schmittgen, 2001).
Statistics
Statistical analysis of qPCR data was performed on ΔCt values using commercially available software (IBM, SPSS Statistics 22). Repeated measures ANOVA, followed by Bonferroni post hoc testing was performed P < 0.05 was considered statistically significant.
Immunohistochemistry
3D reconstructed epidermis was formalin-fixed and processed for routine histology. 6 μm paraffin sections were stained with antibodies using an indirect immunoperoxidase (Vectastain, Vector Laboratories) or immunofluorescence technique. Details on antibodies are presented in Table S2.
Transepidermal water loss measurement
Transepidermal water loss (TEWL) was determined according to condenser-chamber method (Aquaflux AF200, Biox Systems, London, UK), as described before (Niehues et al., 2016).
In vitro outside-in dye penetration
1 mM Lucifer Yellow (LY, Sigma Aldrich) was added to the upper chambers of 3D reconstructed epidermis in 12-well dishes for one hour. Immunohistological sections were counterstained with 4′,6-diamidino-2-phenylindole (DAPI, Boehringer Mannheim) and assessed with fluorescence microscopy for LY penetration. Validation of this model is described in Niehues et al. (Niehues et al., 2016).
Hydrocortisone diffusion analysis
Stratum corneum (SC) isolation was performed according to the procedure as earlier described 20,21. In short, SC was isolated from ex vivo ski or 3D epidermal equivalents and used in the diffusion set-up was described previously (Mojumdar et al., 2014) using 0.34 mg/ml hydrocortisone in acetate buffer (pH 5.0) as diffusion fluid with 2–2,5 ml/hour flow speed. Samples were collected for a period of 15 hours, with a sampling time of 60 minutes. Hydrocortisone content was analyzed by UPLC (Acquity UPLC-UV system, Milford, USA) at 243 nm and analysis was performed using Masslynx v4.1 for peak integration, calculation of the concentration and flux determination, which is represented as flux in μg/cm2/hour.
Expression and synthesis of LCE proteins
The cloning of LCE genes has been described previously16. LCE fusion-proteins of glutathione S-transferase (GST) were produced according to standard procedures (Frangioni and Neel, 1993). Fusion-proteins were thrombin-cleaved to liberate the LCE protein from GST. LCE3B-GST fusion-protein did not show any antibacterial activity, but after GST-removal, the LCE protein clearly displayed antimicrobial activity. As pure LCE proteins were difficult to obtain, most likely due to their extremely high cysteine content, LCE3A,B,C proteins were synthesized by solid phase peptide synthesis, purified to 85–90% purity by reverse-phase HPLC and characterized by electrospray mass spectroscopy (Pepmic, Suzhou, China). LCE proteins were delivered in reduced form. Reducing agents and other low molecular weight components were removed by spin column dialysis (Amicon Ultra Centrifugal Filters 3K, Merck Millipore) and LCE proteins were extensively dialyzed against 10mM sodium phosphate buffer (PB, pH 7.4).
Bacteria culture
Diverse bacterial strains were grown on Columbia agar with 5% sheep blood (Becton, Dickinson and Co.) and single colonies were used to inoculate cultures in brain heart infusion medium (Mediaproducts BV, Groningen, The Netherlands) overnight at 37°C. Bacterial cultures were diluted 102 times in BHI medium and allowed to grow for another 2.5 hours to reach exponential growth (except for P. acnes). For strains and specifications of all bacterial strains see Table S3.
Antimicrobial assay
Bacteria were harvested from exponential growth cultures by centrifugation (2100 g, 5 min), washed with PB, and resuspended in PB at a concentration of 104 ~ 105 CFU/ml. Bacterial suspensions were exposed to LCE peptides in an assay volume of 100 μl, for 2 hours at 37°C in a 96 wells microplate (Greiner Bio-one). The suspensions were serially diluted in steps of ten and 10 μl of each dilution was plated on 5% sheep blood containing Columbia agar plates overnight at 37°C. Antimicrobial effects were determined by counting CFU.
