ISSN: 2206-7418Nanotheranostics
Nanotheranostics 2018; 2(1):96-105. doi:10.7150/ntno.22960 This issue Cite
Research Paper
Two-Color-Based Nanoflares for Multiplexed MicroRNAs Imaging in Live Cells
1. State Key Laboratory of Chemo/Biosensing and Chemometrics, College of Chemistry and Chemical Engineering, Key Laboratory for Bio-Nanotechnology and Molecular Engineering of Hunan Province, Hunan University, Changsha, China.
2. State Key Laboratory of Medical Genetics & School of Life Science, Central South University, Changsha, China.
Abstract

MicroRNAs (miRNAs) have become an ideal biomarker candidate for early diagnosis of diseases. But various diseases involve changes in the expression of different miRNAs. Therefore, multiplexed assay of miRNAs in live cells can provide critical information for our better understanding of their roles in cells and further validating of their function in clinical diagnoses. Simultaneous detection of multiple biomarkers could effectively improve the accuracy of early cancer diagnosis. Here, we develop the two-color-based nanoflares for simultaneously detecting two distinct miRNA targets inside live cells. The nanoflares consist of gold nanoparticles (AuNPs) functionalized with a dense shell of recognition sequences hybridized to two short fluorophore-labeled DNA molecules, termed “flares”. In this conformation, the close proximity of the fluorophore to the AuNPs surface leads to quenching of the fluorescence. However, when target miRNAs bind to the recognition sequence, the concomitant displacement of the flare can be detected as a corresponding increase in fluorescence. The results demonstrate that the two-color-based nanoflares can simultaneously detect miR-21 and miR-141 expression levels in various live cancer cells successfully. Compared to the traditional single-color-based nanoflares, the two-color-based nanoflares could offer more reliable and practical information for cancer detection, improving the accuracy of early disease diagnosis.
Keywords: two-color-based nanoflares, multiplexed microRNAs imaging, living cells
Introduction
MicroRNAs (miRNAs) are a group of small, single-stranded, endogenous noncoding RNAs that can regulate the expression of target genes and play significant roles in physiologic and pathologic processes.1-4 In particular, there is accumulating evidences that the dysregulated expression of miRNA is associated with various human diseases, such as cancers.5-7 Thus, miRNAs have become an ideal class of biomarker candidate for clinical diagnosis. Meanwhile, Multiple different miRNAs collaboratively regulate important cellular events at the same time, and thus, it is important to simultaneously monitor different miRNAs in cells.8 In addition, recent studies have identified that a change in the cellular status is typically related to a simultaneous change in the level of several miRNAs.9 Detection of multiplexed miRNAs is of great significance in understanding biological functions of miRNA and early diagnosis of cancers, meanwhile, simultaneous detection of multiple miRNAs brings new opportunities for improving the accuracy of early cancer detection over the single-marker assay.10,11 Unfortunately, the most commonly used methods of miRNA detection, such as qRT-PCR, Northern blot and microarrays, cannot be used in living cells. Accurate in detecting relative miRNA in bulk samples, are incapable of detecting cell-to-cell variations, they require pooling miRNA from lysates acquired from groups of cells.12-19 Therefore, a major challenge exists to accurately detect relative miRNA levels in individual live cells. Fluorescence analysis provides new possibilities for measurement and visualization of biomarkers in living cells owing to the simplicity and high sensitivity.11
Fluorescence in situ hybridization (FISH) has been the dominate detection method for detecting of intracellular miRNA.20,21 However, the requirement of cell fixation in FISH limits the application of such miRNA detection methods in live cells. Recently, it has attracted intense research interest for the development of new methodologies to meet the purpose of sensing intracellular miRNAs.22-28 For example, the Ju group employed the nucleic-acid adsorbed carbon nitride nanosheets for detecting intracellular miRNA-18a in living HepG2 cells.26 Huh and co-workers used hyaluronic-acid-based nanocontainers containing miR-34a beacons as probes for imaging miR-34a in MCF-7 cancer cells.27 Zhu group utilized an electrostatic nucleic acid AuNP nanoassembly and a MNAzyme-funtionalized gold nanorod nanodevice for imaging intracellular miRNAs.28 Simultaneous detection and imaging of multiple intracellular biomarkers hold great promise for early cancer detection.29 However, it still remains one of the major challenges. It is necessary to develop a quicker, simpler and more accurate approach to sense multiplexed microRNAs in live cells.30
One approach to meeting the above challenges termed nanoflares, which have previously been shown to be useful for detecting mRNA targets in live cells.31-33 Herein, we develop the two-color-based nanoflares for simultaneously detecting two distinct miRNA targets inside a living cell. As shown in Fig. 1, the nanoflares consists of gold nanoparticles (AuNPs) functionalized with a dense shell of recognition sequences hybridized to two short fluorophore-labeled DNA molecules, termed “flares”. In this conformation, the close proximity of the fluorophore to the AuNP surface leads to quenching of the fluorescence. However, when target miRNAs bind to the recognition sequence, the concomitant displacement of the flare can be detected as a corresponding increase in fluorescence. This two-color-based nanoflares could provide an easy, noninvasive, instantaneous and intuitive way to imaging multiplex miRNAs in living cells.30
Materials and Methods
Reagents and instruments
Trisodium citrate was obtained from Sinopharm Chemical Reagent Co., Ltd. (China). Chloroauric acid (HAuCl4·4H2O) was obtained from Shanghai Chemical Reagent Company (Shanghai, China). MgCl2, NaCl and KCl were purchased from China National Pharmaceutical Group Corporation (Shanghai, China). All other reagents were of analytical grade. 3-(4,5-Dimethylthiazol-2-yl)-2-diphenyltetrazolium bromide (MTT) were purchased from Sinopharm Chemical Reagent Co., Ltd. (China). Deoxyribonuclease I (DNase I) was purchased from Sangon Biotechnology Co., Ltd (Shanghai, China). All aqueous solutions were prepared using ultrapure water (≥18 MΩ, Milli-Q, Millipore). All oligonucleotides (Table 1) were synthesized and HPLC purified by Sangon Biotechnology Co., Ltd (Shanghai, China).
