Abstract
Free full text

LIVER WHOLE SLIDE IMAGE ANALYSIS FOR 3D VESSEL RECONSTRUCTION
Abstract
The emergence of digital pathology has enabled numerous quantitative analyses of histopathology structures. However, most pathology image analyses are limited to two-dimensional datasets, resulting in substantial information loss and incomplete interpretation. To address this, we have developed a complete framework for three-dimensional whole slide image analysis and demonstrated its efficacy on 3D vessel structure analysis with liver tissue sections. The proposed workflow includes components on image registration, vessel segmentation, vessel cross-section association, object interpolation, and volumetric rendering. For 3D vessel reconstruction, a cost function is formulated based on shape descriptors, spatial similarity and trajectory smoothness by taking into account four vessel association scenarios. An efficient entropy-based Relaxed Integer Programming (eRIP) method is proposed to identify the optimal inter-frame vessel associations. The reconstructed 3D vessels are both quantitatively and qualitatively validated. Evaluation results demonstrate high efficiency and accuracy of the proposed method, suggesting its promise to support further 3D vessel analysis with whole slide images.
1. INTRODUCTION
Whole slide images scanned from high resolution digital scanners provide rich information about morphological and functional characteristics of biological systems. As a result, such image modality provides insights on the underlying mechanisms of disease onset and progression. Although numerous image analysis methods have been proposed to analyze microscopy images [1, 2], they mainly focus on 2D biological structure analysis with substantial information loss. This presents challenges to such applications involving 3D modeling and analysis. In liver disease diagnosis, for instance, 3D structural changes in liver vessels and their 3D spatial relationships are essential for better understanding disease pathological evolution and progression [3]. A large set of approaches on vessel structure analysis have been proposed, ranging from vessel segmentation [4], structure tracking [5], to 3D vasculature visualization [6]. However, these methods are developed for radiology image analysis. Applying them to whole slide images directly is not feasible, as pathology images have much higher resolutions and phenotypic variations.
