Abstract
Free full text

Pannexins, a family of gap junction proteins expressed in brain
Associated Data
Abstract
Database search has led to the identification of a family of proteins, the pannexins, which share some structural features with the gap junction forming proteins of invertebrates and vertebrates. The function of these proteins has remained unclear so far. To test the possibility that pannexins underlie electrical communication in the brain, we have investigated their tissue distribution and functional properties. Here, we show that two of these genes, pannexin 1 (Px1) and Px2, are abundantly expressed in the CNS. In many neuronal cell populations, including hippocampus, olfactory bulb, cortex and cerebellum, there is coexpression of both pannexins, whereas in other brain regions, e.g., white matter, only Px1-positive cells were found. On expression in Xenopus oocytes, Px1, but not Px2 forms functional hemichannels. Coinjection of both pannexin RNAs results in hemichannels with functional properties that are different from those formed by Px1 only. In paired oocytes, Px1, alone and in combination with Px2, induces the formation of intercellular channels. The functional characteristics of homomeric Px1 versus heteromeric Px1/Px2 channels and the different expression patterns of Px1 and Px2 in the brain indicate that pannexins form cell type-specific gap junctions with distinct properties that may subserve different functions.
Gap junctions are collections of intercellular channels that, in vertebrates, are formed by connexins, a multigene family of which 20 members have been identified in humans (1). It is generally accepted that gap junctions between neurons represent the anatomical substrate of electrical synapses (reviewed in refs. 2 and 3). Although the incidence of electrical coupling relative to chemical synapses in the adult is relatively low, several studies have demonstrated that different types of interneurons of the hippocampus and neocortex communicate by means of electrical synapses in a cell-specific manner (4-11). These observations suggest that this additional form of intercellular communication is more widespread than previously imagined and delineates independent networks of coupled cells.
Besides the undisputed role of chemical transmission in network oscillations, both computer simulations and electrophysiological recordings have recently emphasized a key role for electrical synapses in the generation of synchronous activity in the hippocampus and neocortex (6, 12-17). The identification of connexin36 (Cx36) as the main neuronal connexin expressed in several areas of the brain (18), suggested that it may be an important component of gap junctions involved in the synchronization of large-scale neuronal networks. This possibility has been directly tested in mice with a targeted ablation of Cx36, which exhibit impaired electrical coupling in several brain regions (15, 19-23). Loss of this gap-junction protein abolishes electrical coupling between hippocampal interneurons and disrupts γ-frequency network oscillations in vitro and in vivo (15, 24). The specificity of this impairment was indicated by the finding that high-frequency rhythms in hippocampal pyramidal cells are unaffected by the lack of Cx36 (15).
These observations raise two possibilities: either a different connexin is specifically deployed throughout the pyramidal cell network or, alternatively, another class of molecules expressed in the mammalian brain forms electrical junctions between pyramidal cells. The latter hypothesis has received theoretical support from the discovery, in the database, of a family of genes for which the name pannexin (Px) has been proposed (25). Because they share structural features with gap junction proteins of invertebrates and vertebrates, we investigated their tissue distribution and analyzed their ability to form functional channels.
Materials and Methods
Molecular Cloning and mRNA Distribution. cDNA clones were obtained by screening a rat hippocampal cDNA library, prepared from postnatal day (P)15 rats with [α-32P]end-labeled oligonucleotides, which were complementary to nucleotides 181-225 and 316-360 of the mouse Px1 ORF, and to nucleotides 199-243 and 334-378 of the human Px2 ORF (25). A probe for Px3 was generated by PCR by using the oligonucleotide pair derived from nucleotides 569-592 and 1059-1082 of the rat Px3 ORF, which was identified in the database. The tissue distribution of pannexin gene expression was investigated by reacting blots containing rat poly(A)+ RNA (Rat MTN blots I and II, catalog nos. 7764-1 and 7795-1, respectively; Clontech) with [α-32P]dCTP-labeled probes derived either from the entire ORF of Px1 and Px2, or with a fragment derived from the β-actin transcript (supplied along with the blots). Radioactive (26) and nonradioactive (27) in situ hybridization experiments were performed essentially as described. [α-35S]dATP-end-labeled oligonucleotides corresponded to nucleotides 181-225 and 334-378 of the mouse Px1 and human Px2 coding sequence, whereas the entire rat ORF was used to generate digoxygenin-labeled sense and antisense riboprobes.
