Advertisement
Research ArticleNeuroscience Free access | 10.1172/JCI66827
FcγRIIb mediates amyloid-β neurotoxicity and memory impairment in Alzheimer’s disease
Tae-In Kam,1 Sungmin Song,1 Youngdae Gwon,1 Hyejin Park,1 Ji-Jing Yan,2 Isak Im,3 Ji-Woo Choi,4 Tae-Yong Choi,5 Jeongyeon Kim,6 Dong-Keun Song,2 Toshiyuki Takai,7 Yong-Chul Kim,3 Key-Sun Kim,4 Se-Young Choi,5 Sukwoo Choi,6 William L. Klein,8 Junying Yuan,9 and Yong-Keun Jung1
1Global Research Laboratory, School of Biological Sciences/Bio-Max Institute, Seoul National University, Seoul, Republic of Korea. 2Department of Pharmacology, College of Medicine, Institute of Natural Medicine, Hallym University, Chunchon, Republic of Korea. 3Department of Life Science, Gwangju Institute of Science and Technology, Gwangju, Republic of Korea. 4Center for Neural Science, Brain Science Institute, Korea Institute of Science and Technology, Seoul, Republic of Korea. 5Department of Physiology, Dental Research Institute, Seoul National University School of Dentistry, Seoul, Republic of Korea. 6School of Biological Sciences, Seoul National University, Seoul, Republic of Korea. 7Department of Experimental Immunology, Tohoku University, Sendai, Japan. 8Department of Neurobiology and Physiology, Northwestern University, Chicago, Illinois, USA. 9Department of Cell Biology, Harvard Medical School, Boston, Massachusetts, USA.
Address correspondence to: Yong-Keun Jung, School of Biological Sciences, Seoul National University, 599 Gwanak-ro, Gwanak-gu, Seoul 151-747, Republic of Korea. Phone: 82.2.880.4401; Fax: 82.2.873.7524; E-mail: [email protected].
Authorship note: Tae-In Kam and Sungmin Song contributed equally to this work.
Find articles by Kam, T. in: JCI | PubMed | Google Scholar
1Global Research Laboratory, School of Biological Sciences/Bio-Max Institute, Seoul National University, Seoul, Republic of Korea. 2Department of Pharmacology, College of Medicine, Institute of Natural Medicine, Hallym University, Chunchon, Republic of Korea. 3Department of Life Science, Gwangju Institute of Science and Technology, Gwangju, Republic of Korea. 4Center for Neural Science, Brain Science Institute, Korea Institute of Science and Technology, Seoul, Republic of Korea. 5Department of Physiology, Dental Research Institute, Seoul National University School of Dentistry, Seoul, Republic of Korea. 6School of Biological Sciences, Seoul National University, Seoul, Republic of Korea. 7Department of Experimental Immunology, Tohoku University, Sendai, Japan. 8Department of Neurobiology and Physiology, Northwestern University, Chicago, Illinois, USA. 9Department of Cell Biology, Harvard Medical School, Boston, Massachusetts, USA.
Address correspondence to: Yong-Keun Jung, School of Biological Sciences, Seoul National University, 599 Gwanak-ro, Gwanak-gu, Seoul 151-747, Republic of Korea. Phone: 82.2.880.4401; Fax: 82.2.873.7524; E-mail: [email protected].
Authorship note: Tae-In Kam and Sungmin Song contributed equally to this work.
Find articles by Song, S. in: JCI | PubMed | Google Scholar
1Global Research Laboratory, School of Biological Sciences/Bio-Max Institute, Seoul National University, Seoul, Republic of Korea. 2Department of Pharmacology, College of Medicine, Institute of Natural Medicine, Hallym University, Chunchon, Republic of Korea. 3Department of Life Science, Gwangju Institute of Science and Technology, Gwangju, Republic of Korea. 4Center for Neural Science, Brain Science Institute, Korea Institute of Science and Technology, Seoul, Republic of Korea. 5Department of Physiology, Dental Research Institute, Seoul National University School of Dentistry, Seoul, Republic of Korea. 6School of Biological Sciences, Seoul National University, Seoul, Republic of Korea. 7Department of Experimental Immunology, Tohoku University, Sendai, Japan. 8Department of Neurobiology and Physiology, Northwestern University, Chicago, Illinois, USA. 9Department of Cell Biology, Harvard Medical School, Boston, Massachusetts, USA.
Address correspondence to: Yong-Keun Jung, School of Biological Sciences, Seoul National University, 599 Gwanak-ro, Gwanak-gu, Seoul 151-747, Republic of Korea. Phone: 82.2.880.4401; Fax: 82.2.873.7524; E-mail: [email protected].
Authorship note: Tae-In Kam and Sungmin Song contributed equally to this work.
Find articles by Gwon, Y. in: JCI | PubMed | Google Scholar
1Global Research Laboratory, School of Biological Sciences/Bio-Max Institute, Seoul National University, Seoul, Republic of Korea. 2Department of Pharmacology, College of Medicine, Institute of Natural Medicine, Hallym University, Chunchon, Republic of Korea. 3Department of Life Science, Gwangju Institute of Science and Technology, Gwangju, Republic of Korea. 4Center for Neural Science, Brain Science Institute, Korea Institute of Science and Technology, Seoul, Republic of Korea. 5Department of Physiology, Dental Research Institute, Seoul National University School of Dentistry, Seoul, Republic of Korea. 6School of Biological Sciences, Seoul National University, Seoul, Republic of Korea. 7Department of Experimental Immunology, Tohoku University, Sendai, Japan. 8Department of Neurobiology and Physiology, Northwestern University, Chicago, Illinois, USA. 9Department of Cell Biology, Harvard Medical School, Boston, Massachusetts, USA.
Address correspondence to: Yong-Keun Jung, School of Biological Sciences, Seoul National University, 599 Gwanak-ro, Gwanak-gu, Seoul 151-747, Republic of Korea. Phone: 82.2.880.4401; Fax: 82.2.873.7524; E-mail: [email protected].
Authorship note: Tae-In Kam and Sungmin Song contributed equally to this work.
Find articles by Park, H. in: JCI | PubMed | Google Scholar
1Global Research Laboratory, School of Biological Sciences/Bio-Max Institute, Seoul National University, Seoul, Republic of Korea. 2Department of Pharmacology, College of Medicine, Institute of Natural Medicine, Hallym University, Chunchon, Republic of Korea. 3Department of Life Science, Gwangju Institute of Science and Technology, Gwangju, Republic of Korea. 4Center for Neural Science, Brain Science Institute, Korea Institute of Science and Technology, Seoul, Republic of Korea. 5Department of Physiology, Dental Research Institute, Seoul National University School of Dentistry, Seoul, Republic of Korea. 6School of Biological Sciences, Seoul National University, Seoul, Republic of Korea. 7Department of Experimental Immunology, Tohoku University, Sendai, Japan. 8Department of Neurobiology and Physiology, Northwestern University, Chicago, Illinois, USA. 9Department of Cell Biology, Harvard Medical School, Boston, Massachusetts, USA.
Address correspondence to: Yong-Keun Jung, School of Biological Sciences, Seoul National University, 599 Gwanak-ro, Gwanak-gu, Seoul 151-747, Republic of Korea. Phone: 82.2.880.4401; Fax: 82.2.873.7524; E-mail: [email protected].
Authorship note: Tae-In Kam and Sungmin Song contributed equally to this work.
Find articles by Yan, J. in: JCI | PubMed | Google Scholar
1Global Research Laboratory, School of Biological Sciences/Bio-Max Institute, Seoul National University, Seoul, Republic of Korea. 2Department of Pharmacology, College of Medicine, Institute of Natural Medicine, Hallym University, Chunchon, Republic of Korea. 3Department of Life Science, Gwangju Institute of Science and Technology, Gwangju, Republic of Korea. 4Center for Neural Science, Brain Science Institute, Korea Institute of Science and Technology, Seoul, Republic of Korea. 5Department of Physiology, Dental Research Institute, Seoul National University School of Dentistry, Seoul, Republic of Korea. 6School of Biological Sciences, Seoul National University, Seoul, Republic of Korea. 7Department of Experimental Immunology, Tohoku University, Sendai, Japan. 8Department of Neurobiology and Physiology, Northwestern University, Chicago, Illinois, USA. 9Department of Cell Biology, Harvard Medical School, Boston, Massachusetts, USA.
Address correspondence to: Yong-Keun Jung, School of Biological Sciences, Seoul National University, 599 Gwanak-ro, Gwanak-gu, Seoul 151-747, Republic of Korea. Phone: 82.2.880.4401; Fax: 82.2.873.7524; E-mail: [email protected].
Authorship note: Tae-In Kam and Sungmin Song contributed equally to this work.
Find articles by Im, I. in: JCI | PubMed | Google Scholar
1Global Research Laboratory, School of Biological Sciences/Bio-Max Institute, Seoul National University, Seoul, Republic of Korea. 2Department of Pharmacology, College of Medicine, Institute of Natural Medicine, Hallym University, Chunchon, Republic of Korea. 3Department of Life Science, Gwangju Institute of Science and Technology, Gwangju, Republic of Korea. 4Center for Neural Science, Brain Science Institute, Korea Institute of Science and Technology, Seoul, Republic of Korea. 5Department of Physiology, Dental Research Institute, Seoul National University School of Dentistry, Seoul, Republic of Korea. 6School of Biological Sciences, Seoul National University, Seoul, Republic of Korea. 7Department of Experimental Immunology, Tohoku University, Sendai, Japan. 8Department of Neurobiology and Physiology, Northwestern University, Chicago, Illinois, USA. 9Department of Cell Biology, Harvard Medical School, Boston, Massachusetts, USA.
