Abstract
We have analyzed 5,088 bacterial 16S rRNA gene sequences from the distal intestinal (cecal) microbiota of genetically obese ob/ob mice, lean ob/+ and wild-type siblings, and their ob/+ mothers, all fed the same polysaccharide-rich diet. Although the majority of mouse gut species are unique, the mouse and human microbiota(s) are similar at the division (superkingdom) level, with Firmicutes and Bacteroidetes dominating. Microbial-community composition is inherited from mothers. However, compared with lean mice and regardless of kinship, ob/ob animals have a 50% reduction in the abundance of Bacteroidetes and a proportional increase in Firmicutes. These changes, which are division-wide, indicate that, in this model, obesity affects the diversity of the gut microbiota and suggest that intentional manipulation of community structure may be useful for regulating energy balance in obese individuals.
Keywords: energy balance/obesity, host-microbial interactions, intestinal bacterial diversity, ob/ob mice, phylogenetics
The 10 trillion to 100 trillion microorganisms that populate our adult intestines benefit us in a number of ways (1). One benefit is that they allow us to extract calories from otherwise indigestible common polysaccharides in our diet. This benefit occurs because components of the microbiota are able to adaptively deploy a large array of glycoside hydrolases and polysaccharide lysases that we humans do not encode in our genome (2, 3) (http://afmb.cnrs-mrs.fr/CAZY/). Furthermore, studies using germ-free and colonized normal and knockout mice fed a standard, polysaccharide-rich rodent-chow diet indicate that this mutualistic host-microbe relationship allows the extracted energy to be stored in adipocytes through a pathway that involves microbial regulation of the intestinal epithelial expression of fasting-induced adipocyte protein (Fiaf), a circulating inhibitor of lipoprotein lipase (LPL) (4). Microbial fermentation of dietary polysaccharides to monosaccharides and short-chain fatty acids in the distal gut and their subsequent absorption stimulate de novo synthesis of triglycerides in the liver. Microbial suppression of Fiaf in the gut epithelium results in reduced levels of this circulating LPL inhibitor, increased LPL activity in adipocytes, and enhanced storage of liver-derived triacyglycerols in fat cells (4).
Although the root cause of obesity is excess caloric intake compared with expenditure, differences in gut microbial ecology between humans may be an important factor affecting energy homeostasis; i.e., individuals predisposed to obesity may have gut microbial communities that promote more efficient extraction and/or storage of energy from a given diet, compared with these communities in lean individuals. This hypothesis raises a number of basic questions about gut microbial ecology in humans and mice. For example, how do the distal-gut microbiotas of the two hosts compare? Does kinship play an important role in the composition of the microbial community? Does adiposity affect community structure, and, if so, at what taxonomic level do these effects occur, and do they reflect a heretofore unappreciated form of homeostatic feedback between the microbiota and host energy balance?
Although information is limited, a current conceptualization of bacterial diversity in the human gut is that there is a restricted suite of highly adapted bacteria, likely inherited from the immediate family and, possibly, filtered by host genotype (5). Studies are needed to characterize the rules controlling microbial diversity in the human gut. Remarkably, a comprehensive enumeration of the gut microbiota has not yet been reported for Mus musculus, even though this mammalian species provides a very attractive model for systematically exploring the roles of host genotype, maternal exposure, diet, and energy balance on intestinal microbial ecology. Therefore, in this report, we use C57BL/6 mice, homozygous for a mutation in the leptin gene (ob/ob) that produces a stereotyped, fully penetrant obesity phenotype (6, 7), and their lean ob/+ and +/+ siblings, to show that microbial-community composition in the distal intestine changes at a division-wide level in response to increasing adiposity. This finding provides another perspective about the link between the gut microbiota and host energy balance.