Oligonucleotide sequences in this work.
| Oligo | Sequences (5'-3') |
|---|---|
| Recognition sequence 21 | TCAACATCAGTCTGATAAGCT-T10-SH |
| Recognition sequence 141 | CCATCTTTACCAGACAGTGTTA-T10-SH |
| FAM-Flare | FAM -TAGCTTATCAGACTG (15 bases) FAM -TAGCTTATCAGACT (14 bases) FAM -TAGCTTATCAGAC (13 bases) FAM -TAGCTTATCAGA (12 bases) FAM -TAGCTTATCAG (11 bases) |
| Cy5-Flare | Cy5 -TAACACTGTCTGGTA (15 bases) Cy5 -TAACACTGTCTGGT (14 bases) Cy5 -TAACACTGTCTGG (13bases) Cy5 -TAACACTGTCTG (12 bases) Cy5 -TAACACTGTCT (11 bases) |
| miRNA21(DNA target) one-base mismatched miRNA21 | TAGCTTATCAGACTGATGTTGA TAGCATATCAGACTGATGTTGA |
| miRNA141(DNA target) one-base mismatched miRNA-41 | TAACACTGTCTGGTAAAGATGG TAACTCTGTCTGGTAAAGATGG |
| miR-21 forward | ACACTCCAGCTGGGTAGCTTATCAGACTGA |
| miR-21 reverse | CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGTCAACATC |
| miR-141 forward | ACACTCCAGCTGGGTAACACTGTCTGGT |
| miR-141 reverse | CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGCCATCTTT |
| U6 forward | CTCGCTTCGGCAGCACA |
| U6 reverse | AACGCTTCACGAATTTGCGT |
The transmission electron microscopic (TEM) images were obtained on a JEM-2100 transmission electron microscope (JEOL Ltd., Japan). The UV-vis absorption spectra were obtained with a Biospec-nano UV-vis spectrophotometer (Japan). The fluorescence spectra were obtained on a Hitachi F-7000 fluorescence spectrometer (Japan). The confocal microscope measurements were obtained using an Olympus laser-scanning confocal microscope (Tokyo, Japan). The Flow cytometry analysis was gained from Gallios machine (Beckman Coulter, USA). Cell incubation was done using a humidified HF90 CO2 incubator (Shanghai Lishen Scientific Equipment Co. Ltd.). All pH measurements of buffer were performed with Thermo Scientific Orion 3 Star pH-meter (Waltham, MA, USA). Centrifuge was performed with Beckman Coulter Allegra 25R centrifuge (Brea, CA, USA).
Schematic of the two-color-based nanoflares for miRNAs detection in live cell.

Preparation and characterization of AuNPs and nanoflares
The 13 nm AuNPs were synthesized using the sodium citrate reduction method.34 Before the experiments, all glassware was cleaned in aqua regia (HCl/HNO3, 3:1), rinsed with H2O, and then oven-dried. Next, 100 mL of 0.01% HAuCl4 was heated to boiling with vigorous stirring and 3.5 mL trisodium citrate (1%) was added under stirring. Then the solution color turned from pale yellow to colorless and finally to burgundy. Boiling was continued for an additional 10 min. After the heating source was removed, the colloid was stirred until the solution reached room temperature. Then it was filtered through a 0.45 μm Millipore membrane filter. Transmission electron microscopy (TEM) images and UV absorption indicated the particle sizes are 13 nm ± 2 nm. The concentration of AuNPs was determined by measuring their extinction at 519 nm (ε = 2.7×108 L mol-1 cm-1). The prepared AuNPs were stored at 4 oC.
