Advertisement
Research ArticleVirology Free access | 10.1172/JCI93472
Anthrax toxin receptor 1 is the cellular receptor for Seneca Valley virus
Linde A. Miles,1 Laura N. Burga,2 Eric E. Gardner,1 Mihnea Bostina,2,3 John T. Poirier,1,4 and Charles M. Rudin1,4,5
1Molecular Pharmacology Program and Department of Medicine, Memorial Sloan Kettering Cancer Center, New York, New York, USA.
2Department of Microbiology and Immunology and
3Otago Centre for Electron Microscopy, University of Otago, Dunedin, New Zealand.
4Department of Medicine, Memorial Sloan Kettering Cancer Center, New York, New York, USA.
5Department of Medicine, Weill Cornell Medical College, New York, New York, USA.
Address correspondence to: Charles M. Rudin, Memorial Sloan Kettering Cancer Center, 300 E. 66th Street, RM1203, New York, New York, 10065, USA. Phone: 646.888.4527; Email: [email protected]. Or to: John T. Poirier, Memorial Sloan Kettering Cancer Center, 415 E. 68th Street, RM1701, New York, New York 10065, USA. Phone: 646.888.3588; Email: [email protected].
Find articles by Miles, L. in: JCI | PubMed | Google Scholar
1Molecular Pharmacology Program and Department of Medicine, Memorial Sloan Kettering Cancer Center, New York, New York, USA.
2Department of Microbiology and Immunology and
3Otago Centre for Electron Microscopy, University of Otago, Dunedin, New Zealand.
4Department of Medicine, Memorial Sloan Kettering Cancer Center, New York, New York, USA.
5Department of Medicine, Weill Cornell Medical College, New York, New York, USA.
Address correspondence to: Charles M. Rudin, Memorial Sloan Kettering Cancer Center, 300 E. 66th Street, RM1203, New York, New York, 10065, USA. Phone: 646.888.4527; Email: [email protected]. Or to: John T. Poirier, Memorial Sloan Kettering Cancer Center, 415 E. 68th Street, RM1701, New York, New York 10065, USA. Phone: 646.888.3588; Email: [email protected].
Find articles by Burga, L. in: JCI | PubMed | Google Scholar
1Molecular Pharmacology Program and Department of Medicine, Memorial Sloan Kettering Cancer Center, New York, New York, USA.
2Department of Microbiology and Immunology and
3Otago Centre for Electron Microscopy, University of Otago, Dunedin, New Zealand.
4Department of Medicine, Memorial Sloan Kettering Cancer Center, New York, New York, USA.
5Department of Medicine, Weill Cornell Medical College, New York, New York, USA.
Address correspondence to: Charles M. Rudin, Memorial Sloan Kettering Cancer Center, 300 E. 66th Street, RM1203, New York, New York, 10065, USA. Phone: 646.888.4527; Email: [email protected]. Or to: John T. Poirier, Memorial Sloan Kettering Cancer Center, 415 E. 68th Street, RM1701, New York, New York 10065, USA. Phone: 646.888.3588; Email: [email protected].
Find articles by Gardner, E. in: JCI | PubMed | Google Scholar
1Molecular Pharmacology Program and Department of Medicine, Memorial Sloan Kettering Cancer Center, New York, New York, USA.
2Department of Microbiology and Immunology and
3Otago Centre for Electron Microscopy, University of Otago, Dunedin, New Zealand.
4Department of Medicine, Memorial Sloan Kettering Cancer Center, New York, New York, USA.
5Department of Medicine, Weill Cornell Medical College, New York, New York, USA.
Address correspondence to: Charles M. Rudin, Memorial Sloan Kettering Cancer Center, 300 E. 66th Street, RM1203, New York, New York, 10065, USA. Phone: 646.888.4527; Email: [email protected]. Or to: John T. Poirier, Memorial Sloan Kettering Cancer Center, 415 E. 68th Street, RM1701, New York, New York 10065, USA. Phone: 646.888.3588; Email: [email protected].
Find articles by Bostina, M. in: JCI | PubMed | Google Scholar
1Molecular Pharmacology Program and Department of Medicine, Memorial Sloan Kettering Cancer Center, New York, New York, USA.
2Department of Microbiology and Immunology and
3Otago Centre for Electron Microscopy, University of Otago, Dunedin, New Zealand.
4Department of Medicine, Memorial Sloan Kettering Cancer Center, New York, New York, USA.
5Department of Medicine, Weill Cornell Medical College, New York, New York, USA.
Address correspondence to: Charles M. Rudin, Memorial Sloan Kettering Cancer Center, 300 E. 66th Street, RM1203, New York, New York, 10065, USA. Phone: 646.888.4527; Email: [email protected]. Or to: John T. Poirier, Memorial Sloan Kettering Cancer Center, 415 E. 68th Street, RM1701, New York, New York 10065, USA. Phone: 646.888.3588; Email: [email protected].
Find articles by Poirier, J. in: JCI | PubMed | Google Scholar
1Molecular Pharmacology Program and Department of Medicine, Memorial Sloan Kettering Cancer Center, New York, New York, USA.
2Department of Microbiology and Immunology and
3Otago Centre for Electron Microscopy, University of Otago, Dunedin, New Zealand.
4Department of Medicine, Memorial Sloan Kettering Cancer Center, New York, New York, USA.
5Department of Medicine, Weill Cornell Medical College, New York, New York, USA.
Address correspondence to: Charles M. Rudin, Memorial Sloan Kettering Cancer Center, 300 E. 66th Street, RM1203, New York, New York, 10065, USA. Phone: 646.888.4527; Email: [email protected]. Or to: John T. Poirier, Memorial Sloan Kettering Cancer Center, 415 E. 68th Street, RM1701, New York, New York 10065, USA. Phone: 646.888.3588; Email: [email protected].
Find articles by Rudin, C. in: JCI | PubMed | Google Scholar
Published June 26, 2017 - More info
J Clin Invest. 2017;127(8):2957–2967. https://doi.org/10.1172/JCI93472.
Copyright © 2017, American Society for Clinical Investigation
Received: February 16, 2017; Accepted: May 4, 2017
-
Abstract
Seneca Valley virus (SVV) is an oncolytic picornavirus with selective tropism for neuroendocrine cancers. It has shown promise as a cancer therapeutic in preclinical studies and early-phase clinical trials. Here, we have identified anthrax toxin receptor 1 (ANTXR1) as the receptor for SVV using genome-wide loss-of-function screens. ANTXR1 is necessary for permissivity in vitro and in vivo. However, robust SVV replication requires an additional innate immune defect. We found that SVV interacts directly and specifically with ANTXR1, that this interaction is required for SVV binding to permissive cells, and that ANTXR1 expression is necessary and sufficient for infection in cell lines with decreased expression of antiviral IFN genes at baseline. Finally, we identified the region of the SVV capsid that is responsible for receptor recognition using cryoelectron microscopy of the SVV-ANTXR1-Fc complex. These studies identify ANTXR1, a class of receptor that is shared by a mammalian virus and a bacterial toxin, as the cellular receptor for SVV.
