Abstract
Free full text

Genome-Wide Analysis of Evolutionary Markers of Human Influenza A(H1N1)pdm09 and A(H3N2) Viruses May Guide Selection of Vaccine Strain Candidates
Associated Data
Abstract
Here we analyzed whole-genome sequences of 3,969 influenza A(H1N1)pdm09 and 4,774 A(H3N2) strains that circulated during 2009–2015 in the world. The analysis revealed changes at 481 and 533 amino acid sites in proteins of influenza A(H1N1)pdm09 and A(H3N2) strains, respectively. Many of these changes were introduced as a result of random drift. However, there were 61 and 68 changes that were present in relatively large number of A(H1N1)pdm09 and A(H3N2) strains, respectively, that circulated during relatively long time. We named these amino acid substitutions evolutionary markers, as they seemed to contain valuable information regarding the viral evolution. Interestingly, influenza A(H1N1)pdm09 and A(H3N2) viruses acquired non-overlapping sets of evolutionary markers. We next analyzed these characteristic markers in vaccine strains recommended by the World Health Organization for the past five years. Our analysis revealed that vaccine strains carried only few evolutionary markers at antigenic sites of viral hemagglutinin (HA) and neuraminidase (NA). The absence of these markers at antigenic sites could affect the recognition of HA and NA by human antibodies generated in response to vaccinations. This could, in part, explain moderate efficacy of influenza vaccines during 2009–2014. Finally, we identified influenza A(H1N1)pdm09 and A(H3N2) strains, which contain all the evolutionary markers of influenza A strains circulated in 2015, and which could be used as vaccine candidates for the 2015/2016 season. Thus, genome-wide analysis of evolutionary markers of influenza A(H1N1)pdm09 and A(H3N2) viruses may guide selection of vaccine strain candidates.
Introduction
Human influenza A viruses impact public health and the global economy by causing annual epidemics and occasional pandemics. The viruses infect 5–10% of adults and 20–30% of children annually. Approximately 17% of infected people visit physicians, and 2% are hospitalized (Hayward et al. 2014). The average annual rate of influenza-associated hospitalizations and deaths is around 25 and 6 per 100,000 infected people, respectively (Lozano et al. 2012; Goldstein et al. 2015).
Influenza vaccines have been developed to protect the general population and especially people at risk (young children, pregnant women, health professionals, and adults over 65 years of age) from developing severe disease (Osterholm et al. 2012). However, vaccines need to be reformulated each year owing to the rapid viral antigenic evolution (Plotkin et al. 2002; Smith et al. 2004; Lee et al. 2015a). Twice a year, the World Health Organization (WHO) provides recommendations on influenza strains that should be included in vaccines for the upcoming influenza season, for southern and northern hemispheres (www.who.int/influenza/vaccines/virus/recommendations/en/). For example, in February 2014, the WHO has recommended to include A/California/7/2009(H1N1pdm09)-like, A/Texas/50/2012(H3N2)-like, and B/Massachusetts/2/2012(Yamagata)-like viruses in trivalent vaccines for the 2014/2015 influenza season for the northern hemisphere. However, overall vaccine effectiveness against laboratory-confirmed influenza associated with medically attended acute respiratory infections was as low as 23% for 2014/2015 (Flannery et al. 2015). Similarly, vaccine effectiveness was moderate for other seasons and varied from year to year (Simpson et al. 2013; Broberg et al. 2015; Flannery et al. 2015; Rondy et al. 2015; Valenciano et al. 2015). This could be owing to antigenic differences between the circulating influenza viruses and the vaccine strains recommended by the WHO (Broberg et al. 2015).
The WHO recommendations are based on phylogenetic and immunological analyses of influenza viruses from previous seasons (http://apps.who.int/gb/pip/pdf_files/Fluvaccvirusselection.pdf?ua=1). However, the phylogenetic approaches are often limited to the analysis of the nucleotide sequences of viral hemagglutinin (HA) and neuraminidase (NA) (Plotkin et al. 2002; Smith et al. 2004; Drummond et al. 2006; Strelkowa and Lassig 2012; Bhatt et al. 2013; Bedford et al. 2014; Lee et al. 2015a; Tharakaraman and Sasisekharan 2015). However, newer studies increasingly utilized whole-genome sequences of influenza viruses, shedding new light on virus evolution and aiding vaccine strain selection process (Holmes et al. 2005; Nelson and Holmes 2007; Du et al. 2012; Luksza and Lassig 2014; Neher et al. 2014; Steinbruck et al. 2014; Lee et al. 2015b; Neher and Bedford 2015; Vijaykrishna et al. 2015).
Here, we present a simple approach for reliable real-time tracking and prediction of viral evolution based on whole-genome sequences of influenza A(H1N1)pdm09 and A(H3N2)viruses, which could further improve vaccine strain selection process and, thereby, enhance vaccine efficacy. In particular, we analyzed the evolution of human influenza A viruses utilizing thousands of available whole-genome sequences of influenza A(H1N1)pdm09 and A(H3N2) strains representing different geographic regions. We identified hundreds mutation sites in influenza genomes. We noticed that mutations at several sites resulted in amino acid changes, which were maintained in influenza proteins for a relatively long time. We named these characteristic variants as evolutionary markers, as they seemed to contain valuable information regarding the viral evolution. Furthermore, we showed that A(H1N1)pdm09 and A(H3N2) viruses acquired their own sets of these markers. We also searched for these markers in corresponding vaccine strains and found many mismatches between vaccine strains and circulating influenza viruses. We also applied our approach to identify recent influenza A(H1N1)pdm09 and A(H3N2) strains that contain all the emerging evolutionary markers. We propose to use these strains as vaccine candidates for the upcoming influenza season. Thus, our evolutional analysis may improve the selection process of vaccine strain candidates, enhance vaccine efficacy, and supplement disease management.
Materials and Methods
The complete sets of influenza A(H1N1)pdm09 and A(H3N2) virus genome segments were retrieved from the Influenza Virus Resource (IVR; http://www.ncbi.nlm.nih.gov/genomes/FLU/FLU.html) and the Global Initiative on Sharing Avian Influenza Data (GISAID; http://platform.gisaid.org/) databases.
Nucleotide sequences were translated into protein sequences. Protein sequences of each strain were concatenated. To construct a reference-free multiple sequence protein alignment, the sequences were aligned using Muscle v3.7 software under default parameters (Edgar 2004). Distances were calculated using the Jukes–Cantor algorithm, and phylogenetic trees were constructed using the neighbor-joining method in Geneious 8.0 software (Kearse et al. 2012). Sequences of vaccine A/California/07/2009(H1N1) and A/Brisbane/10/2007(H3N2) strains were included in the alignments and phylogenetic trees.
Sites for amino acids changes were identified in concatenated protein sequences, and frequencies (F) of substitutions at these sites were calculated for 2009–2015 using A/California/07/2009 and A/Brisbane/10/2007 as references for A(H1N1)pdm09 and A(H3N2) influenza strains, respectively. Frequencies of changes were also calculated yearly (1.7.20xx–31.6.20xy). Substitutions that have difference in frequencies (ΔF = Fmax − Fmin) of more than 0.1 were selected and named evolutionary markers. These markers were also analyzed every three months in viral sub-populations and ΔFs were calculated. We also estimated false discovery rates for these changes, as described previously (Storey 2002).
