Abstract
Free full text

Overview of the voltage-gated sodium channel family
Abstract
Selective permeation of sodium ions through voltage-dependent sodium channels is fundamental to the generation of action potentials in excitable cells such as neurons. These channels are large integral membrane proteins and are encoded by at least ten genes in mammals. The different sodium channels have remarkably similar functional properties, but small changes in sodium-channel function are biologically relevant, as underscored by mutations that cause several human diseases of hyperexcitability.
Voltage-gated sodium channels play an essential role in the initiation and propagation of action potentials in neurons and other electrically excitable cells such as myocytes and endocrine cells [1,2]. When the cell membrane is depolarized by a few millivolts, sodium channels activate and inactivate within milliseconds. Influx of sodium ions through the integral membrane proteins comprising the channel depolarizes the membrane further and initiates the rising phase of the action potential. The voltage-gated sodium channel is a large, multimeric complex, composed of an α subunit and one or more smaller β subunits [3]. The ion-conducting aqueous pore is contained entirely within the α subunit, and the essential elements of sodium-channel function - channel opening, ion selectivity and rapid inactivation - can be demonstrated when α subunits are expressed alone in heterologous cells. Coexpression of the β subunit is required for full reconstitution of the properties of native sodium channels, as these auxiliary subunits modify the kinetics and voltage-dependence of the gating (that is, opening and closing) of the channel. Although different sodium channels have broadly similar functional characteristics, small differences in properties do distinguish different isoforms and contribute to their specialized functional roles in mammalian physiology and pharmacology. Sodium channels that are not voltage-gated also exist in biology; the epithelial sodium channels (EnaC) of the EnaC/degenerin (DEG) gene family mediate sodium transport in epithelia and other cell types and are structurally unrelated to the voltage-gated sodium channels. We use the term 'sodium channel' here to mean 'voltage-gated sodium channel'.
Sodium channel subunits and structure
Sodium-channel proteins in the mammalian brain are composed of a complex of a 260 kDa α subunit in association with one or more auxiliary β subunits (β1, β2 and/or β3) of 33-36 kDa [3] (Figure (Figure1).1). Nine α subunits (Nav1.1-Nav1.9) have been functionally characterized, and a tenth related isoform (Nax) may also function as a Na+ channel. The primary sequence predicts that the sodium channel α subunit folds into four domains (I-IV), which are similar to one another and contain six α-helical transmembrane segments (S1-S6). In each of the domains, the voltage sensor is located in the S4 segments, which contain positively charged amino-acid residues in every third position. A re-entrant loop between helices S5 and S6 is embedded into the transmembrane region of the channel to form the narrow, ion-selective filter at the extracellular end of the pore. The wider intracellular end of the pore is formed by the four S6 segments. Small extracellular loops connect the transmembrane segments, with the largest ones connecting the S5 or S6 segments to the membrane re-entrant loop. Larger intracellular loops link the four homologous domains. Large amino-terminal and carboxy-terminal tail domains also contribute to the internal face of the sodium channel. This view of sodium channel architecture has been largely confirmed by biochemical, electrophysiological, and structural experiments.
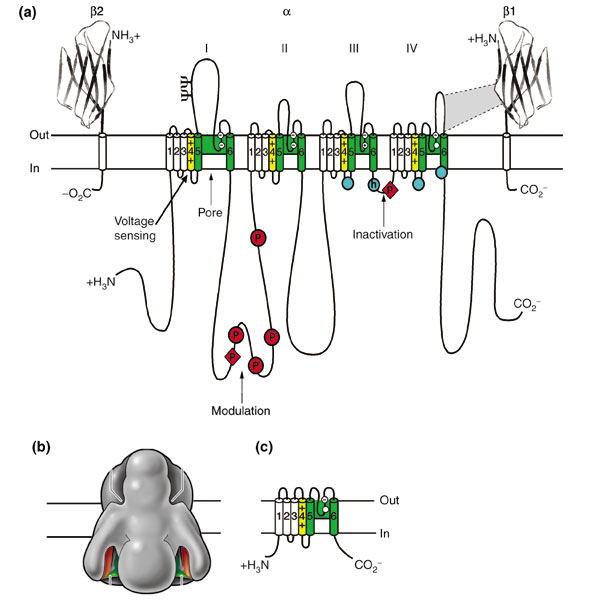
Structure of voltage-gated sodium channels. (a) Schematic representation of the sodium-channel subunits. The α subunit of the Nav1.2 channel is illustrated together with the β1 and β2 subunits; the extracellular domains of the β subunits are shown as immunoglobulin-like folds, which interact with the loops in the α subunits as shown. Roman numerals indicate the domains of the α subunit; segments 5 and 6 (shown in green) are the pore-lining segments and the S4 helices (yellow) make up the voltage sensors. Blue circles in the intracellular loops of domains III and IV indicate the inactivation gate IFM motif and its receptor (h, inactivation gate); P, phosphorylation sites (in red circles, sites for protein kinase A; in red diamonds, sites for protein kinase C); ψ, probable N-linked glycosylation site. The circles in the re-entrant loops in each domain represent the amino acids that form the ion selectivity filter (the outer rings have the sequence EEDD and inner rings DEKA). (b) The three-dimensional structure of the Nav channel α-subunit at 20 Å resolution, compiled from electron micrograph reconstructions. Adapted from [6]. (c) Schematic representation of NaChBac, the bacterial voltage-gated sodium channel.