-
Introduction
Seneca Valley virus (SVV) is the prototype member of the Senecavirus genus within the family Picornaviridae (1). SVV is a clade I strain of Senecavirus A (SVA) that has been shown to selectively infect and lyse cancers with neuroendocrine features, including a subset of small-cell lung cancer (SCLC) and pediatric neuroendocrine solid tumors (1–4). These cancers constitute a major cause of morbidity and mortality — SCLC alone is responsible for approximately 30,000 deaths annually in the US (5). Previous studies in preclinical mouse models and early-phase clinical trials confirmed the safety and potential efficacy of SVV as a novel cancer treatment, but clinical development of SVV has been hampered by a lack of understanding of host dependency factors (2, 3, 6–9).
Picornavirus tropism is restricted to permissive cells by the requirement for host expression of a viral receptor. Picornavirus receptors are typically immunoglobulin superfamily (IgSF) proteins that consist of a single transmembrane domain and 2–5 Ig domains (10). In all cases, the extreme amino-terminal Ig domain of the receptor interacts with the viral capsid in a canyon that surrounds the 5-fold axis. While IgSF receptors are most common, non-IgSF receptors and attachment factors are also documented and include transmembrane proteins as well as carbohydrates such as sialic acid and heparan sulfate (11, 12). Sialic acid acts as a receptor or attachment factor for a number of viruses and has been proposed as a component of the SVV receptor. Although enzymatic removal or blocking of sialic acid with lectins modestly reduces SVV infection in pediatric glioma models, the definitive identity of the SVV receptor has remained elusive (13).
Despite a requirement for receptor expression, this alone may not be sufficient for productive infection in all contexts. Mammalian viruses have varying degrees of sensitivity to innate cellular antiviral responses (14). The potency of cellular antiviral responses is similarly variable in normal and cancerous cells (14, 15). SCLC in particular has been shown to frequently lack key components of MHC class I antigen presentation as well as to have decreased expression of immune stimulatory cytokines (16–18). Both result in decreased tumor cell recognition and removal by the immune system. In the absence of innate immune defect, exogenous expression of the appropriate receptor may be insufficient for permissivity and is likely a shared requirement for successful replication of many oncolytic viruses (19).
In this study, we performed 2 genome-wide loss-of-function screens followed by gene expression analysis of publicly available cancer cell line data to identify essential host dependency factors of SVV infection.
-
Results
Genome-wide loss-of-function screens identify ANTXR1 as essential for SVV infection. The pooled genome-scale CRISPR knockout (GeCKO) v2 human single-guide RNA (sgRNA) library targets over 19,000 genes within the human genome and has the ability to efficiently knock out genes using the Cas9 DNA nuclease (20–24). Because of the high efficiency of gene disruption in haploid cells, we performed a genome-wide screen in one of the only known human haploid cell lines, HAP1, which we found to be permissive to SVV at relatively high MOI (refs. 25, 26, and Supplemental Figure 1A; supplemental material available online with this article; https://doi.org/10.1172/JCI93472DS1). HAP1-Cas9 cells were transduced with pooled GeCKO lentivirus library and challenged with an SVV infection at an MOI of 1,000 virus particle/cell (vp/cell), which resulted in more than 90% cell death (Figure 1A and Supplemental Figure 1A). Genomic DNA was extracted from the expanded surviving cell population and subsequently analyzed by high-throughput sequencing to determine changes in sgRNA representation in comparison with controls. Representation of nontargeting control sgRNAs was maintained from the plasmid pools until the end of the screen; however, notable changes were observed in targeting sgRNAs, reflecting loss of sgRNAs that target essential genes (Supplemental Figure 1, B–E). The most significantly enriched sgRNAs in the SVV selected pool were found to target the ANTXR1 gene, which encodes anthrax toxin receptor 1 (27). ANTXR1 and the testis expressed 2 gene (TEX2) were the only genes with multiple sgRNAs significantly enriched in the SVV-resistant sample (Figure 1B). Highly enriched sgRNAs were individually tested for the ability to confer resistance to SVV in HAP1 cells (Figure 1C). We observed 6 sgRNAs targeting 3 different genes that conferred SVV resistance after gene knockout in HAP1 cells, including 3 independent sgRNAs targeting the ANTXR1 gene. Both enriched sgRNAs targeting the TEX2 gene conferred resistance as well as 1 sgRNA targeting the nuclear receptor gene NR2C2.
 Figure 1
Figure 1Identification of ANTXR1 as host dependency factor for SVV. (A) Depiction of genome-scale CRISPR knockout (GeCKO) screen workflow. After lentiviral transduction of the sgRNA library, transduced cells were selected by puromycin. Cells were then challenged with SVV to select for resistant cells. (B) The screen identified ANTXR1 (blue) and TEX2 (red) as the most significant hits. Nontargeting control sgRNAs are highlighted in black. Log fold change (logFC) in selected over control pools is indicated on the vertical axis as a function of the average log counts per million reads (logCPM). (C) HAP1 cells were transduced with individual sgRNAs identified from the HAP1 GeCKO screen. Cell viability was assayed in the absence (light gray) or presence (black) of SVV. Each bar corresponds to the average of n = 6 replicates with error bars representing SD. Dashed lines indicate parental HAP1 cell viability in the absence and presence of SVV. (D) Individual sgRNAs identified in the 25 H446 GeCKO screen colonies were transduced into parental H446 cells. Cell viability was tested in the absence (light gray) or presence (black) of SVV. Parental H446 cell viability in the absence and presence of SVV is indicated with dashed lines. Each bar corresponds to the average of n = 6 replicates with error bars representing SD.
To validate our results in a cell line of immediate relevance to neuroendocrine cancers, we repeated the GeCKO screen in the highly SVV-permissive H446 SCLC cell line. GeCKO lentivirus-transduced H446-Cas9 cells were challenged with SVV at an MOI of 1 vp/cell, which resulted in more than 99% cell death. The percentage of surviving cells after SVV infection was much lower than in the HAP1 screen, allowing isolation of individual cell colonies, instead of a pooled population. Genomic DNA from each colony was extracted, and individual sgRNAs were identified by Sanger sequencing (Table 1). In 23 of 25 resistant colonies (92%), sgRNAs targeting ANTXR1 were present, and comprised 3 independent sgRNAs targeting ANTXR1. Each sgRNA identified in the H446 screen was tested individually in a secondary screen of parental H446 cells for the ability to confer SVV resistance (Figure 1D). All 3 ANTXR1-targeting sgRNAs identified in the screen were able to confer resistance to SVV; however, no other candidate sgRNAs altered SVV permissivity in parental H446 cells.
ANTXR1 is necessary for permissivity in neuroendocrine cancer cell lines. We assessed the genomic sequence of ANTXR1 in clones isolated from the H446 screen and found that all 5 ANTXR1-KO clones contained insertions or deletions (indels) in exon 2 of the ANTXR1 gene. These indels would lead to a frameshift and premature stop codon, predicted to result in a truncated ANTXR1 protein (Table 2). We also confirmed the loss of SVV permissivity in ANTXR1-KO clones using a cell viability assay with parental H446 cells and nonpermissive A549 cells as positive and negative controls, respectively (Figure 2A). After a 72-hour incubation period with SVV, we observed a significant loss of viability with increasing MOI of SVV in parental H446 cells. All ANTXR1-KO lines as well as A549 cells showed no significant change in cell viability with MOI of SVV over 5 logs higher than effective exposures for parental H446 cells, indicative of highly SVV-resistant cells.