The thiol-modified oligonucleotides were reduced by Tris (2-carboxyethyl) phosphine hydrochloride (TCEP·HCl) for about 1 hour and then mixed separately with corresponding flare strands, heated to 95 oC and maintained 5 min, then slowly cooled to room temperature. Two dye-modified flare duplexes (flares labelled with FAM or Cy5) were mixed and were added to 13 nm gold colloids at a concentration of 3 μM of oligonucleotide per 1 mL of 10 nM colloid and kept 16 h at 4 oC. After 16 h, Phosphate buffer (0.2 M; pH =7.4) was added to the mixture to achieve a 0.01 M phosphate concentration, the NaCl (4.0 M) concentration of the mixture was slowly increased to 0. 3 M from 0.1 M over an eight-hour period. The solution containing the nanoflares was centrifuged (13,000 rpm, 30 min) and resuspended in phosphate buffered saline (PBS; 137 mM NaCl, 10 mM Phosphate, 2.72 mM KCl, pH 7.4) three times to produce the purified AuNPs and resuspended in desired solution, stock solution at 4 oC. The nanoflares were diluted to certain concentration for use in all subsequent experiments. The concentration of AuNPs were determined by measuring their extinction at 524 nm (ε = 2.7×108 L mol-1 cm-1).
Optimization experimental conditions
To achieve optimal experimental conditions in vitro, we investigate the fluorescence change of different flares (11, 12, 13, 14, 15 bases) at various temperatures. Then, to refine the hybridization time. We treated the nanoflares with miRNA-21 target or miRNA-141 target for time monitoring 1h. The fluorescence of FAM and Cy5 were measured, respectively.
Quantitation of DNA duplexes on the AuNPs
DNA sequences loaded on AuNPs were quantitated according to the published protocol.35 Mercaptoethanol (20 mM) was added to the nanoflares solution, which was incubated overnight with shaking at room temperature. Released DNA probes were then separated via centrifugation and the fluorescence was measured with a fluorescence spectrometer. The fluorescence of FAM labelled flare was excited at 488 nm and measured at 520 nm and the fluorescence of Cy5 labelled flare was excited at 635 nm and measured at 662 nm. The fluorescence was converted to molar concentrations of flares by interpolation from a standard linear calibration curve that was prepared with known concentrations of flares with identical buffer pH, ionic strength and ME concentrations. The average and standard deviation were obtained by 3 parallel experiments in each trial. By dividing molar concentrations of each flare by the original nanoflares concentration, we calculated that there was 75±2 flares per nanoflares (40±1 FAM labelled flares; 35±1 Cy5 labelled flares).
Fluorescence experiments
For analyte detection, the nanoflares were diluted to the concentration of 3 nM in Tris-HCl buffer and treated with different concentration of the miRNA-21 targets or miRNA-141 targets (0, 2, 5, 10, 25, 50, 100, 200 nM). After incubation for 40 min at 37 oC, the fluorescence was monitored at appropriate excitation wavelengths. The fluorescence of FAM was excited at 488 nm and measured at 520 nm, the fluorescence of Cy5 was excited at 635 nm and measured at 662 nm. The nanoflares had a linear range of 0-25 nM, and the detection limits were calculated from the derived calibration curve (>13 standard deviations), as defined by IUPAC. All experiments were repeated at least three times.
Specificity experiments
To evaluate the specificity of nanoflares (3 nM) for the target miRNA-21 (100 nM) and miRNA-141 (100 nM) against other three negative control miRNAs (miRNA-200b, let 7d, and miRNA-429) with the concentration of 200 nM. Then, the nanoflares treated with no target, one-base mismatched target and target, respectively. All experiments were repeated at least three times.
Nuclease stability
Two groups nanoflares were diluted to 3 nM in tris-HCl at 37 oC. After allowing the sample to equilibrium, 1.3 μL DNase I (2U/L) was added to one group, and then time scanning 1 h to monitor the fluorescence intensity of the sample by F7000. 200 nM targets were added to above two groups to incubate for 40 min at 37 oC, respectively, the fluorescence was measured at appropriate excitation wavelengths after the solution was cooled to room temperature.
Cell culture and MTT assay
HeLa cells (human cervical cancer cell line), 22Rv1(human prostate cancer cells line), SMMC-7721 cells (human hepatocyte cell line) and LOVE-1 cells (human colon cancer cell line) were grown in RPMI 1640 medium supplemented with 10% inactivated fetal bovine serum and100 U/mL 1% antibiotics penicillin/streptomycin solution and maintained at 37 oC in a 100% humidified atmosphere containing 5% CO2 at 37 oC.
LOVE-1 cells were dispersed within replicate 96-well microtiter plates to a total volume of 200 μL well-1. Plates were maintained at 37 oC in 5% CO2/95% air incubator for 24 h. The nanoflares were added to each well after the original medium was removed. LOVE-1 cells were incubated with unmodified AuNPs (1 nM), nanoflares (1 nM and 5 nM) for 6 h, 12 h, 24 h and 48 h. Then 100 μL MTT solutions (0.5 mg mL-1 in PBS) were added to each well. After 4 h, the remaining MTT solution was removed, and 150 μL of DMSO was added to each well to dissolve the formazan crystals. The absorption was measured at 490 nm with a RT 6000 microplate reader.
