Abstract
Free full text

Integrative, Multi-modal Analysis of Glioblastoma Using TCGA Molecular Data, Pathology Images and Clinical Outcomes
Abstract
Multi-modal, multi-scale data synthesis is becoming increasingly critical for successful translational biomedical research. In this paper, we present a large-scale investigative initiative on glioblastoma, a high-grade brain tumor, with complementary data types using in silico approaches. We integrate and analyze data from The Cancer Genome Atlas Project on glioblastoma that includes novel nuclear phenotypic data derived from microscopic slides, genotypic signatures described by transcriptional class and genetic alterations, and clinical outcomes defined by response to therapy and patient survival. Our preliminary results demonstrate numerous clinically and biologically significant correlations across multiple data types, revealing the power of in silico multi-modal data integration for cancer research.
I. Introduction
With rapid technological advances in acquiring data from diverse platforms in cancer research, numerous large scale datasets have become available, providing high resolution views and multi-faceted descriptions of biological systems. Such efforts include those in brain tumor research by The Cancer Genome Atlas (TCGA) [1], and the Repository of Molecular Brain Neoplasia Data (REMBRANDT) [2], which have collected large volumes of multi-modal data from complementary platforms on patients with diffuse glioma. As manual processing of this large-scale data is both error-prone and intractably time-consuming, recent investigations have either primarily focused on in silico experiments that interrogate these datasets or use them to generate or corroborate hypotheses.
In the In Silico Brain Tumor Research Center (ISBTRC), one of the six National Cancer Institute (NCI) funded In Silico Research Centers of Excellence1, we explore novel approaches and develop tools for integrative multi-scale, multi-modal data analysis of diffuse gliomas. Our current research has focused on potential relations across tumor genomic and gene expression profiles, complex nuclear morphometric features, neuro-imaging, and clinical outcomes. By conducting complementary, multi-scale in silico experiments, we aim not only to improve prognostic capabilities, but also to develop a better understanding of biological underpinnings that drive the rapid progression of these devastating diseases [3].
As a first step towards seamless data integration for improved diagnosis and stratification, we describe our methodology for correlating nuclear morphometric features derived from digitized microscopic images of glioblastoma with 1) genetic alterations, 2) transcriptional subtypes, and 3) treatment response and patient survival.
We hypothesize that digitized pathology images contain rich and as yet untapped biological information trapped in morphologic features that can be resolved by image analysis to provide correlations with genetic alterations and patient prognosis. In this paper, we present results correlating computer-generated nuclear morphometry from large-scale microscopic images with survival, treatment response, and clinically relevant molecular characterizations. The results demonstrate the potential of multi-modal data integration within the setting of large-scale in silico research.
II. Data set and analysis infrastructure
The overall framework for data analysis, management, integration, and computation infrastructure is illustrated in Fig. 1, where nuclear morphometric features from microscopic images, molecular signatures, clinical outcomes, and neuroimaging annotations from the same cohort of patients are stored in a database for large-scale multi-modal data query, integration, and analysis.
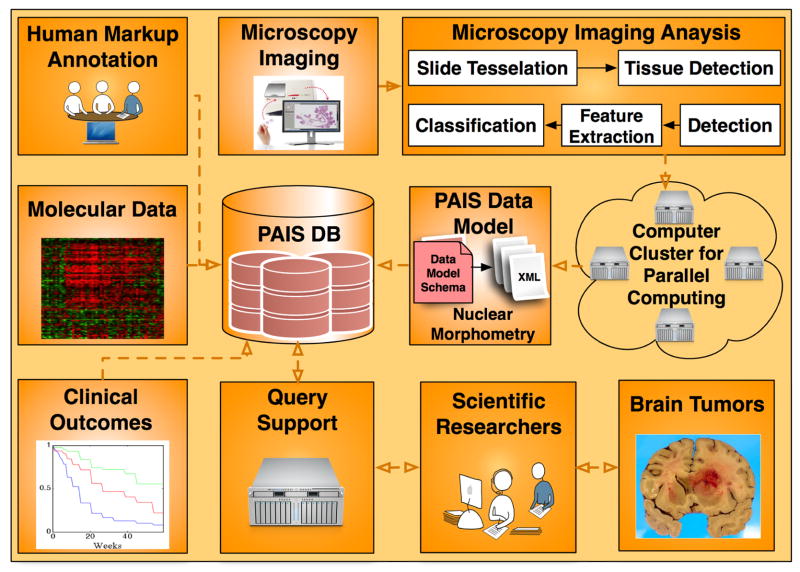
The overall architecture for the In Silico brain tumor research is presented. All data sources, i.e. microscopy imaging features, molecular data, and clinical outcomes are captured by PAIS database with which scientific researchers can query for multi-data integration results.