Supplementary Material
Acknowledgments
We thank Heiman Wertheim (Department of Medical Microbiology, Radboudumc) for providing us with the bacterial strains used in this study. This research was funded by a TOP grant (91211052) of the Netherlands Organization for Health Research and Development (ZonMw) awarded to JS and PZ, and by a personal grant (to PZ) from the National Psoriasis Foundation/USA. Furthermore, this study was supported by NIH R01 grants AR042742, AR050511, AR054966, AR062382, and AR065183 to JE. JE is supported by the Ann Arbor Veterans Affairs Hospital. He is also supported by the Dawn and Dudley Holmes Memorial Fund and the Babcock Endowment Fund to the Department of Dermatology at the University of Michigan.
ABBREVIATIONS
- AMP
antimicrobial peptide
- CE
cornified envelope
- CFU
colony forming units
- EDC
epidermal differentiation complex
- EM
electron microscopy
- eQTL
expression quantitative trait loci
- GST
glutathione S-transferase
- GWAS
genome wide association studies
- hBD2
human beta-defensin 2
- IL
interleukin
- LCE
late cornified envelope
- LCE3B/C-del
the 32 kb deleted fragment comprising LCE3B and 3C
- LY
Lucifer Yellow
- NN
normal skin
- PB
sodium phosphate buffer
- PN
psoriasis uninvolved skin
- PP
psoriasis lesional skin
- SC
stratum corneum
- SNP
single nucleotide polymorphism
- TCEP
tris(2-carboxyethyl) phosphine
- TEM
transmission electron microscopy
- TEWL
transepidermal water loss
- Th1/17
T helper 1/17
Footnotes
CONFLICT OF INTEREST
The authors declare no conflict of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bassaganyas L, Riveira-Munoz E, Garcia-Aragones M, Gonzalez JR, Caceres M, Armengol L, et al. Worldwide population distribution of the common LCE3C-LCE3B deletion associated with psoriasis and other autoimmune disorders. BMC Genomics. 2013;14:261. doi: 10.1186/1471-2164-14-261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergboer JG, Tjabringa GS, Kamsteeg M, van Vlijmen-Willems IM, Rodijk-Olthuis D, Jansen PA, et al. Psoriasis risk genes of the late cornified envelope-3 group are distinctly expressed compared with genes of other LCE groups. Am J Pathol. 2011;178:1470–7. doi: 10.1016/j.ajpath.2010.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergboer JG, Zeeuwen PL, Schalkwijk J. Genetics of psoriasis: evidence for epistatic interaction between skin barrier abnormalities and immune deviation. J Invest Dermatol. 2012;132:2320–31. doi: 10.1038/jid.2012.167. [DOI] [PubMed] [Google Scholar]
- Candi E, Schmidt R, Melino G. The cornified envelope: a model of cell death in the skin. Nat Rev Mol Cell Biol. 2005;6:328–40. doi: 10.1038/nrm1619. [DOI] [PubMed] [Google Scholar]
- Capon F, Semprini S, Chimenti S, Fabrizi G, Zambruno G, Murgia S, et al. Fine mapping of the PSORS4 psoriasis susceptibility region on chromosome 1q21. J Invest Dermatol. 2001;116:728–30. doi: 10.1046/j.1523-1747.2001.01311.x. [DOI] [PubMed] [Google Scholar]