Address correspondence to: Yong-Keun Jung, School of Biological Sciences, Seoul National University, 599 Gwanak-ro, Gwanak-gu, Seoul 151-747, Republic of Korea. Phone: 82.2.880.4401; Fax: 82.2.873.7524; E-mail: [email protected].
Authorship note: Tae-In Kam and Sungmin Song contributed equally to this work.
Find articles by Choi, J. in: JCI | PubMed | Google Scholar
1Global Research Laboratory, School of Biological Sciences/Bio-Max Institute, Seoul National University, Seoul, Republic of Korea. 2Department of Pharmacology, College of Medicine, Institute of Natural Medicine, Hallym University, Chunchon, Republic of Korea. 3Department of Life Science, Gwangju Institute of Science and Technology, Gwangju, Republic of Korea. 4Center for Neural Science, Brain Science Institute, Korea Institute of Science and Technology, Seoul, Republic of Korea. 5Department of Physiology, Dental Research Institute, Seoul National University School of Dentistry, Seoul, Republic of Korea. 6School of Biological Sciences, Seoul National University, Seoul, Republic of Korea. 7Department of Experimental Immunology, Tohoku University, Sendai, Japan. 8Department of Neurobiology and Physiology, Northwestern University, Chicago, Illinois, USA. 9Department of Cell Biology, Harvard Medical School, Boston, Massachusetts, USA.
Address correspondence to: Yong-Keun Jung, School of Biological Sciences, Seoul National University, 599 Gwanak-ro, Gwanak-gu, Seoul 151-747, Republic of Korea. Phone: 82.2.880.4401; Fax: 82.2.873.7524; E-mail: [email protected].
Authorship note: Tae-In Kam and Sungmin Song contributed equally to this work.
Find articles by Choi, T. in: JCI | PubMed | Google Scholar
1Global Research Laboratory, School of Biological Sciences/Bio-Max Institute, Seoul National University, Seoul, Republic of Korea. 2Department of Pharmacology, College of Medicine, Institute of Natural Medicine, Hallym University, Chunchon, Republic of Korea. 3Department of Life Science, Gwangju Institute of Science and Technology, Gwangju, Republic of Korea. 4Center for Neural Science, Brain Science Institute, Korea Institute of Science and Technology, Seoul, Republic of Korea. 5Department of Physiology, Dental Research Institute, Seoul National University School of Dentistry, Seoul, Republic of Korea. 6School of Biological Sciences, Seoul National University, Seoul, Republic of Korea. 7Department of Experimental Immunology, Tohoku University, Sendai, Japan. 8Department of Neurobiology and Physiology, Northwestern University, Chicago, Illinois, USA. 9Department of Cell Biology, Harvard Medical School, Boston, Massachusetts, USA.
Address correspondence to: Yong-Keun Jung, School of Biological Sciences, Seoul National University, 599 Gwanak-ro, Gwanak-gu, Seoul 151-747, Republic of Korea. Phone: 82.2.880.4401; Fax: 82.2.873.7524; E-mail: [email protected].
Authorship note: Tae-In Kam and Sungmin Song contributed equally to this work.
Find articles by Kim, J. in: JCI | PubMed | Google Scholar
1Global Research Laboratory, School of Biological Sciences/Bio-Max Institute, Seoul National University, Seoul, Republic of Korea. 2Department of Pharmacology, College of Medicine, Institute of Natural Medicine, Hallym University, Chunchon, Republic of Korea. 3Department of Life Science, Gwangju Institute of Science and Technology, Gwangju, Republic of Korea. 4Center for Neural Science, Brain Science Institute, Korea Institute of Science and Technology, Seoul, Republic of Korea. 5Department of Physiology, Dental Research Institute, Seoul National University School of Dentistry, Seoul, Republic of Korea. 6School of Biological Sciences, Seoul National University, Seoul, Republic of Korea. 7Department of Experimental Immunology, Tohoku University, Sendai, Japan. 8Department of Neurobiology and Physiology, Northwestern University, Chicago, Illinois, USA. 9Department of Cell Biology, Harvard Medical School, Boston, Massachusetts, USA.
Address correspondence to: Yong-Keun Jung, School of Biological Sciences, Seoul National University, 599 Gwanak-ro, Gwanak-gu, Seoul 151-747, Republic of Korea. Phone: 82.2.880.4401; Fax: 82.2.873.7524; E-mail: [email protected].
Authorship note: Tae-In Kam and Sungmin Song contributed equally to this work.
Find articles by Song, D. in: JCI | PubMed | Google Scholar
1Global Research Laboratory, School of Biological Sciences/Bio-Max Institute, Seoul National University, Seoul, Republic of Korea. 2Department of Pharmacology, College of Medicine, Institute of Natural Medicine, Hallym University, Chunchon, Republic of Korea. 3Department of Life Science, Gwangju Institute of Science and Technology, Gwangju, Republic of Korea. 4Center for Neural Science, Brain Science Institute, Korea Institute of Science and Technology, Seoul, Republic of Korea. 5Department of Physiology, Dental Research Institute, Seoul National University School of Dentistry, Seoul, Republic of Korea. 6School of Biological Sciences, Seoul National University, Seoul, Republic of Korea. 7Department of Experimental Immunology, Tohoku University, Sendai, Japan. 8Department of Neurobiology and Physiology, Northwestern University, Chicago, Illinois, USA. 9Department of Cell Biology, Harvard Medical School, Boston, Massachusetts, USA.
Address correspondence to: Yong-Keun Jung, School of Biological Sciences, Seoul National University, 599 Gwanak-ro, Gwanak-gu, Seoul 151-747, Republic of Korea. Phone: 82.2.880.4401; Fax: 82.2.873.7524; E-mail: [email protected].
Authorship note: Tae-In Kam and Sungmin Song contributed equally to this work.
Find articles by Takai, T. in: JCI | PubMed | Google Scholar
1Global Research Laboratory, School of Biological Sciences/Bio-Max Institute, Seoul National University, Seoul, Republic of Korea. 2Department of Pharmacology, College of Medicine, Institute of Natural Medicine, Hallym University, Chunchon, Republic of Korea. 3Department of Life Science, Gwangju Institute of Science and Technology, Gwangju, Republic of Korea. 4Center for Neural Science, Brain Science Institute, Korea Institute of Science and Technology, Seoul, Republic of Korea. 5Department of Physiology, Dental Research Institute, Seoul National University School of Dentistry, Seoul, Republic of Korea. 6School of Biological Sciences, Seoul National University, Seoul, Republic of Korea. 7Department of Experimental Immunology, Tohoku University, Sendai, Japan. 8Department of Neurobiology and Physiology, Northwestern University, Chicago, Illinois, USA. 9Department of Cell Biology, Harvard Medical School, Boston, Massachusetts, USA.
Address correspondence to: Yong-Keun Jung, School of Biological Sciences, Seoul National University, 599 Gwanak-ro, Gwanak-gu, Seoul 151-747, Republic of Korea. Phone: 82.2.880.4401; Fax: 82.2.873.7524; E-mail: [email protected].
Authorship note: Tae-In Kam and Sungmin Song contributed equally to this work.
Find articles by Kim, Y. in: JCI | PubMed | Google Scholar
1Global Research Laboratory, School of Biological Sciences/Bio-Max Institute, Seoul National University, Seoul, Republic of Korea. 2Department of Pharmacology, College of Medicine, Institute of Natural Medicine, Hallym University, Chunchon, Republic of Korea. 3Department of Life Science, Gwangju Institute of Science and Technology, Gwangju, Republic of Korea. 4Center for Neural Science, Brain Science Institute, Korea Institute of Science and Technology, Seoul, Republic of Korea. 5Department of Physiology, Dental Research Institute, Seoul National University School of Dentistry, Seoul, Republic of Korea. 6School of Biological Sciences, Seoul National University, Seoul, Republic of Korea. 7Department of Experimental Immunology, Tohoku University, Sendai, Japan. 8Department of Neurobiology and Physiology, Northwestern University, Chicago, Illinois, USA. 9Department of Cell Biology, Harvard Medical School, Boston, Massachusetts, USA.
Address correspondence to: Yong-Keun Jung, School of Biological Sciences, Seoul National University, 599 Gwanak-ro, Gwanak-gu, Seoul 151-747, Republic of Korea. Phone: 82.2.880.4401; Fax: 82.2.873.7524; E-mail: [email protected].
Authorship note: Tae-In Kam and Sungmin Song contributed equally to this work.
Find articles by Kim, K. in: JCI | PubMed | Google Scholar
1Global Research Laboratory, School of Biological Sciences/Bio-Max Institute, Seoul National University, Seoul, Republic of Korea. 2Department of Pharmacology, College of Medicine, Institute of Natural Medicine, Hallym University, Chunchon, Republic of Korea. 3Department of Life Science, Gwangju Institute of Science and Technology, Gwangju, Republic of Korea. 4Center for Neural Science, Brain Science Institute, Korea Institute of Science and Technology, Seoul, Republic of Korea. 5Department of Physiology, Dental Research Institute, Seoul National University School of Dentistry, Seoul, Republic of Korea. 6School of Biological Sciences, Seoul National University, Seoul, Republic of Korea. 7Department of Experimental Immunology, Tohoku University, Sendai, Japan. 8Department of Neurobiology and Physiology, Northwestern University, Chicago, Illinois, USA. 9Department of Cell Biology, Harvard Medical School, Boston, Massachusetts, USA.