A complete three-dimensional structure of the sodium channel is not yet available; however, limited but interesting structural information on the sodium channel is beginning to emerge. The solution structure of the inactivation gate on the intracellular loop between domains III and IV (Figure (Figure1)1) has been determined by nuclear magnetic resonance (NMR) [4]. This analysis predicts a rigid α-helical structure, preceded by two turns that position a key hydrophobic sequence motif (the IFM motif, Ile-Phe-Met) such that it can interact with and block the inner mouth of the pore (Figure (Figure2).2). The global fold of the sodium-channel pore region is predicted to be largely similar to that of potassium channels, whose structure has been solved by X-ray crystallography [5]. In this hypothetical structural view, the S5 and S6 helices of each sodium-channel domain are arrayed in four-fold symmetry around the aqueous ion-conduction pathway and the pore is lined mostly by S6 segments and the re-entrant loops that form the ion-selectivity filter. Direct structural determination at 19 Å resolution by cryo-electron microscopy and image reconstruction techniques [6] show sodium channels as having a bell-like shape (when viewed from the side), with a four-fold symmetry of transmembrane masses and large intracellular and extracellular masses through which several inlets and outlets allow aqueous access (Figure (Figure1b).1b). Unexpectedly, this image of sodium channels predicts a central pore that does not directly connect the intracellular and extracellular spaces but instead splits into four branches. Moreover, it suggests that there are four peripherally located transmembrane pores of unknown function, possibly acting as gating pores for voltage-sensor movement.
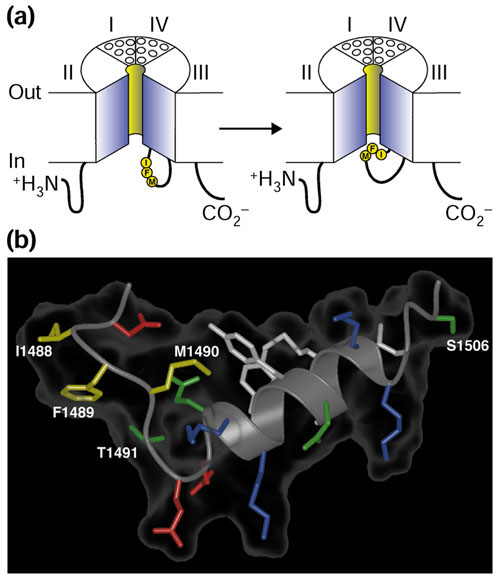
Mechanism of inactivation of sodium channels. (a) The hinged-lid mechanism. The intracellular loop connecting domains III and IV of the sodium channel is depicted as forming a hinged lid with the critical phenylalanine (F1489) within the IFM motif shown occluding the mouth of the pore during the inactivation process. The circles represent the transmembrane helices. (b) Three-dimensional structure of the central segment of the inactivation gate, as determined by multidimensional NMR. Side chains of the critical IFM motif residues (I1488, F1489 and M1490) are shown in yellow, and those of T1491, which is important for inactivation, and S1506, which is a protein-kinase-C-dependent phosphorylation site, are also indicated. Adapted from [4].
Comparison of the primary structures of the auxiliary β subunits with those of other proteins reveals a probable structural relationship to the family of proteins that contain immunoglobulin-like folds [7]. The extracellular domains of these type I single-membrane-spanning proteins are predicted to fold in a similar manner to the myelin protein Po, whose immunoglobin-like fold is known to be formed by a sandwich of two β sheets held together by hydrophobic interactions [8].
Evolutionary history and sodium channel genes
Sodium channels were the first members of the ion channel superfamily to be discovered; the superfamily also includes voltage-gated K+ channels, voltage-gated Ca2+ channels, Trp-related channels (a diverse family permeable to various cations) and cyclic-nucleotide-gated channels [2]. But in evolution, the sodium channel family is the most recent of the voltage-gated ion channels to have arisen, having evolved from similarly structured Ca2+ channels that contain four homologous domains. Ca2+ channels, in turn, probably arose by two rounds of gene duplication from the ancestors of K+ and cyclic-nucleotide-gated channels, which form functional channels as tetramers of single-domain subunits. The appearance of four-domain sodium channels coincided with evolution of metazoans that had specialized neurons [2,9]. Although 'classical' four-domain sodium channels have not been identified from prokaryotes, a recent report has identified a bacterial sodium channel in (Figure (Figure1c)1c) the salt-loving bacterium Bacillus halodurans [10]. This single-domain voltage-dependent sodium channel, which also has sequence and pharmacological characteristics resembling those of Ca2+ channels, opens up the intriguing possibility that it is similar to the ancestral single-domain channel from which the Ca2+ and Na+ channels have arisen.