- Chandra A, Lahiri A, Senapati S, Basu B, Ghosh S, Mukhopadhyay I, et al. Increased Risk of Psoriasis due to combined effect of HLA-Cw6 and LCE3 risk alleles in Indian population. Sci Rep. 2016;6:24059. doi: 10.1038/srep24059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu H, Pazgier M, Jung G, Nuccio SP, Castillo PA, de Jong MF, et al. Human alpha-defensin 6 promotes mucosal innate immunity through self-assembled peptide nanonets. Science. 2012;337:477–81. doi: 10.1126/science.1218831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Cid R, Riveira-Munoz E, Zeeuwen PL, Robarge J, Liao W, Dannhauser EN, et al. Deletion of the late cornified envelope LCE3B and LCE3C genes as a susceptibility factor for psoriasis. Nat Genet. 2009;41:211–5. doi: 10.1038/ng.313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Guzman Strong C, Conlan S, Deming CB, Cheng J, Sears KE, Segre JA. A milieu of regulatory elements in the epidermal differentiation complex syntenic block: implications for atopic dermatitis and psoriasis. Hum Mol Genet. 2010;19:1453–60. doi: 10.1093/hmg/ddq019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Docampo E, Rabionet R, Riveira-Munoz E, Escaramis G, Julia A, Marsal S, et al. Deletion of the late cornified envelope genes, LCE3C and LCE3B, is associated with rheumatoid arthritis. Arthritis Rheum. 2010;62:1246–51. doi: 10.1002/art.27381. [DOI] [PubMed] [Google Scholar]
- Frangioni JV, Neel BG. Solubilization and purification of enzymatically active glutathione S-transferase (pGEX) fusion proteins. Anal Biochem. 1993;210:179–87. doi: 10.1006/abio.1993.1170. [DOI] [PubMed] [Google Scholar]
- Gallo RL, Hooper LV. Epithelial antimicrobial defence of the skin and intestine. Nat Rev Immunol. 2012;12:503–16. doi: 10.1038/nri3228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genetic Analysis of Psoriasis C, the Wellcome Trust Case Control C. Strange A, Capon F, Spencer CC, Knight J, et al. A genome-wide association study identifies new psoriasis susceptibility loci and an interaction between HLA-C and ERAP1. Nat Genet. 2010;42:985–90. doi: 10.1038/ng.694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genomes Project C. Abecasis GR, Altshuler D, Auton A, Brooks LD, Durbin RM, et al. A map of human genome variation from population-scale sequencing. Nature. 2010;467:1061–73. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harder J, Bartels J, Christophers E, Schroder JM. A peptide antibiotic from human skin. Nature. 1997;387:861. doi: 10.1038/43088. [DOI] [PubMed] [Google Scholar]
- Harder J, Schroder JM, Glaser R. The skin surface as antimicrobial barrier: present concepts and future outlooks. Exp Dermatol. 2013;22:1–5. doi: 10.1111/exd.12046. [DOI] [PubMed] [Google Scholar]
- Hollox EJ, Huffmeier U, Zeeuwen PL, Palla R, Lascorz J, Rodijk-Olthuis D, et al. Psoriasis is associated with increased beta-defensin genomic copy number. Nat Genet. 2008;40:23–5. doi: 10.1038/ng.2007.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huffmeier U, Bergboer JG, Becker T, Armour JA, Traupe H, Estivill X, et al. Replication of LCE3C-LCE3B CNV as a risk factor for psoriasis and analysis of interaction with other genetic risk factors. J Invest Dermatol. 2010;130:979–84. doi: 10.1038/jid.2009.385. [DOI] [PubMed] [Google Scholar]
- Jackson B, Tilli CM, Hardman MJ, Avilion AA, MacLeod MC, Ashcroft GS, et al. Late cornified envelope family in differentiating epithelia--response to calcium and ultraviolet irradiation. J Invest Dermatol. 2005;124:1062–70. doi: 10.1111/j.0022-202X.2005.23699.x. [DOI] [PubMed] [Google Scholar]
