Abstract
Objective
To identify novel genetic loci that predispose to early-onset myasthenia gravis (EOMG) applying a two-stage association study, exploration, and replication strategy.Methods
Thirty-four loci and one confirmation loci, human leukocyte antigen (HLA)-DRA, were selected as candidate genes by team members of groups involved in different research aspects of MG. In the exploration step, these candidate genes were genotyped in 384 EOMG and 384 matched controls and significant difference in allele frequency were found in eight genes. In the replication step, eight candidate genes and one confirmation loci were genotyped in 1177 EOMG patients and 814 controls, from nine European centres.Results
ALLELE FREQUENCY DIFFERENCES WERE FOUND IN FOUR NOVEL LOCI: CD86, AKAP12, VAV1, B-cell activating factor (BAFF), and tumor necrosis factor-alpha (TNF-α), and these differences were consistent in all nine cohorts. Haplotype trend test supported the differences in allele frequencies between cases and controls. In addition, allele frequency difference in female versus male patients at HLA-DRA and TNF-α loci were observed.Interpretation
The genetic associations to EOMG outside the HLA complex are novel and of interest as VAV1 is a key signal transducer essential for T- and B-cell activation, and BAFF is a cytokine that plays important roles in the proliferation and differentiation of B-cells. Moreover, we noted striking epistasis between the predisposing VAV1 and BAFF haplotypes; they conferred a greater risk in combination than alone. These, and CD86, share the same signaling pathway, namely nuclear factor-kappaB (NFκB), thus implicating dysregulation of proinflammatory signaling in predisposition to EOMG.Free full text

VAV1 and BAFF, via NFκB pathway, are genetic risk factors for myasthenia gravis
Associated Data
Abstract
Objective
To identify novel genetic loci that predispose to early-onset myasthenia gravis (EOMG) applying a two-stage association study, exploration, and replication strategy.
Methods
Thirty-four loci and one confirmation loci, human leukocyte antigen (HLA)-DRA, were selected as candidate genes by team members of groups involved in different research aspects of MG. In the exploration step, these candidate genes were genotyped in 384 EOMG and 384 matched controls and significant difference in allele frequency were found in eight genes. In the replication step, eight candidate genes and one confirmation loci were genotyped in 1177 EOMG patients and 814 controls, from nine European centres.
Results
Allele frequency differences were found in four novel loci: CD86, AKAP12, VAV1, B-cell activating factor (BAFF), and tumor necrosis factor-alpha (TNF-α), and these differences were consistent in all nine cohorts. Haplotype trend test supported the differences in allele frequencies between cases and controls. In addition, allele frequency difference in female versus male patients at HLA-DRA and TNF-α loci were observed.
Interpretation
The genetic associations to EOMG outside the HLA complex are novel and of interest as VAV1 is a key signal transducer essential for T- and B-cell activation, and BAFF is a cytokine that plays important roles in the proliferation and differentiation of B-cells. Moreover, we noted striking epistasis between the predisposing VAV1 and BAFF haplotypes; they conferred a greater risk in combination than alone. These, and CD86, share the same signaling pathway, namely nuclear factor-kappaB (NFκB), thus implicating dysregulation of proinflammatory signaling in predisposition to EOMG.
Introduction
Myasthenia gravis (MG) is a relatively rare heterogeneous autoimmune neuromuscular disorder, clinically characterized by weakness and fatigability of skeletal and extraocular muscles.1 MG is a T-cell-dependent antibody-mediated disease, caused by autoantibodies directed against acetylcholine receptor (AChR) in over 80% of patients.2 Patients with early-onset MG (EOMG; disease onset before age 50) form a particularly well-defined subgroup, with a disease prevalence in Europeans of about 10–20 in 100,000 and a strong female bias.3–5 Pathogenesis in this subgroup involves not only the muscle AChR as the target of the autoimmune response, but also the thymus, which very frequently shows lymph node-like infiltrates.6 EOMG is rarely inherited within a family, but occurrence of other autoimmune and immune-mediated diseases among myasthenia patients' relatives is relatively common.7 The concordance rate in MG twins is estimated to be 35% in monozygotic twins, compared with 4–5% in dizygotic twins,8 supporting genetic contribution to disease pathogenesis. The current hypothesis is that EOMG is a rare complex disease associated with numerous genetic contributors each having a small effect.9
Human leukocyte antigen ancestral haplotype (HLA-AH8.1) has been consistently associated with EOMG in Caucasian.4,10 The HLA-AH8.1 haplotype, that tightly associates the various alleles of HLA-A1, B8, DR3 genes, shows an exceptionally strong tendency to be inherited together, making it hard to distinguish the contribution of each individual gene. Remarkably, HLA-AH8.1 haplotype has been associated with several autoimmune diseases, such as celiac, thyroid diseases, and systemic lupus erythematosus, but not with the late-onset MG, supporting the hypothesis of shared genetic risk factors for several autoimmune diseases.11,12 In addition, polymorphisms in AChR alpha (CHRNA1) have been shown to confer increased risk to very early-onset MG.13
In the past 5 years, important biologic discoveries have come from genome-wide association studies (GWAS) aimed at unbiased detection of variants at loci predisposing to complex traits, such as cardiac, autoimmune diseases, and psychiatric disorders.14 Because most such variants have modest effects, they can only achieve genome-wide significance in well-powered GWAS with thousands of samples. A candidate gene approach is a valuable alternative, as it needs only a few hundred samples to highlight even modest genotype-risk associations, with the caveats that it requires some knowledge of disease pathophysiology, knowledge that may be biased.
To identify additional susceptibility loci, we undertook a two-stage study on 34 candidate genes and one confirmation loci and eight genes in the replication stage, of which five showed significant associations as described below.
Materials and Methods
Study design
The study was designed to identify candidate loci in the exploration stage and check them in a larger group of patients and controls in the replication stage (Fig.(Fig.1).1). We estimated that the study had 0.8 power to detect odds ratios (OR) of ≥2 in the exploration stage and 1.5 in the replication stage, respectively.