A. Microscopy Imaging Data
Digital microscopy is rapidly emerging as a tool for establishing pathologic diagnosis, evaluating treatment efficacies and performing morphologic research. In distinction to traditional visual review of histological sections, which introduces human bias and remains largely qualitative [4], a computer-based analysis of virtual microscopic slides can be systematic, objective, efficient, and complete [5][6][7]. Moreover, many features in a microscopic image can be identified and analyzed by computer algorithms but not by human observers. Thus, imaging data from histologic slides contains rich phenotypic information that can potentially be exploited to yield clinically meaningful results.
In our research, we have used the microscopic images from TCGA project on glioblastomas (GBMs), which are WHO grade IV astrocytic neoplasms that are rapidly progressive and ultimately fatal. All digitized slides are Haematoxylin and Eosin (H&E) stained permanent sections that were formalin-fixed and paraffin-embedded. In aggregate, 428 whole slides associated with 162 patients are included. All were scanned at 20x magnification with a high-resolution, high-throughput digitized scanner. The overall storage size of the complete image data set for study is about 175Gbytes with JPEG compression ratio of 5.11. The image resolution is up to 63922 × 45753 pixels.
B. “Omics” Data
Phenotypic data derived from digitized images was correlated with TCGA molecular data, providing insight to underlying biological mechanisms and potentially uncovering therapeutic targets within a morphologic class. Each TCGA sample was characterized by multiple molecular platforms including gene (mRNA) and microRNA expression, DNA copy number variation, DNA sequence and DNA methylation.
A recent study of TCGA GBMs defined four transcriptional subtypes: proneural (PN), neural (NR), classical (CL) and mesenchymal (MS) [8]. Each subtype is defined by a characteristic gene expression profile and genetic alterations, including mutations and chromosomal changes (amplification/deletion). For our study, transcriptional subtypes were either obtained from the supplementary information in an earlier work [8] or determined with Prediction Analysis of Microarray (PAM) software version 2.21 using RMA normalized Affymetrix HT-HGU133 mRNA expression platform data. A sample expression average was computed for samples with multiple corresponding arrays. Unlogged expression was filtered to remove probes with a fold change less than 1.5 or an expression range less than 20.
Somatic mutations and chromosome alterations (amplification or deletion) for genes CDKN2A, EGFR, IDH1, NF1, PDGFRA, TP53, and PTEN, have been provided by the Memorial Sloan-Kettering Cancer Center (MSKCC)2. Mutational status from 205 samples was available. Copy number variation data from the same set consists of a consensus derived from a combination of platforms (Agilent, Affymetrix SNP 6, Illumina) together with methods (RAE[9], GISTIC[10], GTS[11]) for identifying regions of genomic aberration likely to drive cancer pathogenesis. Copy number alterations are represented by homozygous deletion, hemizygous deletion, neutral change, gain, and high-level amplification.
C. Clinical Outcomes
Clinical data on patient age, chemo- and radiotherapies, and survival was downloaded from the TCGA portal3.
D. Computational Infrastructure
High resolution digitized pathology images are extremely large, with some occupying several gigabytes even in compressed form. The TCGA dataset include hundreds of pathology images and presents a significant computational challenge for analysis. To expedite processing, we partitioned each whole slide image into non-overlapping regions of 4096 × 4096 pixels to permit parallel analysis. This choice balances between memory requirements and the loss of microanatomy due to tiling. Larger regions have physical memory constraints. Smaller ones place a greater fraction of nuclei on region boundaries resulting in their loss during analysis. To scale up the analysis component of the architecture, we process images with a large-scale, high-performance computation infrastructure where a cluster of computer nodes executes jobs simultaneously. This configuration currently consists of seven Dell 1950 1U rack mount units. Each unit is configured with Dual Xeon E5420 CPUs running with four cores at 2.5Ghz for a total of eight cores per node.