We also compared evolutionary markers in circulating influenza viruses and vaccine strains recommended by the WHO for corresponding seasons focusing on markers, which had F > 0.5 during season covered by vaccine. We also analyzed evolutionary markers that were prevalent (F > 0.5) in A(H1N1)pdm09 and A(H3N2) viral populations in 2015, in influenza vaccine strains recommended by the WHO for 2015/2016 season. In addition, we identified influenza A strains, which possess all the identified markers of circulating influenza strains from 2015 and could be included in the vaccine for 2015/2016. References to influenza vaccine strain candidates were retrieved from the WHO Web site (www.who.int/influenza/vaccines/virus/recommendations/en/).
The sites for evolution markers were mapped onto available influenza A virus protein structures, retrieved from the Protein Data Bank (PDB; supplementary table S1). PyMol was used to visualize the sites on the protein structures and generate structural images (www.pymol.org).
The mutation rates of influenza A viruses were calculated by dividing the average number of substitutions in viral proteins occurring every year, by the total number of amino acids in the concatenated protein sequences. The resulting numbers were multiplied by 100 to express mutation rates in percentages.
Results
Evolutionary Markers of Influenza A(H1N1)pdm09 Viruses
We retrieved 3,969 whole-genome sequences of human influenza A(H1N1)pdm09 viruses, which circulated in the world from 1.5.2009 to 30.9.2015. We translated nucleotide sequences into viral HA, M1, M2, NA, NP, NS1, NS2, PA, PA-X, PB1, and PB2 protein sequences. Protein sequences of each strain were concatenated, and concatenated sequences were aligned. The concatenated protein sequence of vaccine influenza A/California/07/2009 strain, which is one of the earliest A(H1N1)pdm09 isolates, was used as a reference in the alignment (fig. 1A). Analysis of aligned sequences revealed 481 sites for amino acid changes in 4,807-amino acid-long protein sequences, indicating that approximately 0.18% of the amino acids of influenza A(H1N1)pdm09 virus changed every year.
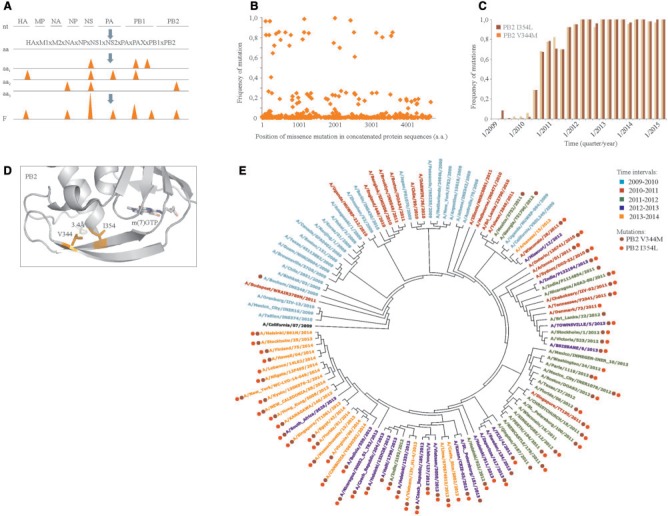
Evolutionary markers of influenza A(H1N1)pdm09 viruses. (A) Schematic representation of the process of identification of changes at amino acid level and their frequencies in the influenza A(H1N1)pdm09 viruses that circulated in the world during 2009–2015. (B) A scatter plot showing frequencies and distribution of these changes in concatenated viral protein sequence of the A(H1N1)pdm09 viruses from 2009 to 2015. (C) Frequencies of I354L and V344M substitutions in PB2 were analyzed in three-month intervals and plotted against virus collection dates. (D) Zoom into structure of PB2:m(7)GTP complex (PDB ID: 4PB6) showing position of 354 and 344 amino acids and their hydrophobic interaction. The distance between interacting residues is shown. (E) Phylogenetic tree constructed using A(H1N1)pdm09 strains that were circulating in the world during 2009–2015. Approximately 20 strains per year representing different geographies and collection times were used in the phylogenetic analysis. Strains were color-coded based on the year of virus collection. Full circles indicate strains that carry I354L and V344M substitutions in PB2.
We calculated frequencies of different amino acids changes at 481 sites. The frequency of each change was plotted against its position in concatenated protein sequences (fig. 1B). We then calculated frequencies of these changes yearly (supplementary table S2), and computed differences between maximal and minimal frequencies, taking into account the number of available sequences for each year. The majority of changes had ΔF ≤ 0.1, suggesting that these changes were introduced as a result of random drift, and, therefore, they were discarded from further analysis. However, there were 61 changes with ΔF > 0.1, indicating that these changes were subjected to selective evolution pressure. We defined these changes as evolutionary markers. Interestingly, these markers were found in 11 influenza proteins, in contrast to random changes, which resided mainly in HA and NA. The contribution of these markers into global mutation rate of A(H1N1)pdm09 viruses was as low as 13%.
We next analyzed the dynamics of acquisition of evolutionary markers by influenza A(H1N1)pdm09 viruses using three-month intervals, taking into account the number of available sequences for each period. We found that some of these markers showed similar evolutionary patterns. For example, the I354L mutation of PB2 was acquired in April and the V344M mutation of PB2 was acquired in May 2009 in the United States (fig. 1C). Strains with both mutations were detected in June 2010, in Singapore. Since August 2010, both mutations were found in viruses circulating in Thailand, Australia, and, later on, in Russia, Canada, Denmark, Greece, China, and the United Kingdom. Since October 2011, more than 90% of A(H1N1)pdm09 strains carried these mutations. The similar evolutionary pattern of these two mutations could be explained by structural and functional constraints: sites for both mutations are located in PB2 cap-binding domain, and both markers can participate in formation of hydrophobic interactions between two antiparallel β-strands, which are involved in cap binding (fig. 1D) (Pautus et al. 2013; Reich et al. 2014). We also mapped these markers on phylogenetic trees that were generated using concatenated protein sequences of selected A(H1N1)pdm09 strains (fig. 1E). Thus, the acquisition of these and other markers or their combinations could be beneficial for global evolution of influenza A(H1N1)pdm09 viruses. Interestingly, 29 of 62 identified mutations or their combinations were associated previously with viral fitness or virulence (Uraki et al. 2013; Elderfield et al. 2014; Sun et al. 2014; Mishel et al. 2015).
Evolutionary Markers of Influenza A(H3N2) Virus
We analyzed 4,774 whole-genome sequences of human influenza A(H3N2) viruses that circulated in the world from 1.1.2009 to 30.9.2015. We translated the nucleotide sequences into viral HA, M1, M2, NA, NP, NS1, NS2, PA, PB1, and PB2 proteins. We concatenated the protein sequences and aligned them. We calculated the total number of mutation sites using a sequence of the vaccine A/Brisbane/10/2007 strain as a reference (fig. 2A). We found 533 sites for amino acid substitutions in 4,465-amino acid-long concatenated protein sequences, indicating that approximately 0.21% of the amino acids of influenza A(H3N2) virus changed annually.