- Jansen PA, Rodijk-Olthuis D, Hollox EJ, Kamsteeg M, Tjabringa GS, de Jongh GJ, et al. Beta-defensin-2 protein is a serum biomarker for disease activity in psoriasis and reaches biologically relevant concentrations in lesional skin. PLoS One. 2009;4:e4725. doi: 10.1371/journal.pone.0004725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston A, Xing X, Swindell WR, Kochkodan J, Riblett M, Nair RP, et al. Susceptibility-associated genetic variation at IL12B enhances Th1 polarization in psoriasis. Hum Mol Genet. 2013;22:1807–15. doi: 10.1093/hmg/ddt034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan CT, Cao L, Roberson ED, Pierson KC, Yang CF, Joyce CE, et al. PSORS2 is due to mutations in CARD14. Am J Hum Genet. 2012;90:784–95. doi: 10.1016/j.ajhg.2012.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kudryashova E, Quintyn R, Seveau S, Lu W, Wysocki VH, Kudryashov DS. Human defensins facilitate local unfolding of thermodynamically unstable regions of bacterial protein toxins. Immunity. 2014;41:709–21. doi: 10.1016/j.immuni.2014.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehrer RI, Barton A, Daher KA, Harwig SS, Ganz T, Selsted ME. Interaction of human defensins with Escherichia coli. Mechanism of bactericidal activity. J Clin Invest. 1989;84:553–61. doi: 10.1172/JCI114198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Tsoi LC, Swindell WR, Gudjonsson JE, Tejasvi T, Johnston A, et al. Transcriptome analysis of psoriasis in a large case-control sample: RNA-seq provides insights into disease mechanisms. J Invest Dermatol. 2014;134:1828–38. doi: 10.1038/jid.2014.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichtenstein A, Ganz T, Selsted ME, Lehrer RI. In vitro tumor cell cytolysis mediated by peptide defensins of human and rabbit granulocytes. Blood. 1986;68:1407–10. [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) Method. Methods. 2001;25:402–8. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Lowes MA, Suarez-Farinas M, Krueger JG. Immunology of psoriasis. Annu Rev Immunol. 2014;32:227–55. doi: 10.1146/annurev-immunol-032713-120225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu X, Guo J, Zhou X, Li R, Liu X, Zhao Y, et al. Deletion of LCE3C_LCE3B is associated with rheumatoid arthritis and systemic lupus erythematosus in the Chinese Han population. Ann Rheum Dis. 2011;70:1648–51. doi: 10.1136/ard.2010.148072. [DOI] [PubMed] [Google Scholar]
- McFadden JP, Baker BS, Powles AV, Fry L. Psoriasis and streptococci: the natural selection of psoriasis revisited. Br J Dermatol. 2009;160:929–37. doi: 10.1111/j.1365-2133.2009.09102.x. [DOI] [PubMed] [Google Scholar]
- Mojumdar EH, Helder RW, Gooris GS, Bouwstra JA. Monounsaturated fatty acids reduce the barrier of stratum corneum lipid membranes by enhancing the formation of a hexagonal lateral packing. Langmuir. 2014;30:6534–43. doi: 10.1021/la500972w. [DOI] [PubMed] [Google Scholar]
- Moser C, Weiner DJ, Lysenko E, Bals R, Weiser JN, Wilson JM. beta-Defensin 1 contributes to pulmonary innate immunity in mice. Infect Immun. 2002;70:3068–72. doi: 10.1128/IAI.70.6.3068-3072.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nair RP, Duffin KC, Helms C, Ding J, Stuart PE, Goldgar D, et al. Genome-wide scan reveals association of psoriasis with IL-23 and NF-kappaB pathways. Nat Genet. 2009;41:199–204. doi: 10.1038/ng.311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nestle FO, Kaplan DH, Barker J. Psoriasis. N Engl J Med. 2009;361:496–509. doi: 10.1056/NEJMra0804595. [DOI] [PubMed] [Google Scholar]
- Niehues H, Schalkwijk J, van Vlijmen-Willems IM, Rodijk-Olthuis D, van Rossum MM, Wladykowski E, et al. Epidermal equivalents of filaggrin null keratinocytes do not show impaired skin barrier function. J Allergy Clin Immunol. 2016 doi: 10.1016/j.jaci.2016.09.016. [DOI] [PubMed] [Google Scholar]