In this paper, we present an automated framework for 3D vessel reconstruction with whole slide images of liver tissue sections. Due to the overwhelming number of vessels and large variations in vessel shapes, it is challenging to associate vessel cross-sections from different image slides in a biologically meaningful way. To address this, we formulate a cost function based on shape descriptors, spatial similarity and trajectory smoothness by taking into account four vessel association scenarios. Optimal vessel associations are achieved when the aggregated cost function is minimized by an entropy-based Relaxed Integer Programming (eRIP) algorithm that is efficient to converge to the global optimal solution. The proposed 3D vessel analysis framework is generic and can be readily applied to studies using other organ tissues.
2. METHODS
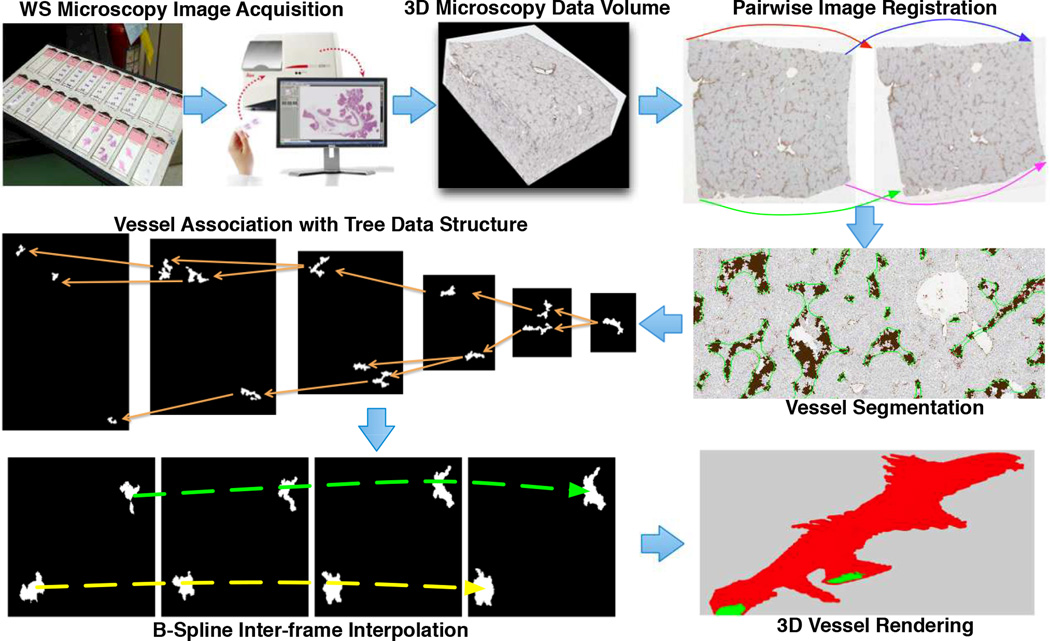
We illustrate in Figure 1 the overall working schema: image acquisition, registration, segmentation, vessel cross-section association, interpolation, and volumetric rendering. Our dataset includes 12 whole slide images of liver tissue sections [3] from the University of Leeds. Tissue sections are stained with dual chromogen Immunohistochemistry (IHC). Each whole-slide image has a typical resolution of 75k × 65k pixels, with the physical resolution 2.508e-1µm per pixel at the base level. Inter-slide physical resolution is 50 µm. We analyze images down-sampled from the base level by 64:1.
2.1. Image registration
In order to accurately reconstruct and characterize vessels in 3D space, we first register sequential microscopy images into the same reference space. The registration process consists of rigid and non-rigid steps. For rigid registration, the scale, 2D rotation matrix, and translation vector are computed by phase correlation in a log polar coordinate system [3]. For non-rigid registration, the whole image is partitioned into a set of evenly spaced blocks that are aligned separately with a rigid block matching approach. A cubic B-Spline transformation [7] is estimated by a regularized least squared difference minimizing method [3] to approximate a set of point translations derived from the rigid block registration. In aggregate, non-rigid spatial changes at 8 × 8 = 64 knots are computed. This non-rigid transform is applied to the rigidly transformed image to compensate for the non-rigid residual. Given these 64 pairs of displacements {k(x(i),y(j)) = (kx(x(i),y(j)),ky(x(i),y(j))), i, j = 1,2, …,8} at locations of {(x(i),y(j))}, the non-rigid displacement at a given location (p,q) can be computed as
For a given point (p,q), we construct two 4 × 4 matrices
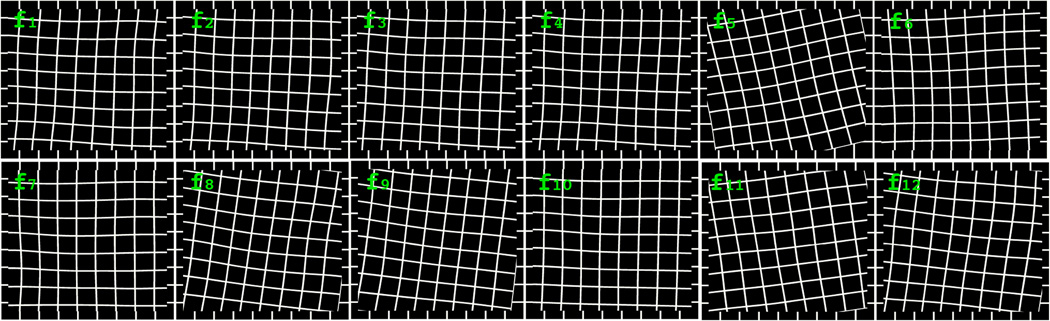
The final displacement at location (p,q) is the summation of rigid and non-rigid displacement. Our results show the median and maximum absolute displacements in x-y pairs are (10.40,9.58) and (139.58,103.66) in pixels. We demonstrate the registration mappings of 12 images with deformed grids in Figure 2.