Materials and Methods
Animals. C57BL/6J ob/+ mothers and their ob/ob, ob/+, and +/+ offspring were raised under a 12-h light cycle, in a specified pathogen-free state. Weaning and adult mice were fed PicoLab chow diet (Purina) ad libitum. All experiments involving mice were performed under protocols approved by the Washington University Animal Studies Committee. All animals were killed at the same time of day.
PCR Amplification of 16S rRNA Genes. Ceca were recovered immediately after the mice were killed. The contents of each intact cecum was recovered by manual extrusion and was frozen immediately (-80°C) until use. A frozen aliquot (≈100 mg) of each sample was added to tubes containing 500 μl of extraction buffer (200 mM Tris, pH 8.0/200 mM NaCl/20 mM EDTA), 210 μl of 20% SDS, 500 μl of phenol:chloroform:isoamyl alcohol (24:24:1), and 500 μl of 0.1-mm-diameter zirconia/silica beads (BioSpec Products, Bartlesville, OK). Microbial cells were disrupted mechanically at 23°C with a bead beater (BioSpec Products; instrument set on high for 2 min). Isolated DNA was fractionated by electrophoresis through 1% agarose gels, and the dominant band was purified by using the MiniElute Gel Extraction kit (Qiagen).
For each mouse, five replicate 25-μl PCRs were performed, each containing 100-200 ng of purified genomic DNA, 100 mM Tris (pH 8.3), 500 mM KCl, 20 mM MgSO4, 200 μM dNTPs, 200 μM concentration of the bacteria-specific primer 8F (5′-AGAGTTTGATCCTGGCTCAG-3′), 200 μM concentration of the universal primer 1391R (5′-GACGGGCGGTGWGTRCA-3′) (8), 1 M betaine, 800 μg/ml BSA, and 1 unit of Taq polymerase (Invitrogen). Cycling conditions were 94°C for 2 min, followed by 20 cycles of 94°C for 1 min, 55°C for 45 sec, and 72°C for 2 min, with a final extension period of 20 min at 72°C. Replicate PCRs were pooled. The resulting 1.3-kb amplicons were gel-purified by using the Qiagen extraction kit and subcloned into TOPO TA pCR4.0, followed by transformation into Escherichia coli TOP10 (Invitrogen). Extraction controls (no cecal material added) did not produce detectable PCR products or colonies. For each mouse, 384 colonies containing cloned amplicons were processed for sequencing. Plasmid inserts were sequenced bidirectionally by using vector-specific primers.
Sequence Assembly, Alignment, and blast. 16S rRNA gene sequences were edited and assembled into consensus sequences [phred and phrap software packages, aided by the program xplorseq (9)]. Sequences that did not assemble were discarded. Bases with phrap quality scores of <20 were trimmed. The final data set (n = 5,088 sequences, arb alignment and tree available at http://gordonlab.wustl.edu/mice; sequence designations listed in Table 1, which is published as supporting information on the PNAS web site) was compared with the 47,210-sequence Ribosomal Database Project (RDP-II) release 9 (10) and with an 11,831-sequence human colonic bacterial data set (11) by using the program blast (12).
Consensus sequences were aligned to a data set of >10,000 small-subunit rRNA sequences (an augmented version of the database described in ref. 13) in the arb software package [www.arb-home.de (14)] with a combination of the arb autoaligner and manual curation. Hypervariable regions of the 16S rRNA molecule were ignored by using the filter “lanemaskPH,” provided with the arb database (13). Aligned sequences were added to the arb neighbor-joining tree (based on pairwise distances with Olsen correction) with the parsimony insertion tool. Sequences with internal regions of poor quality leading to alignment problems were excluded from further analysis.
Mouse Community Clustering with unifrac. A phylogenetic tree, containing only the 16S rRNA sequences from this study, was exported from arb and annotated to indicate the mouse of origin for each sequence. Information in the phylogenetic tree was used to measure the difference between bacterial communities in mice by using the unifrac metric. The total branch length was calculated for each mouse's bacterial community represented in the tree. For each possible pair of mice, the fraction of the total branch length shared by the two communities and unique to each community (the unifrac) was then determined. unifrac measures the degree of divergence between 16S rRNA sequences, when comparing communities, without assignment of sequences to phylotypes (assignment is a necessary step when traditional community ecology indices are used and can blur distinctions between sequences by treating them as equal categories).