Confocal fluorescence imaging
All cells were plated on 35-mm confocal laser culture dishes for 24 h. Then the nanoflares (3 nM) were respectively delivered into the cell at 37 oC in 5 % CO2 for 6 h. The cells were examined by confocal laser scanning microscopy (CLSM) with different laser transmitters. The miRNA-21 was recorded by FAM in green channel with 488 nm excitation, miRNA-141 was recorded by Cy5 in red channel with 633 nm excitation. The fluorescence images were presented after processing by image proplus 6.0 software and Image J version 1.38x software.
Flow cytometric assay and qRT-PCR
LOVE-1, 22Rv1, HeLa and SMMC-7721 cells were incubated with nanoflares. After 6 h, the cells were washed to remove the redundant particles. Cells were then detached from culture dishes using Trypsin-EDTA Solution. The solution containing treated cells was centrifuged (2000 rpm, 4 min) and resuspended in PBS three times. Flow cytometric assay was performed using Beckman Coulter Gallios machine.
Total cellular RNA was extracted from cells using Trizol reagent (Sangon Co. Ltd., Shanghai, China) according to the indicated protocol. The cDNA samples were prepared by using the reverse transcription (RT) reaction with AMV First Strand cDNA Synthesis Kit (BBI, Toronto, Canada). qPCR analysis of miRNAs were performed with SG Fast qPCR Master Mix (2X) (BBI), according to the indicated protocol on an Light Cycler480 Software Setup (Roche). The primers used in this experiment are shown in Table 1. We evaluated all the data with respect to the miRNA expression by normalizing to the expression of U6 and using the 2-∆∆Ct method.
Results and discussion
We designed the two-color-based nanoflares using 13-nm AuNPs (Fig. S1), since the size particle is an efficient quencher and does not efficiently scatter visible light,36 which is important for designing optical probes with minimal interference. The UV/Vis absorption spectra indicated that the maximum absorption of the AuNPs was at 519 nm and that it was red-shifted to 524 nm for the nanoflares (Fig. S2), which confirmed that the AuNPs were successfully functionalized with flare duplexes. But the length of flares is very important for nanoflares. If the length is short, it was instable hybridization with high background fluorescence. Conversely, it is difficult to displacement by targets. So, we study the different lengths of flares. As shown in Fig. S3, the signal-to-noise ratio increased and then up to plateau with increasing of length, while the background signals increased and then decreased. The results revealed that both of 14 and 15 bases have good signal-to-noise ratio, but 15 bases with more low background and stability than 14 bases, so we chose 15 bases as the below experiment. To demonstrate the working principle of the two-color-based nanoflares, we chose miRNA-141 and miRNA-21 as the model targets. miRNA-141 is an epithelial-associated miRNA expressed in a wide range of common human cancers including breast, lung, colon, and prostate cancer.14 miRNA-21 is reported as an oncogene and antiapoptotic indicator and is upregulated in many cancer cell lines.37-39 To obtain optimal experimental conditions, we study the kinetics of nanoflares treated with target (Fig. S4), the results suggested that nanoflares complete reaction with targets in 40 min. Equal concentrations of miRNA-141 and miRNA-21 targeted DNA strands were hybridized to their flares, respectively. Then, the complex of DNA duplex was immobilized as a dense mixed monolayer on the AuNPs surface via gold-thiol bond formation. The resultant nanoflares were then purified by centrifugation. Different fluorophore labels, FAM and Cy5, were respectively used for miRNA-21 and miRNA-141 flares to allow independent detection for each target. Quantification of DNA surface loading by fluorescence reveals that each AuNP contains approximately 75±2 flares (40±1 FAM labeled flares targeting miRNA-21, 35±1 Cy5 labeled flares targeting miRNA-141) (Fig. S5).
Fluorescence spectra for the nanoflares (3 nM) in the presence of various concentrations (0, 2, 5, 10, 25, 50, 100 and 200 nM) of miRNA-21 or miRNA-141 target. The miRNA-21 and miRNA-141 flares are labeled with FAM and Cy5, respectively. (a) Fluorescence spectra corresponding to FAM signal with excitation at 488 nm. (b) Fluorescence spectra corresponding to Cy5 signal with excitation at 635 nm.

To investigate the behavior of the two-color-based nanoflares in a cell free system, the fluorescence signals of the flares were monitored by adding of different concentration of DNA synthetic targets (Fig. 2) at appropriate excitation wavelengths. When the synthetic DNA targets were added respectively, the related fluorescence signals increased at wavelengths corresponding to the DNA flares with increasing concentration of the DNA targets from 0 to 200 nM, thus suggesting that the hybridization of the nanoflares and DNA targets can result in fluorescence recovery and that the change of fluorescence intensity is associated with the concentration of the DNA targets. The detection limits of the nanoflares were calculated to be 0.9 nM for miRNA-21 and 1.2 nM for miRNA-141, respectively.