Functional Expression in Xenopus Oocytes. The coding sequence of each pannexin was subcloned into the pBSxG expression vector (28). In vitro transcription, biochemical analysis, preparation of Xenopus oocytes, and RNA injection were performed as described elsewhere (29). Metabolic labeling of oocytes indicated that all three pannexin RNAs directed the synthesis of specific polypeptide bands, whose electrophoretic mobility was similar to that of the in vitro translated constructs (data not shown).
For physiological analysis, cells were injected with a total volume of 40 nl of either an antisense oligonucleotide (3 ng per cell) to suppress the endogenous Xenopus Cx38 (30), or a mixture of antisense plus the specified RNA (20-80 ng per cell). The ability of pannexins to form hemichannels was assessed in single oocytes 2-4 days after RNA injection, by using a two-electrode voltage clamp. To investigate whether Px1 and Px2 could functionally interact, oocytes were coinjected with Px1 RNA (40-80 ng per cell) together with the specified amounts of RNAs encoding either Px2 or the W77R mutation of human Cx26, which is devoid of functional activity (31). To analyze whether pannexins formed intercellular channels, oocytes were stripped of the vitelline envelope 1-2 days after RNA injection, and paired for 24-48 h before measuring junctional conductance with a dual-voltage clamp. The setup, hardware, and software used for electrophysiological measurements and data analysis were as described (29, 32).
Statistical Analysis. Results are shown as mean ± SEM. An independent experiment is defined as a series of data obtained with oocytes isolated from one animal. Comparisons between two populations of data were made by using Student's unpaired t test. P values of 0.01 or less were considered to be significant.
Results
Structure and Organization of the Pannexin Genes. Analysis of the cDNA sequences for Px1, Px2, and Px3 identified ORFs encoding proteins with calculated molecular masses of 48,072, 73,270 and 44,976 Da, respectively. The sequences of all three proteins predict, as for connexins, four transmembrane domains and cytoplasmic N and C termini. A hallmark of gap-junction-forming proteins is the presence of conserved, regularly spaced cysteine residues located on the two extracellular loops. Whereas the connexins contain three such residues, pannexins contain only two, thus resembling, in this respect, innexins, the invertebrate constituents of intercellular channels (33) (see also Fig. 5 A and B, which is published as supporting information on the PNAS web site). A comparison of the cDNAs to the mouse genomic sequence (obtained from the Ensembl database; www.ensembl.org) resulted in the determination of the exon-intron structure of the three mouse pannexin genes (Fig. 1A). Considerable variability was found in the organization and length of the three gene loci, the protein-coding regions could be assigned to five, three, and four exons, respectively, for the Px1, Px2, and Px3 genes.

Gene organization and mRNA expression in rodents. (A) The loci of the three pannexins in the mouse genome, indicating their exon (numbered boxed regions) and intron structure, are displayed. Within each exon, nucleotides contributing to the presumed protein sequence for each pannexin are shaded. (B) Northern blot analysis was performed on rat poly(A)+ RNA (lanes 1-16: adrenal gland, bladder, eye, spinal cord, thyroid, stomach, prostate, large intestine, testis, kidney, skeletal muscle, liver, lung, spleen, brain, and heart, respectively). The two filters were hybridized with probes for each of the three pannexins and exposed for 16 h at -70°C. The Px1 probe hybridized to a 2.2-kb mRNA that was detectable in several organs, including spinal cord and brain. The 3.5-kb Px2 was most abundant in spinal cord and brain and was also present in other organs. A less prominent 2.5-kb transcript was observed in some organs. Px3 mRNA was observed only in skin (data not shown).
Distribution of Pannexin mRNA. Northern blots indicated that Px1 and Px2 transcripts were coexpressed in many tissues, including eye, thyroid, prostate, kidney, liver, and CNS (Fig. 1B). Px2, in particular, was expressed at relatively higher levels in the brain and spinal cord (see Fig. 6, which is published as supporting information on the PNAS web site). By contrast, Px3 transcripts could only be detected in the skin, which was found, by RT-PCR, to be devoid of Px1 and Px2 mRNA (data not shown).