Address correspondence to: Yong-Keun Jung, School of Biological Sciences, Seoul National University, 599 Gwanak-ro, Gwanak-gu, Seoul 151-747, Republic of Korea. Phone: 82.2.880.4401; Fax: 82.2.873.7524; E-mail: [email protected].
Authorship note: Tae-In Kam and Sungmin Song contributed equally to this work.
Find articles by Choi, S. in: JCI | PubMed | Google Scholar
1Global Research Laboratory, School of Biological Sciences/Bio-Max Institute, Seoul National University, Seoul, Republic of Korea. 2Department of Pharmacology, College of Medicine, Institute of Natural Medicine, Hallym University, Chunchon, Republic of Korea. 3Department of Life Science, Gwangju Institute of Science and Technology, Gwangju, Republic of Korea. 4Center for Neural Science, Brain Science Institute, Korea Institute of Science and Technology, Seoul, Republic of Korea. 5Department of Physiology, Dental Research Institute, Seoul National University School of Dentistry, Seoul, Republic of Korea. 6School of Biological Sciences, Seoul National University, Seoul, Republic of Korea. 7Department of Experimental Immunology, Tohoku University, Sendai, Japan. 8Department of Neurobiology and Physiology, Northwestern University, Chicago, Illinois, USA. 9Department of Cell Biology, Harvard Medical School, Boston, Massachusetts, USA.
Address correspondence to: Yong-Keun Jung, School of Biological Sciences, Seoul National University, 599 Gwanak-ro, Gwanak-gu, Seoul 151-747, Republic of Korea. Phone: 82.2.880.4401; Fax: 82.2.873.7524; E-mail: [email protected].
Authorship note: Tae-In Kam and Sungmin Song contributed equally to this work.
Find articles by Choi, S. in: JCI | PubMed | Google Scholar
1Global Research Laboratory, School of Biological Sciences/Bio-Max Institute, Seoul National University, Seoul, Republic of Korea. 2Department of Pharmacology, College of Medicine, Institute of Natural Medicine, Hallym University, Chunchon, Republic of Korea. 3Department of Life Science, Gwangju Institute of Science and Technology, Gwangju, Republic of Korea. 4Center for Neural Science, Brain Science Institute, Korea Institute of Science and Technology, Seoul, Republic of Korea. 5Department of Physiology, Dental Research Institute, Seoul National University School of Dentistry, Seoul, Republic of Korea. 6School of Biological Sciences, Seoul National University, Seoul, Republic of Korea. 7Department of Experimental Immunology, Tohoku University, Sendai, Japan. 8Department of Neurobiology and Physiology, Northwestern University, Chicago, Illinois, USA. 9Department of Cell Biology, Harvard Medical School, Boston, Massachusetts, USA.
Address correspondence to: Yong-Keun Jung, School of Biological Sciences, Seoul National University, 599 Gwanak-ro, Gwanak-gu, Seoul 151-747, Republic of Korea. Phone: 82.2.880.4401; Fax: 82.2.873.7524; E-mail: [email protected].
Authorship note: Tae-In Kam and Sungmin Song contributed equally to this work.
Find articles by Klein, W. in: JCI | PubMed | Google Scholar
1Global Research Laboratory, School of Biological Sciences/Bio-Max Institute, Seoul National University, Seoul, Republic of Korea. 2Department of Pharmacology, College of Medicine, Institute of Natural Medicine, Hallym University, Chunchon, Republic of Korea. 3Department of Life Science, Gwangju Institute of Science and Technology, Gwangju, Republic of Korea. 4Center for Neural Science, Brain Science Institute, Korea Institute of Science and Technology, Seoul, Republic of Korea. 5Department of Physiology, Dental Research Institute, Seoul National University School of Dentistry, Seoul, Republic of Korea. 6School of Biological Sciences, Seoul National University, Seoul, Republic of Korea. 7Department of Experimental Immunology, Tohoku University, Sendai, Japan. 8Department of Neurobiology and Physiology, Northwestern University, Chicago, Illinois, USA. 9Department of Cell Biology, Harvard Medical School, Boston, Massachusetts, USA.
Address correspondence to: Yong-Keun Jung, School of Biological Sciences, Seoul National University, 599 Gwanak-ro, Gwanak-gu, Seoul 151-747, Republic of Korea. Phone: 82.2.880.4401; Fax: 82.2.873.7524; E-mail: [email protected].
Authorship note: Tae-In Kam and Sungmin Song contributed equally to this work.
Find articles by Yuan, J. in: JCI | PubMed | Google Scholar
1Global Research Laboratory, School of Biological Sciences/Bio-Max Institute, Seoul National University, Seoul, Republic of Korea. 2Department of Pharmacology, College of Medicine, Institute of Natural Medicine, Hallym University, Chunchon, Republic of Korea. 3Department of Life Science, Gwangju Institute of Science and Technology, Gwangju, Republic of Korea. 4Center for Neural Science, Brain Science Institute, Korea Institute of Science and Technology, Seoul, Republic of Korea. 5Department of Physiology, Dental Research Institute, Seoul National University School of Dentistry, Seoul, Republic of Korea. 6School of Biological Sciences, Seoul National University, Seoul, Republic of Korea. 7Department of Experimental Immunology, Tohoku University, Sendai, Japan. 8Department of Neurobiology and Physiology, Northwestern University, Chicago, Illinois, USA. 9Department of Cell Biology, Harvard Medical School, Boston, Massachusetts, USA.
Address correspondence to: Yong-Keun Jung, School of Biological Sciences, Seoul National University, 599 Gwanak-ro, Gwanak-gu, Seoul 151-747, Republic of Korea. Phone: 82.2.880.4401; Fax: 82.2.873.7524; E-mail: [email protected].
Authorship note: Tae-In Kam and Sungmin Song contributed equally to this work.
Find articles by Jung, Y. in: JCI | PubMed | Google Scholar
Authorship note: Tae-In Kam and Sungmin Song contributed equally to this work.
Published June 10, 2013 - More info
J Clin Invest. 2013;123(7):2791–2802. https://doi.org/10.1172/JCI66827.
© 2013 The American Society for Clinical Investigation
Received: February 6, 2013; Accepted: April 11, 2013
-
Abstract
Amyloid-β (Aβ) induces neuronal loss and cognitive deficits and is believed to be a prominent cause of Alzheimer’s disease (AD); however, the cellular pathology of the disease is not fully understood. Here, we report that IgG Fcγ receptor II-b (FcγRIIb) mediates Aβ neurotoxicity and neurodegeneration. We found that FcγRIIb is significantly upregulated in the hippocampus of AD brains and neuronal cells exposed to synthetic Aβ. Neuronal FcγRIIb activated ER stress and caspase-12, and Fcgr2b KO primary neurons were resistant to synthetic Aβ-induced cell death in vitro. Fcgr2b deficiency ameliorated Aβ-induced inhibition of long-term potentiation and inhibited the reduction of synaptic density by naturally secreted Aβ. Moreover, genetic depletion of Fcgr2b rescued memory impairments in an AD mouse model. To determine the mechanism of action of FcγRIIb in Aβ neurotoxicity, we demonstrated that soluble Aβ oligomers interact with FcγRIIb in vitro and in AD brains, and that inhibition of their interaction blocks synthetic Aβ neurotoxicity. We conclude that FcγRIIb has an aberrant, but essential, role in Aβ-mediated neuronal dysfunction.
-
Introduction
The pathological features of Alzheimer’s disease (AD), including memory loss and neuronal degeneration, are believed to be associated with the accumulation of amyloid-β (Aβ) in the brain (1, 2), but the mechanism underlying this pathogenesis remains uncertain. Of particular note, the membrane proteins responsible for Aβ neurotoxicity and memory impairment have not yet been identified, though there are several Aβ-binding proteins thought to be involved in AD pathology, including receptors for advanced glycation end-products (RAGE), Aβ-binding alcohol dehydrogenase (ABAD), and cellular prion protein (PrPc) (3–5). RAGE was originally reported to interact with Aβ in neurotoxicity and was found to act in the blood-brain barrier as a transporter of Aβ into the brain (3, 6). ABAD is an intracellular binding partner of Aβ, acting on mitochondria (4). PrPc was recently reported to act as a receptor for the Aβ oligomer, but this is still controversial and needs to be further clarified (5, 7–10). Despite a growing number of reports, the identification of a plausible membrane receptor responsible for Aβ neurotoxicity and memory impairment has yet to be made. Therefore, identification of such a neuronal receptor will be important and greatly beneficial for understanding and controlling AD pathology.
We previously isolated a cytosolic mediator of Aβ neurotoxicity, E2-25K/Hip-2, based on a global analysis of the genes expressed in the primary cortical neurons exposed to Aβ1-42 (11, 12). Using the same approach, we found that Aβ1-42 strongly upregulated the expression of FcγRIIb in the cortical neurons. FcγRIIb was initially identified as a type of IgG receptor, which binds to IgG immune complexes containing relevant antigens and is mainly expressed in B cells, macrophages, and neutrophils (13). In B cells, FcγRIIb acts to inhibit B cell receptor–mediated (BCR-mediated) immune responses and plays a key role in preventing autoimmunity (14). Thus, Fcgr2b KO mice display an elevated humoral immune response and are highly susceptible to autoimmune disease in the B6 strain (15, 16). Also, a mutation replacing Ile232 with Thr (I232T) in the transmembrane domain impairs the ability of FcγRIIb to function as an inhibitory receptor and leads to autoimmunity (14). Further, it was proposed that BCR-independent aggregation of FcγRIIb induces apoptosis in B cells (14). Recently, it was reported that FcγRIIb is also expressed in Purkinje cells and regulates cerebellar functions in the brain (17). Although there are a growing number of reports showing the expression and function of IgG-binding Fcγ receptors (FcγRs) in the CNS (18, 19), the neuronal function of FcγRIIb in the brain is largely unknown.