Study subjects
Following local Ethics Committee approval and informed consent signed, we recruited: (1) Caucasian patients with ocular and generalized MG with anti-AChR antibodies from nine European centres (Table(Table1)1) according to these inclusion criteria: males and females diagnosed with MG onset between ages 15 and 50, without thymoma and positive for anti-AChR antibodies15; (2) healthy age-matched Caucasian controls from six of the same centres, without any other neurological or autoimmune diseases. Samples were recruited from nine European centres, France, Italy, Poland, United Kingdom, Greece, Spain, Germany, Hungary, and Norway. About 1300 anti-AChR-positive EOMG patients were genotyped for the current study, 384 for the exploration stage, and additional 912 for the replication stage. However, about 10% of samples were excluded due to poor DNA quality, genotype quality, or population certification issues. In total, 1177 patients (240 males and 937 females) and 814 controls were used for the statistical analysis (Table(Table1).1). The recently published GWAS and the current study shared DNA (those from the United Kingdom, Norway, and some from France); hence, the two studies have some overlap.5
Table 1
European centres involved in DNA sample collection for MG patients and matched controls.
| Centre | Country | Male | Female | Total | |||
|---|---|---|---|---|---|---|---|
| EOMG | Controls | EOMG | Controls | EOMG | Controls | ||
| INSERM, Paris | France | 80 | 136 | 270 | 152 | 350 | 288 |
| INNCB, Milano | Italy | 44 | 78 | 120 | 90 | 164 | 168 |
| Oslo University Hospital | Norway | 28 | 38 | 115 | 154 | 143 | 192 |
| Medical University of Warsaw | Poland | 22 | 24 | 146 | 24 | 168 | 48 |
| Hellenic Pasteur Institute, Athens | Greece | 16 | 11 | 48 | 53 | 64 | 64 |
| Tübingen University Medical Center | Germany | 14 | 0 | 34 | 0 | 48 | – |
| University of Pecs, Pecs | Hungary | 12 | 0 | 36 | 6 | 48 | 6 |
| Weatherall Institute for Molecular Medicine, Oxford | U.K. | 15 | 0 | 129 | 0 | 144 | – |
| Hospital University of Bellvitge, Barcelona | Spain | 9 | 18 | 39 | 30 | 48 | 48 |
| Total | 240 | 305 | 937 | 510 | 1177 | 814 | |
Candidate gene selection and SNP selection
Thirty-four loci, in addition to HLA-DRA, a known susceptibility locus, were selected as candidate genes by team members of groups involved in different research aspects of MG for more than two decades in the framework of a MG European network project (FIGHT-MG). The selection was focused on immune genes, but also included MG-specific candidates, such as the AChR subunits. Both the discovery and replication genotypes of the current study were completed by the time of the EOMG GWAS publication in December 2012,5 hence, the candidate gene selection was not influenced by the GWAS results. To reduce genotyping costs, a small subset of informative SNPs (tag SNPs) was selected using Tagger and JMP-Genomics software.16 Hence, 384 (Table S1) and 95 (Table S2) SNPs were genotyped during the exploration and replication stages, respectively.
Genotype methodology and quality control
Genotyping was carried out using Illumina GoldenGate assays for the exploration stage at the Genomics Core Facility (Technion, Haifa, Israel), and the Biomark system (Fluidigm, South San Francisco, CA) for the replication stage at Biological Services (Weizmann Institute for Science, Rehovot, Israel), according to the manufacturer's protocols.17,18 SNP assays were designed by Illumina or by Fluidigm SNPtype Assays. Subjects with <90% successful genotyping or deviating from the expected European clustering were excluded from the analysis, about 10% of the samples. SNPs with minor allele frequencies (MAF) less than 0.05, or not in Hardy–Weinberg equilibrium, or with GenCall score <0.5 were also excluded from the analysis, about 30% of SNPs.19 Population substructure was determined using principal component analysis and 24 SNPs used to infer population ancestry (genomic control SNPs) (Table(Table22).20 The FST score, that is, the contribution by each centre (the S subscript) to the total genetic variance (the T subscript), was about 0.02, which is as expected for a European population, and implies a high degree of similarity among cohorts.21
Table 2
Gene symbol and number of SNPs selected for EOMG association test.
| Symbol | No. SNP | Symbol | No. SNP | Symbol | No. SNP |
|---|---|---|---|---|---|
| AIRE | 4 | CHRNE | 4 | IL6 | 9 |
| AKAP12 | 15 | CXCL13 | 18 | IL7R | 7 |
| C2 | 4 | FGF11 | 2 | IRF3 | 3 |
| CCL21 | 4 | FOXP3 | 9 | LTA | 5 |
| CD40 | 7 | HLA-DRA | 16 | MUSK | 30 |
| CD40LG | 14 | IFIH1 | 8 | MYD88 | 3 |
| CD80 | 24 | IFNB1 | 2 | BAFF | 11 |
| CD86 | 15 | IFNG | 3 | VAV1 | 25 |
| CFB | 11 | IL17A | 9 | TLR3 | 8 |
| CHRNA1 | 6 | IL17RA | 16 | TSLP | 5 |
| CHRNB1 | 5 | IL18 | 9 | TNF | 8 |
| CHRND | 2 | IL2RA | 30 | European ancestry marker SNPs | 24 |
In bold loci selected for the replication stage.
Statistical analyses
We used JMP-Genomics V6 software (SAS Institute, Cary, NC) to compare allele frequencies between cases and controls; differences were considered significant when P-values were <0.05 after false discovery rate (FDR) adjustment. ORs and 95% confidence intervals (CI) were calculated for statistically significant SNPs. Linkage disequilibrium (LD) blocks and SNP haplotypes were inferred using the Expectation–Maximization algorithm, as implemented in JMP-Genomics. Haplotype trends and SNP↔SNP interactions were tested using the same software.
Results
Analysis of the exploration stage genotype data identified significant associations in EOMG with eight of the 34 genes tested, and confirmed the known associations at HLA-DRA.4,10 However, no difference in allele frequency between EOMG and controls were found for SNPs in AChR subunits or for SNPs in the genomic region of the autoimmune regulator (AIRE).13 It is possible that some of the candidate genes may still be found to associate with EOMG if tested in larger study, because of the limited statistical power of the exploration stage. Complete analyses of the combined exploration and replication cohorts identified 10 SNPs, in six genes, showing significant associations (Table(Table3):3): HLA class II DR α chain (HLA-DRA); tumor necrosis factor-α (TNF-α), both part of the extended HLA-AH8.1 haplotype; A kinase (PRKA) anchor protein 12 (AKAP12 or gravin); CD86 antigen (CD28 antigen ligand 2, B7-2 antigen); vav 1 guanine nucleotide exchange factor (VAV1); TNF ligand superfamily member 13B (TNFSF13B), also known as B-cell activating factor (BAFF). Analysis of one representative SNP per gene shows differences from the controls that were consistently in the same direction and with similar delta values in each European Center for SNPs in: HLA-DRA; TNF-α; BAFF; VAV1 (Fig.(Fig.2).2). In particular, the consistent 4–5% differences between EOMG and controls in MAFs at BAFF (despite the marginally significant P-value of 0.045 for the trend test) suggest that this is most likely a true association that requires further investigation. The differences at CD86 and AKAP12 were less consistent between centres, possibly due to their low MAFs.
