Abstract
Free full text

ComPRePS: An Automated Cloud-based Image Analysis tool to democratize AI in Digital Pathology
Abstract
Artificial intelligence (AI) has extensive applications in a wide range of disciplines including healthcare and clinical practice. Advances in high-resolution whole-slide brightfield microscopy allow for the digitization of histologically stained tissue sections, producing gigapixel-scale whole-slide images (WSI). The significant improvement in computing and revolution of deep neural network (DNN)-based AI technologies over the last decade allow us to integrate massively parallelized computational power, cutting-edge AI algorithms, and big data storage, management, and processing. Applied to WSIs, AI has created opportunities for improved disease diagnostics and prognostics with the ultimate goal of enhancing precision medicine and resulting patient care.
The National Institutes of Health (NIH) has recognized the importance of developing standardized principles for data management and discovery for the advancement of science and proposed the Findable, Accessible, Interoperable, Reusable, (FAIR) Data Principles1 with the goal of building a modernized biomedical data resource ecosystem to establish collaborative research communities. In line with this mission and to democratize AI-based image analysis in digital pathology, we propose ComPRePS: an end-to-end automated Computational Renal Pathology Suite which combines massive scalability, on-demand cloud computing, and an easy-to-use web-based user interface for data upload, storage, management, slide-level visualization, and domain expert interaction. Moreover, our platform is equipped with both in-house and collaborator developed sophisticated AI algorithms in the back-end server for image analysis to identify clinically relevant micro-anatomic functional tissue units (FTU) and to extract image features.
1. INTRODUCTION
In this work, we present ComPRePS, an open-source, cloud-based computational tool for data upload, storage, and digital histology visualization integrated with server-based AI algorithms to perform complex image analysis tasks. Our tool offers various segmentation methods to detect clinically relevant structures as well as a pipeline to extract features of clinical import. In addition, our platform has the goal of reconciling AI-based image analysis and digital pathology for clinical application. Our publicly available tool and software create the opportunity for research communities to contribute to more reliable, accurate, and explainable AI-based image analysis methods.
Digital and computational pathology is becoming increasingly popular and being adopted by institutions globally2,3 with the promise of improving diagnostics and prognostics across organ systems. Requirements for successful implementation include addressing data management and storage of many large-scale digitized slides. In addition, high computational demand for extracting and analyzing relevant information from each WSI remains a significant bottleneck for wider adoption. With ComPRePS, we developed a platform with an intuitive browser-based user interface (UI) to improve a pathologist’s interaction with WSIs and apply AI to augment clinical decision making. A few noteworthy features in ComPRePS include: (1) enabling users to upload image data with linked metadata while maintaining the highest standard of security and confidentiality, and (2) managing user data in an AI-ready format by maintaining inter-operability between popular digital pathology management such as xml, json, csv, xlsx etc. for downstream applications. In this paper, by demonstrating the segmentation task of FTUs from kidney biopsy WSIs using ComPRePS, we will describe the end-to-end pipeline to automate a time-consuming process that to date has been performed manually by an expert. We hope that ComPRePS will be the catalyst at shifting the paradigm in the research community by creating a collaborative platform where data, AI models, codes, and software are public and accessible to any user globally regardless of skill level. Our user-friendly platform enables end users including clinicians and pathologists to perform analyses on their own very large images without any AI/software training,4 allowing all users to contribute to ever more powerful AI models to solve some of the most difficult healthcare challenges.
2. METHODS
2.1. ComPRePS User-Interface (UI)
HistomicsUI is the front-end of our tool where WSIs are organized, managed using Girder,5 a free data management platform with visualization using Digital Slide Archive (DSA).6 The user interface (UI) is accessible via the University of Florida Athena instance7 of DSA (credentials: username=public, password=public) from any browser, no desktop installation required. Once the WSI is opened in the HistomicsUI, the user can interact with the digital slide as in any stand-alone WSI viewer with the ability to pan and zoom in order to view structures of interest in appropriate detail. In addition, the user can display color-coded annotation layers overlaid on the WSI itself.