 Figure 2
Figure 2Knockout of ANTXR1 leads to the loss of SVV permissivity. (A) Three of the H446 ANTXR1-KO cell lines were challenged with increasing MOIs of SVV for 72 hours. Cell viability was determined via AlamarBlue. Parental H446 cells and the nonpermissive NSCLC cell line A549 were used as positive and negative controls, respectively. Each data point represents the average of n = 6 replicates with error bars representing SD. (B) Permissive SCLC and pediatric cancer cell lines were transduced with an sgRNA targeting ANTXR1. Parental (light gray) and ANTXR1-KO (black) cells were challenged with SVV-GFP and analyzed by flow cytometry. Each bar represents the average of n = 3 replicates with error bars representing SD. Unpaired 2-sided t tests were used to determine statistical significance. (C) Parental H446 cells (top), H446 ANTXR1-KO mCherry cells (bottom), or 1:1 mixture of parental/ANTXR1-KO mCherry cells (middle) were challenged with SVV-GFP. White arrows indicate adjacent SVV-GFP–infected parental H446 and uninfected ANTXR1-KO mCherry cells. Scale bars: 100 μm. Images representative of 3 independent experiments. (D) Parental H446 tumors (light green/dark green), H446 ANTXR1-KO mCherry tumors (pink/red), and 1:1 mixture of parental/ANTXR1-KO mCherry tumors (light purple/dark purple) were challenged with WT SVV-001 or PBS vehicle. Tumor volumes were measured every other day. Each data point corresponds to the average of n = 4–5 tumors with error bars representing SD. (E) Tumors were excised at the experiment endpoint and analyzed by flow cytometry. Each bar represents the average of n = 4–5 tumors with error bars representing SD. A 2-way ANOVA test with multiple comparisons was used to determine statistical significance. ****P ≤ 0.0001.
To determine whether ANTXR1 is essential for SVV infection in additional neuroendocrine cancer cell lines, we generated ANTXR1-KO lines in the SCLC cell lines H446, LX22cl, and H82, as well as HAP1 and the SVV-permissive pediatric cancer cell lines Y79 and TC-71. Each ANTXR1-KO line was challenged with an infectious SVV reporter virus that expresses GFP within the viral polyprotein (SVV-GFP) (3, 28). Cells were analyzed by flow cytometry using the corresponding parental cell line as a positive control (Figure 2B). In all cases ANTXR1-KO profoundly decreased SVV-GFP infection by at least 70% in the KO cell lines compared with the corresponding parental lines. ANTXR1 gene knockout leads to a loss of SVV permissivity in permissive cell lines of multiple tumor types.
Some viruses are able to increase viral spread into bystander cells without the use of their canonical receptor, but it is currently unknown whether SVV has this capability (29). We further examined whether bystander cells lacking ANTXR1 expression could be infected by neighboring cells through cell-cell spread in a mixed cell population containing both parental and ANTXR1-KO cells. We first created an H446 ANTXR1-KO clone that stably expressed the mCherry fluorescent protein (ANTXR1-KO mCherry). We then cocultured parental H446 and ANTXR1-KO mCherry cells at a 1:1 cell number ratio and challenged cells with SVV-GFP using pure parental and ANTXR1-KO mCherry cultures as controls (Figure 2C). As expected, only single-GFP-positive (GFP+) or single-mCherry-positive (mCherry+) cells but not dual-positive GFP+mCherry+ cells were observed in the admixed cell culture. Additionally, we engrafted immunodeficient nude mice with parental H446 cells, ANTXR1-KO mCherry cells, or a 1:1 admixture of parental and ANTXR1-KO mCherry cells (Figure 2D) and challenged with SVV-001. Parental H446 tumors completely regressed upon administration of SVV-001, whereas the 1:1 parental/ANTXR1-KO mCherry tumor cohort showed only an initial delay in tumor progression. Tumors that progressed in the 1:1 parental/ANTXR1-KO mCherry SVV-001 cohort were significantly enriched in mCherry+ cells, consistent with elimination of the parental H446 population from the tumor (Figure 2E). As expected, ANTXR1-KO mCherry tumors were unaffected by the administration of SVV-001 compared with control. These results confirm that ANTXR1 expression is required for eradication of H446 xenograft tumors in vivo.
Defects in innate immune signaling are required for SVV replication. We next sought to determine whether ANTXR1 expression level in cell lines is predictive of permissivity using publicly available gene expression data of the 1,037 cell lines in the Cancer Cell Line Encyclopedia (CCLE) (30). We first determined an expression cutoff based on the distribution of expression in the CCLE (31). Approximately 37% of cell lines fell below the expression cutoff (Figure 3A). Of the cell lines in the CCLE, 81 have been previously assessed for permissivity (1, 2). Of these lines, biased toward inclusion of neuroendocrine cancer lines, 20 were found to be permissive. ANTXR1 expression was significantly associated with permissivity (P = 0.0023, Fisher’s exact test). Most strikingly, none of the 20 permissive cell lines lacked expression of ANTXR1, supporting the hypothesis that ANTXR1 is a required host dependency factor for SVV infection.
 Figure 3
Figure 3ANTXR1 expression is significantly associated with permissivity. (A) Scaled log2 ANTXR1 gene expression of permissive (top), nonpermissive (middle), and all (1,037) cell lines in the CCLE (bottom). ANTXR1 expression was significantly associated with permissivity (P = 0.0023, Fisher’s exact test). Inset table summarizes the findings of cell lines confirmed to be permissive (P) or nonpermissive (NP) and their ANTXR1 expression status as ANTXR1-expressing (ANTXR+) or nonexpressing (ANTXR1–). (B) An enrichment score was calculated for each cell line tested and for each of the 7 significant gene sets. Enrichment scores are highly correlated between the gene sets given their overlapping gene constituents. Permissivity is indicated in the column label. (C) An enrichment barcode plot depicting the negative enrichment of type I IFN signaling genes in permissive cell lines (q = 0.0046). The t statistic for each gene in the set is plotted as a vertical black bar. Significantly upregulated genes fall within the red region, while significantly downregulated genes fall within the blue region. (D) Sample-wise enrichment scores were calculated for the top enriched gene set and plotted based on the histology of the tumor of origin. SCLC and neuroblastoma stand out as lacking genes involved in type I IFN signaling. (E) Receiver operating characteristics of ANTXR1 (AUC = 0.66), IFN enrichment score (AUC = 0.80), or a combination of the 2 predictors (AUC = 0.89).
While ANTXR1 expression appears to be a requirement for SVV permissivity, the CCLE data set suggests that it is not sufficient: 42 of 62 ANTXR1-expressing cell lines analyzed for permissivity (67.7%) are nonpermissive, corresponding to a low positive predictive value of 0.32. We were therefore prompted to identify predictive gene expression differences between ANTXR1-expressing permissive and nonpermissive classes. We used competitive gene set enrichment to identify significantly differentially expressed gene sets from the Reactome database (32, 33). We identified 7 gene sets, all of which were significantly downregulated in permissive cell lines expressing ANTXR1 (Figure 4B). The most significant gene set was INTERFERON_ALPHA_BETA_SIGNALING, in which 34 of 44 genes (77%) were significantly downregulated in permissive cell lines. The enrichment for this gene set (q = 0.0046) can be visualized in Figure 3C. A sample-wise analysis of gene expression was performed to see whether the gene set enrichment we observed was driven by cell lines derived from a particular tumor histology (ref. 34 and Figure 3B). We found that lack of expression of these gene sets was enriched among SCLC and neuroblastoma cell lines (Figure 3D). To determine whether this expression signature was operant in vivo, we performed a similar analysis of human tumor xenograft data from the Pediatric Preclinical Testing Program (PPTP) and found that permissivity to SVV is concordant with downregulation of IFN signaling at baseline (Supplemental Figure 2, A and B, and refs. 2, 35). An analysis of the receiver operating characteristics indicates that the best-performing predictor consisting of a combination of ANTXR1 expression and type I IFN enrichment score results in an area under the curve (AUC) of 0.89 (Figure 3E). Moreover, the combined predictor performs significantly better than either predictor individually (P < 0.005). Taken together, these results suggest that robust SVV replication requires both expression of the cellular receptor ANTXR1 and downregulation of expression of antiviral IFN signaling genes at baseline.