2.2. Image segmentation
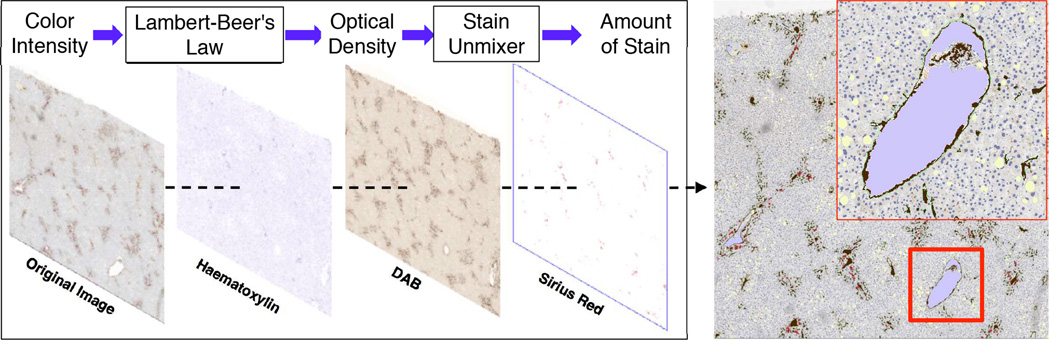
We next segment vessels in each registered image frame. First, we deconvolve each IHC stain component from the original color image. Color deconvolution [8] can be realized by Lambert-Beer’s law on the relationship between the intensity of light entering a specimen Ii and that through a specimen Io as: Io = Iie(−Ab) where A and b are the amount of stain and the absorption factor, respectively. The resulting Optical Density (OD) is defined as: OD = −log(Io/Ii). We define the un-mixer
2.3. Inter-frame vessel association
After blood vessel segmentation, inter-frame vessel grouping and filtering are performed. Vessel grouping is done by an image dilation operation [10] to make small vessel pieces cohesive to their source vessels. Candidates are then chosen among the grouped vessels by size for further analysis. We characterize the selected vessels by shape, spatial relationship and vessel trajectory smoothness. For our dataset, we consider four distinct vessel association cases: one-to-one (growing), one-to-two (bifurcating), one-to-none (disappearing) and none-to-one (appearing). The resulting cost functions are formulated as follows:
One-to-one (vessel extension to the next frame):
One-to-two (vessel bifurcation):
One-to-none (vessel disappearance):
None-to-one (new vessel emergence):
where
Additionally, function g(·), d(·) and b(·) are defined as:
where
We assume there are m1 and m2 vessel objects in frame t and t + 1 respectively, and r possible associations between these two frames. We solve the frame-by-frame vessel association problem by a Relaxed Integer Programming (RIP) framework [11]. The optimal vessel associations can be achieved as follows:
where c is a r × 1 vector with each entry representing the cost of one vessel object association; M is a r × (m1 + m2) binary matrix with Mij = 1 if and only if the i-th object association involves the j-th element from the union of m1 vessel objects in frame t and m2 in frame t + 1; (MT x)i is the i-th element of (MTx) and the constraint (MTx)i ≤ 1 guarantees that each vessel object in a given frame can be selected at most once in the result; A is a matrix composed with the first m1 columns in M; 1 is a m1 × 1 vector with all entries being 1s.This equality constraint guarantees that the sum of association probabilities for any given vessel object from frame t is one. The optimal solution x is a r × 1 binary vector where xi = 1 indicates the i-th association is included in the optimal solution.
We propose an efficient entropy-based Relaxed Integer Programming (eRIP) mechanism to find the best vessel association with RIP as a building block (Algorithm 1). In eRIP, we first solve RIP for one possible solution xRIP. If the entropy of xRIP is sufficiently low, we take it as the final result. Otherwise, we switch to Integer Programming (IP) by replacing the constraint 0 ≤ xj ≤ 1 in RIP with xj ![[set membership]](/http/europepmc.org/corehtml/pmc/pmcents/x2208.gif) {0,1}.