2.2. ComPRePS AI pipeline for FTU segmentation
Our browser-accessible, cloud-based, user-friendly tool enables clinicians, pathologists, biologists and computational scientists to independently perform analyses with the goal of reducing their segmentation burden. To achieve this goal, our custom segmentation tool is based off of Facebook AI Research’s Detectron2 library to perform complex panoptic segmentation tasks of 6 kidney-specific WSI FTUs.8 Our segmentation pipeline can be used for training new models as well as generating predicted annotations with new user uploaded data.
For model training, we leverage the workflow of the Detectron2 library9 and build our customized pipeline for FTU segmentation and classification, which implements convenient functions, methods and attributes by making use of a panoptic feature pyramidal architecture from input image patches. Fixed-size training patches are randomly chopped and extracted from training samples and fed into the training process. Our training data consisted of 190 WSIs with H&E, PAS, trichrome, Jones’ Methenamine Silver staining, including examples of diabetic nephropathy (n=53), lupus nephritis (n=39), and transplant surveillance needle core biopsies (11), as well as 58 large sub-WSIs manually cropped from healthy portions of 33 reference kidneys, 23 small sub-WSIs of 5 H&E needle core biopsies, 2 small sub-WSIs from a silver-stained biopsy, and 4 sub-WSIs from a trichrome biopsy. Each WSI used for training had 6 associated annotation layers performed manually by domain experts: non-globally sclerotic and globally sclerotic glomeruli, cortical and medullary interstitium, tubules, arteries/arterioles.10 Our tool also has 2 training pipelines to segment peritubular capillaries (PTC) and interstitial fibrosis and tubular atrophy (IFTA).
After the training is completed and validated on a hold-out dataset not participated in training, the trained model becomes available in ComPRePS model zoo to be applied on a prediction workflow for segmenting FTUs on new user-uploaded WSIs. The model prediction workflow is also built by abstracting the Detectron2 library functions and methods which follow first by cropping the large input WSI into small sized patches, generating segmentation masks for each patch and finally stitching all mask patches together on the WSI level. The resulting segmentation masks are saved as JSON annotations which can be seamlessly visualized in the ComPRePS UI overlaying on the WSI.
Each segmentation pipelines are deployed in ComPRePS as a docker image including codes and dependencies by following best software engineering practices so our developed AI pipelines can be inter-operable on different servers and deployed in a quick fashion. Our user-friendly UI enables the user to select various hyper-parameters for AI tuning in order to customize training and prediction workflows according to users choice. All the pre-trained models for various FTU segmentation are saved in our default open-sourced model zoo which are ready for our users to consume directly and apply in their data. Each generated segmentation masks are displayed automatically as different color coded layers for easy visualization and quality evaluation directly on ComPRePS UI, no additional tool or application is needed. In addition, ComPRePS UI allows users to edit FTU mask boundaries as needed which are automatically saved in the JSON files. In our knowledge, currently there is no digital pathology tool available in the market which performs end-to-end (E2E) computational AI based image analysis that performs the following: First, enabling a novice user such as a bench scientist, clinician or nephropathologist to do the entire E2E slide visualization, FTU segmentation and mask generation, quality evaluation and editing AI generated masks according to their liking in a single software platform without the need of any additional third-party add-on/applications. Second, reinforcing the mission of democratizing healthcare in all over the world by harnessing the transformative power of AI in patient care with open-sourced software/code and pre-trained models for community access and feedback.
2.3. ComPRePS Pathomic Feature Extraction from Segmented FTUs
Following segmentation, the feature extraction pipeline extracts pathologist-informed morphometric features from segmented FTUs (non-globally sclerotic and globally sclerotic glomeruli, tubules and arteries/arterioles) using image analysis techniques. Extracted features are summarized as follows and can be extracted into a downloadable spreadsheet format for: Glomeruli: Glomerular area, mesangial area, computed mesangial fraction and bounding box coordinates for each glomerulus. Tubules: Average tubular basement membrane thickness, average cell thickness, luminal fraction and bounding box coordinates. Arteries/arterioles: Arterial area and bounding box coordinates for each segmented artery/arteriole. Each FTU feature sets have been chosen evaluating their clinical significance by consulting with several nephro-pathologists. We are currently developing an additional morphometric feature extraction pipeline that segments 3 sub-compartments (nuclei, PAS+/eosinophilic areas, and luminal space) within each FTU and extracts sub-compartment specific features.