 Figure 4
Figure 4Re-expression of ANTXR1 reconstitutes SVV permissivity. Cells were cotransfected with the ANTXR1-HA and mCherry expression plasmids (A, B, and D). (A) An H446 ANTXR1-KO cell line was transfected, then challenged with SVV-GFP. Scale bars: 100 μm. Images representative of 3 independent experiments. (B) H446 and LX22cl ANTXR1-KO cell lines were transfected, challenged with SVV-GFP, and analyzed by flow cytometry. mCherry-transfected parental (P) and ANTXR1-KO cells were used as positive and negative controls, respectively. Each bar represents the average of n = 3 replicates with error bars representing SD. A 2-way ANOVA test with multiple comparisons was used to determine statistical significance. (C) ANTXR1-HA protein expression was confirmed by Western blot. Blot representative of 2 independent Westerns. (D) H69 and H146 cells were transduced with a Dox-inducible ANTXR1-HA expression lentivirus. Parental and ANTXR1-expressing cells were incubated in absence or presence of 1 μg/ml doxycycline for 72 hours, challenged with SVV-GFP, and analyzed by flow cytometry. Each bar represents the average of n = 3 replicates with error bars representing SD. A 2-way ANOVA test with multiple comparisons was used to determine statistical significance. (E) ANTXR1-HA protein expression in the presence of doxycycline was confirmed by Western blot. Blot representative of 2 independent Westerns. ****P ≤ 0.0001
We have previously shown, using gene expression data from SCLC cell lines and patient-derived xenograft models, that SVV permissivity is correlated to the differential gene expression of 2 neurogenic transcription factors, NEUROD1 and ASCL1 (3). High NEUROD1/ASCL1 gene expression ratios were enriched in SVV permissive cell lines and therefore were suggested as a classifier for SCLC subtype and SVV permissivity. Interestingly, these cell lines are enriched for downregulation of antiviral IFN signaling genes, potentially explaining our prior observations (Supplemental Figure 2C).
We further sought to test whether an SVV infection in ANTXR1-expressing cells could be affected by the presence of an active IFN pathway. We performed 1-step single growth kinetic assays with SVV-GFP (MOI = 0.1 vp/cell) in parental H446, H446 ANTXR1-KO, and DMS79 cells, which express ANTXR1 but have higher gene expression of the IFN pathway components (Supplemental Figure 2D). H446 ANTXR1-KO or DMS79 were significantly impaired in viral production compared with parental H446 cells. These results suggest that high baseline expression of IFN pathway components in ANTXR1-expressing cells can attenuate an SVV infection.
Re-expression of ANTXR1 rescues SVV permissivity. To test the specificity of the ANTXR1 sgRNAs, we evaluated whether exogenous re-expression of ANTXR1 could rescue permissivity to SVV in ANTXR1-KO cells. We cotransfected 3 H446 ANTXR1-KO lines with an ANTXR1-HA expression plasmid and an mCherry fluorescent protein expression plasmid and challenged the cells with SVV-GFP 16 hours after transfection (Figure 4A and Supplemental Figure 3). Compared with untransfected ANTXR1-KO cells that did not show any GFP+ cells, ANTXR1-KO cells transfected with the ANTXR1-HA expression plasmid consistently showed GFP+ cells, indicative of a productive SVV-GFP infection and rescue of SVV permissivity. To further test the importance of ANTXR1 expression in permissive cells, we cotransfected the H446 and LX22cl ANTXR1-KO lines with the ANTXR1-HA and mCherry expression plasmids, and subsequently incubated the cells with SVV-GFP. Cells were analyzed by flow cytometry and gated to select for transfected cells (mCherry+). Compared with parental mCherry+GFP+ cells, we observed a significant decrease in the mCherry+GFP+ population in ANTXR1-KO H446 and LX22cl cells that was rescued upon transfection with the ANTXR1-HA expression plasmid (Figure 4B). Expression of the ANTXR1-HA fusion protein was confirmed in each ANTXR1-HA–transfected cell line by immunoblot using an HA tag–specific antibody (Supplemental Figure 3 and Figure 4C). Re-expression of ANTXR1 protein in ANTXR1-KO cell lines is sufficient to rescue SVV permissivity.
Exogenous expression of ANTXR1 is sufficient to induce SVV permissivity. We next sought to determine whether expression of ANTXR1 protein was sufficient to increase the permissivity of the nonpermissive SCLC cell lines H69 and H146, which do not express the gene. After transduction with a doxycycline-inducible ANTXR1-HA expression lentivirus, we incubated parental and ANTXR1-expressing H69 and H146 cells in the presence or absence of 1 μg/ml doxycycline for 72 hours, challenged with SVV-GFP, and analyzed by flow cytometry (Figure 4D). Parental H69 and H146 cells and ANTXR1-transduced cells, in the absence of doxycycline, showed GFP+ populations under 1.5%, as expected. Upon doxycycline treatment, both ANTXR1-transduced H69 and H146 cells showed a significant increase in SVV-GFP–infected cells to 7.46% ± 0.17% and 18.3% ± 0.20%, respectively. Expression of the ANTXR1-HA protein in the doxycycline-induced cells was confirmed by Western blot (Figure 4E). These data confirm that exogenous expression of ANTXR1 is sufficient to induce permissivity in SVV-resistant SCLC cell lines that lack endogenous ANTXR1.
ANTXR1 interacts directly with SVV. As ANTXR1 is a transmembrane protein and required for SVV infection in various permissive SCLC cell lines, we sought to determine whether ANTXR1 interacts directly with SVV. We used an ANTXR1-Fc chimera or a control isotype IgG1-Fc protein for coimmunoprecipitation (co-IP) studies. After incubation of Fc-bead complexes with SVV, all bound proteins were eluted and analyzed by Western blot using SVV rabbit antisera (Figure 5A). In all serially diluted ANTXR1-Fc samples incubated with SVV, we observed viral protein bands as well as a decrease in intensity of the bands corresponding to a decrease of bound ANTXR1-Fc protein. We did not detect any SVV protein bands in samples incubated with the IgG1-Fc isotype control or samples not incubated with SVV. After confirming a direct interaction, we repeated the ANTXR1-Fc chimera co-IP studies in the presence of increasing amounts of sodium chloride (NaCl) to investigate the strength of the interaction in vitro under high ionic strength (Figure 5B). The intensity of viral protein bands did not change significantly with increasing salt concentration up to 2 M NaCl. As ANTXR1 has high sequence similarity to the high-affinity anthrax receptor, ANTXR2, we performed the co-IP with the ANTXR2-Fc chimeric protein using the ANTXR1-Fc protein as a positive control (27). We did not observe any bands corresponding to viral proteins in ANTXR2-Fc samples incubated with SVV, indicating the absence of an interaction between the extracellular domain of ANTXR2 and SVV (Figure 5C). Bands corresponding to viral protein were observed only in ANTXR1-Fc samples. Our results indicate that ANTXR1, and not ANTXR2, can directly interact with SVV in a high-affinity and stable interaction.