E. Pathology Image Data Representation and Management
Digital microscopy images contain a tremendous array of micro-anatomic structures, which collectively characterize specimens phenotypically. In a study with hundreds or thousands of high-resolution images, millions of nuclear morphometric features need to be represented and curated in a systematic manner such that they can be efficiently queried for correlative investigations. In addition, image analysis using either multiple algorithms or multiple parameter sets can further increase the size of data to be recorded. As a result, information models are needed to organize and represent virtual slide-related image, annotation, mark-up and feature information. To address these challenges, we developed the Pathology Analytical and Imaging Standards (PAIS) model to support flexible, efficient, and semantically enabled data representations for pathology image analysis and characterization4. We also implemented a relational database realization of PAIS using IBM DB2 Enterprise Edition 9.7.3 with its spatial extender. The current database runs on PowerEdge T410 Linux server with four quadcore CPUs, 16GB memory, and a 7200 rpm hard drive.
PAIS makes it possible to represent and share data generated from pathology images. More importantly, it is useful tool for scientific discovery through its powerful query support, including those that are metadata-based, spatially based or semantically based [12][13]. Further, we incorporate related molecular data and clinical information into PAIS database to provide integrative queries.
III. Integrated multi-modal data analysis
We next present our methodologies for high throughput microscopic image analysis and multi-modal data integration.
A. Microscopy Imaging Analysis
We developed a suite of image analysis tools for segmenting and characterizing nuclei. To reliably identify nuclei, we applied the fast hybrid grayscale reconstruction algorithm to images for normalizing background regions degraded by artifacts arising from tissue preparation and scanning [18]. This operation substantially separates the foreground from the normalized background and allows recognition of nuclei by simple thresholding. Overlapped nuclei were subsequently separated with the watershed method.
We then extracted a complementary set of features for each identified nucleus to obtain phenotypic signatures of GBMs. These features fall under four primary headings: nuclear morphometry, region texture, intensity and gradient statistics, as summarized in Fig. 2 (a) [14]. Since specific nuclear features have traditionally been used to distinguish types of gliomas, morphometric features (such as the degree of elongation, and size) are included. Nuclear texture information is captured by multiple descriptors, as it varies across nuclei due to the content and clumping of chromatin. Features relevant to nuclear intensity and intensity gradient are included as well. All nuclear features are computed with the grayscale image channel converted from the original color image. Additionally, we applied the same set of texture and gradient features to “cytoplasm” regions surrounding nuclei. Since the true cellular borders of glioma cells cannot be resolved on H&E stained images, cytoplasm refers to a fixed-distance radius surrounding a nucleus. In practice, we dilated the nuclear regions with an eight-pixel margin to identify this space. Fig. 2 (b) presents a small image region where glioma nuclei and cytoplasm regions are depicted. Features derived from cytoplasm are computed with the grayscale image channel as well as the isolated channels for H&E stain signals separated by a color deconvolution algorithm [15]. As the cytoplasm space is obtained by dilating the nuclear regions, its morphologic features are not calculated. Cytoplasm features are then combined with nuclear features for better representation. In aggregate, 74 features extracted from nuclei and proximal cytoplasm describe the morphology and texture characteristics of each nucleus and its neighboring area.
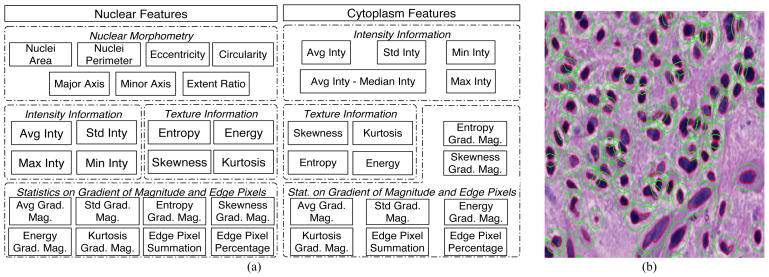
(a) Features computed from nuclear and “cytoplasm” regions are summarized; (b) A typical image region is overlaid with nuclear (red) and “cytoplasm” (green) boundaries identified by computer algorithms.