Here, we report that FcγRIIb plays a critical role in the Aβ neurotoxicity and memory impairment that appear to be relevant to AD pathogenesis. FcγRIIb deficiency prevented Aβ-induced inhibition of long-term potentiation (LTP) and synaptic dysfunction and rescued the memory impairments in an AD mouse model, demonstrating that FcγRIIb is essential for Aβ1-42-induced neurotoxicity.
-
Results
Induction of FcγRIIb expression in neurons incubated with Aβ and in AD brains. We first analyzed the expression of FcγRIIb in the brains of AD patients using anti-human FcγRIIb antibody (EP888Y), which did not show any cross-reactivity with FcγRIIa and FcγRIIIb on Western blot analysis (Supplemental Figure 1A; supplemental material available online with this article; doi: 10.1172/JCI66827DS1). FcγRIIb was detected in the brains, and we found its expression level to be significantly increased in the hippocampal regions of the AD patients, while the level of NeuN was slightly reduced compared with normal control or mild cognitive impairment (MCI) patients (Figure 1, A and B, and Supplemental Table 1). Increased neuronal expression of FcγRIIb was also observed in the NeuN-positive hippocampal neurons of AD patients (Figure 1C and Supplemental Figure 2A). Interestingly, intraneuronal Aβ visualized by Nu-1 antibody, which detected Aβ-derived diffusible ligands (ADDLs), including monomeric and oligomeric forms (20), colocalized with the immunoreactivity of FcγRIIb in the hippocampal neurons of AD patients (Figure 1D). Western blot analysis identified FcγRIIb expression in the mouse brains and the lack of its expression in Fcgr2b KO mouse brains (Supplemental Figure 1B). Furthermore, FcγRIIb was expressed in most parts of the brain, especially in the cortex, hippocampus, and cerebellum (Supplemental Figure 2, B and C). Compared with age-matched WT mice, the level of FcγRIIb was increased in the cortex of the 6- and 17-month-old AD-model mice (J20 line), which express familial AD mutant amyloid precursor protein (hAPP) and show learning and memory deficits (21), while the FcγRIIb level was not significantly changed in the 2-month-old J20 mice (Supplemental Figure 2D).
 Figure 1
Figure 1Increased expression of FcγRIIb in the neurons of AD brains. (A) Increased expression of FcγRIIb in AD patients. Hippocampal homogenates from normal, MCI (Braak III), and AD patients with Braak stage V/VI were analyzed by Western blotting. PHF1 antibody detecting phospho-tau was used as a marker for AD. (B) Levels of FcγRIIb (left) and NeuN (right) were quantified by densitometric measurement. Values are the mean ± SEM. *P < 0.05; **P < 0.005; ***P < 0.001; 2-tailed t test. (C) Immunohistochemical detection of FcγRIIb in the NeuN-positive hippocampal neurons of AD patients. Tissue samples from normal and AD patients (n = 3) were immunostained using anti-FcγRIIb and anti-NeuN antibodies and examined under a confocal microscope. Scale bars: 20 μm. (D) Immunohistochemical detection of FcγRIIb and Aβ in the hippocampus of AD patients. Tissue samples were immunostained using anti-FcγRIIb and anti-Aβ antibodies and then examined under a confocal microscope. Scale bars: 10 μm. (E) Immunoblot analysis showing FcγRIIb induction by Aβ1-42 in neuronal cells. Primary cortical neurons and SH-SY5Y cells were incubated with 5 μM Aβ1-42 for 36 hours, and cell extracts were analyzed with Western blotting using anti-FcγRIIb antibody (left, 2.4G2; right, EP888Y). The signals on the blots were quantified by densitometric analysis (bottom). (F) Increase in Fcgr2b mRNA by Aβ1-42 in primary cortical neurons. Neurons cultured from WT and Fcgr2b KO mice were incubated with 5 μM Aβ1-42 for 36 hours, after which total RNA was isolated for RT-PCR analysis with synthetic primers (top). Levels of mRNA in WT neurons were quantified by densitometric measurement (bottom). Values are the mean ± SD (n = 3). *P < 0.05; 2-tailed t test.
Western blot analysis showed that FcγRIIb expression was markedly increased by Aβ1-42 in the primary cultured neurons and SH-SY5Y neuroblastoma cells (Figure 1E). Interestingly, among FcγRs, it was Fcgr2b that was induced by Aβ1-42 in the primary cortical neurons, as examined by RT-PCR analysis (Figure 1F). Thus, we investigated the molecular mechanism by which FcγRIIb expression is regulated by Aβ1-42. We first found that Aβ1-42 activated c-Jun N-terminal protein kinase (JNK) and its downstream transcription factor, c-Jun, in WT cortical neurons, but not in Fcgr2b KO neurons (Supplemental Figure 3A). In addition, the JNK inhibitor (SP600125), but not the ERK1/2 inhibitor (U0126), suppressed Aβ-triggered FcγRIIb expression in both mRNA and protein levels in SH-SY5Y cells (Supplemental Figure 3B) and inhibited an Aβ-triggered increase in the reporter activity that was driven by the FCGR2B promoter (Supplemental Figure 3C). Moreover, the JNK inhibitor suppressed Aβ1-42-induced neurotoxicity in both primary cortical neurons and HT22 mouse hippocampal cells (Supplemental Figure 3, D and E). Together with the previous report showing that the AP-1 complex regulates FCGR2B gene expression through direct interaction of c-Jun with the FCGR2B promoter (22), our results suggest that Aβ1-42 regulates FCGR2B gene expression at the transcriptional level via JNK-c-Jun.
Neurotoxic role of FcγRIIb in Aβ signaling. Given that the oligomeric form is reportedly one of the primary neurotoxic Aβ species (23–25) and that neuronal Aβ is apparently seen in AD (26, 27), we examined the role of FcγRIIb in Aβ neurotoxicity. We prepared the toxic oligomers and confirmed their species using SDS PAGE and atomic force microscopy. Oligomeric species of Aβ were formed (Figure 2A, left) as described in ADDLs (20) and contained nonfibrillar and globular particles (Figure 2A, right). Notably, unlike WT neurons, cultured hippocampal neurons from Fcgr2b KO mice were quite resistant to the neurotoxicity of synthetic Aβ1-42 containing soluble oligomers (Figure 2B and Supplemental Figure 4A). Resistance to Aβ neurotoxicity was also observed in cultured Fcgr2b KO cortical neurons and Fcgr2b knockdown (KD) B103 rat neuroblastoma cells (data not shown). Conversely, ectopic expression of FcγRIIb induced significant HT22 cell death, while an I232T FcγRIIb mutant failed to do so and, rather, suppressed Aβ neurotoxicity (Figure 2C). These results suggest that FcγRIIb is required for Aβ neurotoxicity in cultured cells and that the neurotoxic signaling triggered by Aβ might be shared with the immune inhibitory activity of FcγRIIb.
 Figure 2
Figure 2FcγRIIb is required for Aβ neurotoxicity. (A) Characterization of Aβ1-42 oligomers. The prepared Aβ1-42 oligomers were identified by SDS-PAGE (left) and atomic force microscopy (right). Scale bars: 0.5 μm. (B) Fcgr2b KO neurons are resistant to Aβ toxicity. Primary hippocampal neurons (5 DIV) were incubated with Aβ1-42 oligomers for 2 days, after which the relative neuronal viability was determined by MTT assay. Data are the mean ± SD (n = 6). ***P < 0.001, 1-way ANOVA. (C) FcγRIIb I232T mutant potently inhibits Aβ toxicity. HT22 cells transfected and incubated with or without Aβ1-42. Bars depict the incidence of cell death. Values are the mean ± SD (n = 3). *P < 0.05; ***P < 0.001, unpaired t test (top). Expression of constructs was identified by Western blotting (bottom). (D) FcγRIIb sensitizes Aβ-induced cell death. HT22 cells transfected with the indicated constructs were incubated for 48 hours with or without CHO-CM, 7PA2-CM, or 1 μM of synthetic Aβ1-42. Values are the mean ± SD (n = 3). *P < 0.05; **P < 0.005, unpaired t test. NS, not significant. (E and F) Inhibition of Aβ1-42-induced FcγRIIb expression and neuronal death by the addition of purified hFcγRIIb-ED protein. SH-SY5Y cells (E) and primary cortical neurons (F) were incubated for 48 hours with Aβ1-42 with or without 50 μg/ml of hFcγRIIb-ED protein. Cell extracts were subjected to Western blot analysis (E); cell viability was determined using Calcein-AM (F). Values are the mean ± SD (n = 4).
Many studies have used synthetic Aβ1-42 to induce neuronal death in vitro (9, 28, 29). However, the concentration of synthetic Aβ is high compared with Aβ oligomers found in the human brain, although AD brains have locally and highly concentrated Aβ near the plaques. Thus, to assess whether low-dose soluble Aβ contributes to FcγRIIb-mediated neurotoxicity, we used cell-derived, naturally secreted Aβ oligomers (24). Immunoprecipitation analysis confirmed the presence of soluble Aβ monomers and oligomers in the conditioned medium (CM) of Chinese hamster ovary (CHO) cells that stably expressed human APP bearing the V717F familial AD mutation (7PA2 cells) (Supplemental Figure 4B). The concentration of soluble Aβ1-42 in 7PA2-CM was estimated by ELISA to be 200 pg/ml (Supplemental Figure 4C). There was no significant incidence of neurotoxicity when HT22 cells were incubated with 7PA2-CM only (Figure 2D). On the other hand, 7PA2-CM sensitized the HT22 cells, which were transfected with and thus overexpressed FcγRIIb, resulting in cell death. Similarly, synthetic Aβ was also sufficiently potent to enhance FcγRIIb-mediated cell death (Figure 2D). In addition, when HT22 cells expressing FcγRIIb were cocultivated with 7PA2 cells, the incidence of HT22 cell death was enhanced (Supplemental Figure 4D).