We used two permutations of the unifrac metric. The first permutation analyzes 16S rRNA sequences present in a sample, without regard to their abundance. The second permutation weights branch lengths, based on the relative abundance of sequences from each mouse. Employing the two permutations separately, we clustered bacterial communities from each animal using the unweighted-pair-group method with an arithmetic mean. The robustness of specific nodes in the tree, with respect to the presence or absence of each mouse and sampling coverage, was tested by jackknifing. For both unifrac permutations, we conducted principal-coordinates analysis and then used ANOVA of the first two principal coordinates to determine the effect of the mother's identity and ob genotype on community composition.
We also tested the effect of genotype on the abundance of the two most highly represented divisions of Bacteria, the Bacteroidetes and Firmicutes, controlling for kinship and gender. Sequences belonging to each division were tallied in arb, and the percentage of each division represented in each mouse was calculated.
All data were checked for normality with probability plots before ANOVA. ANOVA was conducted with a model-comparison approach (15) by using the program proc glm and contrast codes in the software package sas (Version 8, SAS Institute, Cary, NC). We used a cutoff of P ≤ 0.05 to indicate a significant effect for type II SS (equal cell size) and type III SS (unequal cell size).
Taxon Bins and Chao-Jaccard Abundance-Based Similarity Indices. A distance matrix was generated in arb, excluding the hypervariable regions of the 16S rRNA molecule because they cannot be aligned. Sequences were binned into taxa, based on a pairwise sequence identity, with lower-end cutoffs of 86-99%. We used the Chao-Jaccard abundance-based similarity index (16) to compare all possible pairs of mice at each taxon level (i.e., from 86% to 99% identity) by using the program estimates (Version 7.5; developed by R. K. Colwell) (http://purl.oclc.org/estimates).
Phylogenetic Analysis of the Cyanobacteria. GenBank rRNA sequence entries related to the putative cyanobacterial sequences from this study were identified using blast, and their phylogenetic relationships were tested by using reference taxa from 15 previously described phylogenetic divisions (17), chosen to represent the breadth of the bacterial domain (for the full list see Supporting Methods, which is published as supporting information on the PNAS web site). Maximum likelihood (ML), maximum parsimony (MP), and Bayesian inference (BI) were used to infer trees and estimate clade support. Heuristic searches were performed (program raxml, Version V; ref. 18) with ML and MP (no. of repetitions = 100) to find the best trees. Both methods yielded the same topology. To test the robustness of inferred topologies, posterior probabilities were determined by a Bayesian Markov chain Monte Carlo method implemented in the program mr bayes v3.0 (19). One million generations were run, and the trees and model parameters were sampled every 100 generations. The posterior probability stabilized after 170,000 generations, so all parameter estimates before generation 170,100 were omitted. In addition, bootstrap analyses were performed with minimal evolution [with K2P, HKY85, general time reversible (GTR), and ML modes of evolution] by using the program paup* 4.0 (http://paup.csit.fsu.edu) with data sets containing 75 or 375 taxa. We used the GTR+Γ+I model of evolution, chosen by log-likelihood testing with the program modeltest (20). Bootstrap support of ≥70% and posterior probability of ≥90% were considered to identify supported nodes.
Results and Discussion
Dominance of the Firmicutes and Bacteroidetes in the Distal Intestines of Mice. The offspring of C57BL/6J ob/+ matings and their three mothers were studied. Mothers 1 and 3 (M1 and M3) were siblings from different litters. M2 produced two litters, the second one immediately after the first. Newborn mice were raised together with their mother and littermates until they were weaned, during the third postnatal week. The mice were then housed alone in microisolator cages until 8 weeks of age, when they were killed. Mothers were killed at 1 year, 4 months after their litters were killed. Siblings and mothers were fed the same standard, polysaccharide-rich rodent-chow diet.