Voltage-dependent sodium channel genes have been identified in a variety of animals, including flies, leeches, squid and jellyfish, as well as mammalian and non-mammalian vertebrates [9,11]. The biophysical properties, pharmacology, gene organization, and even intron splice sites of invertebrate sodium channels are largely similar to the mammalian sodium channels, adding further support to the idea that the primordial sodium channel was established before the evolutionary separation of the invertebrates from the vertebrates [12].
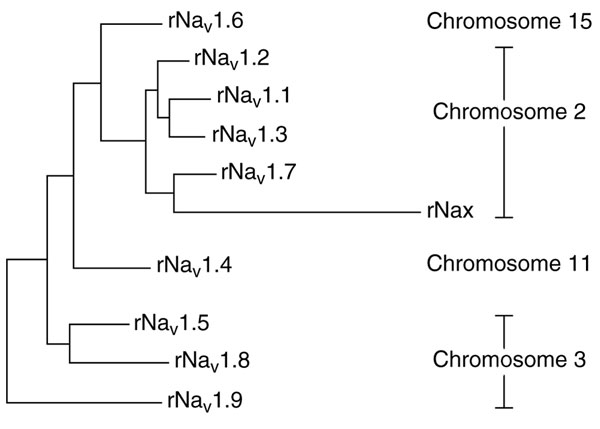
Vertebrate α-subunit genes, especially those encoding the human and rodent sodium channels, are the best characterized to date. These sodium channels are greater than 50% identical in amino-acid sequence in the transmembrane and extracellular domains and are considered to be of one subfamily. According to the convention of the International Union of Pharmacologists, the nomenclature of sodium channels (for example, Nav1.1) consists of the chemical symbol of the principal permeating ion (Na) and the principal physiological regulator (voltage, subscript) followed by a number indicating the gene subfamily and a decimal that separates the number assigned to specific channel isoforms [13]. At least 20 exons encode each of the nine sodium channel α-subunit proteins. In an evolutionary analysis, the genes fall into four groups, and members of each group map to the same chromosome segment (Figure (Figure3).3). The four chromosome segments containing sodium channel α-subunit genes are paralogous, and each also contains a cluster of Hox genes, which encode transcription factors involved in the control of developmental patterning [12]. Genes encoding sodium channels Nav1.1, Nav1.2, Nav1.3, and Nav1.7 are located on chromosome 2 in both human and mouse, and these channels share similarities in sequence, biophysical characteristics, the ability to be blocked by nanomolar concentrations of the neurotoxin tetrodotoxin, and broad expression in neurons [13]. A second cluster of genes encoding the channels Nav1.5, Nav1.8, and Nav1.9 is located on human chromosome 3p21-24 and to the orthologous region of chromosome 3 in mouse [13]. Although they are approximately 75% identical in amino-acid sequence to the group of channels on chromosome 2, these closely related sodium channels have amino-acid changes that confer varying degrees of tetrodotoxin resistance. In Nav1.5, the principal cardiac isoform, a single amino-acid change from phenylalanine to cysteine in the pore region of domain I is responsible for 200-fold reduction in tetrodotoxin sensitivity [14] relative to the channels encoded on chromosome 2. At the corresponding position in channels Nav1.8 and Nav1.9 the residue is serine, and this change results in even greater resistance to tetrodotoxin [15]. The latter two channels are preferentially expressed in peripheral sensory neurons [16,17]. The other two isoforms, Nav1.4 (which is expressed in skeletal muscle) and Nav1.6 (which is highly abundant in the central nervous system), have greater than 85% sequence identity and similar functional properties to the chromosome-2-encoded channels, including tetrodotoxin sensitivity in the nanomolar concentration range. Despite these similarities, phylogenetic analysis by parsimony suggests a more distant evolutionary relationship from the chromosome-2-encoded channels (Figure (Figure3),3), consistent with their distinct chromosomal localizations - Nav1.4 on human chromosome 11 (mouse chromosome 17) and Nav1.6 on human chromosome 15 (mouse 12) [11,13].
A tenth sodium channel, Nax, whose gene is located near the sodium channels of chromosome 2, is more divergent in sequence and has been suggested to be evolutionarily more distant [11]. Key differences in functionally important regions of the voltage sensor and the inactivation gate, and lack of detectable function upon expression in heterologous cells, suggest that Nax may not function as a voltage-dependent sodium channel. Indeed, Nax knockout mice have deficits in sensing of extracellular salt levels in the hypothalamus [18], indicating that Nax may act as a salt sensor rather than as a voltage-activated channel.
The chromosomal locations of the β subunits are known in humans - the β1 gene maps to 19q13, whereas β2 and β3 are located on 11q22-23. Although β3 appears to be more similar to β1 in sequence and in function, it appears to be closer phylogenetically to β2 [19].