In situ hybridization studies demonstrated widespread expression of both transcripts in many brain regions, including cortex, striatum, olfactory bulb, hippocampus, thalamus, and cerebellum (Fig. 2 A and B). On closer inspection at the cellular level, a differential distribution of Px1 and Px2 mRNA was apparent. In hippocampus, for example, both Px1 and Px2 were expressed in the pyramidal cell layer and in individual neurons in the stratum oriens and stratum radiatum (Fig. 2 C and D). Based on their location, the scattered neurons (NeuN positive, data not shown) can be inferred to be GABAergic interneurons. By contrast, in the cerebellum, Px1 expressing cells were abundant in the white matter where Px2 expression was absent (Fig. 2 E and F; note, however, the high Px2 labeling in the Purkinje cell layer). The labeling of Px1 expressing cells in the white matter was not restricted to the cerebellum but was also observed in other white-matter structures (e.g., corpus callosum and fimbria fornix), which, similarly, were also devoid of Px2 expression (data not shown).
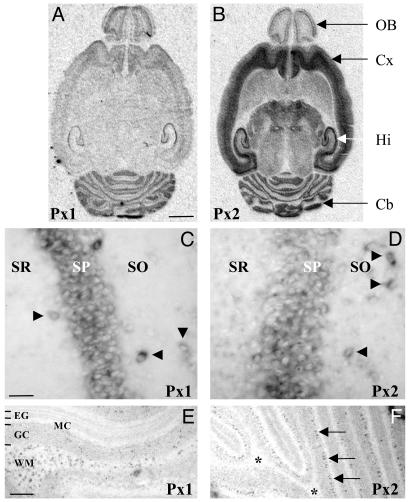
Expression of Px1 and Px2 mRNA in the brain. (A and B) The distribution of transcripts encoding Px1 and Px2 was determined by radioactive in situ hybridization in horizontal brain sections obtained from rats at P15. X-ray autoradiograms illustrate a partially overlapping expression profile and indicate that they are abundant in the olfactory bulb (OB), cortex (Cx), hippocampus (Hi), and cerebellum (Cb). No signal was detected in parallel competition experiments with an excess of unlabeled probe (data not shown). (Scale bar, 2.5 mm.) (C-F) Nonradioactive in situ hybridization demonstrating that high expression of Px1 (C) and Px2 (D) was detected in the stratum pyramidalis (SP) of the hippocampus and in individual neurons (arrowheads) in the stratum oriens (SO) and stratum radiatum (SR). By contrast, in the cerebellum, there was a strong labeling of Px1-expressing cells (E) in the white matter (WM) where Px2 expression was absent (F, *). Note, however, that the Px2 riboprobe strongly labeled cells in the Purkinje cell layer (F, arrows). No staining was obtained with sense probes (data not shown). EG, external granule cell layer; MC, molecular cell layer; GC, granule cell layer. [Scale bars, 50 μm (C and D) and 250 μm (E and F).]
Functional Expression in Single Xenopus Oocytes. Large, voltage-activated outward currents were consistently induced when oocytes expressing Px1 were stepped to voltages >-20 mV (Fig. 3 A and B). At large positive potentials, Px1 currents reached a peak within 30-60 msec of the imposition of the voltage step and then declined slowly; this rectification becoming more pronounced with increasing positive potentials. By contrast, neither Px2 nor Px3 induced membrane currents above those recorded in controls (Fig. 3 A and B). Furthermore, incubation of oocytes for 10-30 min in carbenoxolone completely suppressed Px1 currents (peak amplitudes at +60 mV were 1,189 ± 170 and 255 ± 47 nA for control medium and 30 μM carbenoxolone, respectively; n = 4) and this effect was fully reversible (peak amplitude at +60 mV after a 30-min recovery period in control medium was 963 ± 239 nA; n = 4).