We then determined the molecular mechanism of FcγRIIb-mediated neuronal death by Aβ1-42. We first found that Aβ- and FcγRIIb-mediated cell death was significantly suppressed by salubrinal, a selective eIF2α dephosphorylation inhibitor, thus protecting against ER stress–mediated neurotoxicity (30) (Supplemental Figure 5, A and B) and suggesting that FcγRIIb mediates Aβ1-42 neurotoxicity through ER stress. Consistently, Aβ1-42-induced upregulation of the typical ER stress markers, GRP78 and CHOP, observed in WT cortical neurons was markedly suppressed in Fcgr2b KO cortical neurons (Supplemental Figure 5C). Conversely, overexpressed FcγRIIb, but not the I232T FcγRIIb mutant, induced similar ER stress responses in HT22 cells (Supplemental Figure 5D). Of particular note, Aβ1-42-induced activation of caspase-12, a key player in ER stress- and Aβ-mediated apoptosis (12, 31), was abrogated in Fcgr2b KO cortical neurons (Supplemental Figure 5C). Furthermore, ectopic expression of the caspase-12 active site mutant (C298S) significantly suppressed FcγRIIb-mediated HT22 cell death, while caspase-12 significantly enhanced it (Supplemental Figure 5E). These observations suggest that homoaggregation of FcγRIIb by Aβ1-42 transduces the toxic signal into neurons to induce ER stress, which in turn activates caspase-12, leading to neuronal death.
We then assessed the possible role of FcγRIIb as a membrane receptor of Aβ1-42 in Aβ1-42 neurotoxicity. As observed in cultured primary neurons, incubation of SH-SY5Y cells with Aβ1-42 increased the expression of FcγRIIb (Figure 2E). On the other hand, the addition of the purified ectodomain (hFcγRIIb-ED) hFcγRIIb protein to culture medium abolished the induction of FcγRIIb expression and JNK activation triggered by Aβ1-42. In parallel, the addition of the FcγRIIb-ED protein also inhibited cell death of primary rat cortical neurons, which were exposed to Aβ1-42 (Figure 2F). Thus, the blockade of Aβ neurotoxicity by hFcγRIIb-ED protein on the extracellular side led us to examine whether FcγRIIb is able to bind to Aβ1-42.
Interaction between FcγRIIb and Aβ1-42. To determine the mode of action of FcγRIIb, we addressed the direct interaction between FcγRIIb and Aβ both in vitro and in vivo. We first performed a set of in vitro binding assays using the purified hFcγRIIb-ED protein. We used soluble fractions of cell-derived Aβ and synthetic Aβ to exclude the nonspecific precipitation of aggregated Aβ. In an in vitro pulldown assay, we found that monomeric and oligomeric forms of both cell-derived Aβ and synthetic Aβ1-42 were coimmunoprecipitated with FcγRIIb-ED protein (Figure 3A and Supplemental Figure 6, A and B), though boiling of the samples for Western blotting converted the oligomeric forms of Aβ1-42 into monomers. Interestingly, FcγRIIb-ED protein bound more effectively to the oligomeric forms of Aβ1-42, a pathogenic form of Aβ, than to Aβ1-40 (Supplemental Figure 6C). It is not yet clear whether the differing oligomeric states of these Aβ peptides cause this difference in binding. We further investigated the direct interaction of Aβ with FcγRIIb and the kinetics of this interaction using surface plasmon resonance (SPR) analysis (Figure 3B). Interestingly, Aβ1-42 potently interacted with hFcγRIIb-ED (Kd = 5.67 × 10–8 M), while human IgG1 (hIgG1) bound to hFcγRIIb-ED with relatively low affinity (Kd = 9.43 × 10–6 M) (Figure 3B and Supplemental Figure 6D).
 Figure 3
Figure 3Interaction of FcγRIIb with Aβ1-42. (A) In vitro binding of FcγRIIb-ED protein to cell-derived Aβ. Purified GST or hFcγRIIb-ED protein was incubated with CHO- or 7PA2-CM for 6 hours and then immunoprecipitated with anti-GST or anti-FcγRIIb antibody. The precipitates were analyzed by Western blotting. L.C., light chain. (B) Direct interaction of FcγRIIb-ED with Aβ1-42 in SPR analysis. BSA or oligomeric Aβ1-42 was immobilized and the interactions with hFcγRIIb-ED were analyzed using Biacore 3000. (C) Interaction of FcγRIIb with oligomeric Aβ1-42 in AD patients. Hippocampal extracts from AD patients and controls (C) were subjected to immunoprecipitation assays using IgG or Nu-1 antibody, and the precipitates were analyzed with Western blotting. Asterisk indicates nonspecific signal, which differs in size from the Aβ monomer. (D) Computational simulation showing the structure of the FcγRIIb-Aβ1-42 complex. The N terminus of Aβ1-42 and IgG-binding region of hFcγRIIb were handled using the Affinity program. (E) Inhibition of Aβ1-42-induced reporter activation of the FcγRIIb-ED/CD40-Luc chimera by Aβ1-9 or FcγRIIb107-114. Synthetic peptides spanning the predicted interaction regions of FcγRIIb and Aβ1-42 and their mutants are represented (top). The reporter assay was performed with or without synthetic peptides with a molar ratio of 1:3 (bottom). (F) Binding of Aβ1-42 to the Ig2 domain of FcγRIIb. Domain structures of FcγRIIb and its deletion constructs are presented (top). HEK293T cells were transfected with the constructs and treated with 1 μM Aβ1-42 for 1 hour. Cell lysates were subjected to immunoprecipitation analysis using Aβ antibody or preimmune (Pre), and the precipitates were analyzed with Western blotting. Expression of each FcγRIIb protein was checked in whole cell lysates. IP, immunoprecipitation.
Using immunoprecipitation assays, we also detected in vivo interaction between FcγRIIb and monomeric and oligomeric forms of Aβ in the hippocampal extracts of AD patients (Figure 3C). In contrast, this interaction was not observed in the control tissues. We used the tissues showing similar expression of FcγRIIb to exclude the effect of different amounts of FcγRIIb protein. The reverse immunoprecipitation assay using FcγRIIb antibody was unsuccessful, probably because the amount of the precipitated Aβ was too low to be detected. The interaction between Aβ1-42 and HA-tagged FcγRIIb was also assessed in cultured cells. We found that synthetic Aβ oligomers and FcγRIIb were coimmunoprecipitated in FcγRIIb-expressing cell lines (Supplemental Figure 6, E and F). We further examined the interaction with reporter assays using a FcγRIIb(ED)-CD40 chimera. The chimera consisted of the extracellular domain of FcγRIIb for Aβ binding and the transmembrane and cytosolic domains of CD40 for nuclear factor-κB (NF-κB) activation (Supplemental Figure 7A). In the reporter assay, we observed that Aβ1-42 treatment increased the luciferase activity, which is under the control of NF-κB DNA elements, by about 4 fold in NIH3T3 cells expressing FcγRIIb(ED)-CD40, but not in cells expressing CD40 (Supplemental Figure 7B). Apparently, Aβ1-42 binds to FcγRIIb-ED to activate NF-κB, further supporting the notion that FcγRIIb binds to extracellular Aβ.
We were eager to understand the mode of interaction between FcγRIIb and Aβ1-42, but Aβ1-42 was readily aggregated and resisted crystallization. Oligomeric structures of Aβ1-42 and Aβ1-40 were computationally predicted to show that the hydrophilic N termini of Aβ1-42 and Aβ1-40 are exposed, whereas C termini bind together to form oligomers (32). In particular, the N terminus of Aβ1-42 is more extended and flexible than that of Aβ1-40 (32), which may provide a hint as to why Aβ1-42 is more toxic to neurons than Aβ1-40 (33). Moreover, the neurotoxicity of oligomeric Aβ isolated from AD patients is protected by Aβ N-terminal antibodies, but not by Aβ C-terminal antibodies (25). Our observations that FcγRIIb interacted with Aβ1-42 better than Aβ1-40 led us to examine whether the extended N-terminus of Aβ1-42 and the IgG-binding region of FcγRIIb contributed to Aβ neurotoxicity. Using in silico modeling of the structure of each domain (34, 35), the mode of interaction was predicted (Figure 3D) and the interaction specificity was addressed. The N-terminal region of Aβ1-42 was predicted to bind to the Ig-like domain 2 of FcγRIIb, and 4phenylalanine of Aβ1-42 and 111tryptophan of FcγRIIb were important in this interaction.