3. RESULTS
The current list of image analysis and AI pipelines available in ComPRePS are shown in Fig.1 top panel. After selecting the desired pipeline, the user can launch either a training or prediction job by first populating the tab with hyper-parameters of interest. As an example, for the multi-compartment segmentation pipeline which segments multiple FTUs in a single pipeline: (1) user selects the image, image folder and pre-trained model and submits the job (Fig.1 lower panel), (2) upon job completion, 6 annotation layers corresponding to cortical and medullary interstitium, non-globally and globally sclerotic glomeruli, tubules, and arteries/arterioles are added under the annotation tab automatically with different colors and can be visualized by toggling the eye icon next to each layer (Fig.1 lower panel). The segmentation output from our pipeline produces accurate and precise segmentation results of kidney FTUs (Fig.2 panels a-h) which are also verified by various nephropathologists from various institutions. ComPRePS UI enables users to perform quality evaluations of the generated annotations and correct them if necessary by using various editing tools.
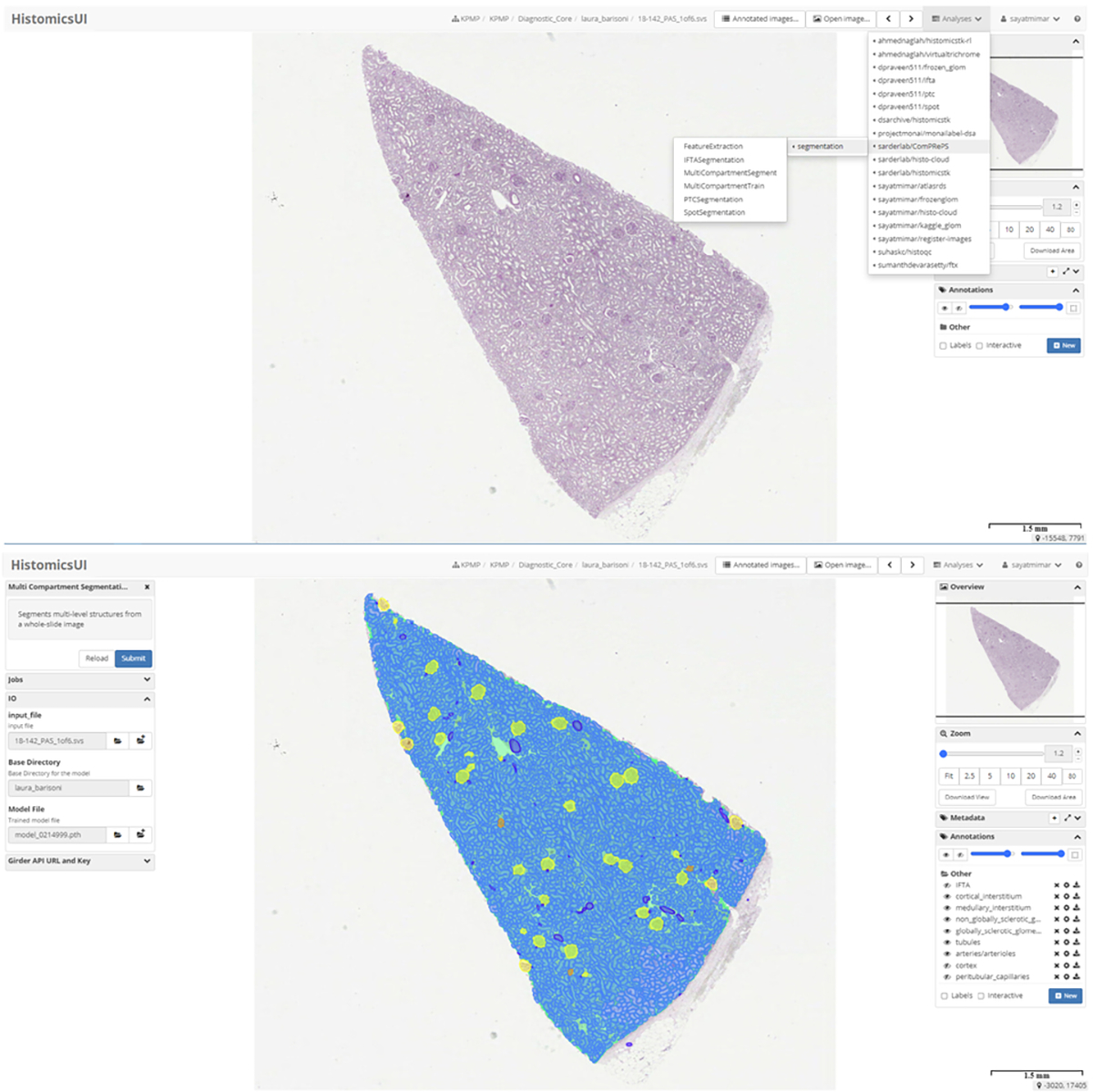
HistomicsUI and ComPRePS with all publicly available plugins. Multi compartment segmentation plugin shown on the top panel. The job can be run after selecting the image file, image directory and the pre-trained model. Once the job is run, annotations are added under the annotations tab and can be visualized by turning on/off the eye icon as shown in the lower panel.