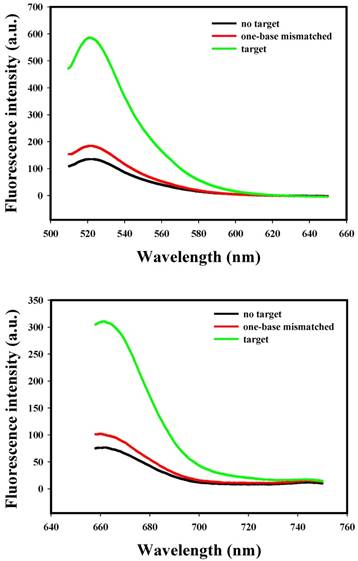
To evaluate the specificity of nanoflares, we conducted the following experiments. As shown in Fig. 3, despite the overdose of the negative control miRNAs (miR-200b, let-7d, miR-429), the negligible fluorescence intensity change was exhibited compared with that of the no target, however, when the miRNA-21 or miRNA-141 were added, significant increasing of fluorescence intensity was observed. The results demonstrated that the nanoflares have high specificity. Moreover, the results in Fig. 4 suggested that the nanoflares were efficient at signaling the presence of a specific target and also could be used to distinguish the target with a single base mismatch.
Nuclease stability is a key property of probes for detection applications in living cells. enzyme deoxyribonucleaseI (DNaseI) was used to investigate the nuclease stability of the nanoflares. The result revealed that there was not obviously degraded of the nanoflares treated with DNaseI compared to of that without DNaseI (Fig. S6). Interestingly, when target DNA were respectively added to the nanoflares and nanoflares/DNaseI, the fluorescence intensity in both solutions were enhanced greatly after hybridization, which prove that the fluorescence recovery was due to the hybridization of the nanoflares and target DNA instead of nuclease degradation.
To evaluate the cytotoxicity of the nanoflares, we performed an MTT assay in LOVE-1 cell line as an example. The absorption of MTT at 490 nm is dependent upon the degree of activation of the cells. The cell viability is then expressed by the ratio of the absorption of the cells incubated with the nanoflares to that of the cells incubated with the culture medium only. The results indicated that the unmodified AuNPs (1 nM) and the nanoflares (1 nM and 5 nM) showed almost no cytotoxicity or side effects in living cells (Fig. S7), and confirmed that the nanoflares could be used in intracellular detection.
To evaluate that the nanoflares are capable of sensing two-color-based miRNAs in living cells. Fluorescence response of the nanoflares for cellular miRNA-21 and miRNA-141 were performed using HeLa cells (human cervical cancer cell line), 22Rv1 cells (human prostate cancer cells line), LOVE-1 cells (human colon cancer cell line) and SMMC-7721 cells (human hepatocyte cell line) as models. Previous reports have shown that miRNA-21 is relative high expression levels in HeLa cells, while miRNA-141 is overexpression in 22Rv1 cells and LOVE-1 cells, and low expression level in 7721 cells.40 To test whether the nanoflares can remain stable for a long time to realize the detection of target in living cells. We incubated the nanoflares with LOVE-1 cells (high expression with miR-141) and SMMC-7721 cells (low expression with miR-141) as a control signal for Cy5, at the same time, HeLa cell (high expression with miR-21) and 22Rv1 (low expression with miR-21) as a control signal for FAM with various time from 2 h to 10 h and then imaged by a confocal microscope, respectively (Fig. S8).
Selectivity of the nanoflares for detecting miRNA-21 and miRNA-141 (100 nM) against other control miRNAs (200 nM).

Selectivity of the nanoflares for detecting target and one-base mismatched target. (a) Fluorescence spectra corresponding to FAM signal with excitation at 488 nm. (b) Fluorescence spectra corresponding to Cy5 signal with excitation at 635 nm.
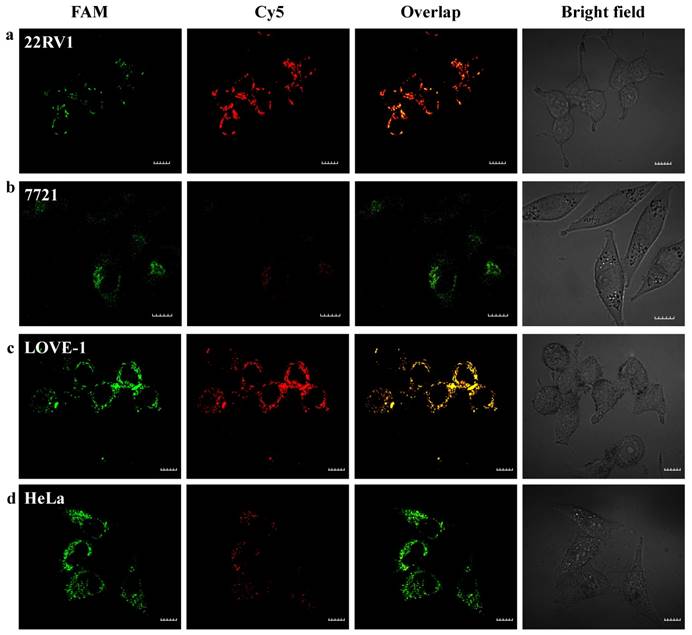
The red fluorescence intensity in LOVE-1 cells gradually increased with the incubation time and reached saturation at 6 h, while no obvious red fluorescence intensity in SMMC-7721 cells observed. These results confirm that the nanoflares are capable of application in cellular environment for at least 10 h. Here, we chose 6 h as the incubation time. Next, we applied the nanoflares for simultaneous detection of two-color-based miRNAs in the above different cells. As shown in Fig. 5, strong green and red fluorescence signals for the two miRNAs were observed under confocal microscope in LOVE-1 after incubation with nanoflares. Interestingly, the green fluorescence signal was strong, while red fluorescence signal was faint in HeLa cell, indicating that the expression of miRNA-21 was higher than that of miRNA-141 in HeLa cell, However, the result was reverse with observed in 22Rv1 cell. There was no obvious red fluorescence signal in SMMC-7721 cell, revealing that the SMMC-7721 cell has a lower miRNA-141 expression level than miRNA-21.