Table 3
Significantly associated SNPs with allele frequencies and odds ratio estimate.
| SNP_ID | Locus_ID | MAFs frequency | FDR corrected negLog10 P-value | Odds ratio and 95% CIs | ||||
|---|---|---|---|---|---|---|---|---|
| Controls | EOMG | Genotype | Trend | OR | Lower | Upper | ||
| rs3129882 | HLA-DRA | 0.48 | 0.42 | 2.58 | 2.76 | 0.78 | 0.68 | 0.89 |
| rs2239806 | HLA-DRA | 0.14 | 0.32 | 32.00 | 31.59 | 3.07 | 2.56 | 3.68 |
| rs3129888 | HLA-DRA | 0.18 | 0.14 | 1.75 | 2.42 | 0.72 | 0.60 | 0.87 |
| rs1800629 | TNF | 0.13 | 0.33 | 32.00 | 32.00 | 3.53 | 2.92 | 4.25 |
| rs1124264 | AKAP12 | 0.15 | 0.09 | 5.40 | 6.19 | 0.56 | 0.69 | 0.46 |
| rs17080949 | AKAP12 | 0.07 | 0.15 | 5.43 | 7.31 | 1.88 | 1.53 | 2.32 |
| rs9282641 | CD86 | 0.08 | 0.04 | 5.35 | 5.35 | 0.45 | 0.61 | 0.34 |
| rs347033 | VAV1 | 0.26 | 0.19 | 3.95 | 4.70 | 0.68 | 0.58 | 0.80 |
| rs4807102 | VAV1 | 0.28 | 0.24 | 1.16 | 1.59 | 0.80 | 0.69 | 0.93 |
| rs9514827 | BAFF | 0.33 | 0.29 | 1.16 | 1.30 | 0.82 | 0.95 | 0.71 |
Columns 5 and 6 show P-values for genotype and additive trend tests after FDR correction, expressed as negative log10 values, higher numbers reflecting stronger significance. ORs estimates the protective (values <1) or the predisposing (values >1) contributions to disease risk with 95% CIs.
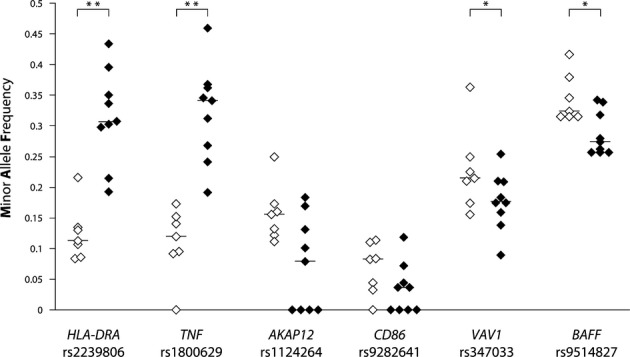
Analysis of MAFs across populations from different European centres for EOMG (♦) and controls (◊). Horizontal lines indicate medians, *P<0.05; **P<0.01 by Mann–Whitney test.
Previous reports suggest an interaction between gender and HLA-alleles. Our data clearly confirm that the associations were even stronger at both the HLA-DRA and TNF-α loci in the 937 females, but were still significant in the 240 males (Fig.(Fig.3),3), with ORs of ~4 and ~2, respectively. We found no such gender differences at the other four loci.
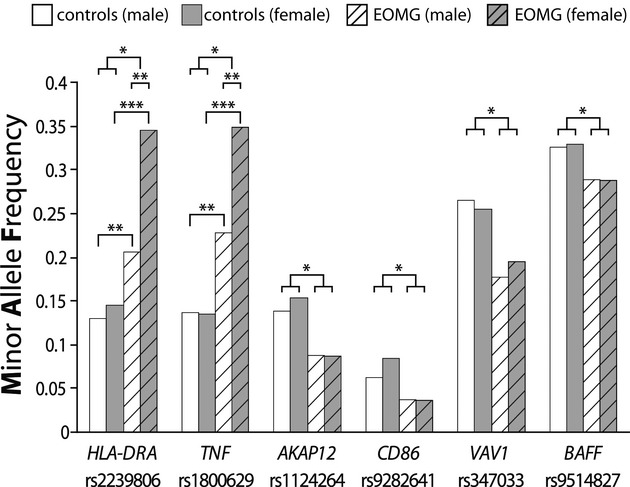
MAFs for six associated genes by gender in patients and controls: *P<0.05; **P<0.01, ***P<0.001 by χ2 tests.
“LD blocks” or “haplotypes” are often more informative than single SNPs, because they are more likely to include causative as well as marker variants. When we defined such haplotypes and compared their frequencies (Table(Table4),4), we noted the expected LD and frequency differences between the three SNPs spanning the 1 Mb in the HLA-AH8.1 haplotype between the TNF-α promoter and the HLA-DRA region. The 90 Kbp genomic region on chromosome 19 that harbors VAV1 has a fragmented pattern of haplotype blocks, each with ≤2 SNPs, and several other SNPs located between them (Fig.(Fig.4).4). The LD block between rs4807102 and rs3786688 creates three possible haplotypes (H1, H2, and H4); of these, H4 was less frequent in EOMG cases (P = 0.0015), but the others showed no differences (Table(Table4).4). In BAFF, three SNPs in the promoter and 5′ coding regions are in LD. They create four possible haplotypes (H2, H3, H4, and H6); of these, H2 was less frequent in EOMG than control subjects (P = 0.005; TableTable4,4, Fig.Fig.5).5). Overall, the haplotype trend test supports the differences in allele frequencies between cases and controls shown in TableTable3,3, but none showed stronger effects than its top-ranking component SNP.
