Abstract
Free full text

OpenSlide: A vendor-neutral software foundation for digital pathology
Abstract
Although widely touted as a replacement for glass slides and microscopes in pathology, digital slides present major challenges in data storage, transmission, processing and interoperability. Since no universal data format is in widespread use for these images today, each vendor defines its own proprietary data formats, analysis tools, viewers and software libraries. This creates issues not only for pathologists, but also for interoperability. In this paper, we present the design and implementation of OpenSlide, a vendor-neutral C library for reading and manipulating digital slides of diverse vendor formats. The library is extensible and easily interfaced to various programming languages. An application written to the OpenSlide interface can transparently handle multiple vendor formats. OpenSlide is in use today by many academic and industrial organizations world-wide, including many research sites in the United States that are funded by the National Institutes of Health.
INTRODUCTION
Anatomic pathology stands at the threshold of a major transformation. Starting in the late 20th century and continuing into the 21st century, digital slide technology has evolved from early attempts to manually digitize the entire glass slide to sophisticated slide formats and robotic scanning devices. The online storage, Internet-based access and archival preservation of digital slides (also known as “whole-slide images”) overcome numerous areas of inefficiency in the conventional workflow of anatomic pathology. For example, they eliminate the need for physical transport and archival storage of glass slides. They also enable within-hospital-network and Internet-based sharing of digital slides for a variety of purposes ranging from diagnoses and quality control to medical research. In addition, there are numerous advances in computer vision and machine learning that enable automated or semi-automated analysis and comparison of digital slides.
For example, an area ripe for the taking is what pathologists affectionately refer to as “wallpaper matching” − namely, comparison of an unknown diagnostic entity encountered under the microscope to entities described and/or photographed in textbooks. This is a long and arduous process with conventional textbooks, hampered by the fact that color imagery is minimal in printed literature because it increases printing costs. Further, only small regions of entire glass slides can be shown in textbooks because their format is too small. If an entire glass slide were to be photographed at ×40 objective (×400 magnification equivalent), the size of the image would roughly be the size of a football field! Using digital slides in conjunction with deep-zoom visualization software and a content-based image search tool called Diamond,[1,2] we hope to circumvent this problem. This would enable pathologists to leverage Internet-based software to greatly improve the speed and accuracy of analysis and to rapidly identify archival material that is comparable to current material.
An important non-technical challenge when transitioning to digital slides will be the ability and willingness of experienced pathologists who are comfortable with glass slides to embrace new technology. Digital pathology will need to offer some benefits (such as improved accuracy, speed or convenience) to practicing pathologists to encourage them to become more proficient with the new workflows. These changes will take time to percolate through the system, possibly a decade or even longer. Newly-trained pathologists who are comfortable with digital slides from the beginning of their careers will accelerate this process of transformation.
One technical challenge is the difficulty of processing digital slides using standard image processing tools and libraries. These are typically designed for images that can be comfortably uncompressed into memory. Unfortunately, digital slides are enormous relative to image data such as personal photographs. Even after aggressive compression, file sizes of hundreds of megabytes to many gigabytes are common in this domain. At full resolution, images routinely exceed memory sizes and often occupy tens of gigabytes when uncompressed. These images are typically multi-resolution, with only a small amount of image data being relevant at any particular resolution and viewpoint.
A second technical challenge is that each vendor implements its own digital slide formats, libraries and viewers. Vendors typically do not document their formats. Even when there is documentation, important details are omitted or inaccurate. Because a vendor's library or viewer is the only way to view a particular digital slide, doctors and researchers suffer from long-term vendor lock-in − they are unable to take advantage of improvements offered by other vendors. In 2010, a Digital Imaging and Communications in Medicine (DICOM) standard for digital slides was released (supplement 145).[3,4] This standard was based on experience with the DICOM standard for visible light images (supplement 15, released in 1999)[5] and the efforts of DICOM Working Group 26.[6] Although the DICOM standard may eventually alleviate the problem of format proliferation, there is no evidence of this yet. Realistically, it may be many years before this standard is widely supported by all vendors. Even after widespread adoption, each vendor may continue to preferentially advocate its own proprietary format and merely provide slow export/import functionality for conversion to/from the DICOM standard. Further, unless an extensive effort of format conversion is undertaken, access to archived digital slides in pre-DICOM formats will remain an important obstacle.