Molecular basis of function
Classical work by Hodgkin and Huxley [1] defined the three key features of sodium channels: voltage-dependent activation, rapid inactivation, and selective ion conductance. Building on this foundation, more recent structure-function studies using molecular, biochemical and electrophysiological techniques have provided us with a good understanding of the molecular basis of sodium-channel function. Critical to this were the neurotoxins tetrodotoxin and saxitoxin, whose pore-blocking properties were exploited to purify the sodium-channel proteins and to reveal the amino-acid residues involved in the outer pore and in the selectivity filter. The outer pore is formed by the re-entrant loops between transmembrane segments S5 and S6 of each domain. Two important amino acids in analogous positions in all four domains are thought to form the negatively charged outer and inner rings that serve as a receptor site for pore-blockers and the selectivity filter (Figure (Figure1a).1a). Mutations of these residues have significant effects on binding of tetrodotoxin and saxitoxin [20], and also have marked effects on the selectivity of permeation of organic and inorganic monovalent cations through the sodium channel [21,22]. The most convincing evidence comes from a study by Heinemann et al. [23], who produced a calcium-selective sodium channel by mutating the inner ring residues (DEKA in the single-letter amino-acid code) to their counterparts in calcium channels (EEEE).
As is the case for other voltage-gated ion channels, the voltage-dependence of activation of sodium channels derives from the outward movement of charged residues as a consequence of an altered electric field across the membrane [1]. The S4 segments of each homologous domain serve as the voltage sensors for activation. They are composed of repeated motifs of one positively charged residue followed by two hydrophobic residues, potentially creating a helical arrangement of positive charges through the membrane. Upon depolarization, the outward movement of the S4 helices and their concurrent rotation initiates a conformational change that opens the sodium-channel pore. This 'sliding helix' [24] or 'helical screw' [25] model is supported by strong evidence [3]. For example, neutralization of the key positively charged residues in S4 markedly reduces the voltage-dependence of gating [26]. The outward gating movement of the S4 segments has also been detected directly by the fact that, when some residues in these helices are substituted with cysteines, extracellular sulfhydryl reagents react with them only after channel activation [27-29].
Inactivation of the sodium channel is a critical process that occurs within milliseconds of channel opening. In the generally accepted model of this process, the highly conserved intracellular loop that connects domains III and IV serves as an inactivation gate, much like a hinged lid, that binds to the intracellular pore of the channel to inactivate it (Figure (Figure2a).2a). Intracellular perfusion of proteases [30] or intracellular application of antibodies that recognize this loop, but not antibodies to other intracellular structures [31], prevents fast inactivation. The 'latch' of the inactivation gate is formed by three key hydrophobic residues (IFM; Figure Figure2),2), and peptides containing this motif can restore inactivation to sodium channels that have a mutated inactivation gate [32]. Mutations of the key phenylalanine residue (Phe1489) to various hydrophilic residues impair inactivation to varying degrees. Furthermore, if it is replaced with a cysteine residue, covalent modification of the cysteine is prevented when the inactivation gate is closed [33]. Structural determination and NMR analysis of the core portion of the inactivation gate reveal a rigid α-helical structure flanked by the IFM motif [4] (Figure (Figure2b).2b). In this structure, the side-chain of Phe1489 points away from the core of the peptide on the same face as a nearby threonine (Thr1491), another critical residue for inactivation [33]. In contrast to the short IFM motif that forms the inactivation gate, the receptor for the inactivation gate on the body of the channel may be composed of multiple hydrophobic residues near the intracellular mouth of the pore. Scanning mutagenesis experiments implicate hydrophobic residues in intracellular loops S4-S5 of domains III and IV, as well as the intracellular end of the S6 transmembrane segments of these domains, as components of the inactivation gate receptor [3].
The exciting discovery of a bacterial sodium channel (NaChBac; Figure Figure1c),1c), consisting of a single domain of six transmembrane α-helical segments [10], potentially provides new tools for the study of the sodium channel structure-function relationships. The NaChBac channel apparently forms a homotetramer that is a functional voltage-gated sodium channel. It has a 100-fold slower rate of inactivation than the Nav channels (because it has no equivalent of the inactivation gate). Furthermore, its sodium selectivity filter appears to be symmetric, formed by four glutamate residues in key positions in all four pore loops, resembling that of the Ca2+ channel. This bacterial sodium channel will provide a simpler framework than the mammalian channels for mutagenesis experiments to test hypotheses regarding the structure and function of voltage-gated sodium channels. In addition, the small size of the channel may make it easier to obtain crystal-structure information, as has been the case for the pore region of the bacterial potassium channel (KcsA) [5].