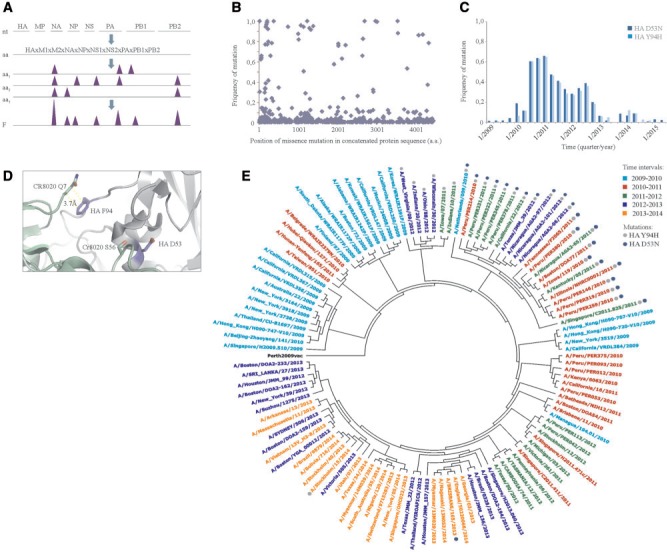
Evolutionary markers of influenza A(H3N2) viruses. (A) Schematic representation of the process of identification of changes at amino acid level and their frequencies in the influenza A(H3N2) viruses that circulated in the world during 2009–2015. (B) A scatter plot showing frequencies and distribution of amino acid substitutions in concatenated viral protein sequence of the A(H3N2) viruses from 2009 to 2015. (C) Frequencies of D53N and Y94H in HA were analyzed in three-month intervals and plotted against virus collection dates. (D) Zoom into structure of HA:CR8020 antibody complex (PDB ID: 3SDY) showing position of sites for amino acid changes (D53N and Y94H) in HA and interacting residues of CR8020. The distance between interacting residues of HA and antibody is shown. (E) Phylogenetic tree constructed using A(H3N2) strains that were circulating in the world during 2009–2015. Approximately 20 strains per year representing different geographies and isolation times were selected. Strains were color-coded based on the year of circulation. Full circles indicate strains that carry D53N and Y94H in HA. Numbering of markers in HA is based on the protein sequence without the signal peptide.
We calculated frequencies of different changes at 533 sites. The frequency of each change was plotted against its position in concatenated protein sequences (fig. 2B). We also calculated frequencies of these changes for each analyzed year (supplementary table S3). We compared mutation frequencies between different years and found that 68 substitutions had ΔF > 0.1. These substitutions were found in 10 protein sequences, in contrast to random changes, which resided mainly in HA and NA protein sequences. Moreover, the contribution of these markers into global mutation rate of A(H3N2) viruses can be calculated at 13%, which is similar to that of A(H1N1)pdm09 viruses.
Many influenza A(H3N2) markers had similar evolutionary patterns (fig. 2C). For example, D53N and Y94H in HA (numbering is based on the protein sequence without the signal peptide) were identified already in influenza A(H3N2) strains circulated in 2009, and their prevalence in the viral population increased up to and through July 2010 and then gradually decreased (numbering of mutations is based on the HA protein without the signal peptide). Interestingly, these markers are located in HA epitope that is recognized by stem-specific CR8020 antibody. Therefore, they could compromise CR8020 binding to HA and promote virus escape from host immune responses (fig. 2D) (Tharakaraman et al. 2014). We further highlighted these markers on a phylogenetic tree that was constructed using concatenated protein sequences of selected influenza A(H3N2) strains representing different geographic regions and different epidemics (fig. 2E). Altogether, these indicate that the acquisition of these and other markers or their combinations could be beneficial for the viruses’ evolution. Interestingly, at least three identified mutations or their combinations were previously associated with viral fitness, virulence, or pathogenicity (Memoli et al. 2009; Forbes et al. 2012, 2013; Job et al. 2014).
Comparison of Evolutionary Markers of Influenza A(H1N1)pdm09 and A(H3N2) Viruses
We next compared evolutionary markers of A(H1N1)pdm09 and A(H3N2) viruses. We found that both viral subtypes have their own sets of evolutionary markers (supplementary tables S2 and S3). To simplify interpretation of the results, we mapped the marker sites on available influenza A virus protein structures (fig. 3). Our analysis revealed no overlap between the sites. Thus, influenza A(H1N1)pdm09 and A(H3N2) viruses evolved in parallel during 2009–2015, which is in agreement with previous studies. Interestingly, many mutation sites resided on the surface of the viral proteins, suggesting that they could be involved in virus–host interactions, such as antigen–antibody interplay depicted on fig. 2D and described in literature (Ekiert et al. 2009, 2011, 2012; Corti et al. 2011; Koel et al. 2013).
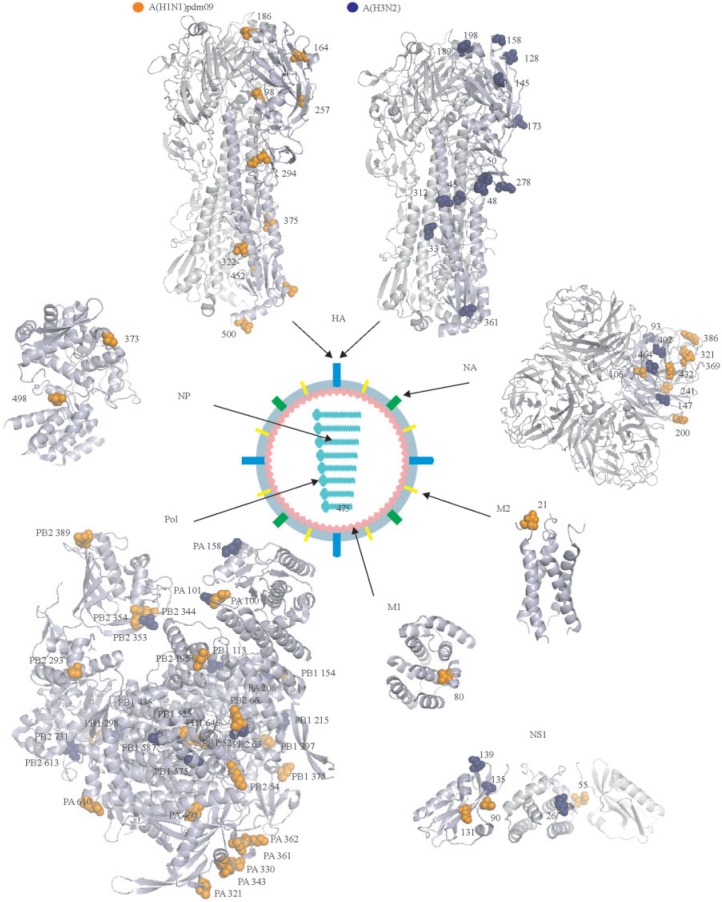
Mapping evolutionary markers of human influenza A(H1N1)pdm09 and A(H3N2) viruses on available virus protein structures. Available three-dimensional structures of individual proteins and the polymerase protein complex of influenza A viruses were used to map the positions of sites for evolutionary markers (PDB IDs: HA – 3LZG and 4FNK, NA – 1IVG, M2 – 2LY0, M1 – 3MD2, pol – 4WSB, NP – 4IRY, and NS1 – 3F5T). Sites of influenza A(H1N1)pdm09 virus are shown in orange, whereas sites of influenza A(H3N2) virus are shown in blue. Numbering starts from Met1 for all proteins except HA. Numbering of markers in HA is based on the protein sequence without the signal peptide. The schematic structure of the influenza A virion is also shown. One monomer in NA tetramer and one monomer in HA trimmer are highlighted with light-blue.