Consistent with previous studies, ob/ob mice consumed 42 ± 4% more chow than their lean ob/+ and +/+ siblings (P < 0.001), resulting in significantly greater body weights (34.5 ± 3.4 g vs. 19.1 ± 1.9 g at 8 weeks; P < 0.001) and epididymal fat-pad weights (1,016 ± 130 mg vs. 130 ± 31 mg; P < 0.001). Body weight, fat-pad weights, and chow consumption were not significantly different among lean ob/+ and +/+ siblings.
DNA was extracted from the cecum of each mouse (n = 19), bacterial 16S rRNA genes were amplified by PCR, and the resulting amplicons were sequenced bidirectionally. There was considerable diversity among the 5,088 rRNA sequences analyzed. By using ≥95% and ≥97% full-sequence identity to delimit a genus and a species, respectively, 64% of the sequences were not assignable to known genera, and only 7% represented previously cultured species.
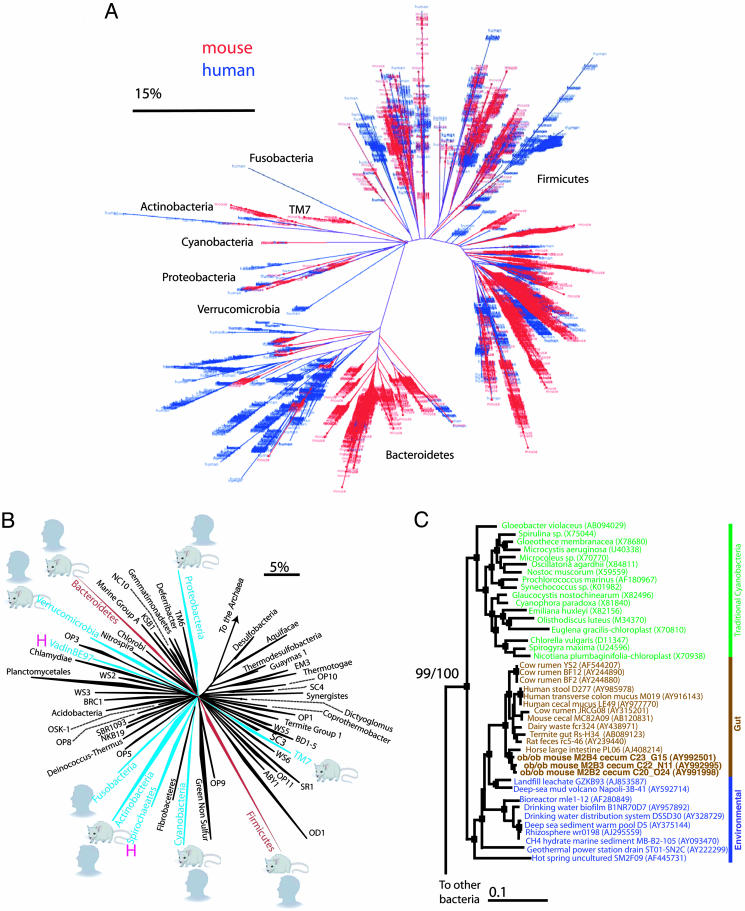
Although 85% of the sequences represent genera that have not been detected in humans (Fig. 1A), there is considerable similarity between human and mouse distal gut microbiotas at the division level (deep evolutionary lineage or superkingdom). Recent analysis of 11,831 bacterial 16S rRNA sequences from three healthy adults (11), combined with a review of 16S rRNAs in GenBank (1), had revealed nine divisions in the human colonic mucosa and feces (Fig. 1B). As in humans, the two most abundant bacterial divisions in mice were the Firmicutes (60-80% of sequences) and the Bacteroidetes (20-40%; also known as the Cytophaga-Flavobacterium-Bacteroides or CFB) (Fig. 1B). Greater than 75% of the Firmicutes are in Clostridium cluster XIVa, a common clade in humans that includes butyrate producers (21) and Eubacterium eligens. The majority (>88%) of the Bacteroidetes belong to Bacteroidetes 4b, which lacks a cultured representative (11). Proteobacteria and Actinobacteria, present at low levels in the human colonic microbiota, and TM7, previously identified in the human mouth (gingival) microbiota, each comprised ≤1% of the mouse cecal bacterial communities.