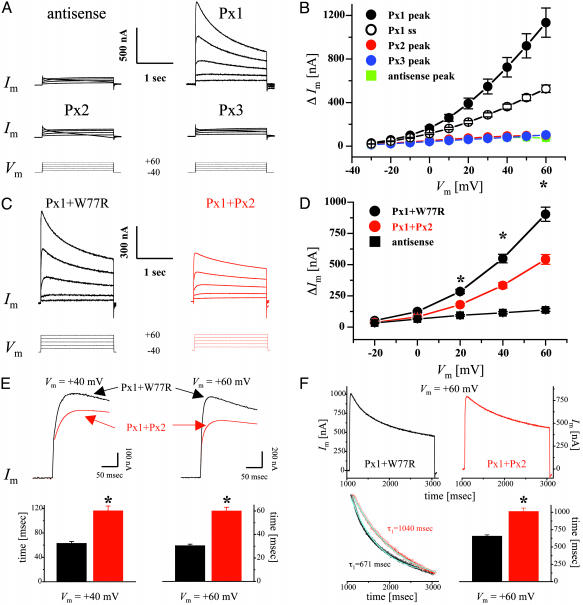
Functional expression of pannexins in single Xenopus oocytes. (A) Whole-cell membrane currents (Im) were measured from single oocytes coinjected with pannexin RNAs and an oligonucleotide antisense to Xenopus Cx38 (see Materials and Methods). Cells were initially clamped at a membrane potential (Vm) of -40 mV, and depolarizing steps lasting 2 sec were applied in 10-mV increments up to +60 mV (bottom traces). For clarity, representative traces are shown only in 20-mV increments. (B) Current-voltage relationships were determined for oocytes injected with either antisense oligonucleotides (blue) or Px1 (black), Px2 (red), and Px3 (green) RNAs plus antisense. Peak current values above holding currents (ΔIm) were calculated and plotted as a function of Vm. Mean values from Px1-injected cells were significantly different (P < 0.01) from those of control oocytes starting at a Vm of -10 mV. For Px1 steady-state currents (open circles), values recorded for 20 msec at the end of the pulse were averaged and plotted as above. Results are shown as mean ± SEM from at least eight independent experiments. Antisense, n = 45; Px1, n = 80; Px2, n = 46; Px3, n = 41. (C-F) Functional interaction of Px1 and Px2 proteins. Antisense-treated oocytes were coinjected with Px1 RNA together with equal amounts of RNAs encoding either Px2 (red traces) or the W77R mutation of human Cx26 (black traces), which is devoid of functional activity (31). (C and D) Coexpression of Px1 and Px2 reduced the amplitude of the outward currents induced by the depolarizing voltage steps (bottom traces). ΔIm recorded from Px1/Px2 (red circles) expressing oocytes was significantly less (*, P < 0.001) than that measured from Px1/W77R cells (black circles). Results are shown as mean ± SEM from four independent experiments. Antisense (n = 39); Px1/W77R (n = 60); Px1/Px2 (n = 67). (E) Px1/Px2 channels exhibit a delayed peak current time. Oocytes were depolarized to +40 mV (top left traces) and +60 mV (top right traces) from a holding potential of -40 mV. Peak currents were reached with a significant delay after the imposition of the voltage step (32 and 68 msec at +60 mV and 62 and 96 msec at +80 mV, for Px1/W77R and Px1/Px2, respectively). The lower panels show the mean ± SEM from three independent experiments for Px1/W77R (n = 45) and Px1/Px2 (n = 50); *, P < 0.001. (F) Px2 slows the kinetics of voltage-dependent closure of Px1 hemichannels. Cells were depolarized to +60 mV from a holding potential of -40 mV (Upper). Px1/Px2 hemichannels (red) gated more slowly than those formed by Px1/W77R (black). The time-dependent decline in Im was well fit by a first-order exponential decay function (Left Lower; cyanide line superposed to the rescaled current traces shown above). (Right Lower) The mean ± SEM from three independent experiments, for Px1/W77R (n = 44) and Px1/Px2 (n = 41); *, P < 0.001.