A third technical challenge is that few vendors provide libraries and viewers for non-Windows platforms. While Windows dominates clinical settings, there is significant use of Linux in medical research and some use of Apple Mac OS X. In addition, the explosive growth of mobile platforms such as Android and iOS tablets and smartphones suggests that these platforms may play important roles in future clinical settings. Some vendors offer multi-platform support through a server-based approach, but this hurts performance by adding a network round-trip delay on every digital slide operation. This makes it suboptimal for high-latency settings such as 3G and 4G cellular networks and unusable in non-networked environments. There are also some open-source projects to read and convert digital slides, such as the LOCI Bio-Formats library[7] and the NYU Virtual Microscope project,[8] but these are limited to a particular programming environment such as Java or to a small number of slide formats.
To address these technical challenges, we have created OpenSlide. This is a C library for reading and manipulating digital slides of diverse vendor formats. This library can be linked to applications written in C or C++, as well as to applications written in other programming languages such as Java and Python that support C bindings. The use of OpenSlide enables clean separation of concerns for an application developer. Rather than dealing with proliferation of vendor-specific formats, the developer need only develop his application using the OpenSlide application programming interface (API). It is OpenSlide's responsibility to deal with the idiosyncrasies of different vendor formats and to translate back and forth at runtime between the OpenSlide API and a specific vendor's data format. In this context, the DICOM standard can be viewed as yet another vendor format. OpenSlide currently handles the following vendor formats:
Aperio SVS
Hamamatsu VMS
Hamamatsu VMU
Leica SCN
3DHISTECH MRXS ("MIRAX")
Trestle TIFF
Generic tiled TIFF.
Since OpenSlide is released as open-source software under the LGPL v2.1 license, the digital pathology community can collaboratively extend it to support new vendor formats. Today, OpenSlide is supported on many Linux distributions, on Windows and on Mac OS X. Although it is not available yet on Android and iOS, we do not foresee any fundamental obstacles to supporting OpenSlide on these platforms.
DESIGN AND IMPLEMENTATION
OpenSlide aims for a good user experience on desktop-class hardware. Its design leverages the fact that although digital slide formats vary widely across vendors, they all address a common set of problems. As a result, vendor formats typically share some broad high-level features:
They pre-compute and store downsampled versions of the full-resolution image for quicker access to lower resolutions
They use various kinds of lossy compression
They are, to varying degrees, optimized for random access
They are effectively unbounded in width and height
They can store slide metadata and additional small images such as thumbnails that serve as annotations
They may not store pixel data for all regions of the image.
OpenSlide is designed to support these features while shielding applications from the details of the various vendor formats. Its vendor-neutral API provides read-only access to digital slides stored in a file system. Table 1 shows the API calls that relate to file access. For brevity and clarity, the prefix “openslide_” is omitted from the calls shown in Tables 1 through 5 and in their discussion throughout this section.
Table 1
Calls to access files

The OpenSlide API provides efficient random access to multiresolution image data using the concept of pyramid levels. Table 2 shows the relevant calls. A digital slide is represented as an ordered list of pyramid levels; level 0 is the highest-resolution level and each subsequent level is a downsampled version of the previous level. In general, no image scaling is performed by the library; the only levels available through the API are those actually stored in the slide file. The centerpiece is the read_region() function, which extracts a rectangular region of a whole-slide image at a particular pyramid level. Image data is returned in uncompressed, premultiplied ARGB (Alpha, Red, Green and Blue) format. Additional calls exist to determine the downsampling factor for each level (relative to level 0) and to determine the next largest level for an arbitrary downsample factor. All image coordinates are specified with respect to the coordinate system of level 0. Pixel data for missing image regions is rendered as transparency.