Analysis of Evolutionary Markers in Vaccine Strains Recommended by WHO for 2010–2014
We searched for identified evolutionary markers in influenza A(H1N1)pdm09 and A(H3N2) vaccine strains recommended by the WHO for past five influenza seasons (supplementary tables S4 and S5). For example, we searched for markers (with F > 0.5) of live influenza strains isolated during 2013–2014 in vaccine strains recommended by WHO for 2013/2014 season. It appeared that only 0–2 of 50 and 19 of 24 evolutionary markers were identified in influenza A(H1N1)pdm09 and A(H3N2) vaccine strains, respectively (table 1). We also found substantial mismatches between evolutionary markers of circulated influenza viruses and vaccine strains recommended for other four seasons. Most importantly, many of these mismatches were located in antibody-binding sites of HA and NA, thereby compromising recognition of circulating influenza strains by antibodies generated in response to corresponding vaccine strains. Thus, the absence of the evolutionary markers in vaccine strains could decrease vaccine efficacy by affecting recognition of circulating viruses by host antibodies developed in response to corresponding vaccine strains.
Table 1
The mismatches between evolutionary markers of circulating influenza viruses and corresponding amino acids in vaccine strains (2010–2014)
| Time interval | A(H1N1)pdm09 vaccine strain | No. of markers | A(H3N2) vaccine strain | No. of markers |
|---|---|---|---|---|
| 2010/2011 | A/California/07/2009 | 0 (11) | A/Perth/16/2009 | 1 (9) |
| A/Wisconsin/15/2009 | 0 (9) | |||
| 2011/2012 | A/California/07/2009 | 0 (14) | A/Perth/16/2009 | 0 (10) |
| 2012/2013 | A/California/07/2009 | 0 (31) | A/Victoria/361/2011 | 16 (22) |
| A/Christchurch/16/2010 | 2 (31) | |||
| A/Brisbane/10/2010 | 2 (31) | |||
| A/California/04/2009 | 0 (31) | |||
| A/Texas/05/2009 | 1 (31) | |||
| A/England/195/2009 | 1 (31) | |||
| A/New York/18/2009 | 1 (31) | |||
| 2013/2014 | A/California/07/2009 | 0 (50) | A/Texas/50/2012 | 19 (24) |
| A/Christchurch/16/2010 | 2 (50) | |||
| A/Brisbane/10/2010 | 2 (50) | |||
| A/California/04/2009 | 0 (50) | |||
| A/Texas/05/2009 | 1 (50) | |||
| A/England/195/2009 | 1 (50) | |||
| A/New York/18/2009 | 1 (50) | |||
| 2014/2015 | A/California/07/2009 | 0 (42) | A/New York/39/2012 | 20 (33) |
| A/Christchurch/16/2010 | 2 (42) | A/Almaty/2958/2013 | 20 (33) | |
| A/Brisbane/10/2010 | 2 (42) | |||
| A/California/04/2009 | 0 (42) | |||
| A/Texas/05/2009 | 1 (42) | |||
| A/England/195/2009 | 1 (42) | |||
| A/New York/18/2009 | 1 (42) |
The numbers in brackets refer to markers present in live viruses, and the numbers in front of the brackets refers to markers in corresponding vaccine strain.
Analysis of Emerging A(H1N1)pdm09 Evolutionary Markers in Corresponding Vaccine Strains Recommended by WHO for 2015/2016, and Identification of Vaccine Strain Candidate for 2015/2016 That Contains all the Evolutionary Markers
We identified 40 evolutionary markers with F > 0.5 in influenza A(H1N1)pdm09 strains circulated during 2015 (fig. 4A). We searched for these markers and found 0 to 2 in vaccine strains recommended by the WHO for 2015/2016 epidemic season. Many of these markers were located at the surface of HA and NA proteins. Moreover, some of them were found at the interfaces between viral glycoproteins and host antibodies (fig. 4B and C). This result indicates that mismatches between evolutionary markers in live viruses and amino acids in vaccine strains could alter recognition of HA and NA of live viruses by anti-HA and anti-NA antibodies generated against vaccine strains. For example, amino acid substitutions at position 375 of HA may alter binding of HB36.3-, HB80.4-, CR6261-, F10-, FI6V3-, and C179-like antibodies, and mismatch at position 353 of NA may interfere with binding of NC41-like antibodies. Thus, we found substantial differences in evolutionary markers between influenza A(H1N1)pdm09 strains circulated in 2015 and vaccine A(H1N1)pdm09 strains recommended by the WHO for 2015/2016 influenza season.

The prevalence of evolutionary markers of live A(H1N1)pdm09 viruses isolated during 2015 in A(H1N1)pdm09 vaccine strains recommended by the WHO and by the authors for 2015/2016 influenza season. (A) Frequencies of 40 selected evolutionary markers of live viruses were plotted against their positions in corresponding viral proteins. The distribution of these markers was analyzed in A(H1N1)pdm09 vaccine strains recommended by the WHO and by the authors (encircled) for 2015/2016 influenza season. Evolutionary markers are shown as black dots. Antibodies that recognize regions of viral proteins where evolutionary markers are located are also shown. (B) Mapping sites for evolutionary markers found in HA of A(H1N1)pdm09 viruses from 2015 on H1 structure (PDB ID: H1-3LZG). Mutation sites and antibodies that recognize corresponding regions on HA are shown (PDB IDs: HA:C05 – 4FP8; HA:S139/1 – 4GMS; HA:CH65 – 3SM5; HA:2D1 – 3LZG; HA:HC45 – 1QFU; HA:HB36.3 – 3R2X; HA:F-HB80.4 – 4EEF; HA:C179 – 5COR; HA:FI6V3 – 3ZTN; HA:CR6261 – 3GBN; HA:F10 – 3FKU). One monomer in HA trimmer is highlighted with light-blue. Numbering of markers in HA is based on the protein sequence without the signal peptide. (C) Mapping sites of evolutionary markers found in NA of A(H1N1)pdm09 viruses from 2015 on N1 structure (PDB ID: N1 – 4B7N). Mutation sites and antibodies that recognize corresponding regions of NA are shown (PDB IDs: NA:NC41 – 1NMB, NA:Mem5 – 2AEP). One monomer in NA tetramer is highlighted with light-blue.
We also searched for influenza strains that contain all 40 evolutionary markers of 2015 live A(H1N1)pdm09 viruses. We identified A/Hawaii/64/2014(H1N1) strain (fig. 4A). This indicates that A/Hawaii/64/2014(H1N1)-like strains could be used as a vaccine candidate for 2015/2016. Importantly, six evolutionary markers of A/Hawaii/64/2014(H1N1) reside in known antibody-binding regions of HA and NA, suggesting that such vaccine candidates could induce better protective immunity against live viruses than the strains recommended by the WHO.