{0,1}.
Algorithm 1
Description of entropy-based Relaxed Integer Programming (eRIP) for Vessel Association
| 1 | Solve the Relaxed Integer Programming (RIP) problem; | |
| 2 | Compute the entropy ex of the solution xRIP from RIP; | |
| 3 | if | ex ≤ e* then |
 | /* e* is a threshold (0 ≤ e* ≤ 1) */ | |
| 4 | x ← xRIP; | |
| 5 | else | |
| 6 |  | Replace the constraint of 0 ≤ xj ≤ 1 with xj ![[set membership]](/http/europepmc.org/corehtml/pmc/pmcents/x2208.gif) {0,1}; {0,1}; |
| 7 | Solve the corresponding Integer Programming (IP) problem for xIP with the updated constraint xj ![[set membership]](/http/europepmc.org/corehtml/pmc/pmcents/x2208.gif) {0,1}; {0,1}; | |
| 8 | x ← xIP; | |
| 9 | end | |
| 10 | return x; | |
3. EXPERIMENTS AND RESULTS
For vessel association, eRIP is proposed to identify the optimal vessel association between adjacent frames. We focus on the largest 25 vessel candidates by size on each slide. Of these candidates, the smallest vessel size is 304.25 ± 104.34 in pixels. At this stage, our working pipeline primarily deals with macro-vessels. The parameters are empirically set as λ1 = 0.58, λ2 = 0.33,
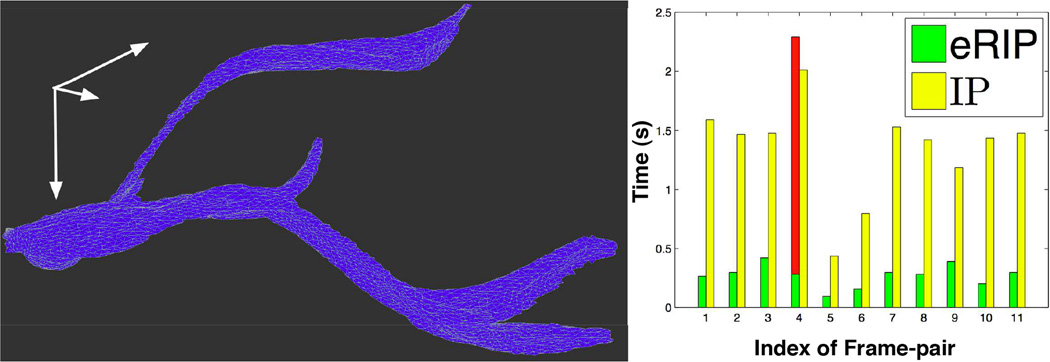
(Left) A 3D close-up view of a representative vessel object; (Right) Time cost comparison between eRIP and IP.