Table 2
Calls relating to image pyramids

The API provides functions for accessing slide metadata and auxiliary images. All other vendor-specific information, such as tile placement and compression formats, is hidden from the application; access to pixel data is only provided in uncompressed, premultiplied ARGB format. This allows OpenSlide to be extended to support new slide formats without changing the external API or the design of applications using the library. As shown in Table 3, OpenSlide exposes slide metadata as a set of properties, each of which is identified by a string and has a string-typed value. OpenSlide exposes generic properties for common slide metadata. Individual vendor drivers also expose properties containing format-specific metadata. In addition, small additional images embedded in the slide file, such as thumbnail or barcode images, are exposed as associated images. Unlike the main slide image, associated images are single-resolution and can only be read in their entirety. Table 4 shows the calls relating to associated images.
Table 3
Calls relating to properties

Table 4
Calls relating to associated images

Calls for error handling and version control are shown in Table 5. If an unrecoverable error such as an I/O error occurs while a slide is being read, the slide handle transitions terminally into an error state. Subsequent invocations of OpenSlide functions on the handle (other than close()) will have no effect; functions that are expected to return values will instead return an error value; read_region() will clear its destination buffer instead of filling it. The get_error() function can be used to check for errors and obtain a string describing the error. This style of error handling allows programs written in C to check for errors only when convenient, without the need to check after every call.
Table 5
Utility calls

OpenSlide is implemented in C99 and uses GLib[9] extensively for memory allocation, data structure manipulation and text file parsing. OpenSlide relies upon LibTIFF,[10] IJG's JPEG library,[11] OpenJPEG,[12] and LibXML2[13] for image reading and processing of the various formats and upon the Cairo graphics library[14] for positioning and rendering slide tiles.
OpenSlide's internal architecture is structured in a manner similar to the device driver model found in operating systems. Application-facing code is linked to vendor-specific code by way of internal constructors and function pointers. When a slide file is first opened, vendor-specific driver code is responsible for parsing the slide's metadata and locating the compressed image data for each region of the slide. Thereafter, read requests are forwarded to the vendor-specific driver, which reads the appropriate compressed data from the slide file and renders it to an ARGB image. To do this, the driver can handle the request itself or – in simple cases – forward it to generic handlers provided by the OpenSlide core.
DIGITAL SLIDE FORMATS
As previously discussed, many vendors provide limited or no public documentation for their slide formats. In some cases documentation is available, but only under disclosure restrictions incompatible with open-source software. As a result, OpenSlide's support for many formats has been implemented via empirical analysis of the raw slide data and a significant amount of trial and error. This is an inherently imperfect and iterative process that relies critically on feedback from the user community. As an OpenSlide driver gets more widely and extensively used, we discover more subtle cases where our understanding of the file format is in error. This leads to modifications that result in a more accurate version of the driver. Over time, the implementation of the driver converges to a stable and accurate version.
This approach is dependent upon the availability of sample slide files for analysis. Because it is often difficult to create these samples without access to a corresponding scanner, we are often dependent on the OpenSlide user community to contribute digital slides in interesting formats. The remainder of this section summarizes the slide formats currently supported by OpenSlide. More detailed documentation, as well as a variety of freely-redistributable slide files, is available on the OpenSlide website.[15]
Aperio SVS
Aperio SVS is a single-file, TIFF-based[16] format with non-standard metadata and a specific internal organization.[17] This format reuses a TIFF field, originally intended for free-form text data, to store structured metadata. Some SVS slides store tiles in JPEG 2000 format, which is not contemplated by the TIFF specification and not supported by LibTiff. For these slides, OpenSlide uses low-level LibTiff routines to extract the tile data and decodes it using OpenJPEG.