Because the in situ hybridization studies revealed coexpression of Px1 and Px2, our subsequent investigations entailed a more detailed functional analysis of these two proteins. To test whether they could form heteromeric channels, currents were recorded from oocytes coexpressing Px1 with Px2 (40-80 ng of RNA per cell) and were found to be significantly reduced with respect to those measured from cells that had been injected with the same amount of Px1 RNA (data not shown). To exclude the possibility that this behavior was merely due to overloading of the synthetic machinery given the difference in the total amount of RNA injected, oocytes were injected with equal amounts of RNA (40-80 ng each per cell) for Px1 and the W77R mutation of human Cx26, which is devoid of functional activity (31). These experiments showed a reduction in current amplitude, suggesting that Px1/Px2 form channels that are different from those composed of Px1 alone (Fig. 3 C and D). Given that Px2 expression in the brain appears to be much stronger than Px1, we also tested whether Px2 could simply function as a dominant-negative partner by coinjecting Px1 and Px2 RNAs at a 1:5 ratio (40:200 ng per cell for Px1:Px2, respectively). Current amplitudes recorded at +60 mV were similar, irrespective of whether Px1 and Px2 were injected at a ratio of 1:1 (591 ± 40 nA; n = 27) or of 1:5 (517 ± 45 nA; n = 20), further indicating that they both interact and form functionally heteromeric channels (see Fig. 7, which is published as supporting information on the PNAS web site). Interestingly, a decrease in current amplitude was not observed when voltage-activated currents were measured from oocytes receiving RNAs for Px1 and Px3, which are not coexpressed in rat tissues (data not shown). Moreover, after the imposition of a voltage step, Px1/Px2 channels reached peak currents with a significant delay, compared with Px1-expressing cells (Fig. 3E), which could result from slower opening or slower closing or both. Finally, analysis of the kinetics of channel closure at the more positive membrane potentials, revealed that currents recorded from Px1/Px2-expressing cells, presumably reflecting heteromeric hemichannels, gated more slowly than those measured from oocytes coinjected with Px1 and W77R RNA, presumably reflecting homomeric Px1 hemichannels (Fig. 3F).
Functional Expression in Paired Xenopus Oocytes. Px1 alone and in combination with Px2 induced the assembly of intercellular channels, whereas Px2 alone failed to do so (Fig. 4A). It should be noted that intercellular channels were consistently detected only from batches of oocytes in which a robust junctional conductance was recorded with homotypic pairs expressing either mouse Cx36 (Fig. 4A) or human Cx26 wild-type (data not shown), which served as positive controls. In this series of experiments, 20 of 23 Px1 pairs and 36 of 42 Px1/Px2 pairs were coupled. Both Px1 and Px1/Px2 pairs displayed a remarkable insensitivity to transjunctional potentials of opposite polarities. Thus, with a driving force ≤ ±60 mV, junctional currents varied linearly with voltage (Fig. 4B), whereas, at higher transjunctional potentials, the conductance of Px1/Px2 channels displayed only a very modest reduction (≈15%) of the initial values (Fig. 4C), which was similar to what was reported for crayfish septate junctions (34) and human Cx31.9 (32). Although it has been reported that junctional currents measured in insect cells are sensitive to changes in membrane potential (35), the relative voltage insensitivity of pannexin intercellular channels with polarization of one cell is a strong indication that polarization of both cells is not likely to affect significantly junctional conductance.

Functional expression of pannexims in paired oocytes. Cells were injected as described in Materials and Methods and were manually paired in homotypic configuration (same construct in both oocytes). (A) Both cells of a pair were initially clamped at -40 mV, and alternating pulses of ±10-20 mV were imposed to one cell. The current delivered to the cell clamped at -40 mV during the voltage pulse is equal in magnitude to the junctional current and can be divided by the voltage to yield the value of junctional conductance (Gj). Pairs of uninjected cells from the different batches of oocytes developed a variable level of junctional currents that exhibited the well known voltage-dependent gating of endogenous Cx38 (42), whereas antisense controls showed negligible coupling, indicating that endogenous currents had been suppressed. Oocyte pairs injected with Px1 either alone or in combination with Px2 (Px1+Px2) developed large junctional currents, whereas homotypic Px2-expressing pairs were uncoupled. Gj values recorded from oocytes expressing the neuronal mouse Cx36 (mCx36) are shown for comparison. Results are shown as the mean ± SEM of the indicated number of oocyte pairs from four to five independent experiments. (B) Px1 (black) and Px1/Px2 (red) intercellular channels exhibit a weak sensitivity to transjunctional voltage (Vj). Junctional currents (Ij) were recorded from oocyte pairs in response to depolarizing Vj steps (bottom traces) applied, from a holding potential of -40 mV, in 20-mV increments. (C) The plot shows the relationship of Vj to steady-state junctional conductance (Gjss), which was measured at the end of the Vj step and normalized to the values recorded at ± 20 mV; Px1/Px2 (•) and mCx36 (□). Data describing the Gj/Vj relationship were fit (smooth cyanide lines) to a Boltzmann equation, whose parameters were in agreement with those reported (43, 44). Results are shown as the mean ± SEM of 7-12 pairs (from four independent experiments) whose Gj was 3.2 ± 0.8 μS and 4.8 ± 1.1 μS for mCx36 and Px1/Px2, respectively. Because of the much larger nonjunctional currents that were present in Px1 homotypic pairs, reliable Gjss/Vj plots with the complete polarization paradigm were difficult to obtain.