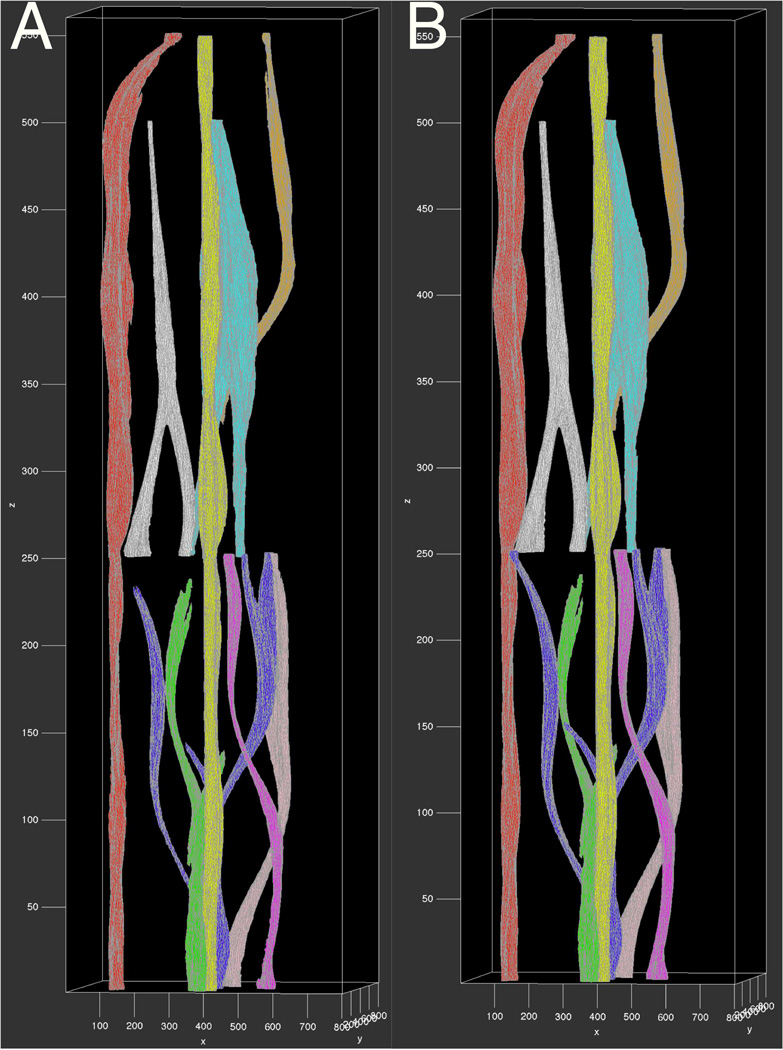
We extensively evaluate our approach with human annotations quantitatively and qualitatively. Table 1 presents the validation results measured by Jaccard coefficient (Jac), precision (Pre), recall (Rec), F1 score (F1) and Hausdorff distance (Haus). The first column shows distinct vessels with their colors identically coded in Figure 4. The best performance assessed by each measure is in bold. Note Vessels in red and yellow are more regular in shape, leading to better agreement between the proposed method and human annotations. 3D vessel rendering results from human annotations in Figure 4 (Right) are used for qualitative assessments. By visual comparisons, 3D vessel structures in Figure 4 (Left) and (Right) are similar. Human annotated vessels are more regular and smooth in shape, whereas machine generated vessels tend to preserve more structural details. Overall, both quantitative measurements and qualitative comparisons suggest a satisfactory concordance between our method and human annotations.
Table 1
Evaluation of the segmentation results (mean, Relative Standard Deviation%). The first column shows vessel color in Figure 4.
| Jac | Pre | Rec | F1 | Haus | |
| R | 0.91,64.73 | 0.94,36.72 | 0.94,36.81 | 0.94,36.69 | 2.81,3.25 |
| Y | 0.92,60.40 | 0.92,37.41 | 0.92,37.53 | 0.92,37.41 | 2.72,3.02 |
| G | 0.80,125.13 | 0.86,41.10 | 0.86,41.06 | 0.86,40.91 | 4.10,5.88 |
| B | 0.80,109.28 | 0.85,41.90 | 0.85,41.69 | 0.85,41.62 | 4.10,5.81 |
| P | 0.84,79.79 | 0.84,39.95 | 0.84,39.82 | 0.84,39.75 | 4.10,5.78 |
| M | 0.81,86.55 | 0.81,40.58 | 0.81,40.45 | 0.81,40.35 | 4.09,6.00 |
| W | 0.87,57.26 | 0.88,35.40 | 0.88,35.52 | 0.88,35.35 | 3.81,3.96 |
| C | 0.81,81.88 | 0.82,37.69 | 0.82,37.80 | 0.82,37.69 | 4.22,3.13 |
| O | 0.86,61.80 | 0.85,37.22 | 0.86,37.33 | 0.86,37.21 | 3.92,3.11 |
In our tests, eRIP and IP produce identical vessel association results. We present the execution time for these two approaches in Figure 5 (Right). The green bars show eRIP execution time, with all values except the fourth less than 0.5 seconds and the majority around 0.3 seconds. By contrast, IP execution time is much longer, with majority around 1.5 seconds (yellow bars). Note that eRIP takes more time than IP for the fourth frame-pair (red bar), as entropy of the solution from RIP is greater than the threshold e* = 0.3. In this case, eRIP automatically switches to IP method. Most xjs from RIP are 0s or 1s in our tests, resulting in less execution time by eRIP overall.