All nuclear and cytoplasmic features associated with a GBM were then summarized into a single vector to represent each patient. To this end, we calculated the first moment of each feature and the second moments of all possible pairs of features [16]. The first moment represents the average value for a specific feature, whereas the second order statistics define relationships between features regarding 1) nuclear morphology, 2) nuclear morphology and nuclear staining, or 3) nuclear morphology/staining and cytoplasmic staining for each patient. The summarization step produces an N(N+3)/2-dimensional feature vector to represent the morphology of each patient in a high dimensional space, where N is equal to 74 in our case. Thus, each patient is represented by a 2849-dimensional imaging signature vector derived by aggregating the features of machine-identified nuclei in the associated microscopic whole-slide images.
This is followed by a consensus clustering procedure to compute the probabilities that signatures of pairwise cases are grouped in the same cluster over 100 independent trials of K-means experiments. This analysis is aimed at uncovering the existence of intrinsic morphological clusters defined by nuclear feature signatures. We set the number of clusters as K=3.
B. PAIS Query Support
Segmentation results and features are stored in the PAIS database. To correlate micro-anatomic morphometry with molecular profiles and clinical outcome, summary statistics on image features need to be computed for each patient. This process involves calculating the mean feature vectors and the feature covariance values of all possible feature pairs over all nuclei in images of each patient. The PAIS database is queried to search for feature pairs and retrieve corresponding feature values. The summary statistics for each image are combined in a separate program to create a single-feature vector for a patient. Queries for the mean, standard deviation, and covariance of feature calculations are supported through IBM DB2 Structured Query Language (SQL) queries with DB2’s built-in aggregation functions: the AVG, STDDEV, and COVARIANCE functions, respectively. An example of PAIS database query for the mean and covariance of three morphometry features, i.e. area, perimeter, and eccentricity, is shown in Fig. 3 where calculation_flat, and patient are two tables storing nuclear morphometry features and patient-slide relationships; pais_uid is the primary key that joins these two tables.
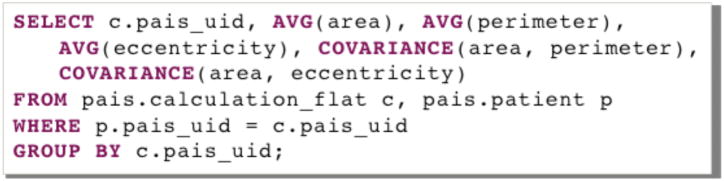
A SQL example is presented to query for mean and covariance nuclear feature vector for each patient in PAIS database.
With the efficient and expressive database query support on morphological signature computation, we are able to correlate nuclear morphometry with clinical outcomes and molecular characterizations and to produce results suggesting a possible relationship across nuclear morphometry, patient survival, and molecular data.
C. Multi-modal Data Correlation
Two methods are used for multi-modal data correlation. The first uses consensus cluster labels to partition patients into three groups and correlate nuclear morphometry signatures with response to treatment and patient survival. This analysis potentially reveals the clinical significance suggested by nuclear morphometry features. The second analysis investigates the relationship of consensus clusters with gene expression subtypes and genetic alterations. The hypergeometric distribution is used to calculate the probability of either a given expression subtype or genetic alteration group being enriched/depleted in a given consensus cluster. This analysis allows us to find those expression subtypes and genetic alteration groups significantly enriched or depleted in a cluster, suggesting a possible relationship between the phenotypic and genomic data of GBMs [16]. We present the hypergeometric probability density function f(x|T,S,K) as in Eq. (1) for x samples of a tumor subtype/genetic alteration group in a consensus cluster with K samples when S out of T samples are expected:
where T is the total population size; S is the number of samples in a given tumor type/genetic alteration group; K is the size of the samples in a given consensus cluster; and x is the number of samples of a given tumor type/genetic alteration group in the given consensus cluster containing K samples. The resulting over- and under-representation p-values can be computed as:
where X is the observed number of samples of a given tumor type/genetic alteration group within the given consensus cluster containing K samples.