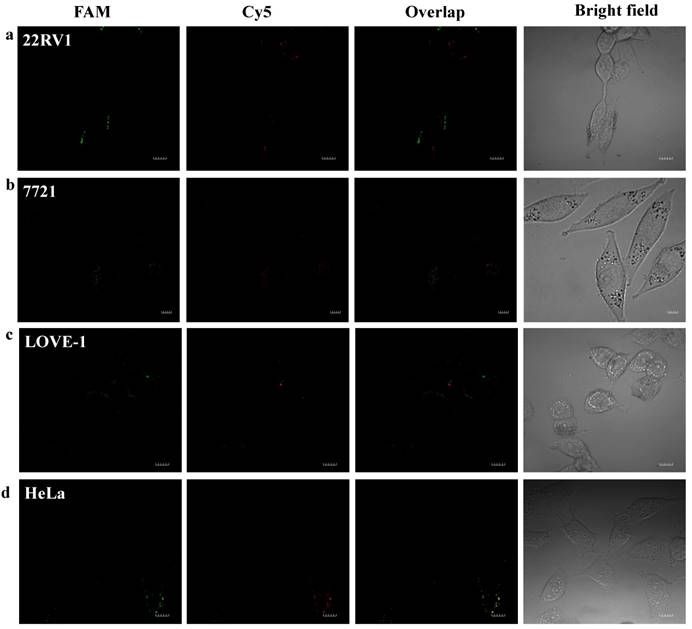
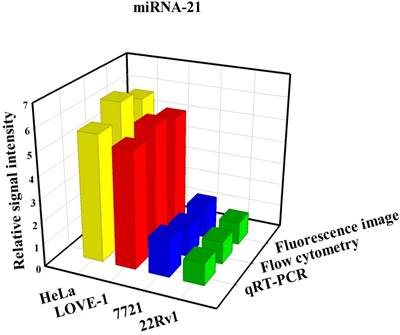
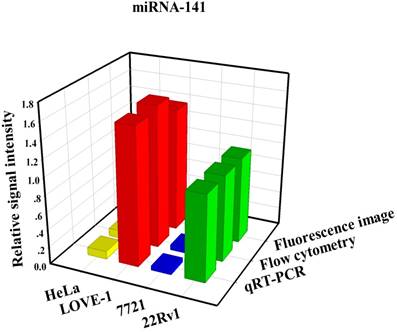
To prove that the fluorescence intensity is due to the nanoflares detecting the target miRNAs and not due to degradation, we treated non-targeting nanoflares with four cell lines as is shown in the Fig. 6, there was no obvious fluorescence intensity, which demonstrated that the fluorescence intensity is due to the nanoflares detecting the target miRNAs and not due to degradation. As is shown in Fig. 7, the flow cytometry analysis (Fig. S9) and qRT-PCR (Fig. S10) also demonstrated that the relative levels of miRNA-21 and miRNA-141 were different in various cell types, which were consistent with the results of the above fluorescence imaging analysis. Therefore, these nanoflares can sense dynamic expression and distribution of tumor-related miRNAs in cancer cells and can be used to distinguish different cell lines, which is an important goal in the field of live cell analysis.
Intracellular fluorescence imaging of miRNA-21 and miRNA-141 under confocal microscope. LOVE-1, 22Rv1, HeLa and SMMC7721 cells were incubated with the nanoflares (3 nM) for 6 h at 37oC. The miRNA-21 was recorded using FAM in the green channel with excitation at 488 nm; miRNA-141 was recorded using Cy5 in the red channel with excitation at 633 nm. Scale bars are 10 µm.
Intracellular fluorescence imaging of miRNA-21 and miRNA-141 under confocal microscope. LOVE-1, 22Rv1, HeLa and SMMC7721 cells were incubated with the non-targeting nanoflares for 6 h at 37 oC. The miRNA-21 was recorded using FAM in the green channel with excitation at 488 nm; miRNA-141 was recorded using Cy5 in the red channel with excitation at 633 nm. Scale bars are 10 µm.