Table 4
Haplotype frequency between cases and controls with confidence interval and P-value.
| Locus ID | H_ID | Haplotype | Haplotype frequencies | χ2 | FDR adjusted P-value | ||
|---|---|---|---|---|---|---|---|
| Controls | EOMG | ||||||
| TNF_HLA-DRA | H1 | rs1800629- rs3129882- rs2239806 | A_A_A | 0.07 | 0.25 | 182.05 | <0.001 |
| TNF_HLA-DRA | H2 | rs1800629- rs3129882- rs2239806 | G_A_A | 0.06 | 0.06 | 0.04 | 0.843 |
| TNF_HLA-DRA | H3 | rs1800629- rs3129882- rs2239806 | G_A_G | 0.36 | 0.25 | 52.57 | <0.001 |
| TNF_HLA-DRA | H4 | rs1800629- rs3129882- rs2239806 | G_G_G | 0.44 | 0.35 | 28.32 | <0.001 |
| AKAP12 | H1 | rs1124264-rs17080949 | C_C | 0.15 | 0.09 | 31.52 | <0.001 |
| AKAP12 | H3 | rs1124264-rs17080949 | T_C | 0.48 | 0.52 | 4.87 | 0.027 |
| AKAP12 | H4 | rs1124264-rs17080949 | T_T | 0.37 | 0.39 | 2.14 | 0.143 |
| CD86 | H1 | rs17203439-rs9282641-rs3792285 | A-C-C | 0.08 | 0.04 | 25.07 | <0.001 |
| CD86 | H2 | rs1800629- rs3129882- rs2239806 | G-A-T | 0.12 | 0.12 | 0.09 | 0.760 |
| CD86 | H3 | rs1800629- rs3129882- rs2239806 | G-C-C | 0.72 | 0.76 | 5.53 | 0.020 |
| CD86 | H4 | rs1800629- rs3129882- rs2239806 | G-C-T | 0.08 | 0.08 | 0.01 | 0.940 |
| VAV1 | H1 | rs4807102-rs3786688 | A_A | 0.37 | 0.40 | 2.72 | 0.099 |
| VAV1 | H2 | rs4807102-rs3786688 | A_G | 0.34 | 0.36 | 0.95 | 0.329 |
| VAV1 | H4 | rs4807102-rs3786688 | G_G | 0.28 | 0.23 | 10.03 | 0.001 |
| BAFF | H2 | rs9514827- rs9514828-rs17564816 | C_T_G | 0.32 | 0.28 | 7.89 | 0.005 |
| BAFF | H3 | rs9514827- rs9514828-rs17564816 | T_C_A | 0.17 | 0.17 | 0.10 | 0.750 |
| BAFF | H4 | rs9514827- rs9514828-rs17564816 | T_C_G | 0.33 | 0.35 | 2.14 | 0.144 |
| BAFF | H6 | rs9514827- rs9514828-rs17564816 | T_T_G | 0.17 | 0.18 | 0.36 | 0.548 |
Locus ID – designations of linked loci haplotype.
Significant haplotypes are in bold

Graphical presentation of the SNP locations and LD structure of VAV1 gene. (A) Chromosome 19 schematic cytogenetic map. (B) Physical map, with exons marked by blocks and SNPs by lines, plus physical coordinates. (C) LD plot. The strength of LD (r2) between all possible pairs of SNPs is indicated by the shade of color, ranging from dark blue (very low) to bright red (very high). (D) Haplotype frequencies of VAV1 in EOMG and controls, and P-value for the comparison between groups. H-ID denotes haplotype ID between rs4808102 and rs3786688 SNPs.

Graphical representation of the SNP locations and LD structure of BAFF gene. (A–C) Chromosome 13 schematic cytogenetic map, physical map and LD plot as described in FigureFigure4.4. (D) Frequencies of haplotypes (H-ID) of the indicated BAFF SNPs in EOMG and controls, plus P-values for differences between them.
Individual SNPs that tag specific genomic regions may predispose independently; alternatively, SNPs from different genomic regions may interact “epistatically”, that is conferring a greater risk in combination than by themselves. We identified a significant interaction between rs2617819 in VAV1 and two SNPs (rs9514827 and rs9514828) in the promoter of region of BAFF at a P-value of 0.03 after correcting for multiple comparisons. Moreover, in carriers of the associating C allele of VAV1 rs2617819, three of the BAFF haplotype combinations (H3, H4, and H6) were increased in EOMG (Fig.(Fig.6).6). These results suggest that epistatic interactions between VAV1 and BAFF alleles confer a higher risk of EOMG than either does alone.
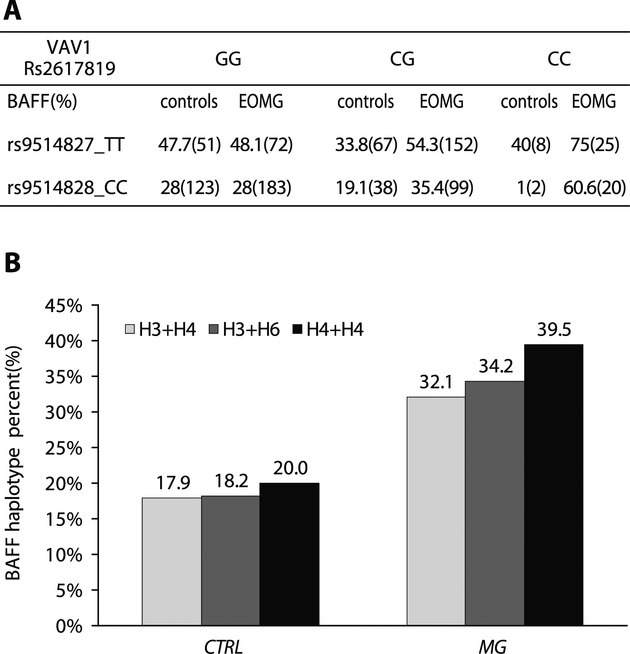
BAFF genotype and haplotype frequencies on the background of VAV1 SNP rs2617819. (A) Percentages and numbers (between brackets) of controls and EOMG patients homozygous for either rs9514827 TT or rs9514828 CC by VAV1 rs2617819 genotype. (B). Percentages of the indicated haplotype combinations in carriers of VAV1 rs2617819 C allele in controls and EOMG.