IV. EXPERIMENTAL RESULTS
With the consensus clustering process, we grouped patients based on the patient-level nuclear morphometry signatures into three clusters, consisting of 70, 10, and 82 patients, respectively. More than 22 million neoplastic nuclei in 428 whole slides from 162 patients were analyzed with the aforementioned image-processing pipeline. We excluded nuclei crossing the tile borders from further analysis, as the number of such nuclei is so small when compared with the enormous number of nuclei completely contained by partitioned regions.
A. Response to Therapy and Survival Analysis
Summary nuclear feature vectors are computed with slides grouped by patients. These patients are further grouped into three consensus clusters based on nuclear morphometry. In Table 1, we present the p-values of the Log-Rank test [17] comparing patient survival to the three consensus clusters. The Log-Rank test between cluster two and three yields statistically significant difference in survival, with longer survivals for patients in cluster three. Additionally, the Kaplan-Meier plot for the three clusters of patients is shown in Fig. 4, where Area Under Curve (AUC) for cluster two (AUC = 296.06) is much smaller than that for clusters one (AUC = 2441.81), and three (AUC = 1302.79). This suggests that patients in cluster two have worse prognosis than those in cluster one and three, although this observation needs to be further validated with a larger number of samples. In Fig. 5, we present the Kaplan-Meier plots of three clusters of patients showing surivals of those treated with either standard and aggressive therapy. The resulting p-values of the Log-Rank tests with patient survivals with regard to response to therapy from cluster one, two and three are 0.00705, 0.158, and 0.000640, respectively. The results suggest that patients in cluster one and three show significantly favorable response to aggressive therapy compared to standard therapy. Cluster two contains a small number of patients and conclusions regarding response to therapy are limited.
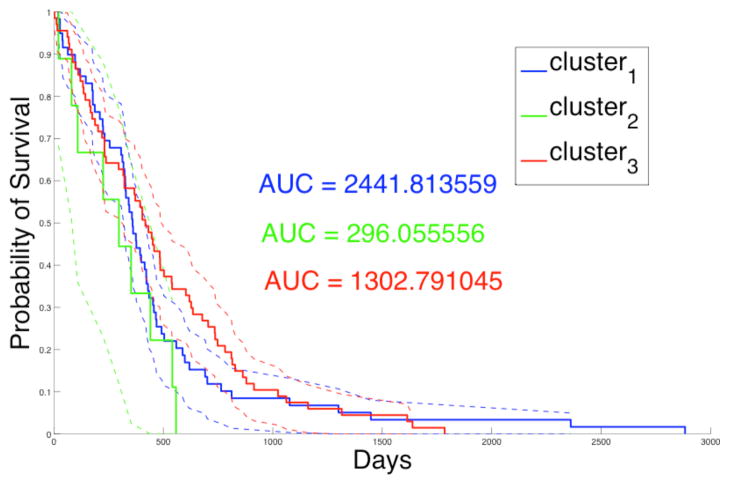
Kaplan-Meier plot for patients of three consensus clusters is presented with AUC values and 95% lower and upper confidence bounds.
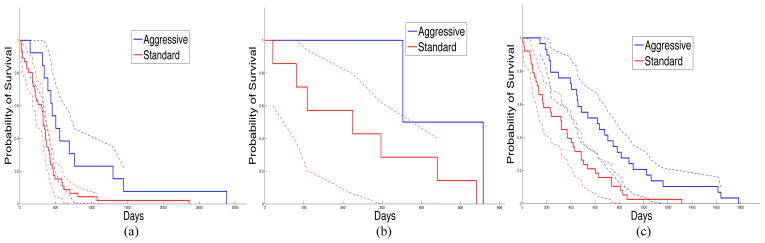
Kaplan-Meier plots for patients treated with aggressive (blue) and standard (red) therapy from (a) cluster 1, (b) cluster 2, and (c) cluster 3, are presented with 95% lower and upper confidence bounds.