Then, to address the interaction specificity, we prepared synthetic peptides derived from Aβ1-42 and FcγRIIb and performed binding competition assays between Aβ1-42 and these peptides using an FcγRIIb(ED)-CD40 reporter. Synthetic peptides derived from the N-terminal region (Aβ1-9 #1; 1DAEFRHDSG9) of Aβ1-42 or from the IgG-binding region (FcγRIIb107-114 #9; 107RCHSWRNK114) of FcγRIIb (36), but not other FcγRIIb95-101 (#8, 95QLVFLEG101) peptides, effectively inhibited the interaction of Aβ1-42 with FcγRIIb in a reporter assay (Figure 3E). In addition, the peptides also suppressed Aβ1-42 neurotoxicity in cultured hippocampal and cortical neurons (Supplemental Figure 8). However, the inhibitory actions of these Aβ1-9 (#1) and FcγRIIb107-114 (#9) peptides were lost upon introduction of mutations into the amino acids that were predicted to be involved in these interactions (4phenylalanine and 7aspartate in Aβ1-9 and 111tryptophan in FcγRIIb107-114) (Figure 3, D and E). We therefore propose that the N terminus of Aβ1-42 is required for the binding of Aβ1-42 to FcγRIIb and for Aβ1-42 neurotoxicity. Further, in immunoprecipitation analysis using hFcγRIIb deletion mutants lacking either 1 of 2 Ig-like domains present in the extracellular region, we found that the hFcγRIIb ΔIg2 mutant lacking the second Ig-like domain failed to interact with Aβ1-42 (Figure 3F). The second Ig-like domain is also known to bind to IgG (13). Collectively, these observations indicate that the second Ig-like domain of hFcγRIIb is critical for interaction with Aβ1-42.
Requirement of FcγRIIb in synaptic dysfunction and memory impairment in AD mice. Many reports have proposed that the oligomeric forms of Aβ1-42 suppress LTP, a measure of synaptic plasticity (24, 37). To explore whether FcγRIIb accounts for Aβ1-42-mediated impairment of synaptic plasticity, we performed extracellular recordings of field excitatory postsynaptic potentials (fEPSPs) in the hippocampal slices of WT or Fcgr2b KO mice. Treatment with synthetic Aβ1-42 (400 nM) inhibited hippocampal LTP after high-frequency stimulation in WT mice (Figure 4, A and B). On the contrary, there was no inhibition of LTP by Aβ1-42 in the Fcgr2b KO hippocampal slices and synaptic response was normal (Figure 4, A and B). We also identified the role of FcγRIIb in LTP inhibition using an AD model mouse. We crossed the AD-mutant hAPP transgenic mice (J20 line) with Fcgr2b KO mice to generate double-transgenic mice (hAPP/KO) and recorded their fEPSPs. Consistently, FcγRIIb deficiency rescued LTP deficits in hAPP mice (Supplemental Figure 9A). Collectively, these observations indicate that FcγRIIb is required for Aβ1-42-induced inhibition of LTP in hippocampal tissue.
 Figure 4
Figure 4Requirement of FcγRIIb in Aβ1-42-mediated inhibition of hippocampal LTP and synaptic dysfunction. (A) Lack of Aβ1-42-induced inhibition of hippocampal LTP in Fcgr2b-deficient mice. WT and Fcgr2b KO hippocampal slices were microinjected with PBS (n = 7 slices from 6 WT mice, n = 7 slices from 5 KO mice) or 400 nM Aβ1-42 (n = 7 slices from 4 WT mice, n = 7 slices from 4 KO mice) for 30 to 50 minutes before HFS. Insets show fEPSP traces of average baseline (black) and after HFS (red). Scale bars: 5 msec and 0.2 mV. (B) Summary bar graphs represent the magnitude of LTP between 50 and 60 minutes. Data are the mean ± SEM. ***P < 0.001, 1-way ANOVA. (C) Abrogation of Aβ-induced synaptic loss in Fcgr2b-deficient neurons. Cultured hippocampal neurons were transfected with pEGFP-N1 and further incubated with CHO- or 7PA2-CM for 5 days. Images represent dendrites of neurons from WT and Fcgr2b KO embryos. Scale bars: 10 μm (left). Quantification of dendrite spine density from 10 randomly chosen neurons measuring 100 μm in length. Bars depict the mean ± SD. ***P < 0.001, 1-way ANOVA (right). (D) Requirement of FcγRIIb-Aβ interaction in Aβ-induced spine density reduction. Primary hippocampal neurons were transfected with pEGFP-N1 and incubated with CHO- or 7PA2-CM for 5 days with or without purified hFcγRIIb-ED protein. Scale bars: 5 μm (left). Average spine density for neurons is indicated as in C. Bars depict the mean ± SD. ***P < 0.001, 1-way ANOVA (right).
Then, we determined whether FcγRIIb contributed to Aβ-induced synaptic loss in cultured hippocampal neurons. After incubation with 7PA2-CM containing soluble Aβ, the neuronal synaptic spine densities were quantified. Compared with the untreated controls, spine density was reduced in the WT hippocampal neurons, but not in the Fcgr2b KO hippocampal neurons (Figure 4C). Similarly, spine density was reduced by incubation with synthetic Aβ1-42 in the WT neurons, but not in the Fcgr2b KO neurons (Supplemental Figure 9B). Furthermore, the addition of hFcγRIIb-ED protein to 7PA2-CM prevented a decrease in the dendritic spine density of cultured neurons (Figure 4D), indicating that the interaction of Aβ with FcγRIIb is required for Aβ-induced synaptic loss. We also found that synaptophysin immunoreactivity was significantly reduced in the CA1 regions of hAPP mice, but not in those of hAPP/Fcgr2b KO mice (Supplemental Figure 9C). Together, these observations suggest that FcγRIIb plays an essential role in Aβ1-42-mediated synaptic loss and dysfunction.
Because the synaptic dysfunction evoked by Aβ provides a molecular and cellular basis for memory deficits and neuropathological features of dementia in AD (38, 39), we investigated the role of FcγRIIb in learning and memory deficits using an AD mouse model. Fcgr2b KO mice did not show any detectable memory impairment compared with WT mice up to 12 months (data not shown). Comparisons of each group’s behavior showed that the defects in spatial working memory of 4- to 6-month-old hAPP mice were significantly improved by FcγRIIb deficiency in a Y-maze test (Figure 5A). In the novel object recognition test, hAPP/Fcgr2b KO mice spent more time exploring the novel object than the other object that they had explored 1 day before, but hAPP mice had no preference for the new object (Figure 5B). Thus, the recognition memory deficit of hAPP mice was also blocked by the removal of FcγRIIb. We further examined passive avoidance memory. During training, step-through latency was similar in each group (data not shown). After training, only the hAPP mice failed to learn and showed impaired task performance, whereas the other 3 groups performed well (Figure 5C). On the other hand, Fcgr2b deficiency did not affect the hyperactivity in hAPP mice that might also be related to memory impairment (40), as reflected in the Y-maze test (Supplemental Figure 10). Because Aβ1-42 and Αβ1-40 levels were similar in the hAPP and hAPP/KO mice (Supplemental Figure 11) and 4- to 6-month-old hAPP mice had not yet shown amyloid plaques (41), we ruled out the possibility that FcγRIIb altered Aβ levels or plaque formation. Together, these results suggest that FcγRIIb is required for learning and memory impairment in the hAPP transgenic mice.
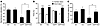 Figure 5
Figure 5FcγRIIb deficiency ameliorates learning and memory deficits in hAPP mice. (A) Suppression of spatial memory defects in hAPP/Fcgr2b-deficient mice. Spatial memory of 4- to 6-month-old WT, Fcgr2b-deficient (KO), hAPP, and hAPP/Fcgr2b-deficient (hAPP/KO) mice was represented as spontaneous alteration on the Y maze (n = 7–18 mice per group; WT, 10 males and 8 females; KO, 4 males and 8 females; hAPP, 4 males and 3 females; hAPP/KO, 10 males and 8 females). Data are the mean ± SEM; *P < 0.05, unpaired t test. (B) Recovered recognition memory impairment in hAPP/Fcgr2b KO mice. Object recognition memory was reflected by the percentage of time each group of mice spent exploring a familiar versus a novel object (n = 9–18 mice per genotype). Values are the mean ± SEM; ***P < 0.001 versus familiar object, paired t test. (C) Prevention of passive avoidance memory defects of hAPP mice by Fcgr2b deficiency (n = 9–13 mice per group). Bars depict the mean ± SEM; *P < 0.05 as indicated by the bracket, unpaired t test.
We then addressed the effects of the inhibitory peptides, Aβ1-9 #1 and FcγRIIb107-114 #9, on memory impairment using i.c.v. injection assays. First, we confirmed the acute memory impairment model triggered by i.c.v.-injected Aβ1-42. When injected with sublethal doses of Aβ1-42 (410 pmol per mouse), the WT mice exhibited significantly impaired spontaneous alteration behavior (Figure 6A), step-through escape latency (Figure 6B), and recognition memory (Figure 6C), as compared with the mice injected with Aβ40-1. However, Fcgr2b KO mice were tolerant to the memory impairment triggered by i.c.v.-injected Aβ1-42 (Figure 6, A–C). Interestingly, when we coinjected Aβ1-42 with either Aβ1-9 (#1) or FcγRIIb107-114 (#9) peptides, Aβ1-42-induced impairments of spontaneous alteration behavior and step-through escape latency were significantly blocked (Figure 6, D and E). However, Aβ1-9 (#7) mutant peptide did not affect those impairments (Figure 6, D and E). Thus, it appears that the interaction of Aβ1-42 with FcγRIIb is important in Aβ1-42-induced neurotoxic activities in vivo such as memory impairment.