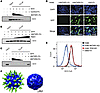 Figure 5
Figure 5SVV interacts directly with ANTXR1. (A) SVV was coimmunoprecipitated with decreasing amounts of an ANTXR1-Fc chimera. Bound proteins were eluted and analyzed by Western blot using an anti-SVV antibody. Input SVV was immunoblotted as a positive control. Blot representative of 2 independent experiments. (B) SVV was coimmunoprecipitated with the ANTXR1-Fc chimera. Washes were performed with increasing concentrations of NaCl up to 2 M. Bound proteins were eluted and analyzed as described in A. Blot representative of 2 independent experiments. (C) SVV was coimmunoprecipitated with the ANTXR1-Fc chimera or decreasing amounts of ANTXR2-Fc chimera and analyzed as described in A. Blot representative of 2 independent experiments. (D) SVV-GFP was preincubated with the ANTXR1-Fc chimera, ANTXR2-Fc chimera, or IgG-Fc isotype control before an 8-hour incubation with parental H446 cells. Cell nuclei were stained with a NucBlue Live ReadyProbe. Scale bars: 100 μm. Images representative of 3 independent experiments. (E) ANTXR1-KO (blue) and TEX2-KO (red) cells were incubated with SVV-Cy5 and analyzed by flow cytometry. Parental H446 (black) and DMS114 (gray) cells were used as positive and negative controls for SVV binding, respectively. Data representative of 2 independent experiments. (F) Cryo-EM density map of SVV capsid (blue) bound to ANTXR1-Fc chimera (green).
Additionally, we investigated which region of the ANTXR1 extracellular domain was essential for the interaction with SVV and was therefore essential for rescue of SVV permissivity in ANTXR1-KO cells. We created an N-terminal deletion series of ANTXR1-HA expression plasmids that deleted increasing regions of the extracellular domain sequence of the ANTXR1 protein while preserving the signal peptide sequence (Supplemental Figure 4A). We then tested the ability of the truncated expression plasmids to rescue SVV permissivity in one of the H446 ANTXR1-KO clones (Supplemental Figure 4B). Unlike full-length ANTXR1 protein, all ANTXR1 truncations were unable to rescue SVV permissivity when challenged with SVV-GFP. We confirmed full-length and truncated ANTXR1-HA protein expression via Western blot (Supplemental Figure 4C). These results suggest there are required residues for the interaction of SVV and ANTXR1 located in the extreme N-terminal region of the ANTXR1 protein.
Soluble ANTXR1-Fc chimera blocks SVV infection in vitro. We next sought to determine whether the interaction between the SVV and ANTXR1-Fc or ANTXR2-Fc chimera could attenuate a cellular SVV infection. We incubated SVV-GFP with the ANTXR1-Fc, ANTXR2-Fc, or IgG1-Fc protein before an overnight incubation with parental H446 cells and subsequent analysis by fluorescence microscopy (Figure 5D). Cells incubated with SVV-GFP and IgG1-Fc or ANTXR2-Fc protein showed high levels of GFP+ cells indicative of a productive SVV infection. Cells incubated with SVV-GFP and ANTXR1-Fc protein showed no detectable GFP+ cells, indicating a substantial lack of SVV-GFP infection in these cells. These results demonstrate that only exogenous ANTXR1 protein and not ANTXR2 protein is able to block a cellular SVV-GFP infection, and further support ANTXR1 as the primary cellular receptor for SVV.
Loss of ANTXR1 protein expression abrogates SVV binding to permissive cells. We sought to determine whether ANTXR1-KO cells had lost the ability to bind SVV. We also assessed the potential role of TEX2, another candidate from the HAP1 screen, in binding SVV. We incubated parental, ANTXR1-KO, and TEX2-KO H446 cells with WT SVV labeled with the fluorophore Cy5 (SVV-Cy5) and analyzed the cells by flow cytometry using the nonpermissive SCLC cell line DMS114 as a negative control for SVV binding (Figure 5E). Parental H446 incubated with SVV-Cy5 showed a high level of fluorescence (mean fluorescence [MF] = 2,373) compared with DMS114 cells incubated with SVV-Cy5 (MF = 425). TEX2-KO H446 cells showed a similar fluorescence profile to parental H446 cells (MF = 2,233), indicating there was no loss of SVV binding ability corresponding to loss of TEX2 protein expression. In contrast, ANTXR1-KO H446 cells showed a markedly diminished fluorescence profile similar to that of the negative control line, DMS114 (MF = 358). Loss of SVV binding was observed only in ANTXR1-KO cells, indicating that not only does ANTXR1 bind directly to SVV based on co-IP data, but it is the major binding determinant for the virus in intact cells.
Cryoelectron microscopy of capsid-receptor complex. We analyzed the complex of SVV bound to the ANTXR1-Fc chimeric protein by cryoelectron microscopy (cryo-EM). Picornaviruses have an icosahedral capsid formed by 60 copies of a protomer consisting of 3 major capsid proteins, VP1, VP2, and VP3, and a fourth much smaller protein, VP4, positioned in the interior of the capsid. Copies of VP1 are assembled around the 5-fold axis, while VP2 and VP3 alternate around the 3-fold axis (36). Our reconstruction matches the existing atomic model of the virus (37) when filtered to 14.5 Å resolution (Figure 5F). Additionally, the map displays the receptor subdomains distributed radially around the 5-fold axis, in crown-like geometry similar to other picornaviruses, such as poliovirus (38), rhinovirus (39), or coxsackieviruses (40). The map revealed the receptor binding quasiperpendicular to the capsid close to the center of the protomer, making contact with all 3 major capsid proteins and centered around the “puff” loop of VP2.
-
Discussion
ANTXR1 functions as 1 of the 2 receptors for the Bacillus anthracis toxin (27). SVV is unique among known viruses in using ANTXR1, or any related protein, as a primary receptor. In contrast to a number of previously identified receptors of other picornaviruses, ANTXR1 is not a member of the immunoglobulin superfamily (IgSF) of receptors (10). Although ANTXR1 shares common features to the IgSF receptors in being a single-pass transmembrane glycoprotein, it may be unique in its role as a receptor to both a mammalian virus and a bacterial toxin.
Interestingly, ANTXR1 was initially discovered as a tumor endothelial marker and is also known as TEM8 (41). We show that ANTXR1 is frequently expressed on the surface of tumor cells compared with normal cells. Efforts to develop a therapeutic antibody targeting ANTXR1 expressed in tumor endothelium have been hampered by cross-reactivity of the antibody to ANTXR2. The exquisite selectivity of SVV for ANTXR1 and the medium-resolution cryo-EM map described here may inform future therapeutic development in the antibody space for both antiangiogenic purposes and a potentially novel target for select neuroendocrine cancers.