Fig. 1.
Bacterial diversity in the distal gut (ceca) of C57BL/6 mice. (A) Phylogenetic tree of 5,088 mouse ceca-associated 16S rRNA sequences reported in this study and 11,831 human colon-associated 16S rRNA sequences from ref. 11. Data from bacteria harvested from both mammalian hosts were obtained by using the same 16S rRNA gene-directed primers and PCR cycle numbers. The bar represents 15% sequence divergence. (B) Phylogenetic tree of the Bacteria showing described divisions (wedges, n = 55). Divisions detected in this study are indicated by the mouse symbol. Divisions detected in a large survey of the human colonic microbiota (11) are indicated by the human-head symbol. “H” denotes additional divisions represented in the human fecal microbiota, as determined from GenBank entries (1). Divisions dominant in mice and humans are colored red, rarer divisions are blue, and undetected divisions are black. The bar indicates changes per nucleotide. (C) Maximum-parsimony tree, showing representative taxa of the Cyanobacteria in green, including chloroplast sequences from Eukaryotes. Sequences detected in the gastrointestinal tracts of animals are in brown, and those detected in other environments are in purple (for additional information about taxa used to construct and root, but not shown in, the tree, see Supporting Methods. Nodes within the tree that are supported [bootstrap values (BT) of >70% and Bayesian posterior probabilities (BPP) of >90%] are indicated by filled squares. BT/BPP is explicitly stated for the basal node. The bar represents 0.1 nucleotide substitutions per site.
A Deep-Branching Clade of the Cyanobacteria in the Guts of Mice and Other Animals. In addition to the divisions described above, we found members of a group of bacteria that had been detected previously in human and other animal guts (Fig. 1C) but not characterized phylogenetically. Our analysis revealed a coherent gut-associated clade rooted deep in the Cyanobacteria, a division whose members perform oxygenic photosynthesis. This group may represent descendants of nonphotosynthetic ancestral cyanobacteria that have adapted to life in animal gastrointestinal tracts. Several sequences from other lightless environments were associated with the gut clade. Sequencing the genomes of these deep-rooting Cyanobacteria could shed light on the great oxygenation event that occurred ≈2.2 billion years ago, when Cyanobacteria profoundly changed the chemistry of the Earth's atmosphere.
Kinship Has a Strong Effect on Microbial Diversity. We next compared bacterial-community composition between mice by using recently developed computational methods that determine the fraction of branch length within a phylogenetic tree that is unique to each animal (unifrac metric; see Materials and Methods). We used two permutations of the unifrac metric: The first considers only taxa that are present, regardless of their abundance; the second weights branch lengths based on the relative abundance of sequences from each mouse (see Materials and Methods, Supporting Results, and Figs. 3 and 4, which are published as supporting information on the PNAS web site and which demonstrate the application of unifrac to the 11,831-member human colonic data set).
The unifrac methods were used to test the effects of kinship and genotype on diversity, in concert with standard multivariate and parametric statistics (principal-coordinates analysis and the unweighted-pair-group method with arithmetic mean and ANOVA applied to principal coordinates). We corroborated the results with an analysis of community similarity that takes into account sampling coverage and “unseen” taxa. This approach, the Chao-Jaccard abundance-based similarity index (16), was applied to taxa delimited by 16S rRNA sequence identity thresholds ranging from 86% to 99%.