 Figure 6
Figure 6Interaction of FcγRIIb with Aβ1-42 is linked to i.c.v. Aβ1-42-induced memory impairment. (A–C) Suppression of i.c.v. Aβ1-42-induced cognitive defects in Fcgr2b-deficient mice. Mice were i.c.v. injected with nontoxic Aβ40-1 or Aβ1-42 (410 pm). One day later, Y-maze (A; **P < 0.005, 1-way ANOVA), passive avoidance (B; n = 10 for each group; *P < 0.05, 1-way ANOVA), and novel object recognition (C; n = 10 for each group) tests were performed as described in the methods. Bars represent the mean ± SEM. (D and E) Suppression of i.c.v. Aβ1-42-induced memory impairment by the peptides inhibiting the interaction between Aβ1-42 and FcγRIIb. WT mice (2-month-old) were i.c.v. injected with Aβ40-1 or Aβ1-42 (410 pm) alone or together with Aβ1-9 (#1), Aβ1-9 mutant (#7), or FcγRIIb107-114 (#9) peptide (molar ratio of 1:3). One day later, the mice were analyzed using Y-maze (D) and passive avoidance (E) tests (n = 8–10). Bars represent the mean ± SEM. **P < 0.005; ***P < 0.001; 1-way ANOVA.
-
Discussion
While it has been believed that the CNS is immune privileged, molecular understanding of the immune system’s role in the progression of or protection against neurodegeneration in the CNS is rapidly growing (42–44). Although IgG is absent or extremely low-level in the CNS, FcγRs also accumulate in the plaques and microglial cells of AD brains (18). Further, one immune inhibitory receptor, CD33, was isolated as an AD risk factor in genome-wide association studies (GWAS) (45). We believe the present report is the first to show FcγRIIb’s role in Aβ neurotoxicity as a binding partner of Aβ1-42 in the CNS. Compared with IgG1, our data show that FcγRIIb bound to Aβ1-42 with higher affinity (Kd = 5.67 × 10–8 M). While the second Ig-like domain of FcγRIIb is required for its binding to both IgG and Aβ1-42, it is likely that there is a subtle difference in their interaction modes. When we injected Aβ1-42 together with IgG1, Aβ-induced impairment of spontaneous alteration behavior and recognition memory was not rescued (data not shown). As seen in other membrane proteins such as RAGE, PrPc, and EphB2 (5, 6, 41), FcγRIIb is not the only binding partner for Aβ oligomers. Along with the essential role of FcγRIIb in Aβ-induced neurotoxicity, synaptic dysfunction, and memory impairment, there is a possibility that FcγRIIb functionally interacts with other Aβ-binding proteins. In addition, the observation that FcγRIIb-ED seems to bind better to the oligomeric forms of Aβ1-42 than those of Aβ1-40 also implies that the receptors have preferential binding affinity for different species and oligomeric forms of Aβ. Thus, as seen in FcγRIIb, it will be interesting to define the critical residues in Aβ1-42 that are involved in the interaction with these proteins and to compare their binding abilities with those of Aβ1-40.
Given an earlier report that homoaggregation of FcγRIIb transduces death signals in B cells (14), another persuasive model may be that oligomeric forms of Aβ1-42 induce similar aggregation of FcγRIIb to generate a neurotoxic signal, including ER stress response and caspase-12 activation. Unlike FcγRIIb, the I232T FcγRIIb mutant, which shows low activity and a decreased affinity of FcγRIIb for lipid rafts (46), failed to induce cell death. On the other hand, Aβ1-42 appeared to bind equally to both FcγRIIb and the I232T mutant in an immunoprecipitation assay (data not shown). These observations imply that the I232T FcγRIIb mutant may act as a decoy receptor to deplete extracellular Aβ or as a dominant negative mutant to suppress FcγRIIb induction. FcγRs have high structural homology in their extracellular domains and need to be analyzed in detail for their expression pattern in the CNS and their ability to bind to Aβ. Among the FcγRs, however, FcγRIIb is the only inhibitory receptor that contains an immunoreceptor tyrosine-based inhibitory motif (ITIM) in its cytoplasmic portion, while the other FcγRs (FcγRI, FcγRIIa, FcγRIII, and FcγRIV) have an immunoreceptor tyrosine-based activating motif (ITAM) (19). We also observed that replacement of the tyrosine residue in the ITIM with phenylalanine (Y273F) caused a loss of FcγRIIb’s proapoptotic activity (data not shown), indicating that FcγRIIb possesses a unique neurotoxic feature among the FcγRs.
The hAPP transgenic mouse (J20 line) shows relatively high expression of the hAPP gene and generates various APP fragments as well as a high level of Aβ. Though the multiple assembly forms of Aβ were presented throughout the lives of the J20 mice (47), various oligomeric Aβs, including Aβ*56 and Aβ trimers, were already detected in the J20 mice before their behavioral defects were observed (48). In J20 mice, more importantly, learning and memory deficits were correlated with certain species of soluble oligomers, such as Aβ*56, but not with plaque load or hAPP and β-CTF levels (49), indicating that Aβ-mediated neurotoxicity is a primary cause of pathology. In addition, Fcgr2b KO mice of the B6 strain are highly susceptible to death due to autoimmunity (15). Although the 129sv/B6 mouse strain may be tolerable to autoimmunity caused by FcγRIIb deficiency (15), AD model mice expressing APP in 129 derivative strains did not show behavioral abnormality in many assessments, including the water maze test (50, 51). Thus, we believe that the J20 mouse showing early learning and memory deficits is a plausible model for demonstrating the role of FcγRIIb in Aβ neurotoxicity in vivo. In our investigation, we used 4- to 6-month-old J20 mice that did not yet show clear-cut pathological endpoints like the formation of plaque burden (52), supporting our proposition that FcγRIIb mediates soluble Aβ–related pathology, such as memory deficits, in these mice. In AD model mice, however, the possibility that FcγRIIb in nonneuronal cells may also contribute to memory impairment through other activity such as neuroinflammation requires further elucidation. Given the previous reports that anti-Aβ antibodies prevented AD-like phenotypes through Fc receptor–mediated phagocytosis in AD model mice (53, 54), immune inhibitory FcγRIIb deficiency may also contribute to rescuing such pathology by eliminating the negative regulation of antibody effector functions.
An acute memory loss model triggered by i.c.v.-injected Aβ1-42 may not be representative for the mental retardation found in AD. Nonetheless, we believe that it is the best model for clarifying the effect of the inhibitory peptides on memory defects. Also, an i.c.v. injection assay of Aβ ADDLs is a useful way to study the role of a subportion or partition of Aβ forms. In this assay, the mice showed no significant differences in their movement, and their acute memory deficits were fully recovered after 3 weeks (data not shown). These observations imply that a single i.c.v. injection of synthetic Aβ1-42 does not interfere with learning and memory nor cause emotional turbulence or physical malaise, and that the synaptic activity impaired by i.c.v-injected Aβ1-42 is not irreversible.
In conclusion, FcγRIIb is responsible for soluble Aβ–associated toxicity, including neuronal death, synaptic dysfunction, and behavioral defects in mice in which the interaction of FcγRIIb with Aβ is critical. Thus, we believe that selective inhibition in the interaction of FcγRIIb with Aβ1-42 can be considered a new therapeutic against the neuropathology caused by Aβ1-42.
-
Methods
Fcg2rb KO and hAPP transgenic mice
Maintenance of and experimentation with WT (C57BL/6), Fcgr2b KO C57BL/6 (16), and hAPP (line J20; The Jackson Laboratory) mice were performed in accordance with the animal care guidelines of Seoul National University.
Preparation of Aβ oligomers
Synthetic Aβ oligomers. The dried synthetic Aβ1-42 peptide (Sigma-Aldrich) was first dissolved in DMSO at 2.2 mM, and then diluted in PBS to obtain a 250-μM stock solution. This solution was incubated at 4°C for at least 24 hours and stored at –80°C until use. Before use, the solution was centrifuged at 12,000 g for 10 minutes and the supernatant was used as an oligomeric Aβ (ADDL). The Aβ preparation was evaluated by neurotoxicity, atomic force microscopy (Park Systems), and Western blotting using anti-Aβ or Nu-1 antibody. For atomic force microscopic analysis, the prepared oligomer solution was aliquoted (20 μl of 5 μM Aβ1-42) and immediately spotted on freshly cleaved mica (Probes). The micas were incubated at room temperature for 10 minutes, rinsed twice with deionized water and then air dried. The images were obtained at scan rates between 0.5 and 1 Hz, with the drive amplitude keeping the contact force at a minimum.
Naturally secreted Aβ oligomers. CHO cells stably expressing human APP751 with the V717F mutation (7PA2 cells; provided by D.J. Selkoe, Harvard Medical School, Cambridge, Massachusetts, USA) were cultured in DMEM with 10% FBS. Cells were grown to approximately 80% confluence, washed with PBS and incubated in serum-free DMEM for approximately 16 hours. The CM was collected and cleared of cells by centrifugation at 230 g for 10 minutes. Supernatants were concentrated at approximately 10-fold using YM-3 Centriprep filters (Millipore), collected as 1-ml aliquots and stored at –80°C until use.
Immunohistochemistry and Western blot analysis
Hippocampal tissues from AD (BraakV-VI), MCI (Braak III), and non-AD patients were provided by the Harvard Brain Tissue Resource Center (McLean Hospital, Belmont, Massachusetts, USA). Immunohistochemical studies of AD brain sections using anti-hFcγRIIb (EP888Y; Novus), anti-NeuN (Chemicon), polyclonal anti-Aβ (Zymed), and 4G8 (Signet Laboratories) antibodies were described previously (55). For Western blotting, anti-mFcγRIIb (2.4G2), anti-GFP, anti–p-c-jun, anti-GRP78, anti-CHOP, anti–caspase-12, anti-synaptophysin (Santa Cruz Biotechnology), anti-pJNK, anti-JNK, anti-pERK, anti-ERK (Cell Signaling Technology), anti–α-tubulin, and anti–β-actin (Sigma-Aldrich) antibodies were purchased. Anti-FcγRIIb antibodies were provided by U. Hammerling (Memorial Sloan Kettering Cancer Center, New York, New York, USA; monoclonal antibody from K9.361 hybridoma) and J. Cambier (University of Colorado Health Sciences Center, Denver, Colorado, USA; rabbit polyclonal), and PHF1 antibody was provided by P. Davies (Albert Einstein College of Medicine, Bronx, New York, USA).