Hamamatsu VMS and VMU
Hamamatsu VMS is a multi-file, JPEG-based[18] format with primarily text-based metadata. Unlike other formats, which split a slide into a two-dimensional array of small tiles encoded with JPEG or JPEG 2000, VMS delivers slide data in a small number of large JPEG images. Support for random access is retrofitted into the JPEG format via a JPEG feature called “restart markers,” originally designed for error recovery. These markers allow the decoder to resynchronize at intervals throughout the image, thus creating “virtual tiles” with a very large aspect ratio (512:1 is not unusual). This aspect ratio forces superfluous decoding of image data well beyond the borders of a region of interest.
IJG's JPEG library does not natively support decoding of individual “virtual tiles,” but can be made to do so via careful restructuring of the image data. The slide metadata records the file offsets corresponding to the first tile in each row; to locate the remainder, OpenSlide scans the slide file in the background. Hamamatsu VMS files contain only two pyramid levels. However, the JPEG algorithm permits efficient downsampling of image data during decode; OpenSlide takes advantage of this feature to generate intermediate slide levels for the convenience of the application.
Hamamatsu VMU is a variant of Hamamatsu VMS, which stores uncompressed 16-bit RGB pixel data. Unlike in VMS, there is no efficient way to synthesize additional downsampled levels, so OpenSlide does not do so.
Leica SCN
Leica SCN is a single-file format based on BigTIFF. Slide metadata is stored in XML format in a TIFF text field. Unlike the other supported formats, SCN provides a pyramidal thumbnail image, though OpenSlide currently provides access to only the highest-resolution level. An SCN file can contain image pyramids for multiple regions of a slide; OpenSlide currently does not support such slides due to API limitations.
3DHISTECH MRXS ("MIRAX")
MRXS is a multi-file format with very complicated metadata in a mixture of text and binary formats. MRXS supports storing image tiles in JPEG, PNG and BMP formats; OpenSlide currently only supports JPEG. Though image tiles are approximately arranged into a grid, each group of (typically) 16 tiles has an individual offset from its “natural” grid position, which is recorded in the metadata.
Unique among the formats supported by OpenSlide, MRXS does not align overlapping tiles before concatenating and downsampling them to produce the lower-resolution levels of the image pyramid. As a result, a single 338 × 254 JPEG image might contain as many as 16,384 subimages, which must be separately extracted and positioned for display at subpixel granularity. This greatly complicates the rendering task.
The MRXS format continues to surprise us. Different versions of the vendor's software produce slightly different variants of the format and we regularly have to modify OpenSlide to support new variants discovered by the user community.
Trestle TIFF
Trestle TIFF is a multi-file format. It consists of a TIFF file containing the image pyramid and a text field with structured metadata, plus additional files containing metadata and thumbnail images. In contrast to standard TIFF, Trestle tiles are individually positioned (as in MRXS) and can overlap. The metadata records the nominal overlaps for a given pyramid level as well as the individual overlaps that apply to particular tiles. OpenSlide currently respects the nominal, but not the individual overlaps. Deviations from the nominal overlaps are typically small so this has not been a major problem in practice.
Generic Tiled TIFF
The standard TIFF format is adequate to support a digital slide containing minimal slide metadata. Such TIFF files can be produced by some scanners and can also be generated using existing open-source tools such as VIPS[19] and ImageMagick.[20] OpenSlide supports tiled TIFF images containing one or more pyramid levels.