Discussion
This study shows significant frequency differences between EOMG and controls in four genes, CD86, AKAP12, VAV1, and BAFF, and the expected strong associations at HLA-DRA and TNF. Three of the genes, CD86, VAV1, and BAFF, use the same NFκB signaling pathway. Moreover, VAV1 and BAFF interact epistatically, conferring a greater risk in combination than by themselves. In addition, we confirmed the stronger association with the HLA-AH8.1 haplotype in females; that with rs1800629 (TNF-308G>A) is of particular interest because of its functional effect on expression levels.4,5,22 By contrast, the role of AKAP12, a cell growth-related protein, in EOMG remains elusive. Autoantibodies against it were noted in EOMG patients more than two decades ago, hence its other name is “gravin,” but it has since been studied more in the context of cell cycle and malignancy than of myasthenia.23–25
There is mounting evidence implicating BAFF in the pathogenesis of several autoimmune diseases, including MG.26 Increased serum levels and expression of BAFF and its receptors have been demonstrated in the thymus of MG patients27. BAFF promotes the survival, growth, and maturation of B-cells, including autoreactive B-cells and rescue B-cells from apoptosis, thereby contributing to autoimmunity.28 Its exact role in EOMG pathogenesis is not clear. If it proves to be more important in disease initiation than progression, it might become a therapeutic target for novel early intervention in EOMG.29
VAV1 is specifically expressed in hematopoietic cells, and is a critical regulator of T-cell cycle progression and proliferation at an early point after convergence of the signaling routes triggered by the T-cell receptor and CD28.30,31 In rats, Vav1 variants control susceptibility to central nervous system (CNS) inflammation by influencing Treg development32 and production of inflammatory cytokines by effector T-cells.33 Several cohorts of patients with multiple sclerosis (MS) have shown associations with VAV1 haplotypes34; we also included these here because of their potential impact on T-cell function. Indeed, one of them showed a protective effect by itself in EOMG, whereas another predisposed synergistically with two adjacent SNPs in the BAFF promoter.
The immune system shows such marked genotypic and phenotypic complexity that it must be fertile soil for epistasis. Elucidating these interactions at functional as well as genetic levels is key to understanding predisposition to autoimmune diseases.35 Epistatic effects have proved to be central in several model organisms and several autoimmune diseases. They can operate at various levels, ranging from direct interactions between proteins, or between regulatory components in different pathways, where strict partitioning is rare. In immune responses, interactions between antigen receptors and their binding partners, between cytokines and their receptors and between signaling molecules in cell activation cascades are all candidates for genetic epistasis. The example that we now show between VAV1 and BAFF probably represents just one of a multitude of similar interactions that influence disease risk.
Activation of the noncanonical NFκB signaling pathway by VAV1 increases the expression of IL-8, BAFF, and Bcl-xL genes and might contribute to EOMG predisposition.30 CD86 and TNF-α, two other EOMG risk factors noted here, also converge on the activation of NFκB cascade. Notably, the choice of candidate genes was based on preselection of immune genes and therefore the involvement of nuclear factor-kappaB (NF-κB) that acts at the crossroads of many signaling pathways retrospectively is not totally surprising. This is also in light of the fact that NFκB plays a crucial role in the establishment of both central tolerance and the peripheral function of Treg cells, and multiple associations have been found between genes involved in NFκB signaling and autoimmune diseases.36 Thus, defective or dysregulated activation of NFκB pathways, as suggested in the present study seem highly plausible contributors to autoreactivity and inflammation in EOMG.
GWAS of MG is a challenge because of its low prevalence and the modest effect sizes anticipated. Nevertheless, a GWAS with more than 600 patients from northern Europe has found that TNFAIP3-interacting protein 1 (TNIP1) is associated with EOMG and confirms the known association with HLA-B08 and protein tyrosine phosphatase nonreceptor 22 (PTPN22).5 TNIP1, also known as ABIN-1, is thought to be involved in ubiquitin-dependent dysregulation of NF-κB signalling37,38 and corepression of nuclear receptors' such as retinoic acid receptors and peroxisome proliferator-activated receptors.39 As by the time of the GWAS publication in December 20125 both the discovery and replication genotypes of the current study were completed, PTPN22 and TNIP1 were not included in the current study as candidate genes. Nevertheless, it is interesting to note that both studies converge on the same signaling pathway, namely NF-κB, in an independent manner.
There is exceptionally strong LD across the entire 4 Mb of the HLA-AH 8.1 haplotype, which is very rich in genes, many with immunological functions. Obviously, with the highly focused SNPs used here, we could not map the association in EOMG precisely. Intriguingly, of the many possible candidate HLA-complex loci, the rs1800629 risk allele in the TNF promoter seems particularly relevant. This functional variant correlates with overexpression and oversecretion of the cytokine in vitro and in vivo22,40 and associates with numerous autoimmune diseases including EOMG.41–44 TNF-α overexpression has been implicated in the development of experimental autoimmune MG onset which is prevented by anti-TNF-α antibodies.45,46 In addition, an autocrine loop involving TNF-α contributes to the production of AKAP12 mRNA in response to inflammation.47 We suggest that overexpression of TNF-α in EOMG patients carrying (-308G>A) may perpetuate the inflammatory response in the thymus, where Treg cell suppressive function is lacking.48 Indeed, TNF-α is increased in both thymic T-cells and sera of EOMG patients, and appears to be involved in the defective regulation observed in these patients.49 Several TNF-α inhibitors are widely used to treat autoimmune diseases such as RA, Crohn's disease, and psoriasis. Hence, if this functional variant proves to be crucial in EOMG patients, these inhibitors might become important alternative options for them too.
Women are disproportionately affected by autoimmune diseases, especially those with onset between puberty and the menopause. The current study confirmed the gender bias at the HLA loci originally noted by Janer et al. and more recently by Gregersen et al.4,5 Interestingly, we also found a very strong gender bias at the TNF loci, that has not been shown previously, and was confirmed in all our nine cohorts. Sex hormones play a pivotal role in the gender bias with a commonly accepted view that androgens are protective. In an animal model for type 1 diabetes, a hormone-dependent differences in microbiota composition in males were found, and these differences were shown to be protective.50 In addition, X-chromosome transcripts were shown to regulate the transcription of autosomal genes, and affect molecular pathways such as DNA methylation, glucose and protein metabolism in a gender-biased manner.51 Last, but not least, numerous HLA-region genes, including HLA-DRA and TNF, contain estrogen response elements.52 Polymorphisms in these response elements might influence transcription in a gender specific manner. Our study highlights the need for more detailed analysis of the effects of sex differences in immune responses.