Analysis of Emerging A(H3N2) Evolutionary Markers in Corresponding Vaccine Strains Recommended by WHO for 2015/2016 and Identification of Vaccine Strain Candidate for 2015/2016 That Contains All the Evolutionary Markers
We also identified 32 evolutionary markers with F > 0.5 in live A(H3N2) strains from 2015. The analysis of these markers in A(H3N2) vaccine strains recommended by the WHO for 2015/2016 epidemic season revealed only 22 markers (fig. 5A). Most of the markers were located on the surfaces of HA and NA (fig. 5B and C). Ten mismatches between evolutionary markers and corresponding amino acids of vaccine strains could alter recognition of viral glycoproteins of live viruses by antibodies generated against vaccine HA and NA. For example, mismatches at position 159 and 160 of HA may lower the affinity of F045-92-like and 2D1-like antibodies that target HA head region. Thus, we found substantial differences in evolutionary markers between live influenza A(H3N2) strains circulated in 2015 and corresponding vaccine strains recommended by the WHO for 2015/2016 influenza season.
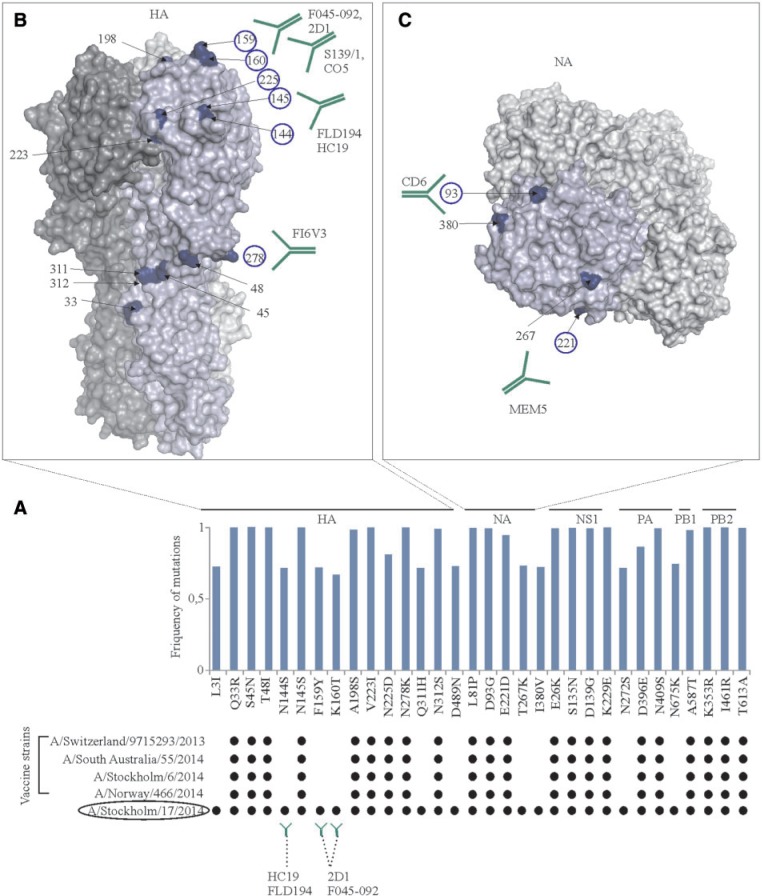
The prevalence of evolutionary markers of live A(H3N2)pdm09 viruses from 2015 in A(H3N2) vaccine strains recommended by the WHO and by the authors for 2015/2016 influenza season. (A) Frequencies of 32 selected evolutionary markers of live viruses were plotted against their positions in corresponding viral proteins. The distribution of these markers were analyzed in A(H3N2) vaccine strains recommended by the WHO and by the authors (encircled) for 2015/2016 influenza season. Evolutionary markers are shown as black dots. Antibodies that recognize regions of viral proteins where evolutionary markers are located are also shown. (B) Mapping sites for evolutionary markers found in HA of A(H3N2) viruses from 2015 on H3 structure (PDB ID: 4FNK). Mutation sites and antibodies that recognize corresponding regions on HA are shown (PDB IDs: A:F045-92 – 4O58, HA:2D1 – 3LZG, HA:S139/1 – 4GMS, HA:C05 – 4FP8, HA:FLD194 – 5A3I, HA:FI6V3 – 3ZTN). One monomer in HA trimmer is highlighted with light-blue. Numbering of markers in HA is based on the protein sequence without the signal peptide. (C) Mapping sites of evolutionary markers found in NA of A(H3N2) viruses from 2015 on N2 structure (PDB ID: N1-1IVG). Mutation sites and antibodies that recognize corresponding regions of NA are shown (PDB IDs: NA:CD6 – 4QNP, NA:Mem5 – 2AEP). One monomer in NA tetramer is highlighted with light-blue.
We also searched for live influenza strain that contain all 32 evolutionary markers. We identified A/Stockholm/17/2014(H3N2) strain (fig. 5A). This indicates that A/Stockholm/17/2014(H3N2)-like strains could be used as vaccine candidates for 2015/2016. Importantly, three evolutionary markers of the A/Stockholm/17/2014(H3N2) strain reside in known antibody-binding sites of HA, suggesting that such vaccine candidates could induce better protective immunity against live viruses than the strains recommended by the WHO. However, clinical trials are needed to validate our predictions.
Discussion
There were many studies that have attempted to track the evolution and evolutionary driving sites in influenza viruses, mostly by identifying those sites under selective pressure, using evolution-aware and statistically rigorous codon models that consider separately the non-synonymous and synonymous substitution rates and base/codon frequencies (Plotkin et al. 2002; Smith et al. 2004; Drummond et al. 2006; Strelkowa and Lassig 2012; Bhatt et al. 2013; Bedford et al. 2014; Lee et al. 2015a; Tharakaraman and Sasisekharan 2015). Moreover, some of these studies utilized whole-genome sequences of influenza viruses and modern software, such as FUBAR and PAML/CODEML (Holmes et al. 2005; Nelson and Holmes 2007; Nunes et al. 2008; Murrell et al. 2013; Vijaykrishna et al. 2015). Phylogenetic trees have also been used in such studies to help differentiate mutations that are gained by individual events and by inheritance in a lineage, and hence to predict the impact of the mutations to the virus survival (Holmes et al. 2005; Smith et al. 2009; Bhatt et al. 2013; Bedford et al. 2014; Steinbruck et al. 2014).
Here we analyzed available primary and tertiary protein structures of human influenza A viruses to better understand their evolution. In particular, we retrieved whole-genome sequences of A(H1N1)pdm09 and A(H3N2) viruses collected during 2009–2015 from public databases, translated them into protein sequences, and identified amino acid changes in these proteins whose frequencies (ΔF) varied in virus populations from one epidemic season to another by ΔF > 0.1. We defined these changes as evolutionary markers. Importantly, these markers were masked by large number of amino acids changes with ΔF ≤ 0.1, that is, substitutions in viral proteins that appeared in relatively small number of strains for short periods.
Some of these markers have emerged in viral sub-populations and remained, whereas others disappeared. Moreover, some of these markers re-emerged. The sites of re-emerged markers may represent hot spots of virus evolution. Such hot spots could be found at positions 235 of HA, 82 of NA, and 330 of PA of A(H1N1)pdm09 or 142 of HA, 129 of NS1, and 613 of PB2 of A(H3N2). However, analysis of longer evolution periods is needed to validate our hypothesis (supplementary tables S2 and S3).