Localization, modulation, and channelopathies
In addition to the differences in cellular and tissue expression, mammalian sodium channels also have differential expression profiles during development and different subcellular localizations, consistent with a distinct role for each channel in mammalian physiology. In rodents, Nav1.3 is highly expressed in fetal nervous tissues, whereas Nav1.1, Nav1.2, and Nav1.6 are abundant in the adult central nervous system. Generally, Nav1.1 and Nav1.3 are localized to the soma of the neuron, where they may control neuronal excitability through integration of synaptic impulses to set the threshold for action potential initiation and propagation to the dendritic and axonal compartments. Evidence from immunocytochemical experiments indicates that Nav1.2 is expressed in unmyelinated axons, where it conducts the action potential [34]. During development, Nav1.6 has been shown to replace Nav1.2 in maturing nodes of Ranvier, the gaps in the myelin sheaths of myelinated axons where action potential conduction takes place [35,36]. Nav1.1, and Nav1.6 are also significantly expressed in the peripheral nervous system (PNS), but the sodium channels that are the most abundantly expressed in the PNS are the three isoforms that have been cloned from sympathetic and dorsal root ganglion neurons, namely Nav1.7, Nav1.8 and Nav1.9. Of these, Nav1.7 is broadly expressed in the PNS and appears to be localized to axons, where it may function in initiating and conducting the action potential [37]. More restricted expression patterns are observed for Nav1.8 and Nav1.9; these channels are differentially expressed in small sensory neurons of the dorsal root and trigeminal ganglia, where they have a key role in the perception of pain [16,38]. Finally, Nav1.4 and Nav1.5 are muscle sodium channels that control the excitability of the skeletal and cardiac myocytes, respectively. Nav1.5 is transiently expressed in developing skeletal muscle but is replaced by Nav1.4 in the adult [19].
Sodium-channel function is known to be regulated in neurons in response to physiological signals (reviewed in [39]). The neurotransmitter dopamine has been shown to modify the firing properties and the input-output properties of medium spiny neurons of the ventral and dorsal striatum, acting through D1-like dopamine receptors and activation of the cAMP pathway to reduce sodium currents and the generation of action potentials. Biochemical studies have shown that purified sodium channels are phosphorylated by the cAMP-dependent protein kinase A at multiple sites in the intracellular loop between domains I and II [39]. In another example of modulation of sodium channels by neural signals, activation of muscarinic acetylcholine receptors by the neurotransmitter acetylcholine inhibits the intrinsic bursts of firing activity of hippocampal CA1 neurons. This effect is mediated in part by protein-kinase-C-dependent phosphorylation of a highly conserved residue (Ser1506 of Nav1.2) in the inactivation gate, which slows sodium-channel inactivation and reduces the peak sodium currents in the neurons. Additional phosphorylation of sites in intracellular loop I-II also influences regulation of sodium channels by protein kinase C [39]. Moreover, activation of tyrosine kinase pathways has also been implicated in sodium-channel modulation [40], as has converse regulation by a receptor tyrosine phosphatase, RPTPβ [41]. Additional modulation of sodium channels by other second messenger systems, including the G-protein subunits Gβγ [42] and calcium/calmodulin [43], is also observed in transfected mammalian cells.
Modulation of sodium currents is undoubtedly important in vivo, although specific deficits that directly affect sodium channel regulation are not yet known. Mutations that subtly alter sodium channel function can lead to human diseases of hyperexcitability. Remarkably, some of these cause changes of only a few percent of the persistent sodium current that results from incomplete inactivation of the mutant sodium channels [44]. Multiple mutations of Nav1.4 that yield hyperactive skeletal sodium channels have been shown to cause hyperkalemic periodic paralysis, a disorder of muscle weakness aggravated by high serum levels of potassium, and paramyotonia congenital, a syndrome of cold- and exercise-induced muscle stiffness [44]. Mutations of Nav1.5 that impair inactivation cause inherited long QT syndrome type 3 and Brugada syndrome [45], both of which lead to an increased risk of ventricular arrhythmias. Recently, a type of epilepsy called generalized epilepsy with febrile seizure plus (GEFS+) type II has been shown to result from gain-of-function mutations of Nav1.1 that enhance the persistent inward sodium current in brain neurons [46,47]. Mutations of the human sodium channel β1 subunit that modify its gating also cause epilepsy, in this case GEFS+ type I [48]. The absolute necessity for intact sodium-channel function in normal physiology is underscored by the finding that knockout mice with specific ablations of Nav1.2, Nav1.5 or Nav1.6 die prematurely [49-51]. In contrast, mutant mice lacking Nav1.8 are viable but have defects in the response to pain [16]. The use of these and other mouse models in which specific sodium channel genes have been targeted should help to delineate the significance of sodium-channel function and its regulation in physiological contexts. The recent resurgence of sodium-channel research, in addition to the large arsenal of information from previous studies, should provide us with a better understanding of the fundamental role of sodium-channel function and dysfunction in excitable cells and may guide us to effective therapies.