EXAMPLE APPLICATIONS
Web-Based Viewer
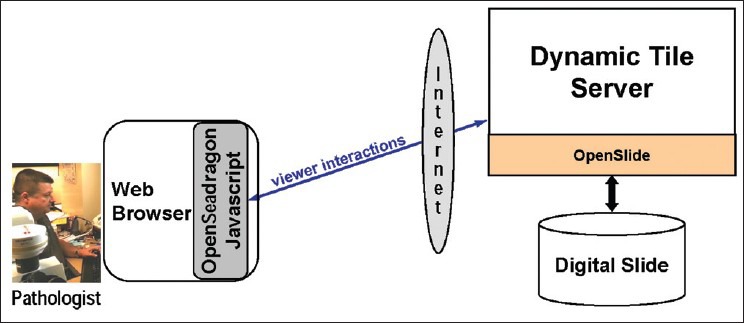
To enable remote viewing of digital slides across the Internet, we have created a web application using the OpenSeadragon AJAX image viewer[21] as shown in Figure 1. The client component of our application is JavaScript that is dynamically loaded into a standard web browser from the web page being browsed. The server component is a web server that returns tiles of specified coordinates and zoom levels on demand. These tiles can be pre-generated using an OpenSlide-based tool and served from the file system or can be produced on demand by a web application that invokes OpenSlide at runtime to generate the tiles. Our experience is that there is no user-perceptible performance difference between dynamically-tiled and statically-tiled viewing sessions. Using dynamic tiling avoids a preprocessing step in the workflow and eliminates the storage overhead of storing tiles. A demo of our viewer is available at http://openslide.org/demo/.
PathFind
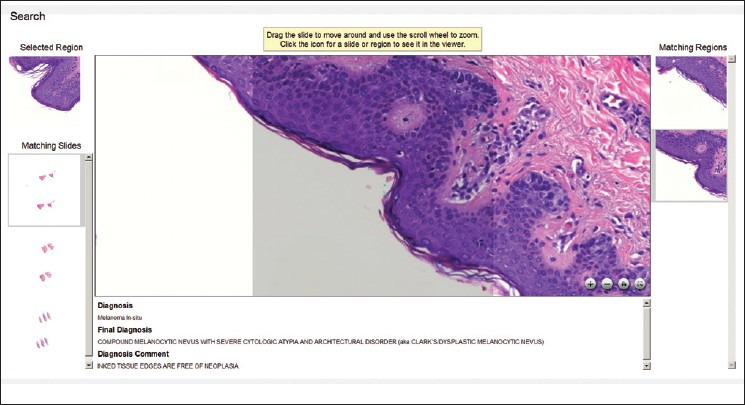
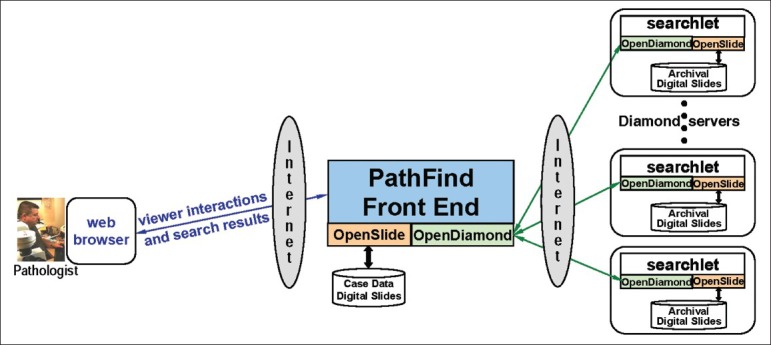
As mentioned earlier, enabling knowledge from previously encountered entities to be brought to bear on an unknown, but similar-looking diagnostic entity is a valuable capability in pathology. Of course, “similar-looking” is expressed in terms of visual attributes that have specific significance to the pathology of the tissue sample being examined. PathFind is an OpenSlide-based tool that accepts a wide range of similarity-detection software called searchlets. Each searchlet is a plugin for the OpenDiamond® platform, which enables interactive, semi-automated, hypothesis-driven searches of complex images. (The OpenDiamond platform was created by the same research group that created OpenSlide.) For example, we have created searchlets for similarity detection of cytologic atypia and pagetoid spread in skin lesions, using a machine-learning technique called Semantic Texton Forests.[22] We have also created searchlets for attributes such as nuclear density using ImageJ,[23] an open-source tool supported by the National Institutes of Health. Figure 2 shows the user interface presented to a pathologist by PathFind. The pathologist can zoom and navigate case data just as he does with a microscope and glass slides today. At any point, he can request a search for archival digital slides that have areas similar to the area currently in view. The results that are returned include entire digital slides as well as specific matching regions within each of those digital slides. Figure 3 shows the end-to-end architecture of the whole system with the roles of OpenSlide and the OpenDiamond platform highlighted. For each result, associated metadata such as anonymized patient information and diagnosis are returned.