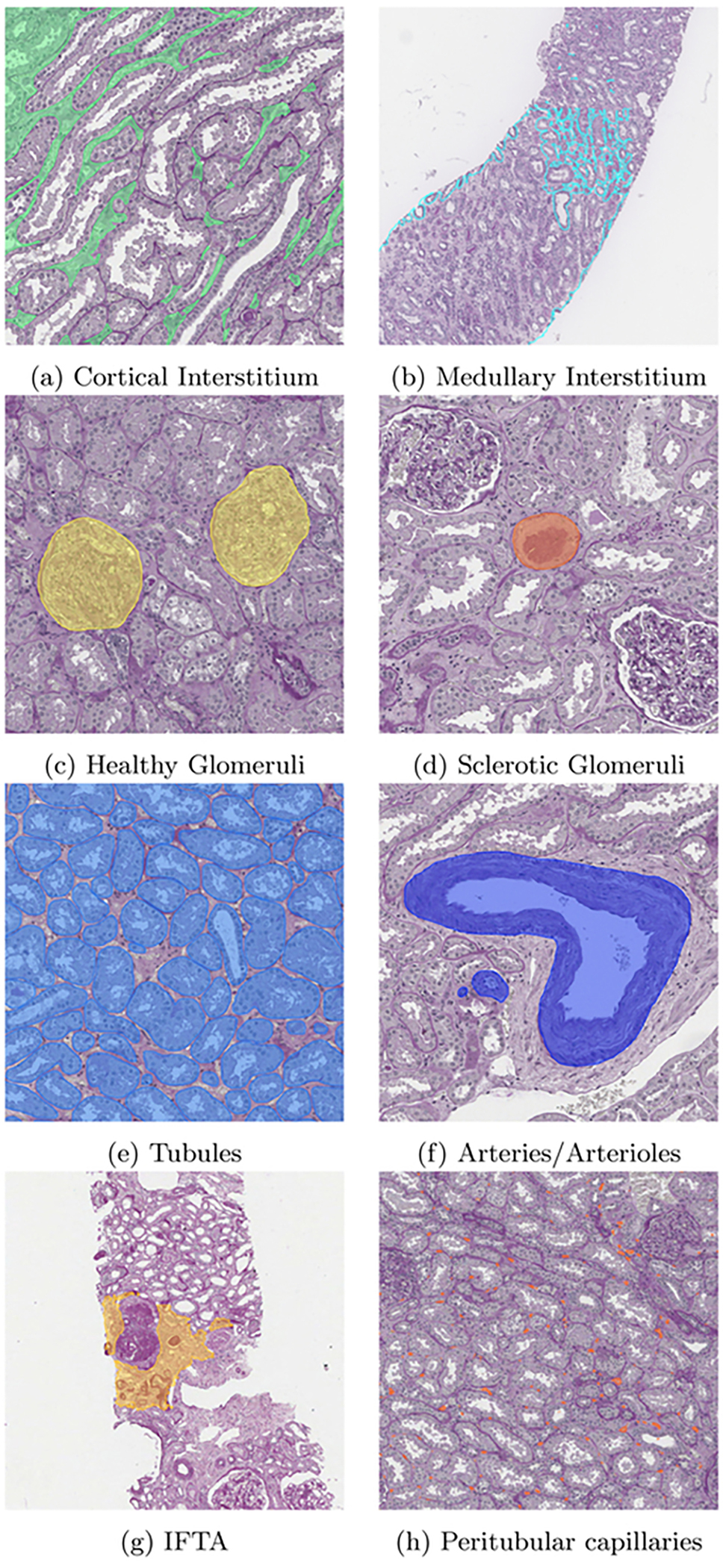
Results of the Multi compartment segmentation model is shown in panels a-f. In panels g and h, IFTA and Peritubular capillaries segmentations are illustrated respectively. Once the job is run, annotations are added under the annotations tab and can be visualized by turning on/off in Histomics UI.