TABLE I
We present P-values from log-rank test with survival data from patients of different nuclear morphometry clusters
| Consensus Cluster | Consensus Cluster(s) | P-value of Log-rank Test |
|---|---|---|
| 1 | (2, 3) | 0.322 |
| 2 | (1, 3) | 0.0719 |
| 3 | (1, 2) | 0.131 |
| 1 | 2 | 0.156 |
| 1 | 3 | 0.222 |
| 2 | 3 | 0.0437 |
B. Correlation with Phenotypic and Genotypic Data
We also investigated whether any of the morphometric clusters was characterized either by a specific gene expression subtype or genetic alteration. We therefore studied the enrichment/depletion relationship between phenotypic and genotypic data. After computing the p-values for over- and under-representations with tumor subtypes in the three consensus clusters, we find that mesenchymal samples are enriched in cluster one with an over-representation p-value of 0.0372. In Fig. 6, the genetic alteration profiles of samples in three clusters are presented for genes of interest for GBMs. With copy number variations, we observe that cluster one is enriched with EGFR amplification (p-value 0.0211) and CDKN2A deletion samples (p-value 0.00586). Cluster two is enrched with PTEN deletion samples with p-value of 0.0244. Additionally, CDKN2A deletion samples are depleted in cluster three with p-value of 0.00958. However, no specific mutations are found to be significantly correlated with the nuclear morphometry clusters.
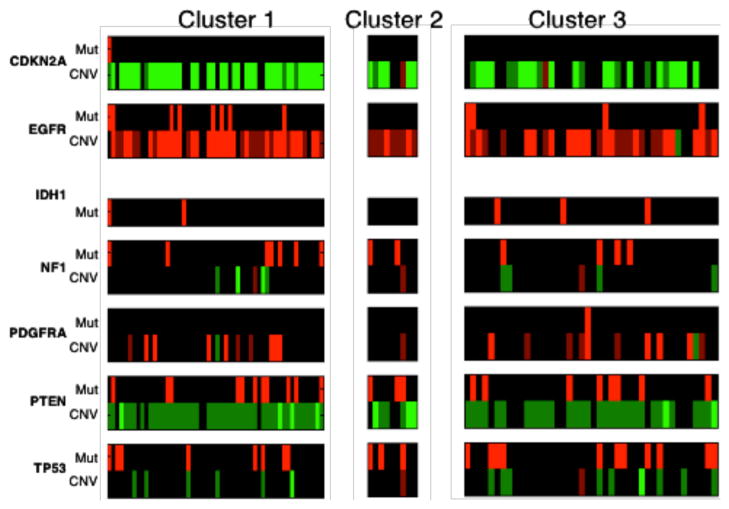
The genetic alteration profiles for three consensus clusters of samples are presented. For copy number data (denoted as CNV), light green, dark green, black, dark red, and light red represent homozygous deletion, hemizygous deletion, neutral change, gain, and high-level amplification. For somatic mutation data (denoted as Mut), red represents mutant.
V. Conclusions
In this letter, we present a large-scale multimodal data correlation study of GBM. Morphological characteristics derived from whole-slide microscopic images are correlated with clinical and molecular data. Results from these analyses revealed a significant survival difference between GBM patients based on the nuclear morphometry cluster of their tumor. This observation suggests a potential for predicting patient outcome based on nuclear morphometry. Our results also suggest that patients within specific nuclear morphometry clusters demonstrate differential therapeutic responses, as the patients in clusters 1 and 3 showed favorable response to aggressive therapy. In a future work, we plan to investigate morphometric features that are most predictive of molecular subtype and clinical behavior. These phenotypic features could then be incorporated into clinical diagnostics.
Acknowledgments
This work was supported by the NCI Contract HHSN261200800001E; TCGA Contract 29 x 55193; NIH 5R01LM009239-04; NHLBI R24 HL085343; and by the Clinical and Translational Science Awards program under PHS Grant UL1RR025008.
Footnotes
1https://wiki.nci.nih.gov/display/ISCRE
2Memorial Sloan-Kettering Cancer Center, http://www.mskcc.org/mskcc/, last access in Mar, 2011
3TCGA portal, http://cancergenome.nih.gov/, last access in Dec 2010
4PAIS wiki: - https://web.cci.emory.edu/confluence/display/PAIS/
Contributor Information
Jun Kong, Center for Comprehensive Informatics, Emory University, Atlanta, GA 30322, USA.
Lee A.D. Cooper, Center for Comprehensive Informatics, Emory University, Atlanta, GA 30322, USA.
Fusheng Wang, Center for Comprehensive Informatics, Emory University, Atlanta, GA 30322, USA.
David A. Gutman, Center for Comprehensive Informatics, Emory University, Atlanta, GA 30322, USA.