References
- Hodgkin AL, Huxley AF. A quantitative description of membrane current and its application to induction and excitation in nerve. J Physiol. 1952;117:500–544. [Abstract] [Google Scholar]
- Hille B. Ion Channels of Excitable Membranes. Sunderland, MA: Sinauer Associates Inc; 2001. [Google Scholar]
- Catterall WA. From ionic currents to molecular mechanisms: the structure and function of voltage-gated sodium channels. Neuron. 2000;26:13–25. [Abstract] [Google Scholar]
- Rohl CA, Boeckman FA, Baker C, Scheuer T, Catterall WA, Klevit RE. Solution structure of the sodium channel inactivation gate. Biochemistry. 1999;38:855–861. [Abstract] [Google Scholar]
- Doyle DA, Cabral JM, Pfuetzner RA, Gulbis JM, Cohen SL, Chait BT, MacKinnon R. The structure of the potassium channel: molecular basis of K+ conduction and selectivity. Science. 1988;280:69–77. [Abstract] [Google Scholar]
- Sato C, Ueno Y, Asai K, Takahashi K, Sato M, Engel A, Fujiyoshi Y. The voltage-sensitive sodium channel is a bell-shaped molecule with several cavities. Nature. 2001;409:1047–1051. [Abstract] [Google Scholar]
- Isom LL, Ragsdale DS, De Jongh KS, Westenbroek RE, Reber BF, Scheuer T, Catterall WA. Structure and function of the beta 2 subunit of brain sodium channels, a transmembrane glycoprotein with a CAM motif. Cell. 1995;83:433–442. [Abstract] [Google Scholar]
- Shapiro L, Doyle JP, Hensley P, Colman DR, Hendrickson WA. Crystal structure of the extracellular domain from P0, the major structural protein of peripheral nerve myelin. Neuron. 1996;17:435–449. [Abstract] [Google Scholar]
- Anderson PAV, Greenberg RM. Phylogeny of ion channels: clues to structure and function. Comp Biochem Physiol B Biochem Mol Biol. 2001;129:17–28. [Abstract] [Google Scholar]
- Ren D, Navarro B, Xu H, Yue L, Shi Q, Clapham DE. A prokaryotic voltage-gated sodium channel. Science. 2001;294:2372–2375. [Abstract] [Google Scholar]
- Goldin AL. Evolution of voltage-gated Na+ channels. J Exp Biol. 2002;205:575–584. [Abstract] [Google Scholar]
- Plummer NW, Meisler MH. Evolution and diversity of mammalian sodium channel genes. Genomics. 1999;57:323–331. [Abstract] [Google Scholar]
- Goldin AL, Barchi RL, Caldwell JH, Hofmann F, Howe JR, Hunter JC, Kallen RG, Mandel G, Meisler MH, Netter YB, et al. Nomenclature of voltage-gated sodium channels. Neuron. 2000;28:365–368. [Abstract] [Google Scholar]
- Satin J, Kyle JW, Chen M, Cribbs LL, Fozzard HA, Rogart RB. A mutant of TTX-resistant cardiac sodium channels with TTX-sensitive properties. Science. 1992;256:1202–1205. [Abstract] [Google Scholar]
- Sivilotti L, Okuse K, Akopian AN, Moss S, Wood JN. A single serine residue confers tetrodotoxin insensitivity on the rat sensory-neuron-specific sodium channel SNS. FEBS Lett. 1997;409:49–52. [Abstract] [Google Scholar]
- Akopian AN, Souslova V, England S, Okuse K, Ogata N, Ure J, Smith A, Kerr BJ, McMahon SB, Boyce S, et al. The tetrodotoxin-resistant sodium current SNS has a specialized function in pain pathways. Nat Neurosci. 1999;2:541–548. [Abstract] [Google Scholar]
- Dib-Hajj S, Black JA, Cummins TR, Waxman SG. NaN/Nav1.9: a sodium channel with unique properties. Trends Neurosci. 2002;25:253–259. [Abstract] [Google Scholar]
- Hiyama TY, Watanabe E, Ono K, Inenaga K, Tamkun MM, Yoshida S, Noda M. Nax channel involved in CNS sodium-level sensing. Nat Neurosci. 2002;5:511–512. [Abstract] [Google Scholar]
- Goldin AL. Resurgence of sodium channel research. Annu Rev Physiol. 2001;63:871–894. [Abstract] [Google Scholar]
- Noda M, Suzuki H, Numa S, Stuhmer W. A single point mutation confers tetrodotoxin and saxitoxin insensitivity on the sodium channel II. FEBS Lett. 1989;259:213–216. [Abstract] [Google Scholar]
- Schlief T, Schonherr R, Imoto K, Heinemann SH. Pore properties of rat brain II sodium channels mutated in the selectivity filter domain. Eur Biophys J. 1996;25:75–91. [Abstract] [Google Scholar]
- Sun YM, Favre I, Schild L, Moczydlowski E. On the structural basis for size-selective permeation of organic cations through the voltage-gated sodium channel. Effect of alanine mutations at the DEKA locus on selectivity, inhibition by Ca2+ and H+, and molecular sieving. J Gen Physiol. 1997;110:693–715. [Europe PMC free article] [Abstract] [Google Scholar]