3.1. Real World Usability Study
As of the time of writing this manuscript, ComPRePS UI and associated AI pipelines such as FTU segmentation and feature extraction have been in active use by pathologists from multiple institutions participating in large National Institute of Health (NIH) consortia; the Kidney Precision Medicine Project (KPMP),11 the Human BioMolecular Atlas Program (HuBMAP),12 to visualize their scanned and uploaded WSIs and to perform quality evaluation and edit AI generated annotations. A formal large-scale usability study is planned for the near future. We invite anyone who would like to participate in the usability study to contact the corresponding author.
4. CONCLUSION
In this work, we present our automated cloud tool, ComPRePS, to perform computational image analysis tasks harnessing the life-altering power of AI to support our mission of democratizing AI in digital pathology including making the technology accessible to clinicians and pathologists so AI based findings can be augmented with human decision making during the E2E life cycle of patient care. In this article, we have reviewed the development and deployment steps of various AI pipelines in ComPRePS to perform automated model training and segmentation on WSIs. The AI pipelines integrated with our user-friendly UI simplifies the utilization process keeping endusers front and center where they can rapidly segment FTUs, generate insights and visualize results without any coding or software development experience. We open-sourced ComPRePS with all our codes and deployed docker images in GitHub and Docker Hub that is scalable to multiple users world-wide and capable to provide helpful service to clinicians and biologists whenever they need it in the clinical pipeline. In addition, ComPRePS allows researchers/practitioners to incorporate their own pre-trained model into the model zoo as well as enable users to further fine-tune their model using our growing database. Our online easily accessible platform, opensource pipelines, and scalable software can help expedite research activity as well as most importantly can make a difference in clinical diagnostics by bringing AI-generated insights to augment the expertise of clinicians and pathologists.
ACKNOWLEDGMENTS
The authors would like to acknowledge the support from the National Institute of Health (NIH) / National institute of Diabetes and Digestive and Kidney Diseases (NIDDK) grants R01 DK114485, R21 DK128668, U01 DK133090, R01 DK129541 and A Computational IMage Analysis Platform (CIMAP) for the Human BioMolecular Atlas Program (HuBMAP) grant OT2 OD033753.
REFERENCES
Full text links
Read article at publisher's site: https://doi.org/10.1117/12.3008469
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
An integrative web-based software tool for multi-dimensional pathology whole-slide image analytics.
Phys Med Biol, 67(22), 09 Nov 2022
Cited by: 2 articles | PMID: 36067783 | PMCID: PMC10039615
Applications of discriminative and deep learning feature extraction methods for whole slide image analysis: A survey.
J Pathol Inform, 14:100335, 14 Sep 2023
Cited by: 3 articles | PMID: 37928897 | PMCID: PMC10622844
Review Free full text in Europe PMC
Browser-based Data Annotation, Active Learning, and Real-Time Distribution of Artificial Intelligence Models: From Tumor Tissue Microarrays to COVID-19 Radiology.
J Pathol Inform, 12:38, 27 Sep 2021
Cited by: 2 articles | PMID: 34760334 | PMCID: PMC8546359
Funding
Funders who supported this work.
NIDDK NIH HHS (4)
Grant ID: R01 DK114485
Grant ID: R21 DK128668
Grant ID: R01 DK129541
Grant ID: U01 DK133090
NIH HHS (1)
Grant ID: OT2 OD033753