4. CONCLUSIONS
In this paper, we present a framework for 3D vessel structure analysis on whole slide images of liver tissue sections. To identify vessel associations, we formulate a cost function with vessel shape descriptors, spatial similarity and trajectory smoothness for four vessel association scenarios. We also propose an eRIP optimization method to efficiently associate vessel objects between adjacent slides. Both quantitative and qualitative evaluation results demonstrate the effectiveness of our 3D vessel reconstruction framework and its promise as a platform for further 3D vessel analysis.
ACKNOWLEDGEMENT
This research is supported in part by grants from National Institute of Health K25CA181503, National Science Foundation ACI 1443054 and IIS 1350885, and CNPq.
REFERENCES
Full text links
Read article at publisher's site: https://doi.org/10.1109/isbi.2015.7163845
Read article for free, from open access legal sources, via Unpaywall:
https://europepmc.org/articles/pmc4578313?pdf=render
Citations & impact
Impact metrics
Citations of article over time
Article citations
An integrative web-based software tool for multi-dimensional pathology whole-slide image analytics.
Phys Med Biol, 67(22), 09 Nov 2022
Cited by: 2 articles | PMID: 36067783 | PMCID: PMC10039615
Efficient 3D Spatial Queries for Complex Objects.
ACM Trans Spat Algorithms Syst, 8(2):14, 12 Feb 2022
Cited by: 1 article | PMID: 36072353 | PMCID: PMC9446285
HISTOPATHOLOGY IMAGE REGISTRATION BY INTEGRATED TEXTURE AND SPATIAL PROXIMITY BASED LANDMARK SELECTION AND MODIFICATION.
Proc IEEE Int Symp Biomed Imaging, 2021:1827-1830, 01 Apr 2021
Cited by: 1 article | PMID: 34548902 | PMCID: PMC8452257
Regional registration of whole slide image stacks containing major histological artifacts.
BMC Bioinformatics, 21(1):558, 04 Dec 2020
Cited by: 4 articles | PMID: 33276732 | PMCID: PMC7718714
Whole Slide Imaging and Its Applications to Histopathological Studies of Liver Disorders.
Front Med (Lausanne), 6:310, 08 Jan 2020
Cited by: 15 articles | PMID: 31970160 | PMCID: PMC6960181
Review Free full text in Europe PMC
Go to all (9) article citations
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
A Framework for 3D Vessel Analysis using Whole Slide Images of Liver Tissue Sections.
Int J Comput Biol Drug Des, 9(1-2):102-119, 01 Jan 2016
Cited by: 6 articles | PMID: 27034719 | PMCID: PMC4809644
A 3D Primary Vessel Reconstruction Framework with Serial Microscopy Images.
Med Image Comput Comput Assist Interv, 9351:251-259, 01 Jan 2015
Cited by: 5 articles | PMID: 26478919 | PMCID: PMC4605571
Improving Algorithm for the Alignment of Consecutive, Whole-Slide, Immunohistochemical Section Images.
J Pathol Inform, 12:29, 03 Aug 2021
Cited by: 3 articles | PMID: 34476109 | PMCID: PMC8378454
3-Dimensional Reconstruction From Histopathological Sections: A Systematic Review.
Lab Invest, 104(6):102049, 19 Mar 2024
Cited by: 1 article | PMID: 38513977
Review
![[open star]](/http/europepmc.org/corehtml/pmc/pmcents/star.gif)