We further clustered the markers into groups based on similarities in their evolution dynamics (amino acid frequency over virus collection time). For example, amino acid changes I354L and V344M of PB2 as well as N321K of PA had similar evolution patterns and therefore can be grouped together (supplementary table S2). Interestingly, I354L and V344M could modulate PB2 activity in snatching caps from host RNAs, whereas N321K of PA was reported to enhance polymerase complex activity in vitro and virus replication in cell culture (Elderfield et al. 2014). Moreover, all three changes could be a part of the adaptation strategy of swine-origin A(H1N1)pdm09 virus to human host, which resulted in virus transition from severe pandemic to mild epidemic mode (Elderfield et al. 2014; Mishel et al. 2015). Also other markers can be divided into groups based on their evolution patterns: D98N and E500K of HA; K166Q and A267T of HA; I34V, K432E of NA, and K361R of PA; N200S of NA, K230R of M1, D21G of M2, and S498N of NP; L90I of NS1 and I397M of PB1; T373A and V425I of NP; E55K of NS1and N29S of NS2; as well as D195N, R293K, and V731I of PB2. The simultaneous increase and reversions of these changes could be reflections of influenza segments being reasonably well-linked.
Similar co-evolving markers can be found in influenza A(H3N2) strains. For example, amino acid changes D53N and Y94H occurred almost simultaneously in HA. These changes could allow the virus to overcome the immunity developed after previous infections or vaccination. Some other influenza A(H3N2) markers within and outside of HA have similar evolution patterns: Q33R and N278K of HA; N144 and Q311H of HA; N145S and V223I of HA; K160T and F159Y of HA; T48I, S45N of HA, and K229E of NS1; T267K of NA and N272S of PA; E421D of NP and E331D of PB1; A587T of PB1, I461V and I63V of PB2, S135N and D139G of NS1, and K88R of NS2; M81I, V560I, K353R, R293K, and A221S of PB2; M129T of NS1; and N92S of NS2. However, the effects of these changes in viral proteins should be evaluated in vitro and in vivo.
Furthermore, we subdivided the evolutionary markers into drivers and passengers based on their time of appearance within these groups (supplementary tables S2 and S3). For example, PB2 I354L was detected few months before PB2 V344M and PA N321K, and thus, can represent a driver in this group of mutations with overall similar evolution dynamics. However, more whole-genome sequences in shorter time intervals would be needed to validate the driver/passenger hypothesis.
We also demonstrated that influenza A(H1N1)pdm09 and A(H3N2) viruses have their own sets of evolutionary markers. But the question remained, whether these viruses can exchange their evolutionary markers via reassortment events. Only a small fraction of the influenza A virus population had reassorted genomes. In particular, A(H1N1)pdm09 viruses acquired PB1 segments from A(H3N2) viruses during 2013 (supplementary fig. S1). But, the ΔF of mutations introduced via reassortment events was less than 0.1; therefore, these mutations should not be considered evolutionary markers. Thus, rare reassortment events did alter the composition of evolutionary markers of influenza A virus subtypes. By contrast, influenza B viruses of Victoria and Yamagata lineages frequently exchange their segments (Matsuzaki et al. 2004; Vijaykrishna et al. 2014; Dudas et al. 2015). The frequent reassortment events complicate genome-wide analysis of evolutionary markers for influenza B virus lineages. Thus, our approach at its current stage is relevant only for influenza A viruses.
We were wondering whether our genome-wide analysis of evolutionary markers of influenza A viruses can improve vaccine strain selection. We analyzed evolutionary markers in vaccine strains recommended by the WHO for corresponding influenza seasons and found only few of them in vaccine strains. Many of these markers were located on the surface of major influenza antigens, HA and NA, and the absence of these markers in vaccine strains could compromise the antigenic properties of vaccine strains, and thus lower vaccine efficacy. Also, many of the markers were located in other viral proteins, and could affect their function and interactions. Furthermore, we identified A/Hawaii/64/2014(H1N1) and A/Stockholm/17/2014(H3N2) strains, which contain all the emerging evolutionary markers, and propose to use these or similar viruses as vaccine candidates for 2015/2016. Thus, our genome-wide analysis of evolutionary markers suggested a potential vaccine candidate that can be a close match with most of the circulating strains, hence providing better protection against influenza A virus infections to the population.
In conclusion, recent advances of next-generation sequencing techniques and increasing number of available whole-genomes sequences of influenza A viruses from different geographic regions can be used for tracking and prediction of virus evolution and for selection of vaccine strain candidates. Here, we developed a simple approach for reliable real-time tracking and prediction of viral evolution based on whole-genome sequences of human influenza A(H1N1)pdm09 and A(H3N2)viruses, which could further improve vaccine strain selection process and, thereby, enhance vaccine efficacy. Our approach may also improve diagnostics and personalized treatment of severe influenza infections. It can also be used in drug development programs as well as in virus surveillance studies, which monitor antiviral drug-resistance. Altogether, these may allow us to better control seasonal influenza outbreaks in near future.
Supplementary Material
Supplementary figure S1 and Supplementary tables S1–S5 are available at Genome Biology and Evolution online (http://www.gbe.oxfordjournals.org/).
Acknowledgments
We gratefully acknowledge the authors, originating and submitting laboratories of the sequences from the GISAID and IVR databases on which this research is based. We also thank Sara Kangaspeska, Triin Lakspere, Petri Jalovaara, Laura Kakkola, Niina Ikonen, Hannimari Kallio-Kokko, Anu Kantele, and Ilkka Julkunen for critical discussion of the manuscript. This work was supported by the University of Helsinki three-year research project grant (465/51/2014), CIMO Fellowship (TM-15-9787), and Jane and Aatos Erkko Foundation.