SlideTutor
OpenSlide has also been used in SlideTutor, an intelligent tutoring system for dermatopathology.[24] Digital slides of the cases to be solved by a student are stored on a server and accessed through OpenSlide by the server-side SlideTutor software. Students access these digital slides over the Internet through a standard web browser. A correct response by a student advances him to the next case. An incorrect response triggers remediation that includes helpful visual and textual information. The results of studies using SlideTutor suggest significant learning gains after just one tutoring session.[25]
Other Use Cases
Several pathology education projects use OpenSlide to decode digital slides. One example is the Cardiac Transplant Endomyocardial Biopsy Acute Cellular Rejection Tutorial[26] from the Society for Cardiovascular Pathology and the Association for the European Cardiovascular Pathology. Another example is the Smart Histology app[27] from Smart In Media. OpenSlide is also used in the backends of online slide management systems such as Simagis Live[28] and Emory's Pathology Image Database System.[29] The VIPS image processing system,[19] a toolkit for analysis and manipulation of very large images, supports reading slide files using OpenSlide.
FUTURE WORK
OpenSlide is an evolving tool and we see many areas for improvement and enhancement:
Supporting new digital slide formats and new variants of existing formats is an ongoing challenge. We would like OpenSlide to ultimately support all major formats, including Hamamatsu NDPI, Olympus VSI, Ventana BIF and DICOM
Language bindings currently exist for Java and Python. We would like to create open-source bindings to additional programming languages such as C# and image-processing toolkits such as MATLAB. GObject bindings would allow OpenSlide to be transparently used by any programming environment that supports GObject Introspection
OpenSlide is designed primarily to support high-resolution brightfield images. As a result, the API supports only three 8-bit color channels per image. Multiple focal planes, z-slices, or time points are not supported. In the future, we would like to extend OpenSlide to support arbitrary numbers of fluorescence channels and physical axes
OpenSlide is designed as infrastructure for third-party applications that wish to support digital slides. While we provide simple viewer programs and tools to exercise the library, including the web-based viewer described earlier, there is no high-quality, open-source viewer application that can easily be installed by end users. Such an application would greatly assist pathologists and researchers wishing to view slide files from Mac OS X or Linux, especially in a multi-vendor environment
The web-based viewer could be extended to support additional features such as annotations, image rotation and synchronized panning/zooming of multiple images. Some of these features already exist in third-party applications built with OpenSlide. In addition, the creation of an embeddable viewer widget for desktop applications, similar to what OpenSeadragon provides in a web browser, would ease third-party development of such applications.
CONCLUSION
Pathology is one of the last medical specialties to be digitized, largely because of the technical problems posed by the capture, storage, transmission, archiving, viewing and annotation of high-resolution, multi-gigabyte, zoomable images. While future improvements in network bandwidth and storage capacity can alleviate these difficulties, their benefit will be limited by the increasing size and complexity of digital slides due to improvements in the resolution, multi-spectral capabilities and other aspects of digital scanner technology. The growth of mobile computing in health-care delivery will exacerbate these challenges because of the need to access digital slides over bandwidth-limited wireless networks from mobile devices with limited screen size. Today's vendor-specific and fragmented approach to digital pathology software offers few opportunities for the latest computer science innovations to be brought to bear on these difficult technical challenges. OpenSlide enables such innovations to be contributed by the best software talent. The merits of specific innovations and their implementations can be evaluated, critiqued and evolved by the entire digital pathology community. We look to a future where this process leads to a vibrant hardware-software ecosystem for digital pathology that fosters interoperability and sharing while preserving ample opportunity for proprietary innovations in hardware and application software.