- Heinemann SH, Terlau H, Stuhmer W, Imoto K, Numa S. Calcium channel characteristics conferred on the sodium channel by single mutations. Nature. 1992;356:441–443. [Abstract] [Google Scholar]
- Catterall WA. Voltage-dependent gating of sodium channels: correlating structure and function. Trends Neurosci. 1986;9:7–10. [Google Scholar]
- Guy HR, Seetharamulu P. Molecular model of the action potential sodium channel. Proc Natl Acad Sci USA. 1986;83:508–512. [Europe PMC free article] [Abstract] [Google Scholar]
- Stuhmer W, Conti F, Suzuki H, Wang XD, Noda M, Yahagi N, Kubo H, Numa S. Structural parts involved in activation and inactivation of the sodium channel. Nature. 1989;339:597–603. [Abstract] [Google Scholar]
- Yang NB, Horn R. Evidence for voltage-dependent S4 movement in sodium channels. Neuron. 1995;15:213–218. [Abstract] [Google Scholar]
- Yang NB, George AL, Jr, Horn R. Molecular basis of charge movement in voltage-gated sodium channels. Neuron. 1996;16:113–122. [Abstract] [Google Scholar]
- Cha A, Ruben PC, George AL, Jr, Fujimoto E, Bezanilla F. Voltage sensors in domains III and IV, but not I and II, are immobilized by Na+ channel fast inactivation. Neuron. 1999;22:73–87. [Abstract] [Google Scholar]
- Armstrong CM. Sodium channel and gating currents. Physiol Rev. 1981;61:644–682. [Abstract] [Google Scholar]
- Vassilev P, Scheuer T, Catterall WA. Inhibition of inactivation of single sodium channels by a site-directed antibody. Proc Natl Acad Sci USA. 1989;86:8147–8151. [Europe PMC free article] [Abstract] [Google Scholar]
- Eaholtz G, Scheuer T, Catterall WA. Restoration of inactivation and block of open sodium channels by an inactivation gate peptide. Neuron. 1994;12:1041–1048. [Abstract] [Google Scholar]
- Kellenberger S, Scheuer T, Catterall WA. Movement of the Na+ channel inactivation gate during inactivation. J Biol Chem. 1996;271:30971–30979. [Abstract] [Google Scholar]
- Westenbroek RE, Merrick DK, Catterall WA. Differential subcellular localization of the RI and RII Na+ channel subtypes in central neurons. Neuron. 1989;3:695–704. [Abstract] [Google Scholar]
- Boiko T, Rasband MN, Levinson SR, Caldwell JH, Mandel G, Trimmer JS, Matthews G. Compact myelin dictates the differential targeting of two sodium channel isoforms in the same axon. Neuron. 2001;30:91–104. [Abstract] [Google Scholar]
- Kaplan MR, Cho MH, Ullian EM, Isom LL, Levinson SR, Barres BA. Differential control of clustering of the sodium channels Nav1.2 and Nav1.6 at developing CNS nodes of Ranvier. Neuron. 2001;30:105–119. [Abstract] [Google Scholar]
- Toledo-Aral JJ, Moss BL, He ZJ, Koszowski AG, Whisenand T, Levinson SR, Wolf JJ, Silos-Santiago I, Halegoua S, Mandel G. Identification of PN1, a predominant voltage-dependent sodium channel expressed principally in peripheral neurons. Proc Natl Acad Sci USA. 1997;94:1527–1532. [Europe PMC free article] [Abstract] [Google Scholar]
- Black JA, Cummins TR, Plumpton C, Chen YH, Hormuzdiar W, Clare JJ, Waxman SG. Upregulation of a silent sodium channel after peripheral, but not central, nerve injury in DRG neurons. J Neurophysiol. 1999;82:2776–2785. [Abstract] [Google Scholar]
- Cantrell AR, Catterall WA. Neuromodulation of Na+ channels: an unexpected form of cellular plasticity. Nat Rev Neurosci. 2001;2:397–407. [Abstract] [Google Scholar]
- Hilborn MD, Vaillancourt RR, Rane SG. Growth factor receptor tyrosine kinases acutely regulate neuronal sodium channels through the src signaling pathway. J Neurosci. 1998;18:590–600. [Abstract] [Google Scholar]
- Ratcliff CF, Qu Y, McCormick KA, Tibbs VC, Dixon JE, Scheuer T, Catterall WA. A sodium channel signaling complex: modulation by associated receptor protein tyrosine phosphatase beta. Nat Neurosci. 2000;3:437–444. [Abstract] [Google Scholar]
- Ma JY, Catterall WA, Scheuer T. Persistent sodium currents through brain sodium channels induced by G protein βγ subunits. Neuron. 1997;19:443–452. [Abstract] [Google Scholar]
- Tan HL, Kupershmidt S, Zhang R, Stepanovic S, Roden DM, Wilde AA, Anderson ME, Balser JR. A calcium sensor in the sodium channel modulates cardiac excitability. Nature. 2002;415:442–447. [Abstract] [Google Scholar]
- Cannon SC. Sodium channel defects in myotonia and periodic paralysis. Ann Rev Neurosci. 1996;19:141–164. [Abstract] [Google Scholar]