Literature Cited
- Bedford T, et al. 2014. Integrating influenza antigenic dynamics with molecular evolution. eLife 3:e01914. [Europe PMC free article] [Abstract] [Google Scholar]
- Bhatt S, et al. 2013. The evolutionary dynamics of influenza A virus adaptation to mammalian hosts. Philos Trans R Soc London Ser B Biol Sci. 368:20120382. [Europe PMC free article] [Abstract] [Google Scholar]
- Broberg E, et al. 2015. Start of the 2014/15 influenza season in Europe: drifted influenza A(H3N2) viruses circulate as dominant subtype. Euro Surveill. 20:21023. [Abstract] [Google Scholar]
- Corti D, et al. 2011. A neutralizing antibody selected from plasma cells that binds to group 1 and group 2 influenza A hemagglutinins. Science 333:850–856. [Abstract] [Google Scholar]
- Drummond AJ, Ho SY, Phillips MJ, Rambaut A. 2006. Relaxed phylogenetics and dating with confidence. PLoS Biol. 4:e88. [Europe PMC free article] [Abstract] [Google Scholar]
- Du X, et al. 2012. Mapping of H3N2 influenza antigenic evolution in China reveals a strategy for vaccine strain recommendation. Nat Commun. 3:709. [Abstract] [Google Scholar]
- Dudas G, Bedford T, Lycett S, Rambaut A. 2015. Reassortment between influenza B lineages and the emergence of a coadapted PB1-PB2-HA gene complex. Mol Biol Evol. 32:162–172. [Europe PMC free article] [Abstract] [Google Scholar]
- Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32:1792–1797. [Europe PMC free article] [Abstract] [Google Scholar]
- Ekiert DC, et al. 2009. Antibody recognition of a highly conserved influenza virus epitope. Science 324:246–251. [Europe PMC free article] [Abstract] [Google Scholar]
- Ekiert DC, et al. 2011. A highly conserved neutralizing epitope on group 2 influenza A viruses. Science 333:843–850. [Europe PMC free article] [Abstract] [Google Scholar]
- Ekiert DC, et al. 2012. Cross-neutralization of influenza A viruses mediated by a single antibody loop. Nature 489:526–532. [Europe PMC free article] [Abstract] [Google Scholar]
- Elderfield RA, et al. 2014. Accumulation of human-adapting mutations during circulation of A(H1N1)pdm09 influenza virus in humans in the United Kingdom. J Virol. 88:13269–13283. [Europe PMC free article] [Abstract] [Google Scholar]
- Flannery B, et al. 2015. Early estimates of seasonal influenza vaccine effectiveness - United States, January 2015. MMWR Morb Mortal Wkly Rep. 64:10–15. [Europe PMC free article] [Abstract] [Google Scholar]
- Forbes N, et al. 2013. Identification of adaptive mutations in the influenza A virus non-structural 1 gene that increase cytoplasmic localization and differentially regulate host gene expression. PLoS One 8:e84673. [Europe PMC free article] [Abstract] [Google Scholar]
- Forbes NE, et al. 2012. Multifunctional adaptive NS1 mutations are selected upon human influenza virus evolution in the mouse. PLoS One 7:e31839. [Europe PMC free article] [Abstract] [Google Scholar]
- Goldstein E, et al. 2015. Estimating the hospitalization burden associated with influenza and respiratory syncytial virus in New York City, 2003–2011. Influenza Other Respir Viruses 9:225–233. [Europe PMC free article] [Abstract] [Google Scholar]
- Hayward AC, et al. 2014. Comparative community burden and severity of seasonal and pandemic influenza: results of the Flu Watch cohort study. Lancet Respir Med. 2:445–454. [Europe PMC free article] [Abstract] [Google Scholar]
- Holmes EC, et al. 2005. Whole-genome analysis of human influenza A virus reveals multiple persistent lineages and reassortment among recent H3N2 viruses. PLoS Biol. 3:e300. [Europe PMC free article] [Abstract] [Google Scholar]
- Job ER, et al. 2014. A single amino acid substitution in the hemagglutinin of H3N2 subtype influenza A viruses is associated with resistance to the long pentraxin PTX3 and enhanced virulence in mice. J Immunol. 192:271–281. [Abstract] [Google Scholar]
- Kearse M, et al. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28:1647–1649. [Europe PMC free article] [Abstract] [Google Scholar]
- Koel BF, et al. 2013. Substitutions near the receptor binding site determine major antigenic change during influenza virus evolution. Science 342:976–979. [Abstract] [Google Scholar]
- Lee AJ, et al. 2015a. Diversifying selection analysis predicts antigenic evolution of 2009 pandemic H1N1 Influenza A virus in humans. J Virol. 89:5427–5440. [Europe PMC free article] [Abstract] [Google Scholar]
- Lee HK, et al. 2015b. Predicting clinical severity based on substitutions near epitope A of influenza A/H3N2. Infect Genet Evol. 34:292–297. [Abstract] [Google Scholar]
- Lozano R, et al. 2012. Global and regional mortality from 235 causes of death for 20 age groups in 1990 and 2010: a systematic analysis for the Global Burden of Disease Study 2010. Lancet 380:2095–2128. [Europe PMC free article] [Abstract] [Google Scholar]
- Luksza M, Lassig M. 2014. A predictive fitness model for influenza. Nature 507:57–61. [Abstract] [Google Scholar]
- Matsuzaki Y, et al. 2004. Genetic diversity of influenza B virus: the frequent reassortment and cocirculation of the genetically distinct reassortant viruses in a community. J Med Virol. 74:132–140. [Abstract] [Google Scholar]
- Memoli MJ, et al. 2009. Recent human influenza A/H3N2 virus evolution driven by novel selection factors in addition to antigenic drift. J Infect Dis. 200:1232–1241. [Europe PMC free article] [Abstract] [Google Scholar]
- Mishel P, et al. 2015. Comparative analysis of whole-genome sequences of influenza A(H1N1)pdm09 viruses isolated from hospitalized and nonhospitalized patients identifies missense mutations that might be associated with patient hospital admissions in Finland during 2009 to 2014. Genome Announc. 3:e00676–15. [Europe PMC free article] [Abstract] [Google Scholar]
- Murrell B, et al. 2013. FUBAR: a fast, unconstrained bayesian approximation for inferring selection. Mol Biol Evol. 30:1196–1205. [Europe PMC free article] [Abstract] [Google Scholar]
- Neher RA, Bedford T. 2015. nextflu: real-time tracking of seasonal influenza virus evolution in humans. Bioinformatics 31:3546–3548. [Europe PMC free article] [Abstract] [Google Scholar]
- Neher RA, Russell CA, Shraiman BI. 2014. Predicting evolution from the shape of genealogical trees. eLife 3:e03568. [Europe PMC free article] [Abstract] [Google Scholar]
- Nelson MI, Holmes EC. 2007. The evolution of epidemic influenza. Nat Rev Genet. 8:196–205. [Abstract] [Google Scholar]
- Nunes B, et al. 2008. Heterogeneous selective pressure acting on influenza B Victoria- and Yamagata-like hemagglutinins. J Mol Evol. 67:427–435. [Abstract] [Google Scholar]
- Osterholm MT, Kelley NS, Sommer A, Belongia EA. 2012. Efficacy and effectiveness of influenza vaccines: a systematic review and meta-analysis. Lancet Infect Dis. 12:36–44. [Abstract] [Google Scholar]
- Pautus S, et al. 2013. New 7-methylguanine derivatives targeting the influenza polymerase PB2 cap-binding domain. J Med Chem. 56:8915–8930. [Abstract] [Google Scholar]