Jingjing Gao, Center for Comprehensive Informatics, Emory University, Atlanta, GA 30322, USA.
Candace Chisolm, Center for Comprehensive Informatics, Emory University, Atlanta, GA 30322, USA.
Ashish Sharma, Center for Comprehensive Informatics, Emory University, Atlanta, GA 30322, USA.
Tony Pan, Center for Comprehensive Informatics, Emory University, Atlanta, GA 30322, USA.
Erwin G. Van Meir, Department of Neurosurgery and Hematology and Medical Oncology, School of Medicine and Winship Cancer Institute, Emory University, Atlanta, GA 30322, USA.
Tahsin M. Kurc, Center for Comprehensive Informatics, Emory University, Atlanta, GA 30322, USA.
Carlos S. Moreno, Center for Comprehensive Informatics, Emory University, Atlanta, GA 30322, USA.
Joel H. Saltz, Center for Comprehensive Informatics, Emory University, Atlanta, GA 30322, USA.
Daniel J. Brat, Center for Comprehensive Informatics, Emory University, Atlanta, GA 30322, USA.
References
Full text links
Read article at publisher's site: https://doi.org/10.1109/tbme.2011.2169256
Read article for free, from open access legal sources, via Unpaywall:
https://europepmc.org/articles/pmc3292263?pdf=render
Citations & impact
Impact metrics
Citations of article over time
Article citations
A deep-learning-based model for assessment of autoimmune hepatitis from histology: AI(H).
Virchows Arch, 15 Jun 2024
Cited by: 0 articles | PMID: 38879691
Biomarkers of Tumor Heterogeneity in Glioblastoma Multiforme Cohort of TCGA.
Cancers (Basel), 15(8):2387, 20 Apr 2023
Cited by: 0 articles | PMID: 37190318 | PMCID: PMC10137245
An integrative web-based software tool for multi-dimensional pathology whole-slide image analytics.
Phys Med Biol, 67(22), 09 Nov 2022
Cited by: 2 articles | PMID: 36067783 | PMCID: PMC10039615
Deep learning features encode interpretable morphologies within histological images.
Sci Rep, 12(1):9428, 08 Jun 2022
Cited by: 10 articles | PMID: 35676395 | PMCID: PMC9177767
Segmentation and Classification in Digital Pathology for Glioma Research: Challenges and Deep Learning Approaches.
Front Neurosci, 14:27, 21 Feb 2020
Cited by: 29 articles | PMID: 32153349 | PMCID: PMC7046596
Go to all (35) article citations
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
An anatomic transcriptional atlas of human glioblastoma.
Science, 360(6389):660-663, 01 May 2018
Cited by: 299 articles | PMID: 29748285 | PMCID: PMC6414061
Bioinformatics and machine learning methodologies to identify the effects of central nervous system disorders on glioblastoma progression.
Brief Bioinform, 22(5):bbaa365, 01 Sep 2021
Cited by: 16 articles | PMID: 33406529
Machine-based morphologic analysis of glioblastoma using whole-slide pathology images uncovers clinically relevant molecular correlates.
PLoS One, 8(11):e81049, 13 Nov 2013
Cited by: 56 articles | PMID: 24236209 | PMCID: PMC3827469
Biobanking: An Important Resource for Precision Medicine in Glioblastoma.
Adv Exp Med Biol, 951:47-56, 01 Jan 2016
Cited by: 2 articles | PMID: 27837553
Review
Funding
Funders who supported this work.
NCATS NIH HHS (1)
Grant ID: UL1 TR000454
NCI NIH HHS (2)
Grant ID: HHSN261200800001E
Grant ID: HHSN261200800001C
NCRR NIH HHS (2)
Grant ID: UL1 RR025008
Grant ID: UL1 RR025008-01
NHLBI NIH HHS (2)
Grant ID: R24 HL085343-05
Grant ID: R24 HL085343
NIBIB NIH HHS (2)
Grant ID: P20 EB000591-03
Grant ID: P20 EB000591
NLM NIH HHS (5)
Grant ID: 5R01LM009239-04
Grant ID: R01 LM009239-04
Grant ID: R01 LM011119
Grant ID: R01 LM009239
Grant ID: R01 LM011119-01