Comparison of miRNA-21 (a) and (b) miRNA-141 levels detected in four kinds of cell lines (HeLa, LOVE-1, SMMC-7721 and 22Rv1) by fluorescence image, flow cytometry and qRT-PCR methods, respectively. Relative signal intensities are normalized to miRNA-21 and miRNA-141 in 22Rv1 cell lines.
Conclusion
In summary, this work shows that the two-color-based nanoflares could be used as a simple, reliable and effective platform for simultaneous detection of various miRNAs in living cells. Since, various diseases and biological processes involve changes in the expression of multiple miRNAs, such a two-color-based assay in living cell can provide critical information for our better understanding of their roles in cells and further validating of their function in clinical diagnoses. Compared to the traditional single-color-based nanoflares, the two-color-based nanoflares could offer more reliable and practical information for cancer detection, improving the accuracy of early disease diagnosis. We anticipate that the nanoflares will benefit drug discovery and early disease diagnostics.
Supplementary Material
Supplementary figures.
Acknowledgements
This work was supported by the Key Project of the National Natural Science Foundation of China (21735002), the Foundation for Innovative Research Groups of NSFC (21521063), the National Natural Science Foundation of Hunan Province (2017JJ2039) and the Fundamental Research Funds for the Central Universities.
Competing Interests
The authors have declared that no competing interest exists.
References
1. He L, Thomson JM, Hemann MT, Hernando-Monge E, Mu D, Goodson S. et al. A microRNA polycistron as a potential human oncogene. Nature. 2005;435:828-33
2. Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6:857-66
3. Rossi JJ. New hope for a microRNA therapy for liver cancer. Cell. 2009;137:990-2
4. Ventura A, Jacks T. MicroRNAs and cancer: short RNAs go a long way. Cell. 2009;136:586-91
5. Jay C, Nemunaitis J, Chen P, Fulgham P, Tong AW. MiRNA profiling for diagnosis and prognosis of human cancer. DNA Cell Biol. 2007;26:293-300
6. Mueller DW, Bosserhoff AK. Role of miRNAs in the progression of malignant melanoma. Br J Cancer. 2009;101:551-6
7. Liang D, Meyer L, Chang DW, Lin J, Pu X, Ye YQ. et al. Genetic variants in microRNA biosynthesis pathways and binding sites modify ovarian cancer risk, survival, and treatment response. Cancer Res. 2010;70:9765-76
8. Ryoo S, Lee J, Yeo J, Na H, Kim Y, Jang H. et al. Quantitative and multiplexed microRNA sensing in living cells based on peptide nucleic acid and nano graphene oxide (PANGO). ACS Nano. 2013;7:5882-91
9. Xi X, Li T, Huang Y, Sun J, Zhu Y, Yang Y. et al. RNA biomarkers: frontier of precision medicine for cancer. Non-coding RNA. 2017;3:9
10. Li N, Chang C, Pan W, Tang B. A multicolor nanoprobe for detection and imaging of tumor-related mRNAs in living cells. Angew Chem Int Ed. 2012;51:7426-30
11. Pan W, Li Y, Wang M. FRET-based nanoprobes for simultaneous monitoring of multiple mRNAs in living cells using single wavelength excitation. Chem Commun. 2016;52:4569-72
12. Liu H, Tian T, Ji D, Ren N, Ge S, Yan M. et al. A graphene-enhanced imaging of microRNA with enzyme-free signal amplification of catalyzed hairpin assembly in living cells. Biosens Bioelectron. 2016;85:909-14
13. Valoczi A, Hornyik C, Varga N, Burgyan J, Kauppinen S, Havelda Z. Sensitive and specific detection of microRNAs by northern blot analysis using LNA-modified oligonucleotide probe. Nucleic Acids Res. 2004;32:e175
14. Varallyay E, Burgyan J, Havelda Z. Detection of microRNAs by northern blot analyses using LNA probes. Methods. 2007;43:140-45
15. Raymond CK, Roberts BS, Garrett-Engele P, Lim LP, Johnson JM. Simple, quantitative primer-extension PCR assay for direct monitoring of microRNAs and short-interfering RNAs. RNA. 2005;11:1737-44
16. Liu CG, Calin GA, Meloon B, Gamliel N, Sevignani C, Ferracin M. et al. An oligonucleotide microchip for genome-wide microRNA profiling in human and mouse tissues. PNAS. 2004;101:9740-44
17. Wanunu M, Dadosh T, Ray V, Jin J, McReynolds L, Drndic M. Rapid electronic detection of probe-specific microRNAs using thin nanopore sensors. Nat Nanotechnol. 2010;5:807-14
18. Yin B, Liu Y, Ye B. One-step, two-color-based fluorescence detection of microRNAs based on duplex-specific nuclease signal amplification. J Am Chem Soc. 2012;134:5064-67