ACKNOWLEDGMENTS
This research was supported by the Clinical and Translational Science Institute of the University of Pittsburgh (CTSI), under National Institutes of Health (NIH) Clinical and Translational Science Award (CTSA) program grants UL1RR024153 and UL1TR000005. We would like to thank Steve Reis and Michelle Broido for their sustained support of this work and Laura Drogowski for her help in the early phases of this project. We gratefully acknowledge and thank the entire OpenSlide community for its continuing contribution of bug reports, patches and slide data. Support for the SCN and MRXS 2.2 formats was contributed by Agelos Pappas. Stephan Lamont contributed initial support for VMU. Hauke Heibel and Marco Feuerstein implemented initial support for Windows and provided valuable feedback. Yves Sucaet contributed sample data in several formats. OpenDiamond is a registered trademark of Carnegie Mellon University.
Footnotes
Available FREE in open access from: http://www.jpathinformatics.org/text.asp?2013/4/1/27/119005
REFERENCES
Articles from Journal of Pathology Informatics are provided here courtesy of Elsevier
Full text links
Read article at publisher's site: https://doi.org/10.4103/2153-3539.119005
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics

Discover the attention surrounding your research
https://www.altmetric.com/details/115628728
Smart citations by scite.ai
Explore citation contexts and check if this article has been
supported or disputed.
https://scite.ai/reports/10.4103/2153-3539.119005
Article citations
The integrated molecular and histological analysis defines subtypes of esophageal squamous cell carcinoma.
Nat Commun, 15(1):8988, 18 Oct 2024
Cited by: 1 article | PMID: 39419971 | PMCID: PMC11487165
From whole-slide image to biomarker prediction: end-to-end weakly supervised deep learning in computational pathology.
Nat Protoc, 16 Sep 2024
Cited by: 1 article | PMID: 39285224
Review
PathEX: Make good choice for whole slide image extraction.
PLoS One, 19(8):e0304702, 29 Aug 2024
Cited by: 0 articles | PMID: 39208135 | PMCID: PMC11361590
Deep learning-based detection and semi-quantitative model for spread through air spaces (STAS) in lung adenocarcinoma.
NPJ Precis Oncol, 8(1):173, 05 Aug 2024
Cited by: 0 articles | PMID: 39103596 | PMCID: PMC11300827
Improving quality control of whole slide images by explicit artifact augmentation.
Sci Rep, 14(1):17847, 01 Aug 2024
Cited by: 0 articles | PMID: 39090284 | PMCID: PMC11294620
Go to all (183) article citations
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
OpenPhi: an interface to access Philips iSyntax whole slide images for computational pathology.
Bioinformatics, 37(21):3995-3997, 01 Nov 2021
Cited by: 2 articles | PMID: 34358287 | PMCID: PMC8570784
Dicom_wsi: A Python Implementation for Converting Whole-Slide Images to Digital Imaging and Communications in Medicine Compliant Files.
J Pathol Inform, 12:21, 11 May 2021
Cited by: 4 articles | PMID: 34267986 | PMCID: PMC8274303
Support system for pathologists and researchers.
J Pathol Inform, 6:34, 23 Jun 2015
Cited by: 0 articles | PMID: 26167378 | PMCID: PMC4485189
Applications and challenges of digital pathology and whole slide imaging.
Biotech Histochem, 90(5):341-347, 15 May 2015
Cited by: 33 articles | PMID: 25978139
Review
Funding
Funders who supported this work.
NCATS NIH HHS (1)
Grant ID: UL1 TR000005
NCRR NIH HHS (1)
Grant ID: UL1 RR024153