- Keating MT, Sanguinetti MC. Molecular and cellular mechanisms of cardiac arrhythmias. Cell. 2001;104:569–580. [Abstract] [Google Scholar]
- Escayg A, MacDonald BT, Meisler MH, Baulac S, Huberfeld G, An-Gourfinkel I, Brice A, LeGuern E, Moulard B, Chaigne D, et al. Mutations of SCN1A, encoding a neuronal sodium channel, in two families with GEFS+2. Nat Genet. 2000;24:343–345. [Abstract] [Google Scholar]
- Lossin C, Wang DW, Rhodes TH, Vanoye CG, George AL., Jr Molecular basis of an inherited epilepsy. Neuron. 2002;34:877–884. [Abstract] [Google Scholar]
- Wallace RH, Wang DW, Singh R, Scheffer IE, George AL, Jr, Phillips HA, Saar K, Reis A, Johnson EW, Sutherland GR, et al. Febrile seizures and generalized epilepsy associated with a mutation in the Na+-channel beta1 subunit gene SCN1B. Nat Genet. 1998;19:366–370. [Abstract] [Google Scholar]
- Burgess DL, Kohrman DC, Galt J, Plummer NW, Jones JM, Spear B, Meisler MH. Mutation of a new sodium channel gene, Scn8a, in the mouse mutant 'motor endplate disease'. Nat Genet. 1995;10:461–465. [Abstract] [Google Scholar]
- Planells-Cases R, Caprini M, Zhang J, Rockenstein EM, Rivera RR, Murre C, Masliah E, Montal M. Neuronal death and perinatal lethality in voltage-gated sodium channel alpha(II)-deficient mice. Biophys J. 2000;78:2878–2891. [Europe PMC free article] [Abstract] [Google Scholar]
- Papadatos GA, Wallerstein PM, Head CE, Ratcliff R, Brady PA, Benndorf K, Saumarez RC, Trezise AE, Huang CL, Vandenberg JI, et al. Slowed conduction and ventricular tachycardia after targeted disruption of the cardiac sodium channel gene Scn5a. Proc Natl Acad Sci USA. 2002;99:6210–6215. [Europe PMC free article] [Abstract] [Google Scholar]
Articles from Genome Biology are provided here courtesy of BMC
Full text links
Read article at publisher's site: https://doi.org/10.1186/gb-2003-4-3-207
Read article for free, from open access legal sources, via Unpaywall:
https://genomebiology.biomedcentral.com/counter/pdf/10.1186/gb-2003-4-3-207
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Discover the attention surrounding your research
https://www.altmetric.com/details/483899
Smart citations by scite.ai
Explore citation contexts and check if this article has been
supported or disputed.
https://scite.ai/reports/10.1186/gb-2003-4-3-207
Article citations
Higher sodium in older individuals or after stroke/reperfusion, but not in migraine or Alzheimer's disease - a study in different preclinical models.
Sci Rep, 14(1):21636, 16 Sep 2024
Cited by: 0 articles | PMID: 39284837 | PMCID: PMC11405707
Potential roles of voltage-gated ion channel disruption in Tuberous Sclerosis Complex.
Front Mol Neurosci, 17:1404884, 26 Aug 2024
Cited by: 0 articles | PMID: 39253727 | PMCID: PMC11381416
Voltage-Gated K+ Channel Modulation by Marine Toxins: Pharmacological Innovations and Therapeutic Opportunities.
Mar Drugs, 22(8):350, 29 Jul 2024
Cited by: 1 article | PMID: 39195466 | PMCID: PMC11355921
Review Free full text in Europe PMC
Genomic analysis of an Ecuadorian individual carrying an SCN5A rare variant.
BMC Cardiovasc Disord, 24(1):388, 27 Jul 2024
Cited by: 0 articles | PMID: 39068398 | PMCID: PMC11282620
Insights into the voltage-gated sodium channel, NaV1.8, and its role in visceral pain perception.
Front Pharmacol, 15:1398409, 23 May 2024
Cited by: 0 articles | PMID: 38855747
Review
Go to all (343) article citations
Other citations
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Voltage-gated sodium channel modulation by scorpion alpha-toxins.
Toxicon, 49(2):142-158, 28 Sep 2006
Cited by: 97 articles | PMID: 17087986 | PMCID: PMC1808227
Review Free full text in Europe PMC
International Union of Pharmacology. XLVII. Nomenclature and structure-function relationships of voltage-gated sodium channels.
Pharmacol Rev, 57(4):397-409, 01 Dec 2005
Cited by: 830 articles | PMID: 16382098
Review
Voltage-gated sodium channels: action players with many faces.
Ann Med, 38(7):472-482, 01 Jan 2006
Cited by: 24 articles | PMID: 17101538
Review
Ion permeation through a voltage- sensitive gating pore in brain sodium channels having voltage sensor mutations.
Neuron, 47(2):183-189, 01 Jul 2005
Cited by: 102 articles | PMID: 16039561

 1
1