19. Wang L, Deng R, Li J. Target-fueled DNA walker for highly selective miRNA detection. Chem Sci. 2015;6:6777-82
20. Kloosterman WP, Wienholds E, de Bruijn E, Kauppinen S, Plasterk RH. In situ detection of miRNAs in Animal embryos using LNA-modifified oligonucleotide probes. Nat Methods. 2006;3:27-29
21. Schneider M, Andersen DC, Silahtaroglu A, Lyngbak S, Kauppinen S, Hansen JL. et al. Cell-specific detection of microRNA expression during cardiomyogenesis by combined in situ hybridization and immunohistochemistry. J Mol Histol. 2011;42:289-99
22. Cheglakov Z, Cronin T, Chuan H, Weizmann Y. Live cell microRNA imaging suing cascade hybridization reaction. J Am Chem Soc. 2015;137:6116-19
23. Min X, Zhang M, Huang F, Lou X, Xia F. Live cell microRNA imaging using exonucleaseIII-aided recycling amplification based on aggregation-induced emission luminogens. ACS Appl Mater Interfaces. 2016;8:8998-9003
24. Wang Y, Yu Z, Zhang Z, Ren R, Zhang S. Orderly nucleic acid aggregates by electrostatic self-assembly in single cells for miRNA detection and visualizing. Analyst. 2016;141:2861-64
25. Yang Y, Huang J, Yang X, He X, Quan K, Xie N. et al. Gold nanoparticle based hairpin-locked-DNAzyme probe for amplified miRNA imaging in living cells. Anal Chem. 2017;89:5850-56
26. Liao X, Wang Q, Ju H. Simultaneous sensing of intracellular microRNAs with a multi-functionalized carbon nitride nanosheet probe. Chem Commun. 2014;50:13604-07
27. Kim E, Yang J, Park J, Kim S, Kim NH, Yook JI. et al. Consecutive targetable smart nanoprobe for molecular recognition of cytoplasmic microRNA in metastatic breast cancer. ACS Nano. 2012;6:8525-35
28. Zhang P, He Z, Chen W, Chen J, Zhao J, Zhu X. et al. In situ amplification of intracellular microRNA with MNAzyme nanodevice for multiplexed imaging, logic operation, and controlled drug release. ACS Nano. 2015;9:789-98
29. Pan W, Zhang T, Yang H, Diao W, Li N, Tang B. Multiplexed detection and imaging of intracellular mRNAs using a four-color nanoprobe. Anal Chem. 2013;85:10581-88
30. Luan M, Li N, Pan W, Yang L, Yu Z, Tang B. Simultaneous detection of multiple targets involved in PI3K/AK pathway for investigating cellular migration and invasion with a multicolor fluorescence nanoprobe. Chem Commun. 2017;53:356-59
31. Seferos DS, Giljohann DA, Hill HD, Prigodich AE, Mirkin CA. Nano-flares: probes for transfection and mRNA detection in living cells. J Am Chem Soc. 2007;129:15477-79
32. Prigodich AE, Seferos DS, Massich MD, Giljohann DA, Lane BC, Mirkin CA. Nano-flares for mRNA regulation and detection. ACS Nano. 2009;3:2147-52
33. Prigodich AE, Randeria PS, Briley WE, Kim NJ, Daniel WL, Giljohann DA. et al. Two-color-based nanoflares: mRNA detection in live cells. Anal Chem. 2012;84:2062-6
34. Grabar KC, Freeman RG, Hommer MB, Natan MJ. Preparation and characterization of Au colloid monolayers. Anal Chem. 1995;67:735-43
35. Demers LM, Mirkin CA, Mucic RC, Reynolds RA, Letsinger RL, Elghanian R. et al. Fluorescence-based method for determining the surface coverage and hybridization efficiency of thiol-capped oligonucleotides bound to gold thin films and nanoparticles. Anal Chem. 2000;72:5535-41
36. Alivisatos A, Johnsson K, Peng X, Wilson T, Loweth C, Bruchez M. et al. Organization of “nanocrystal molecules” using DNA. Nature. 1996;382:609-11
37. Gumireddy K, Young D, Xiong X, Hogenesch J, Huang Q, Deiters A. Small-molecule inhibitors of microRNA miR-21 function. Angew Chem Int Ed. 2008;47:7482-84
38. Ali S, Almhanna K, Chen W, Philip PA, Sarkar FH. Differentially expressed miRNAs in the plasma may provide a molecular signature for aggressive pancreatic cancer. Am J Transl Res. 2010;3:28-47
39. Ali S, Ahmad A, Banerjee S, Padhye S, Dominiak K, Schaffert JM. et al. Gemcitabine sensitivity can be induced in pancreatic cancer cells through modulation of miR-200 and miR-21 expression by curcumin or its analogue CDF. Cancer Res. 2010;70:3606-17
40. Wang Q, Li Q, Yang X, Wang K. Grapheneoxide-gold nanoparticles hybrids-based surface plasmon resonance for sensitive detection of microRNA. Biosens Bioelectron. 2016;77:1001-7
Author contact
![]() Corresponding authors: kmwangedu.cn; jinhuangedu.cn
Corresponding authors: kmwangedu.cn; jinhuangedu.cn
Received 2017-9-23
Accepted 2017-10-30
Published 2018-1-1

 Global reach, higher impact
Global reach, higher impact