- Peterlin BM, Brogie JE, Price DH. 2012. 7SK snRNA: a noncoding RNA that plays a major role in regulating eukaryotic transcription. Wiley Interdiscip Rev RNA 3:92–103. [Europe PMC free article] [Abstract] [Google Scholar]
- Plotkin JB, Dushoff J, Levin SA. 2002. Hemagglutinin sequence clusters and the antigenic evolution of influenza A virus. Proc Natl Acad Sci U S A. 99:6263–6268. [Europe PMC free article] [Abstract] [Google Scholar]
- Reich S, et al. 2014. Structural insight into cap-snatching and RNA synthesis by influenza polymerase. Nature 516:361–366. [Abstract] [Google Scholar]
- Rondy M, et al. 2015. 2012/13 influenza vaccine effectiveness against hospitalised influenza A(H1N1)pdm09, A(H3N2) and B: estimates from a European network of hospitals. Euro Surveill. 20:21011. [Abstract] [Google Scholar]
- Simpson CR, et al. 2013. In Seasonal Influenza Vaccine Effectiveness (SIVE): an observational retrospective cohort study—exploitation of a unique community-based national-linked database to determine the effectiveness of the seasonal trivalent influenza vaccine Southampton (UK). Health Services and Delivery Research, 1.10. [Abstract] [Google Scholar]
- Smith DJ, et al. 2004. Mapping the antigenic and genetic evolution of influenza virus. Science 305:371–376. [Abstract] [Google Scholar]
- Smith GJ, et al. 2009. Origins and evolutionary genomics of the 2009 swine-origin H1N1 influenza A epidemic. Nature 459:1122–1125. [Abstract] [Google Scholar]
- Steinbruck L, Klingen TR, McHardy AC. 2014. Computational prediction of vaccine strains for human influenza A (H3N2) viruses. J Virol. 88:12123–12132. [Europe PMC free article] [Abstract] [Google Scholar]
- Storey JD. 2002. A direct approach to false discovery rates. J Roy Stat Soc B. 64:479–498. [Google Scholar]
- Strelkowa N, Lassig M. 2012. Clonal interference in the evolution of influenza. Genetics 192:671–682. [Europe PMC free article] [Abstract] [Google Scholar]
- Sun Y, et al. 2014. Naturally occurring mutations in the PA gene are key contributors to increased virulence of pandemic H1N1/09 influenza virus in mice. J Virol. 88:4600–4604. [Europe PMC free article] [Abstract] [Google Scholar]
- Tharakaraman K, Sasisekharan R. 2015. Influenza surveillance: 2014–2015 H1N1 “swine”-derived influenza viruses from India. Cell Host Microbe 17:279–282. [Abstract] [Google Scholar]
- Tharakaraman K, et al. 2014. Broadly neutralizing influenza hemagglutinin stem-specific antibody CR8020 targets residues that are prone to escape due to host selection pressure. Cell Host Microbe 15:644–651. [Europe PMC free article] [Abstract] [Google Scholar]
- Uraki R, et al. 2013. Virulence determinants of pandemic A(H1N1)2009 influenza virus in a mouse model. J Virol. 87:2226–2233. [Europe PMC free article] [Abstract] [Google Scholar]
- Valenciano M, et al. 2015. The European I-MOVE Multicentre 2013–2014 Case-Control Study. Homogeneous moderate influenza vaccine effectiveness against A(H1N1)pdm09 and heterogenous results by country against A(H3N2). Vaccine 33:2813–2822. [Abstract] [Google Scholar]
- Vijaykrishna D, et al. 2014. The contrasting phylodynamics of human influenza B viruses. eLife 4:e05055. [Europe PMC free article] [Abstract] [Google Scholar]
- Vijaykrishna D, et al. 2015. The contrasting phylodynamics of human influenza B viruses. eLife 4:e05055. [Europe PMC free article] [Abstract] [Google Scholar]
Articles from Genome Biology and Evolution are provided here courtesy of Oxford University Press
Full text links
Read article at publisher's site: https://doi.org/10.1093/gbe/evv240
Read article for free, from open access legal sources, via Unpaywall:
https://academic.oup.com/gbe/article-pdf/7/12/3472/17924857/evv240.pdf
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics

Discover the attention surrounding your research
https://www.altmetric.com/details/4818782
Smart citations by scite.ai
Explore citation contexts and check if this article has been
supported or disputed.
https://scite.ai/reports/10.1093/gbe/evv240
Article citations
Host adaptive mutations in the 2009 H1N1 pandemic influenza A virus PA gene regulate translation efficiency of viral mRNAs via GRSF1.
Commun Biol, 5(1):1102, 17 Oct 2022
Cited by: 6 articles | PMID: 36253464 | PMCID: PMC9576711
Identifying host-specific amino acid signatures for influenza A viruses using an adjusted entropy measure.
BMC Bioinformatics, 23(1):333, 12 Aug 2022
Cited by: 0 articles | PMID: 35962315 | PMCID: PMC9372975
Antiviral Activity and Mechanisms of Seaweeds Bioactive Compounds on Enveloped Viruses-A Review.
Mar Drugs, 20(6):385, 08 Jun 2022
Cited by: 8 articles | PMID: 35736188 | PMCID: PMC9228758
Review Free full text in Europe PMC
Genome characterization and mutation analysis of human influenza A virus in Thailand.
Genomics Inform, 20(2):e21, 30 Jun 2022
Cited by: 3 articles | PMID: 35794701 | PMCID: PMC9299564
Review Free full text in Europe PMC
Genotypic Variants of Pandemic H1N1 Influenza A Viruses Isolated from Severe Acute Respiratory Infections in Ukraine during the 2015/16 Influenza Season.
Viruses, 13(11):2125, 21 Oct 2021
Cited by: 1 article | PMID: 34834932 | PMCID: PMC8619959
Go to all (14) article citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
Protein structures in PDBe (Showing 24 of 24)
-
(4 citations)
PDBe - 3LZGView structure
-
(2 citations)
PDBe - 4GMSView structure
-
(2 citations)
PDBe - 4FP8View structure
-
(2 citations)
PDBe - 2AEPView structure
-
(2 citations)
PDBe - 4FNKView structure
-
(2 citations)
PDBe - 3ZTNView structure
-
(2 citations)
PDBe - 1IVGView structure
-
(1 citation)
PDBe - 4IRYView structure
-
(1 citation)
PDBe - 3FKUView structure
-
(1 citation)
PDBe - 1NMBView structure
-
(1 citation)
PDBe - 2LY0View structure
-
(1 citation)
PDBe - 4O58View structure
-
(1 citation)
PDBe - 4QNPView structure
-
(1 citation)
PDBe - 4PB6View structure
-
(1 citation)
PDBe - 5CORView structure
-
(1 citation)
PDBe - 4WSBView structure
-
(1 citation)
PDBe - 3F5TView structure
-
(1 citation)
PDBe - 4EEFView structure
-
(1 citation)
PDBe - 3SDYView structure
-
(1 citation)
PDBe - 1QFUView structure
-
(1 citation)
PDBe - 3GBNView structure
-
(1 citation)
PDBe - 5A3IView structure
-
(1 citation)
PDBe - 3R2XView structure
-
(1 citation)
PDBe - 4B7NView structure
Show less
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Evolution of the hemagglutinin expressed by human influenza A(H1N1)pdm09 and A(H3N2) viruses circulating between 2008-2009 and 2013-2014 in Germany.
Int J Med Microbiol, 305(7):762-775, 21 Aug 2015
Cited by: 12 articles | PMID: 26416089
Molecular epidemiology and evolution of A(H1N1)pdm09 and H3N2 virus during winter 2012-2013 in Beijing, China.
Infect Genet Evol, 26:228-240, 07 Jun 2014
Cited by: 14 articles | PMID: 24911284
Genetic diversity of influenza A viruses circulating in Bulgaria during the 2018-2019 winter season.
J Med Microbiol, 69(7):986-998, 01 Jul 2020
Cited by: 3 articles | PMID: 32459617 | PMCID: PMC7481746
[Swine influenza virus: evolution mechanism and epidemic characterization--a review].
Wei Sheng Wu Xue Bao, 49(9):1138-1145, 01 Sep 2009
Cited by: 6 articles | PMID: 20030049
Review