Prior work described by Liu et al. suggested that sialic acid could be a component of the SVV receptor in pediatric glioma models (13). As sialic acids can be found on a number of different carrier molecules, enzymatic removal of sialic acids and competitive blocking by lectins may potentially affect diverse cellular processes or occlude a portion of the virus/receptor binding interface, altering SVV permissivity. Conversely, the data presented here identify ANTXR1, a receptor that is both necessary and sufficient for SVV permissivity in both SCLC and pediatric cancer cells.
Our gene expression analysis suggests that beyond expression of ANTXR1, cancer cell defects in the innate immune response are important determinants of successful SVV replication. We have previously shown that SVV has a preference for the variant subtype of SCLC, with permissivity correlated to a high NEUROD1/ASCL1 gene expression ratio (3). Although these neurogenic transcription factors were not identified in our screens, we have shown that NEUROD1 expression can be used as a correlative indicator of innate immune response pathway activity. These features, together with ANTXR1 expression, may define a category of cancers particularly amenable to treatment with SVV. An improved understanding of how cellular innate immune response pathways dictate permissivity could identify synergistic combination strategies with therapeutic agents targeting these pathways in cancer cells.
The identification of the receptor for a virus is an important first step in understanding its tropism. For SVV, this discovery is additionally germane, as ANTXR1 expression will facilitate the identification of patients who could potentially benefit from SVV virotherapy. SCLC is a highly aggressive and nearly universally lethal cancer, for which few tractable therapeutic targets have been identified. Many large-scale clinical trials, conducted without biomarker selection, have failed to advance the standard of care for this disease (42, 43). Carefully defined biomarkers that would focus novel therapeutic studies on the potentially responsive subset of patients could change this field. The identification of ANTXR1, and possibly suppressed innate immunity, as selection criteria will help define the structure of our subsequent clinical trials.
-
Methods
Human GeCKO v2 library screens. The Human GeCKO v2 library was obtained as 2 half libraries (libraries A and B) in the lentiGuide-Puro plasmid backbone (Addgene plasmid 52962) as a gift from Feng Zhang (Broad Institute, Massachusetts Institute of Technology, Cambridge, MA, USA). The Memorial Sloan Kettering Cancer Center RNAi core facility amplified the pooled libraries by electroporation of Endura electrocompetent cells (Lucigen) as described previously (20, 21). All cell line and virus information is described in the Supplemental Methods. DNA libraries were used to create lentivirus, transduced into H446-Cas9 and HAP1-Cas9 cells, and challenged with SVV-001 as reported in Supplemental Methods. All methods performed to identify and test each sgRNA for importance in SVV permissivity are described in Supplemental Methods.
Cell viability assays and analysis. Twenty-four hours before infection, cells (5.0 × 103) were seeded into black opaque 96-well plates (Corning) in 100 μl media. Plates were infected with serial dilutions of SVV from an MOI of 5,000 vp/cell to 5.0 × 10–5 vp/cell and incubated for 24–72 hours. Each MOI was tested in 3–6 replicate wells with uninfected cells as controls. AlamarBlue cell viability solution was added to each well and incubated at 37°C. Fluorescence emission at 590 nm was obtained after excitation at 565 nm using a Synergy Neo plate reader (BioTek) with wells containing only media as background controls. Background fluorescence values were subtracted and replicate wells averaged to determine average fluorescence and SD for each MOI of SVV. The average fluorescence value at each MOI was divided by average fluorescence value of the control wells to calculate percentage cell viability. Cell viability values and SDs were plotted against MOI of SVV using GraphPad Prism 6 software.
In vivo SVV-001 efficacy. Female athymic nude mice, aged 6–8 weeks, were purchased from Envigo Inc. Mice were engrafted s.c. with a 1:1 mixture of Matrigel (Corning) and either parental H446 cells, H446 ANTXR1-KO mCherry cells, or 1:1 mix of parental/KO mCherry cells in HBSS. Once tumors reached volumes of approximately 100 mm3, mice within each cohort were randomly distributed and administered SVV-001 (1 × 1013 vp/kg) via i.p. injection or PBS, pH 7.4, as vehicle controls. Tumor dimensions were measured with external calipers every 48 hours. Tumor volumes were estimated by the formula V = (L × W2)/2, where L is the length or diameter and W is the width. Calculated tumor averages for each cohort and SDs were plotted using GraphPad Prism 6 software. At the end of the study, mice were euthanized and tumors excised and analyzed by flow cytometry.
ANTXR1 expression experiments. Cells were transiently cotransfected with the ANTXR1-HA and pLenti6 W118-mCherry expression plasmids using polyethylenimine and subsequently harvested for Western blot analysis or challenged with SVV-GFP for flow cytometric analysis. Further detailed experiments are described in Supplemental Methods.
SVV-GFP infections. Unless otherwise stated, cells were seeded in a tissue culture–treated well plate (Corning) 24 hours before infection. Plates were infected with SVV-GFP at an MOI of 5.0 vp/cell and incubated at 37°C for 8 or 16 hours. NucBlue Live ReadyProbe reagent (Invitrogen) was added to each well and incubated at 37°C for 20 minutes. Images of cells were obtained using an EVOS FL Auto fluorescence microscope (Invitrogen). Specific SVV-GFP infections are further detailed in Supplemental Methods.
SVV-Cy5 binding experiments. SVV was incubated with the amine-reactive Cy5 dye (GE Healthcare) in sodium carbonate buffer (pH 9.3) for 1 hour at room temperature. Excess dye was removed by filtration through gel filtration columns (GE Healthcare) in HEPES buffer. Virus aliquots were stored at –80°C. Parental, ANTXR1-KO, and TEX2-KO H446 cells were incubated with SVV-Cy5 for 30 minutes at 37°C in OptiMEM. The nonpermissive SCLC cell line DMS114 was used as a negative control. Cells were then processed and analyzed via flow cytometry as described in Supplemental Methods.
Coimmunoprecipitations. Magnetic Protein G Dynabeads (Invitrogen) were used for all immunoprecipitation experiments. Unless otherwise stated, Dynabeads and Dynabead-protein complexes were washed 3 times with PBS, pH 7.4, supplemented with 0.02% Tween-20 (Sigma-Aldrich). Dynabeads were immobilized for manipulation and washing using a DynaMag magnet (Life Technologies). Proteins were eluted by boiling of Dynabead-protein complexes for 10 minutes at 90°C using RIPA buffer supplemented with NuPAGE sample reducing agent and LDS sample buffer. Details for specific co-IP experiments are reported in Supplemental Methods.
Western blotting. Eluted Dynabead proteins or protein extracts were resolved on a 4%–12% Bis-Tris polyacrylamide gel with MOPS running buffer (Life Technologies) and transferred to a PVDF membrane (Millipore). For co-IP experiments, membranes were blotted with purified rabbit antisera against SVV (Neotropix Inc.) (1). For ANTXR1 transfection cell lysates, membranes were blotted with commercial primary antibodies against the HA tag (Cell Signaling, catalog 3724S) or GAPDH (Santa Cruz Biotechnology, catalog sc-25778) or vinculin (Cell Signaling, catalog 13901) as a loading control. Immunoblotting was performed using HRP-conjugated secondary antibodies (Cell Signaling) and detection by chemiluminescence (GE Life Sciences).