- Niehues H, van Vlijmen-Willems IM, Bergboer JG, Kersten FF, Narita M, Hendriks WJ, et al. Late cornified envelope (LCE) proteins: Distinct expression patterns of LCE2 and LCE3 members suggest non-redundant roles in human epidermis and other epithelia. Br J Dermatol. 2015 doi: 10.1111/bjd.14284. [DOI] [PubMed] [Google Scholar]
- Schroder JM, Harder J. Antimicrobial skin peptides and proteins. Cell Mol Life Sci. 2006;63:469–86. doi: 10.1007/s00018-005-5364-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selsted ME, Ouellette AJ. Mammalian defensins in the antimicrobial immune response. Nat Immunol. 2005;6:551–7. doi: 10.1038/ni1206. [DOI] [PubMed] [Google Scholar]
- Tejasvi T, Stuart PE, Chandran V, Voorhees JJ, Gladman DD, Rahman P, et al. TNFAIP3 gene polymorphisms are associated with response to TNF blockade in psoriasis. J Invest Dermatol. 2012;132:593–600. doi: 10.1038/jid.2011.376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thakoersing VS, van Smeden J, Mulder AA, Vreeken RJ, El Ghalbzouri A, Bouwstra JA. Increased presence of monounsaturated fatty acids in the stratum corneum of human skin equivalents. J Invest Dermatol. 2013;133:59–67. doi: 10.1038/jid.2012.262. [DOI] [PubMed] [Google Scholar]
- Tsoi LC. Transcriptomic studies to characterize the gene expression landscape of psoriasis (GEO dataset GSE63980) 2014. [Google Scholar]
- Tsoi LC, Iyer MK, Stuart PE, Swindell WR, Gudjonsson JE, Tejasvi T, et al. Analysis of long non-coding RNAs highlights tissue-specific expression patterns and epigenetic profiles in normal and psoriatic skin. Genome Biol. 2015a;16:24. doi: 10.1186/s13059-014-0570-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsoi LC, Spain SL, Ellinghaus E, Stuart PE, Capon F, Knight J, et al. Enhanced meta-analysis and replication studies identify five new psoriasis susceptibility loci. Nat Commun. 2015b;6:7001. doi: 10.1038/ncomms8001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsoi LC, Spain SL, Knight J, Ellinghaus E, Stuart PE, Capon F, et al. Identification of 15 new psoriasis susceptibility loci highlights the role of innate immunity. Nat Genet. 2012;44:1341–8. doi: 10.1038/ng.2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valdimarsson H, Thorleifsdottir RH, Sigurdardottir SL, Gudjonsson JE, Johnston A. Psoriasis--as an autoimmune disease caused by molecular mimicry. Trends Immunol. 2009;30:494–501. doi: 10.1016/j.it.2009.07.008. [DOI] [PubMed] [Google Scholar]
- Zeeuwen PL, de Jongh GJ, Rodijk-Olthuis D, Kamsteeg M, Verhoosel RM, van Rossum MM, et al. Genetically programmed differences in epidermal host defense between psoriasis and atopic dermatitis patients. PLoS One. 2008;3:e2301. doi: 10.1371/journal.pone.0002301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeeuwen PL, Ederveen TH, van der Krieken DA, Niehues H, Boekhorst J, Kezic S, et al. Gram-positive anaerobe cocci (GPAC) are underrepresented in the microbiome of filaggrin-deficient human skin. J Allergy Clin Immunol. 2016 doi: 10.1016/j.jaci.2016.09.017. [DOI] [PubMed] [Google Scholar]
- Zuo X, Sun L, Yin X, Gao J, Sheng Y, Xu J, et al. Whole-exome SNP array identifies 15 new susceptibility loci for psoriasis. Nat Commun. 2015;6:6793. doi: 10.1038/ncomms7793. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.