The results revealed that mothers and their offspring shared cecal microbiotas with similar community membership, regardless of their ob genotype. This characteristic held for both generations; i.e., mothers that were siblings (M1 and M3) shared microbiotas of similar composition, and the microbiotas of their offspring (cousins) were similar. Moreover, the composition of the microbiotas of two litters from the same mother (M2) was not significantly different, whereas the composition of the M2 family was significantly different from the M1/M3 family (the effect of kinship is significant to P < 0.001) (Fig. 2A in the text; and see Fig. 5A, which is published as supporting information on the PNAS web site). The differences in composition between the M1/M3 and M2 families occurred at the level of genera (Fig. 2B).
Fig. 2.
Effects of kinship and obesity on gut microbial ecology. (A) Unweighted-pair-group method with arithmetic-mean (UPGMA) tree, based on pairwise differences between the cecal microbial communities of each mouse (UniFrac metric, based on 5,088 sequences). Dark blue, mother 1 and her offspring (M1-); pink, mother 2 and offspring; light blue, M3 and offspring. mothers 1 and 3 are siblings from different litters. Mother 2 had two sequential litters (M2A- and M2B-). All nodes are robust with respect to the specific mice included (jackknife values >0.95 for all nodes, representing the percentage of time the node was present when a randomly chosen mouse was removed from the distance matrix for n = 1,000 replicates). Nodes denoted by a square are robust to sequence number (jackknife values >0.67, representing the number of times the node was present when 200 sequences were randomly chosen from each mouse for n = 100 replicates, excluding mice M2A-1 and M2A-2, which had <200 sequences). (B) Community similarity between the microbiota of mouse pairs belonging to different families (≠ fam), the same family (= fam), different phenotypes [obese or lean, (≠ pheno)], or the same phenotype (= pheno). Community similarity was calculated for taxa (defined by percentage of conserved sequence identities, ranging from 86% to 99%, excluding hypervariable regions of the gene in the alignment) by using the Chao-Jaccard abundance-based similarity index (16). This index estimates the similarity of communities, based on the probability (taking into account community coverage) of a taxon belonging to one community or another or both. Identical communities have an index of 1. Values shown are means for each category. Error bars are standard errors among the similarity indices between all pairs of mice in each category. (C) Proportion of Bacteroidetes and Firmicutes in the cecal microbiota of lean vs. obese mice. Mean values ± SEM are plotted. *, P < 0.05 within the division.
Obesity Correlates with a Shift in the Abundance of Bacteroidetes and Firmicutes. Regardless of family membership, obesity was associated with a large shift in the relative abundance of the specific taxa present (P < 0.01; see Fig. 5B). The change in community structure occurred at the division level. The cecal microbiota of obese mice had a statistically significant 50% reduction in Bacteroidetes, relative to lean mice, and a significantly greater proportion of Firmicutes (P < 0.05, Fig. 2C). The changes were division-wide (there was no specific subgroup that was preferentially lost or amplified within these divisions) and occurred independently of kinship and gender.
Our study included a single runted animal with an ob/ob genotype. Its body mass (20 g) was equivalent to that of lean ob/+ and +/+ siblings, but the ratio of its epididymal fat-pad mass to total body mass was significantly greater (1.6% vs. 0.7% of total body mass; z = 6.64; P < 0.0001). This individual consumed significantly less chow than its lean siblings, yet the relative abundance of Firmicutes (71.2%) and Bacteroidetes (26.1%) was similar to other ob/ob animals, suggesting that alterations in the representation of these divisions cannot be simply ascribed to differences in chow consumption or total body mass.