In conclusion, our findings implicate several genes converging on the proinflammatory NF-κB signaling pathway. Patients DNA samples were assembled from nine different European centres with a total of 1200 samples genotyped, the most robust genetic study that has been conducted to date in MG in terms of number of patients analyzed. With its larger numbers (1177 vs. 649) and wider geographic spread across Europe, our study complements the recent GWAS,5 which could not detect loci with modest ORs, such as VAV1 and BAFF; conversely, the limited number of loci evaluated in the current study did not include PTPN22 and TNIP1 loci. These genetic findings shed light on underlying disease mechanisms in MG and provide targets for the development of novel diagnostic and therapeutic tools for future use in this chronic inflammatory and autoimmune disorder.
Acknowledgments
This work was supported by 7th Framework Program of the European Union FIGHT-MG (grant no. 242210) (A. M., S. B. A., R. M., C. T., A. S., S. T.), the Polish-Norwegian Research Fund Grant PNRF-204-AI-1/07 (A. K. P.), the Sacta-Rashi Foundation (N. A.), the U.K. Medical Research Council (N. W.), the “Association Française contre les Myopathies” (A. S.). The authors would like to thank Liat Linde (The Genomics Core Facility at the Rappaport Faculty of Medicine, Technion) and David Pilzer (Biological services Weizmann Institute) for DNA genotyping, and our many clinical colleagues for collecting patients.
Authorship and Contributorship
Substantial contributions to conception and design: N. A., R. L. P., P. B., S. B. A., R. M., C. T., K. P., S. J. T., A. M. Acquisition of data: N. A., S. H. S., E. G., K. P., Z. Z., M. B., S. J. T., N. W. F. B., C. A., R. M., P. C., L. C. Analysis and interpretation of data: N. A., E. G., R. L. P., P. B., A. S. N. W. S. B. A., A. M. Drafting and revising the article: N. A., A. M., R. L. P. S. B. A., K. P., Z. Z., M. B., S. J. T. Final approval: K. P., Z. Z., M. B., S. J. T., A. M.
Conflicts of Interest
H. F. H., C. T., B. A. L. and A. M. have no conflicts of interest related to this paper. P. B.: speaking fees, MEDA Pharmaceuticals Inc. P. C. L. C., F. B.: no conflict of interest. C. A.: travel support, Merck-Serono, TEVA and Fresenius Medical Care. R. M.: consultancies for Alexion, BioMarin; speaking fees and travel support, Sanofi-Aventis, Merck-Serono, MEDA Pharmaceuticals Inc. A. M.: advisory board and research support, Teva Pharmaceutical Industries Ltd.; Medison Pharma Ltd.; Biogen Idec; Merck Serono; and Bayer Schering Pharma; A. M.: speaking fees and travel support from Teva Pharmaceutical Industries Ltd.; Medison Pharma Ltd.; Biogen Idec; Merck Serono; Bayer Schering Pharma Avanir Pharmaceuticals and Novartis. S. B. A., F. T. and R. L. P. have no conflicts of interest related to this paper.
References
- Meriggioli MN, Sanders DB. Autoimmune myasthenia gravis: emerging clinical and biological heterogeneity. Lancet Neurol. 2009;8:475–490. [Europe PMC free article] [Abstract] [Google Scholar]
- Lindstrom JM, Seybold ME, Lennon VA, et al. Antibody to acetylcholine receptor in myasthenia gravis. Prevalence, clinical correlates, and diagnostic value. Neurology. 1976;26:1054–1059. [Abstract] [Google Scholar]
- Carr AS, Cardwell CR, McCarron PO, et al. A systematic review of population based epidemiological studies in Myasthenia Gravis. BMC Neurol. 2010;10:46. [Europe PMC free article] [Abstract] [Google Scholar]
- Janer M, Cowland A, Picard J, et al. A susceptibility region for myasthenia gravis extending into the HLA-class I sector telomeric to HLA-C. Hum Immunol. 1999;60:909–917. [Abstract] [Google Scholar]
- Gregersen PK, Kosoy R, Lee AT, et al. Risk for myasthenia gravis maps to a (151) Pro–>Ala change in TNIP1 and to human leukocyte antigen-B*08. Ann Neurol. 2012;72:927–935. [Europe PMC free article] [Abstract] [Google Scholar]
- Weiss JM, Cufi P, Bismuth J, et al. SDF-1/CXCL12 recruits B cells and antigen-presenting cells to the thymus of autoimmune myasthenia gravis patients. Immunobiology. 2013;218:373–381. [Abstract] [Google Scholar]
- Mao ZF, Yang LX, Mo XA, et al. Frequency of autoimmune diseases in myasthenia gravis: a systematic review. Int J Neurosci. 2011;121:121–129. [Abstract] [Google Scholar]
- Ramanujam R, Pirskanen R, Ramanujam S, et al. Utilizing twins concordance rates to infer the predisposition to myasthenia gravis. Twin Res Hum Genet. 2011;14:129–136. [Abstract] [Google Scholar]
- Avidan N, Le Panse R, Berrih-Aknin S, et al. Genetic basis of myasthenia gravis – a comprehensive review. J Autoimmun. 2013 10.1016/j.jaut.2013.12.001 [Epub ahead of print] [Abstract] [Google Scholar]
- Vandiedonck C, Beaurain G, Giraud M, et al. Pleiotropic effects of the 8.1 HLA haplotype in patients with autoimmune myasthenia gravis and thymus hyperplasia. Proc Natl Acad Sci USA. 2004;101:15464–15469. [Europe PMC free article] [Abstract] [Google Scholar]
- Candore G, Lio D, Colonna Romano G, et al. Pathogenesis of autoimmune diseases associated with 8.1 ancestral haplotype: effect of multiple gene interactions. Autoimmun Rev. 2002;1:29–35. [Abstract] [Google Scholar]
- Maniaol AH, Elsais A, Lorentzen AR, et al. Late onset myasthenia gravis is associated with HLA DRB1*15:01 in the Norwegian population. PLoS One. 2012;7:e36603. [Europe PMC free article] [Abstract] [Google Scholar]