Flow cytometry analysis. Parental and ANTXR1-KO cell lines were seeded in tissue culture–treated 6-well plates 24 hours before SVV-GFP infection. Cells were infected with SVV-GFP at the 50% tissue culture infective dose (TCID50) for each cell line and incubated at 37°C for 6–16 hours with uninfected cells as controls. Cells were subsequently harvested, processed, and analyzed via flow cytometry on a BD LSR II Flow Cytometer (Becton Dickinson) as reported in Supplemental Methods. Additional gating and analysis were performed with FlowJo analysis software (Tree Star). Analyzed data and SDs were plotted using GraphPad Prism 6 software.
Gene expression analysis. Normalized gene expression data for cancer cell lines was downloaded from the Cancer Cell Line Encyclopedia (CCLE) or from the Pediatric Preclinical Testing Program (PPTP) (30, 35). Custom content descriptor files (CDFs) were used for both gene expression data sets. For CCLE microarray data, we used a CDF corresponding to ENTREZG v15. For PPTP microarray data, which include admixed mRNA of both human and mouse origin, we used a human-specific H-spec CDF (44). To determine the appropriate cutoff for cell lines expressing ANTXR1, local modes in the density distribution of ANTXR1 expression were identified, the lowest of which was designated as nonexpressed. The SD of this peak was then determined and an expression cutoff equal to 10 SDs above the mode was set, based on the work of Zilliox and Irizarry (31). Similar results were obtained using a Gaussian mixture model. Gene expression analysis was performed using the R statistical programming environment and the Bioconductor suite of tools. Differentially expressed genes were identified using LIMMA to fit a linear model to each gene and generate moderated t statistics using an empirical Bayes approach. Gene set enrichment analysis was performed using CAMERA, a purely competitive gene set testing approach (45). Sample-wise enrichment was determined using gene set variation analysis (34). Receiver operating characteristics were calculated using sample-wise enrichment score, ANTXR1 expression, or a combined predictor modeled using logistic regression as predictors.
Cryoelectron microscopy. Equal volumes of virions at 0.2 mg/ml and ANTXR1 at 1 mg/ml were mixed, giving a ratio of approximately 10:1 receptors per binding site. The samples were mixed and kept for 90 minutes at 37°C and transferred on ice for another 90 minutes. Specimens were prepared by application of 3 μl of purified virus on glow-discharged Quantifoil holey carbon grids (Quantifoil Micro Tools GmbH). The excess buffer was blotted and the grid was flash-plunged into liquid ethane using a Leica KF80 cryofixation device (C. Reichert Optische Werke AG). Grids were loaded onto a Gatan 914 Cryoholder. Images were collected on a JEOL JEM2200FS microscope (JEOL Ltd.) operated at 200 kV using minimal dose conditions with an electron dose of approximately 30 electrons/Å2. An in-column omega energy filter was used to improve image contrast by zero-loss filtering with a slit width of 25 eV. Automated data collection was carried out using SerialEM software. The micrographs were recorded at a defocus between 1 and 3 μm, on a 4k × 4k complementary metal oxide semiconductor camera (Tietz Video and Image Processing Systems) at a calibrated magnification of 50,000 corresponding to a pixel size of 3.12 Å.
One thousand seven hundred individual virus particles were selected from micrographs using E2BOXER software (46). Contrast transfer function (CTF) parameters were calculated using CTFFIND3 (47), and micrographs with poor CTF estimates were discarded. Orientation, classification, and refinement were done in Relion (48) using as initial reference a strongly low-pass version of the SVV atomic model (37). By calculating the Fourier shell correlation between 2 halves of the data set, the resolution of the map was estimated to be 14.5 Å. Similarly, a 34-Å reconstruction of the capsid without the receptor bound was calculated from 950 particles. The reconstructed maps were visualized using Chimera (49).
Statistics. Two-way ANOVA tests with multiple comparisons were performed where applicable to determine statistical significance.
Study approval. All animal experiments and procedures were carried out under an animal protocol approved by the Institutional Animal Care and Use Committee at Memorial Sloan Kettering Cancer Center, New York, NY, USA.
-
Author contributions
Authors contributed to this work in the following ways: designing research studies: LAM, LNB, MB, JTP, and CMR; conducting experiments: LAM, LNB, EEG, and JTP; acquiring data: LAM, LNB, and EEG; analyzing data: LAM, LNB, MB, and JTP; and writing the manuscript: LAM, MB, JTP, and CMR.
-
Acknowledgments
We thank Stephen Leppla and Shihui Liu (NIH/NAIAD) for their contribution of plasmids used in this study as well as members of the Rudin laboratory for their critical review of the manuscript. We thank the members of the RNAi core facility at Memorial Sloan Kettering Cancer Center for their technical assistance with the Human GeCKO v2 library. Electron microscopy was performed at the Otago Centre for Electron Microscopy. We also thank Neotropix Inc. and Perceiver Pharmaceuticals LLC for contributions to this work. This study was supported by National Cancer Institute grant T32 CA009243, the Lung Cancer Research Foundation (to LAM), the Burroughs Wellcome Fund, the American Association for Cancer Research, the Caring for Carcinoid Foundation (to JTP and CMR), and National Cancer Institute grant P30 CA008748.
Address correspondence to: Charles M. Rudin, Memorial Sloan Kettering Cancer Center, 300 E. 66th Street, RM1203, New York, New York, 10065, USA. Phone: 646.888.4527; Email: [email protected]. Or to: John T. Poirier, Memorial Sloan Kettering Cancer Center, 415 E. 68th Street, RM1701, New York, New York 10065, USA. Phone: 646.888.3588; Email: [email protected].
-
Footnotes
LAM’s present address is: Human Oncology and Pathogenesis Program, Memorial Sloan Kettering Cancer Center, New York, New York, USA.
Conflict of interest: L.A. Miles, J.T. Poirier, and C.M. Rudin have filed for a patent claiming use of ANTXR1 as a diagnostic biomarker for SVV virotherapy (PCT/US2016/064679).
Reference information: J Clin Invest. 2017;127(8):2957–2967.https://doi.org/10.1172/JCI93472.
-
References
- Reddy PS, et al. Seneca Valley virus, a systemically deliverable oncolytic picornavirus, and the treatment of neuroendocrine cancers. J Natl Cancer Inst. 2007;99(21):1623–1633.
- Morton CL, et al. Initial testing of the replication competent Seneca Valley virus (NTX-010) by the pediatric preclinical testing program. Pediatr Blood Cancer. 2010;55(2):295–303.
- Poirier JT, Dobromilskaya I, Moriarty WF, Peacock CD, Hann CL, Rudin CM. Selective tropism of Seneca Valley virus for variant subtype small cell lung cancer. J Natl Cancer Inst. 2013;105(14):1059–1065.
- Segalés J, Barcellos D, Alfieri A, Burrough E, Marthaler D. Senecavirus A. Vet Pathol. 2017;54(1):11–21.
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2015. CA Cancer J Clin. 2015;65(1):5–29.
- Wadhwa L, Hurwitz MY, Chévez-Barrios P, Hurwitz RL. Treatment of invasive retinoblastoma in a murine model using an oncolytic picornavirus. Cancer Res. 2007;67(22):10653–10656.
- Yu L, et al. A single intravenous injection of oncolytic picornavirus SVV-001 eliminates medulloblastomas in primary tumor-based orthotopic xenograft mouse models. Neuro-oncology. 2011;13(1):14–27.