Prospectus: Contribution of the Population Structure of the Gut Microbiota to Obesity. Together, these results show that the development of obesity in ob/ob mice affects the relative abundance of the major gut bacterial divisions derived from a maternal inoculum. The mechanisms responsible for directing these changes in microbial diversity remain to be defined, although broad division-level effects are not typical of an immune response to bacteria (22).
ob/ob mice are more efficient at harvesting energy from food than are lean +/+ animals (23, 24). The observed alterations in community structure within the distal gut microbiota of ob/ob animals may represent an unheralded contributing factor to their pattern of fuel partitioning. In domestic animals, antibiotics are commonly used to increase the efficiency of conversion of fuel in the diet to body mass (25), although the influence of these antibiotics on gut microbial ecology has not been determined with large-scale molecular phylogenetic surveys.
To date, 73 genomes of members of Firmicutes have been fully or partially sequenced (http://img.jgi.doe.gov). Most sequenced Firmicutes are in the class Bacillus, which is rare in the human or mouse gut microbiota; none is a member of the Clostridium XIV class, which contains the most abundant representatives of this division in the distal guts of mice and humans. Hence, the physiological contributions of Firmicutes to the intestinal ecosystem, and to fuel partitioning, are unclear. The increased ratio of Firmicutes to Bacteroidetes in ob/ob mice may help promote adiposity or, alternatively, could represent a host-mediated adaptive response to limit energy uptake/storage (e.g., by reducing the capacity to ferment polysaccharides). One way to address this question is by comparing gene content in the Firmicutes-enriched cecal microbiota of ob/ob mice with the Bacteroidetes-enriched microbiota of their lean ob/+ and +/+ littermates by using a metagenomics approach (26, 27). The results should provide insights about what metabolic attributes are provided to the host by these division-enhanced communities and how changes in divisional representation may effect energy homeostasis. The idea that gut microbial diversity is linked to obesity deserves exploration in humans, because it may yield new treatment strategies for this growing worldwide threat to our health.
Supplementary Material
Acknowledgments
We thank Sabrina Wagoner, Lucinda Fulton, and Kirk Harris for valuable assistance. This work was supported by grants from the W. M. Keck Foundation and the National Institutes of Health (DK070977 and DK007130).
Author contributions: R.E.L., F.B., and J.I.G. designed research; R.E.L., F.B., and P.T. performed research; C.A.L. and R.D.K. contributed new reagents/analytic tools; R.E.L., F.B., P.T., C.A.L., R.D.K., and J.I.G. analyzed data; and R.E.L., C.A.L., and J.I.G. wrote the paper.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database [accession nos. DQ014552-DQ015671 (mothers) and AY989911-AY993908 (offspring)].
References
- 1.Bäckhed, F., Ley, R. E., Sonnenburg, J. L., Peterson, D. A. & Gordon, J. I. (2005) Science 307, 1915-1920. [DOI] [PubMed] [Google Scholar]
- 2.Sonnenburg, J. L., Xu, J., Leip, D. D., Chen, C. H., Westover, B. P., Weatherford, J., Buhler, J. D. & Gordon, J. I. (2005) Science 307, 1955-1959. [DOI] [PubMed] [Google Scholar]
- 3.Xu, J., Bjursell, M. K., Himrod, J., Deng, S., Carmichael, L. K., Chiang, H. C., Hooper, L. V. & Gordon, J. I. (2003) Science 299, 2074-2076. [DOI] [PubMed] [Google Scholar]
- 4.Bäckhed, F., Ding, H., Wang, T., Hooper, L. V., Koh, G. Y., Nagy, A., Semenkovich, C. F. & Gordon, J. I. (2004) Proc. Natl. Acad. Sci. USA 101, 15718-15723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zoetendal, E. G., Akkermans, A. D. L., Akkermans-van Vliet, W. M., de Visser, J. A. G. M. & de Vos, W. M. (2001) Microb. Ecol. Health Dis. 13, 129-134. [Google Scholar]
- 6.Zhang, Y., Proenca, R., Maffei, M., Barone, M., Leopold, L. & Friedman, J. M. (1994) Nature 372, 425-432. [DOI] [PubMed] [Google Scholar]
- 7.Ingalis, A. M., Dickie, M. M. & Snell, G. D. (1950) J. Hered. 41, 317-318. [DOI] [PubMed] [Google Scholar]
- 8.Lane, D. J. (1991) in Nucleic Acid Techniques in Bacterial Systematics, eds. Stackebrandt, E. & Goodfellow, M. (Wiley, New York), pp. 115-175.