Discussion
The data presented here demonstrate that pannexins, an unexplored family of innexin-like genes expressed in vertebrates, constitute an additional group of gap-junction channel-forming proteins. Electrical recordings from single and paired Xenopus oocytes provided evidence that Px1 forms both hemichannels and intercellular channels alone and in combination with Px2. Based on the high degree of coexpression of Px1 and Px2 in several brain areas, we speculate that they may represent the molecular correlate of a novel class of electrical synapses.
Gap-junction-based intercellular channels have been conserved throughout evolution as the basis of direct cell-cell communication, but vertebrates and invertebrates use two unrelated gene families to accomplish the same task (33). Recently, Panchin et al. (25) identified two innexin-like sequences in the human database and cloned an innexin-like cDNA fragment from mouse fetal brain. Multiple alignments with a representative group of innexins and connexins have revealed that pannexins and connexins belong to clearly distinct families (see Fig. 5 A and B). Comparison between pannexins and innexins, on the other hand, indicates that they are more closely related, and thus can be considered as part of a larger superfamily (25). In a preliminary report, it was shown that injection of RNA encoding the molluscan Px1 ortholog altered the specificity of electrical connections between different classes of neurons in the mollusk Clione limacina (36). However, no information was provided on the channel-forming ability of Px1.
Our experiments demonstrate for the first time, to our knowledge, that pannexins form intercellular channels, and thus may contribute to electrical communication in the nervous system. Pannexin hemichannels exhibit larger currents with increasing depolarizations, as reported for connexins, but peak currents were reached with 10- to 50-fold faster kinetics (29, 37). Pannexin intercellular channels show very weak voltage gating, although, within the range of transjunctional voltages that may occur after the generation of an action potential, this behavior was not different from that of Cx36, the major neuronal connexin. In addition, Px2 could not assemble homomeric channels, but reduced the amplitude and modified the voltage gating kinetics of Px1 hemichannels, suggesting that heteromeric Px1/Px2 channels were present. This behavior is similar to that of the Drosophila innexin, Dm-Inx3, which never forms homomeric channels, but has been shown in coexpression experiments to modify the electrophysiological properties of another innexin, Dm-Inx2 (38). Like a number of innexin and connexins that are unable to form homomeric channels (33, 38, 39), Px3 was not functional in the oocyte system, and, therefore, it is premature to speculate on its function. Considering the likely ability of pannexins to assemble homomeric as well as heteromeric channels, and given their differential distribution in the brain, it is likely that functionally distinct pannexin channels may underlie different tasks.
Electrical synchronization is increasingly recognized as an important mechanism that shapes the activity of neuronal circuits (2, 3, 15, 16, 19, 21, 24). Our studies indicate that pannexins are a family of channel-forming proteins that constitute an additional class of electrical synapses in the vertebrate CNS. In contrast to the neuronal Cx36, which, in the hippocampus, is expressed by interneurons but is conspicuously absent in adult pyramidal cells (15), Px1 and Px2 exhibit high expression levels in interneurons and pyramidal cells alike. Both computer simulations and electrophysiological studies have led to the proposal that axoaxonal coupling between pyramidal cells is necessary for the generation of ultrafast (150-200 Hz) oscillatory activity (14, 40). In addition, Traub et al. (41) have proposed that axonal gap-junction coupling between interneurons may also be relevant for the generation of hippocampal network bursts. Because the identity of these gap-junction proteins has remained elusive so far, it will be interesting to determine whether pannexins are the molecular correlate of these specific forms of interneuronal communication.