- Burke MJ, et al. Phase I trial of Seneca Valley Virus (NTX-010) in children with relapsed/refractory solid tumors: a report of the Children’s Oncology Group. Pediatr Blood Cancer. 2015;62(5):743–750.
- Rudin CM, et al. Phase I clinical study of Seneca Valley Virus (SVV-001), a replication-competent picornavirus, in advanced solid tumors with neuroendocrine features. Clin Cancer Res. 2011;17(4):888–895.
- Rossmann MG, He Y, Kuhn RJ. Picornavirus-receptor interactions. Trends Microbiol. 2002;10(7):324–331.
- Zhou L, Luo Y, Wu Y, Tsao J, Luo M. Sialylation of the host receptor may modulate entry of demyelinating persistent Theiler’s virus. J Virol. 2000;74(3):1477–1485.
- Fry EE, et al. The structure and function of a foot-and-mouth disease virus-oligosaccharide receptor complex. EMBO J. 1999;18(3):543–554.
- Liu Z, et al. Intravenous injection of oncolytic picornavirus SVV-001 prolongs animal survival in a panel of primary tumor-based orthotopic xenograft mouse models of pediatric glioma. Neuro-oncology. 2013;15(9):1173–1185.
- Randall RE, Goodbourn S. Interferons and viruses: an interplay between induction, signalling, antiviral responses and virus countermeasures. J Gen Virol. 2008;89(pt 1):1–47.View this article via: PubMed Google Scholar
- Dunn GP, Koebel CM, Schreiber RD. Interferons, immunity and cancer immunoediting. Nat Rev Immunol. 2006;6(11):836–848.
- Doyle A, et al. Markedly decreased expression of class I histocompatibility antigens, protein, and mRNA in human small-cell lung cancer. J Exp Med. 1985;161(5):1135–1151.
- Yazawa T, et al. Lack of class II transactivator causes severe deficiency of HLA-DR expression in small cell lung cancer. J Pathol. 1999;187(2):191–199.
- Fischer JR, et al. Selective suppression of cytokine secretion in patients with small-cell lung cancer. Ann Oncol. 1995;6(9):921–926.
- Kaufman HL, Kohlhapp FJ, Zloza A. Oncolytic viruses: a new class of immunotherapy drugs. Nat Rev Drug Discov. 2015;14(9):642–662.
- Shalem O, et al. Genome-scale CRISPR-Cas9 knockout screening in human cells. Science. 2014;343(6166):84–87.
- Sanjana NE, Shalem O, Zhang F. Improved vectors and genome-wide libraries for CRISPR screening. Nat Methods. 2014;11(8):783–784.
- Cong L, et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339(6121):819–823.
- Mali P, et al. RNA-guided human genome engineering via Cas9. Science. 2013;339(6121):823–826.
- Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337(6096):816–821.
- Carette JE, et al. Ebola virus entry requires the cholesterol transporter Niemann-Pick C1. Nature. 2011;477(7364):340–343.
- Pillay S, et al. An essential receptor for adeno-associated virus infection. Nature. 2016;530(7588):108–112.
- Bradley KA, Mogridge J, Mourez M, Collier RJ, Young JA. Identification of the cellular receptor for anthrax toxin. Nature. 2001;414(6860):225–229.
- Poirier JT, et al. Characterization of a full-length infectious cDNA clone and a GFP reporter derivative of the oncolytic picornavirus SVV-001. J Gen Virol. 2012;93(pt 12):2606–2613.View this article via: PubMed Google Scholar
- Bergelson JM. Intercellular junctional proteins as receptors and barriers to virus infection and spread. Cell Host Microbe. 2009;5(6):517–521.
- Barretina J, et al. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature. 2012;483(7391):603–607.
- Zilliox MJ, Irizarry RA. A gene expression bar code for microarray data. Nat Methods. 2007;4(11):911–913.
- Milacic M, et al. Annotating cancer variants and anti-cancer therapeutics in reactome. Cancers (Basel). 2012;4(4):1180–1211.
- Croft D, et al. The Reactome pathway knowledge base. Nucleic Acids Res. 2014;42(Database issue):D472–D477.View this article via: PubMed Google Scholar
- Hänzelmann S, Castelo R, Guinney J. GSVA: gene set variation analysis for microarray and RNA-seq data. BMC Bioinformatics. 2013;14:7. View this article via: PubMed Google Scholar
- Neale G, et al. Molecular characterization of the pediatric preclinical testing panel. Clin Cancer Res. 2008;14(14):4572–4583.
- Tuthill TJ, Groppelli E, Hogle JM, Rowlands DJ. Picornaviruses. Curr Top Microbiol Immunol. 2010;343:43–89.View this article via: PubMed Google Scholar
- Venkataraman S, Reddy SP, Loo J, Idamakanti N, Hallenbeck PL, Reddy VS. Structure of Seneca Valley Virus-001: an oncolytic picornavirus representing a new genus. Structure. 2008;16(10):1555–1561.
- Strauss M, Filman DJ, Belnap DM, Cheng N, Noel RT, Hogle JM. Nectin-like interactions between poliovirus and its receptor trigger conformational changes associated with cell entry. J Virol. 2015;89(8):4143–4157.
- Kolatkar PR, Bella J, Olson NH, Bator CM, Baker TS, Rossmann MG. Structural studies of two rhinovirus serotypes complexed with fragments of their cellular receptor. EMBO J. 1999;18(22):6249–6259.
- Organtini LJ, Makhov AM, Conway JF, Hafenstein S, Carson SD. Kinetic and structural analysis of coxsackievirus B3 receptor interactions and formation of the A-particle. J Virol. 2014;88(10):5755–5765.
- Carson-Walter EB, Watkins DN, Nanda A, Vogelstein B, Kinzler KW, St Croix B. Cell surface tumor endothelial markers are conserved in mice and humans. Cancer Res. 2001;61(18):6649–6655.View this article via: PubMed Google Scholar
- Hann CL, Rudin CM. Fast, hungry and unstable: finding the Achilles’ heel of small-cell lung cancer. Trends Mol Med. 2007;13(4):150–157.
- Oze I, et al. Twenty-seven years of phase III trials for patients with extensive disease small-cell lung cancer: disappointing results. PLoS One. 2009;4(11):e7835.
- Isella C, et al. Stromal contribution to the colorectal cancer transcriptome. Nat Genet. 2015;47(4):312–319.
- Wu D, Smyth GK. Camera: a competitive gene set test accounting for inter-gene correlation. Nucleic Acids Res. 2012;40(17):e133.
- Tang G, et al. EMAN2: an extensible image processing suite for electron microscopy. J Struct Biol. 2007;157(1):38–46.
- Mindell JA, Grigorieff N. Accurate determination of local defocus and specimen tilt in electron microscopy. J Struct Biol. 2003;142(3):334–347.
- Scheres SH. RELION: implementation of a Bayesian approach to cryo-EM structure determination. J Struct Biol. 2012;180(3):519–530.
- Pettersen EF, et al. UCSF Chimera — a visualization system for exploratory research and analysis. J Comput Chem. 2004;25(13):1605–1612.
-
Version history
- Version 1 (June 26, 2017): Electronic publication
- Version 2 (August 1, 2017): Print issue publication



Copyright © 2024 American Society for Clinical Investigation
ISSN: 0021-9738 (print), 1558-8238 (online)