- Giraud M, Taubert R, Vandiedonck C, et al. An IRF8-binding promoter variant and AIRE control CHRNA1 promiscuous expression in thymus. Nature. 2007;448:934–937. [Abstract] [Google Scholar]
- Visscher PM, Brown MA, McCarthy MI, et al. Five years of GWAS discovery. Am J Hum Genet. 2012;90:7–24. [Europe PMC free article] [Abstract] [Google Scholar]
- Nicolle MW. Myasthenia gravis. Neurologist. 2002;8:2–21. [Abstract] [Google Scholar]
- de Bakker PI, Yelensky R, Pe'er I, et al. Efficiency and power in genetic association studies. Nat Genet. 2005;37:1217–1223. [Abstract] [Google Scholar]
- Chan M, Chan MW, Loh TW, et al. Evaluation of nanofluidics technology for high-throughput SNP genotyping in a clinical setting. J Mol Diagn. 2011;13:305–312. [Europe PMC free article] [Abstract] [Google Scholar]
- Fan JB, Gunderson KL, Bibikova M, et al. Illumina universal bead arrays. Methods Enzymol. 2006;410:57–73. [Abstract] [Google Scholar]
- Ritchie ME, Liu R, Carvalho BS, et al. Comparing genotyping algorithms for Illumina's Infinium whole-genome SNP BeadChips. BMC Bioinformatics. 2011;12:68. [Europe PMC free article] [Abstract] [Google Scholar]
- Tian C, Gregersen PK, Seldin MF. Accounting for ancestry: population substructure and genome-wide association studies. Hum Mol Genet. 2008;17:R143–R150. [Europe PMC free article] [Abstract] [Google Scholar]
- Nelis M, Esko T, Magi R, et al. Genetic structure of Europeans: a view from the North-East. PLoS One. 2009;4:e5472. [Europe PMC free article] [Abstract] [Google Scholar]
- Elahi MM, Asotra K, Matata BM, et al. Tumor necrosis factor alpha -308 gene locus promoter polymorphism: an analysis of association with health and disease. Biochim Biophys Acta. 2009;1792:163–172. [Abstract] [Google Scholar]
- Gordon T, Grove B, Loftus JC, et al. Molecular cloning and preliminary characterization of a novel cytoplasmic antigen recognized by myasthenia gravis sera. J Clin Invest. 1992;90:992–999. [Europe PMC free article] [Abstract] [Google Scholar]
- Nauert JB, Klauck TM, Langeberg LK, et al. Gravin, an autoantigen recognized by serum from myasthenia gravis patients, is a kinase scaffold protein. Curr Biol. 1997;7:52–62. [Abstract] [Google Scholar]
- Gelman IH. Emerging roles for SSeCKS/Gravin/AKAP12 in the control of cell proliferation, cancer malignancy, and barriergenesis. Genes Cancer. 2010;1:1147–1156. [Europe PMC free article] [Abstract] [Google Scholar]
- Lisak RP, Ragheb S. The role of B cell-activating factor in autoimmune myasthenia gravis. Ann N Y Acad Sci. 2012;1274:60–67. [Abstract] [Google Scholar]
- Kim JY, Yang Y, Moon JS, et al. Serum BAFF expression in patients with myasthenia gravis. J Neuroimmunol. 2008;199:151–154. [Abstract] [Google Scholar]
- Berrih-Aknin S, Ragheb S, Le Panse R, et al. Ectopic germinal centers, BAFF and anti-B-cell therapy in myasthenia gravis. Autoimmun Rev. 2013;12:885–889. [Abstract] [Google Scholar]
- Dalakas MC. Novel future therapeutic options in myasthenia gravis. Autoimmun Rev. 2013;12:936–941. [Abstract] [Google Scholar]
- Tuosto L. NF-kappaB family of transcription factors: biochemical players of CD28 co-stimulation. Immunol Lett. 2011;135:1–9. [Abstract] [Google Scholar]
- Tybulewicz VL, Ardouin L, Prisco A, et al. Vav1: a key signal transducer downstream of the TCR. Immunol Rev. 2003;192:42–52. [Abstract] [Google Scholar]
- Colacios C, Casemayou A, Dejean AS, et al. The p.Arg63Trp polymorphism controls Vav1 functions and Foxp3 regulatory T cell development. J Exp Med. 2011;208:2183–2191. [Europe PMC free article] [Abstract] [Google Scholar]
- Pedros C, Papapietro O, Colacios C, et al. Genetic control of HgCl2-induced IgE and autoimmunity by a 117-kb interval on rat chromosome 9 through CD4 CD45RC(high) T cells. Genes Immun. 2013;14:258–267. [Abstract] [Google Scholar]
- Jagodic M, Colacios C, Nohra R, et al. A role for VAV1 in experimental autoimmune encephalomyelitis and multiple sclerosis. Sci Transl Med. 2009;1:10–21. [Abstract] [Google Scholar]
- Rose AM, Bell LC. Epistasis and immunity: the role of genetic interactions in autoimmune diseases. Immunology. 2012;137:131–138. [Abstract] [Google Scholar]
- Sun SC, Chang JH, Jin J. Regulation of nuclear factor-kappaB in autoimmunity. Trends Immunol. 2013;34:282–289. [Europe PMC free article] [Abstract] [Google Scholar]
- Oshima S, Turer EE, Callahan JA, et al. ABIN-1 is a ubiquitin sensor that restricts cell death and sustains embryonic development. Nature. 2009;457:906–909. [Europe PMC free article] [Abstract] [Google Scholar]
- Wagner S, Carpentier I, Rogov V, et al. Ubiquitin binding mediates the NF-kappaB inhibitory potential of ABIN proteins. Oncogene. 2008;27:3739–3745. [Abstract] [Google Scholar]
- Ramirez VP, Gurevich I, Aneskievich BJ. Emerging roles for TNIP1 in regulating post-receptor signaling. Cytokine Growth Factor Rev. 2012;23:109–118. [Europe PMC free article] [Abstract] [Google Scholar]
- Kroeger KM, Carville KS, Abraham LJ. The -308 tumor necrosis factor-alpha promoter polymorphism effects transcription. Mol Immunol. 1997;34:391–399. [Abstract] [Google Scholar]
- Toonen EJ, Barrera P, Fransen J, et al. Meta-analysis identified the TNFA -308G > A promoter polymorphism as a risk factor for disease severity in patients with rheumatoid arthritis. Arthritis Res Ther. 2012;14:R264. [Europe PMC free article] [Abstract] [Google Scholar]