- 9.Papineau, D., Walker, J. J., Mojzsis, S. J. & Pace, N. R., Appl. Environ. Microbiol., in press. [DOI] [PMC free article] [PubMed]
- 10.Cole, J. R., Chai, B., Farris, R. J., Wang, Q., Kulam, S. A., McGarrell, D. M., Garrity, G. M. & Tiedje, J. M. (2005) Nucleic Acids Res. 33, D294-D296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Eckburg, P. B., Bik, E. M., Bernstein, C. N., Purdom, E., Dethlefsen, L., Sargent, M., Gill, S. R., Nelson, K. E. & Relman, D. A. (2005) Science 308, 1635-1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Altschul, S. F., Madden, T. L., Schäffer, A. A., Zhang, J., Zhang, Z., Miller, W. & Lipman, D. J. (1997) Nucleic Acids Res. 25, 3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hugenholtz, P. (2002) Genome Biol. 3, REVIEWS0003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ludwig, W., Strunk, O., Westram, R., Richter, L., Meier, H., Yadhukumar, Buchner, A., Lai, T., Steppi, S., Jobb, G., et al. (2004) Nucleic Acids Res. 32, 1363-1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Judd, C. M. & McClelland, G. H. (1989) Data Analysis: A Model Comparison Approach (Harcourt Brace Jovanovich, New York).
- 16.Chao, A., Chazdon, R. L., Colwell, R. K. & Shen, T.-J. (2005) Ecol. Lett. 8, 148-159. [Google Scholar]
- 17.Harris, J. K., Kelley, S. T. & Pace, N. R. (2004) Appl. Environ. Microbiol. 70, 845-849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stamatakis, A., Ludwig, T. & Meier, H. (2005) Bioinformatics 21, 456-463. [DOI] [PubMed] [Google Scholar]
- 19.Ronquist, F. & Huelsenbeck, J. P. (2003) Bioinformatics 19, 1572-1574. [DOI] [PubMed] [Google Scholar]
- 20.Posada, D. & Crandall, K. A. (1998) Bioinformatics 14, 817-818. [DOI] [PubMed] [Google Scholar]
- 21.Barcenilla, A., Pryde, S. E., Martin, J. C., Duncan, S. H., Stewart, C. S., Henderson, C. & Flint, H. J. (2000) Appl. Environ. Microbiol. 66, 1654-1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Suzuki, K., Meek, B., Doi, Y., Muramatsu, M., Chiba, T., Honjo, T. & Fagarasan, S. (2004) Proc. Natl. Acad. Sci. USA 101, 1981-1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ferraris, R. P. & Vinnakota, R. R. (1995) Am. J. Clin. Nutr. 62, 540-546. [DOI] [PubMed] [Google Scholar]
- 24.Warwick, B. P. & Romsos, D. R. (1988) Am. J. Physiol. 255, R141-R148. [DOI] [PubMed] [Google Scholar]
- 25.Gustafson, R. H. & Bowen, R. E. (1997) J. Appl. Microbiol. 83, 531-541. [DOI] [PubMed] [Google Scholar]
- 26.Tringe, S. G., von Mering, C., Kobayashi, A., Salamov, A. A., Chen, K., Chang, H. W., Podar, M., Short, J. M., Mathur, E. J., Detter, J. C., et al. (2005) Science 308, 554-557. [DOI] [PubMed] [Google Scholar]
- 27.Handelsman, J., Rondon, M. R., Brady, S., Clardy, J. & Goodman, R. M. (1998) Chem. Biol. 5, R245-R249. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.