Acknowledgments
We thank U. Amtmann for technical assistance, Dr. L. Barrio (Hospital Ramón y Cajal, Madrid) for the gift of the pBSxG plasmid, Dr. M. Dubois-Dalcq (Institut Pasteur) for continuous support, Dr. W. Wisden (University of Heidelberg, Heidelberg) for critical reading of the manuscript, and Dr. R. Traub (State University of New York, Brooklyn, NY) for stimulating discussions. This work and costs of publication were supported by grants from the Schilling Foundation and the Deutsche Forschungsgemeinschaft, Collaborative Research Center 488 (H.M.), and from the Pasteur-Weizmann Joint Research Program (R.B.).
Notes
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: Pxn, pannexin n; Cxn, connexin n.
Data deposition: The sequence reported in this paper has been deposited in the GenBank database [accession nos. AJ557015 (rat Px1 cDNA), AJ557016 (rat Px2 cDNA), and AJ557017 (rat Px3 cDNA)].
References
Articles from Proceedings of the National Academy of Sciences of the United States of America are provided here courtesy of National Academy of Sciences
Full text links
Read article at publisher's site: https://doi.org/10.1073/pnas.2233464100
Read article for free, from open access legal sources, via Unpaywall:
https://europepmc.org/articles/pmc263867?pdf=render
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics

Discover the attention surrounding your research
https://www.altmetric.com/details/101801895
Smart citations by scite.ai
Explore citation contexts and check if this article has been
supported or disputed.
https://scite.ai/reports/10.1073/pnas.2233464100
Article citations
The Ethanolic Extract of Polygala paniculata L. Blocks Panx1 Channels and Reduces Ischemic Brain Infarct in a Dose- and Sex-Dependent Way.
Mol Neurobiol, 13 Sep 2024
Cited by: 0 articles | PMID: 39271622
Pannexin1 Mediates Early-Life Seizure-Induced Social Behavior Deficits.
ASN Neuro, 16(1):2371164, 16 Jul 2024
Cited by: 0 articles | PMID: 39024558 | PMCID: PMC11262470
Inhibition of astroglial hemichannels prevents synaptic transmission decline during spreading depression.
Biol Res, 57(1):39, 12 Jun 2024
Cited by: 0 articles | PMID: 38867288
Ca2+ permeation through C-terminal cleaved, but not full-length human Pannexin1 hemichannels, mediates cell death.
Proc Natl Acad Sci U S A, 121(25):e2405468121, 11 Jun 2024
Cited by: 3 articles | PMID: 38861601
Pannexin1 Channel-Mediated Inflammation in Acute Ischemic Stroke.
Aging Dis, 15(3):1296-1307, 07 May 2024
Cited by: 1 article | PMID: 37196132 | PMCID: PMC11081155
Review Free full text in Europe PMC
Go to all (501) article citations
Other citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
Nucleotide Sequences (3)
- (1 citation) ENA - AJ557017
- (1 citation) ENA - AJ557015
- (1 citation) ENA - AJ557016
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Pharmacological properties of homomeric and heteromeric pannexin hemichannels expressed in Xenopus oocytes.
J Neurochem, 92(5):1033-1043, 01 Mar 2005
Cited by: 322 articles | PMID: 15715654
Pannexin1 and Pannexin2 expression in the developing and mature rat brain.
Brain Res Mol Brain Res, 141(1):113-120, 06 Sep 2005
Cited by: 143 articles | PMID: 16143426
The mammalian pannexin family is homologous to the invertebrate innexin gap junction proteins.
Genomics, 83(4):706-716, 01 Apr 2004
Cited by: 316 articles | PMID: 15028292
Cell-cell communication beyond connexins: the pannexin channels.
Physiology (Bethesda), 21:103-114, 01 Apr 2006
Cited by: 178 articles | PMID: 16565476
Review