- Liu ZH, Ding YL, Xiu LC, et al. A meta-analysis of the association between TNF-alpha -308G>A polymorphism and type 2 diabetes mellitus in Han Chinese population. PLoS One. 2013;8:e59421. [Europe PMC free article] [Abstract] [Google Scholar]
- Skeie GO, Pandey JP, Aarli JA, et al. TNFA and TNFB polymorphisms in myasthenia gravis. Arch Neurol. 1999;56:457–461. [Abstract] [Google Scholar]
- Zelano G, Lino MM, Evoli A, et al. Tumour necrosis factor beta gene polymorphisms in myasthenia gravis. Eur J Immunogenet. 1998;25:403–408. [Abstract] [Google Scholar]
- Duan RS, Wang HB, Yang JS, et al. Anti-TNF-alpha antibodies suppress the development of experimental autoimmune myasthenia gravis. J Autoimmun. 2002;19:169–174. [Abstract] [Google Scholar]
- Wang HB, Li H, Shi FD, et al. Tumor necrosis factor receptor-1 is critically involved in the development of experimental autoimmune myasthenia gravis. Int Immunol. 2000;12:1381–1388. [Abstract] [Google Scholar]
- Yan M, Xia C, Cheng C, et al. The role of TNF-alpha and its receptors in the production of Src-suppressed C kinase substrate by rat primary type-2 astrocytes. Brain Res. 2007;1184:28–37. [Abstract] [Google Scholar]
- Gradolatto A, Nazzal D, Foti M, et al. Defects of immunoregulatory mechanisms in myasthenia gravis: role of IL-17. Ann N Y Acad Sci. 2012;1274:40–47. [Abstract] [Google Scholar]
- Gradolatto A, Nazzal D, Truffault F, et al. Both Treg cells and Tconv cells are defective in the myasthenia gravis thymus: roles of IL-17 and TNF-alpha. J Autoimmun. 2014 ; 10.1016/j.jaut.2013.12.015 [Epub ahead of print] [Abstract] [Google Scholar]
- Yurkovetskiy L, Burrows M, Khan AA, et al. Gender bias in autoimmunity is influenced by microbiota. Immunity. 2013;39:400–412. [Europe PMC free article] [Abstract] [Google Scholar]
- Bermejo-Alvarez P, Rizos D, Lonergan P, et al. Transcriptional sexual dimorphism during preimplantation embryo development and its consequences for developmental competence and adult health and disease. Reproduction. 2011;141:563–570. [Abstract] [Google Scholar]
- Kaur M, Schmeier S, MacPherson CR, et al. Prioritizing genes of potential relevance to diseases affected by sex hormones: an example of myasthenia gravis. BMC Genomics. 2008;9:481. [Europe PMC free article] [Abstract] [Google Scholar]
Articles from Annals of Clinical and Translational Neurology are provided here courtesy of Wiley
Full text links
Read article at publisher's site: https://doi.org/10.1002/acn3.51
Read article for free, from open access legal sources, via Unpaywall:
https://onlinelibrary.wiley.com/doi/pdfdirect/10.1002/acn3.51
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics

Discover the attention surrounding your research
https://www.altmetric.com/details/2276210
Article citations
Positive regulation of Vav1 by Themis controls CD4 T cell pathogenicity in a mouse model of central nervous system inflammation.
Cell Mol Life Sci, 81(1):161, 02 Apr 2024
Cited by: 0 articles | PMID: 38565808 | PMCID: PMC10987373
Preferential X Chromosome Inactivation as a Mechanism to Explain Female Preponderance in Myasthenia Gravis.
Genes (Basel), 13(4):696, 15 Apr 2022
Cited by: 5 articles | PMID: 35456502 | PMCID: PMC9031138
Single-cell RNA-Seq reveals transcriptional heterogeneity and immune subtypes associated with disease activity in human myasthenia gravis.
Cell Discov, 7(1):85, 14 Sep 2021
Cited by: 18 articles | PMID: 34521820 | PMCID: PMC8440681
Estrogen Receptor, Inflammatory, and FOXO Transcription Factors Regulate Expression of Myasthenia Gravis-Associated Circulating microRNAs.
Front Immunol, 11:151, 21 Feb 2020
Cited by: 16 articles | PMID: 32153563 | PMCID: PMC7046803
Associations of BAFF rs2893321 polymorphisms with myasthenia gravis susceptibility.
BMC Med Genet, 20(1):168, 30 Oct 2019
Cited by: 6 articles | PMID: 31666013 | PMCID: PMC6822419
Go to all (12) article citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
SNPs (Showing 15 of 15)
- (9 citations) dbSNP - rs1800629
- (7 citations) dbSNP - rs2239806
- (6 citations) dbSNP - rs3129882
- (6 citations) dbSNP - rs9514827
- (5 citations) dbSNP - rs9514828
- (4 citations) dbSNP - rs17080949
- (4 citations) dbSNP - rs4807102
- (3 citations) dbSNP - rs1124264
- (3 citations) dbSNP - rs17564816
- (2 citations) dbSNP - rs3786688
- (2 citations) dbSNP - rs2617819
- (2 citations) dbSNP - rs9282641
- (1 citation) dbSNP - rs3129888
- (1 citation) dbSNP - rs347033
- (1 citation) dbSNP - rs4808102
Show less
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Genome-Wide Association Study of Late-Onset Myasthenia Gravis: Confirmation of TNFRSF11A and Identification of ZBTB10 and Three Distinct HLA Associations.
Mol Med, 21(1):769-781, 10 Nov 2015
Cited by: 25 articles | PMID: 26562150 | PMCID: PMC4749491
Association with HLA DQ of early onset myasthenia gravis in Southeast Texas region of the United States.
Int J Immunogenet, 38(1):55-62, 25 Nov 2010
Cited by: 18 articles | PMID: 21108743
Risk for myasthenia gravis maps to a (151) Pro→Ala change in TNIP1 and to human leukocyte antigen-B*08.
Ann Neurol, 72(6):927-935, 10 Oct 2012
Cited by: 92 articles | PMID: 23055271 | PMCID: PMC3535539
Autoimmune associations and autoantibody screening show focused recognition in patient subgroups with generalized myasthenia gravis.
Hum Immunol, 74(9):1184-1193, 18 Jun 2013
Cited by: 27 articles | PMID: 23792059