Primary culture and cell viability test
Primary neurons were prepared as described previously (11). Briefly, the hippocampal and cortical neurons were cultured from embryonic day 16 (cortical neurons) and postnatal day 1 (hippocampal neurons), respectively, and incubated for 1 to 2 weeks in neurobasal media containing B27 (Invitrogen). For cell viability assays, apoptotic cells were analyzed by TUNEL assay (Promega), and live cells were detected by MTT assay (Sigma-Aldrich) or staining with Calcein-AM (Molecular Probes) according to the manufacturer’s instructions. In the transient transfection experiments, cell viability was determined based on the morphology of GFP-positive cells under a fluorescence microscope (Olympus) and by trypan blue exclusion assays as described (11).
RT-PCR
Total RNA was purified from mouse tissues with TRIzol reagent (Invitrogen) and used for reverse transcription as described previously (11). Murine hippocampal and cortical neurons were purified using OptiPrep density gradient medium (Sigma-Aldrich) following the manufacturer’s instructions. PCR was performed using synthetic primers for FcγRIIb: FcγRIIb-RT-5′ (5′-CAGCGACCCTGTAGATCTGGGAG-3′) and FcγRIIb-RT-3′ (5′-GCATCCCGAAGCCCTGGATGAAG-3′). Mouse cortical primary neurons were incubated with 5 μM Aβ1-42 for 36 hours, after which total RNA was purified as described above. PCR was performed using the corresponding primers: FcγRI-RT-5′ (5′-CCAGCTTTGGAGATGACATG-3′) and FcγRI-RT-3′ (5′-CGTTGATAAACCATTGTGTG-3′), or FcγRIII-RT-5′ (5′-GGACACCCAGATGTTTCAG-3′) and FcγRIII-RT-3′ (5′-CGGGTC TGCTCCATTTGACAC-3′).
Construction of plasmids
Rat FcγRIIb (rFcγRIIb) cDNA was amplified by PCR from a rat brain cDNA library using synthesized primers (FcγRIIb-5′-EcoRI, 5′-CGCGGAATTCGATGGACAGCAACAGGACT-3′; FcγRIIb-3′-KpnI, 5′-CGGGTACCATAATGTGGTTCTGGTAGTC-3′) and subcloned into pEGFP-N1. The FcγRIIb I232T mutant was generated by PCR using primers containing the corresponding mutation (FcγRIIb [I232T]-5′, 5′-GCTGT-CGCTGGAACTGTAGCTGCC-3′; FcγRIIb [I232T]-3′, 5′-GGCAGCTCAGCAGTTCCAGCGACAGC-3′), and confirmed by DNA sequencing analysis. To construct the rFcγRIIb(ED)-CD40 chimera, the extracellular domain of rFcγRIIb and the transmembrane and cytosolic domains of CD40 were subcloned into pEGFP-N1 using the primers (FcγRIIb-ED-5′-NheI, 5′-GCTAGCGCTATGGACAGCAACAGGACT-3′; FcγRIIb-ED-3′-HindIII, 5′-AAGCTTGGGAGGCAACGAACTGCTGGATTT-3′; CD40-TM+cyto-5′-HindIII, 5′-CCCAAGCTTGGGGCCCTGGTGGTGATCCCCATC-3; and CD40-TM+cyto-3′-KpnI, 5′-CGGGTACCATTCACTGTCTCTCCTGCA C-3′). Caspase-12 GFP and its active site mutant (C298S) have been described previously (12). The promoter (–537 to +43) of the human FcγRIIb (22) was amplified by PCR and subcloned into pGL2-basic vectors.
Measurement of luciferase activity
Cells were harvested and luciferase activity was measured as described previously (56). Briefly, NIH3T3 cells were cotransfected with pNF-κB-luciferase, pβact-lacZ and pEGFP, pTNF receptor1 (TNFR1), pCD40, or pFcγRIIb-CD40, and then incubated for 24 hours with 5 μM Aβ1-42 or 10 ng/ml of murine TNF-α. Cell lysates were prepared using the lysis reagent 5X (Promega), and the reporter activity was determined with a Luciferase Assay System (Promega) using a luminometer (Berthold Technologies). Luciferase activity was normalized to β-galactosidase.
Protein interaction assays
In vitro pulldown. The synthetic Aβ1-42 soluble fraction (supernatant after centrifugation of a 500-μM Aβ1-42 stock solution at 6,000 g for 5 minutes) or 7PA2-CM was incubated with 10 μg hFcγRIIb-ED or GST for 2 hours in binding buffer (50 mM Tris-HCl [pH 7.4], 1 mM DTT, 0.5 mM EDTA, 0.01% Triton X-100, 0.5 mg/ml BSA, 10% [v/v] glycerol, protease inhibitor cocktail). The mixture was then added to protein G beads preincubated with polyclonal anti-FcγRIIb antibody or anti-GST antibody. After incubating for 2 hours, the precipitates were washed 3 times with binding buffer and analyzed by Western blotting using 4G8 antibody.
In vivo pulldown. Human hippocampal brain tissues were homogenized in ice-cold lysis buffer (20 mM Tris-HCL [pH 7.4], 150 mM NaCl, 1% Triton X-100, protease inhibitor cocktail), and the tissue lysates were centrifuged at 12,000 g for 20 minutes at 4°C. Supernatant was then collected, and the protein concentration was determined using the Bradford method (GE Healthcare). Equal amounts (1 mg) of hippocampal lysates from normal control and AD patients’ brains were immunoprecipitated with anti-Aβ antibody (Nu-1) beads at 4°C overnight, then immunoblotted using anti-FcγRIIb antibody (EP888Y).
SPR. Oligomerized Aβ1-42 or hIgG1 was immobilized on a CM5 sensor chip (GE Healthcare) preactivated by 0.05 M 1-ethyl-3-(3-dimethylaminopropyl) carbodiimide (EDC) and 0.2 M N-hydroxysuccinimide (NHS) using amine coupling. The chip was then deactivated with 1 M ethanolamine-HCl, pH 8.5. To analyze the binding kinetics, different concentrations of hFcγRIIb-ED in a running buffer (PBS, pH 7.4) were injected into the sensor chip for 120 seconds at a flow rate of 5 μl per minute, followed by an injection with PBS, and dissociation curves were monitored for 240 seconds. Surface regeneration was achieved by injection of 5 mM of NaOH. The curves obtained from the SPR experiments were analyzed and Kd values were calculated using BIAevaluation software (GE Healthcare). When Aβ1-42 binding to hIgG1 was analyzed, dose-dependent binding was not observed, indicating that nonspecific interaction was negligible (data not shown).
Dendritic spine density analysis
Hippocampal neurons were transfected at 11 days in vitro (DIV) with pEGFP-N1. Two days after transfection, neurons were incubated with the CHO- or 7PA2-CM for 5 days. For treatment with synthetic Aβ1-42, cultured neurons at 13 DIV were transfected with pEGFP-N1 for 3 days and then exposed to 2 μM of Aβ1-42 for 2 days. Cultures were fixed in 4% paraformaldehyde for 10 minutes followed by immunostaining using anti-synaptophysin antibody, if necessary. Images of GFP-expressing cells were acquired by confocal microscopy (Carl Zeiss), and more than 10 randomly chosen neurons measuring 100 μm in length were analyzed in a blind fashion.
ELISA analysis for Aβ quantification
Aβ levels were analyzed using a sandwich ELISA kit (Invitrogen) according to the manufacturer’s instructions. Briefly, whole hemibrains were microdissected and the hippocampus was isolated. Tissues were homogenized in 10 volumes of ice-cold guanidine buffer (5 M guanidine-HCl/50 mM Tris-Cl, pH 8.0) and mixed for 3 hours at room temperature. The brain homogenates were further diluted 1:10 with cold reaction buffer (5% BSA, 0.03% Tween-20, and 5 mM EDTA in PBS supplemented with protease inhibitor cocktail), then centrifuged at 16,000 g and 4°C for 20 minutes. The diluents were mixed 1:1 with standard dilution buffer in the assay kit.
Tissue slice preparation and electrophysiological recordings
WT C57BL/6 and Fcg2rb KO mice (2–3 months of age) for the Aβ/LTP assay or WT, Fcg2rb KO, hAPP, and hAPP/Fcg2rb KO mice (9–13 months of age) for LTP were anesthetized with isoflurane and decapitated. Brains were rapidly removed and placed in an ice-cold modified artificial cerebrospinal fluid (aCSF) solution [175 mM sucrose, 20 mM NaCl, 3.5 mM KCl, 1.25 mM NaH2PO4, 26 mM NaHCO3, 1.3 mM MgCl2 and 11 mM D-(+)-glucose]. Coronal slices (350 μm, Aβ/LTP), including hippocampus, or transverse hippocampal slices (400 μm, LTP) were cut using a vibroslicer (NVSL; World Precision Instruments) and incubated in normal aCSF solution [120 mM NaCl, 3.5 mM KCl, 1.25 mM NaH2PO4, 26 mM NaHCO3, 1.3 mM MgCl2 and 11 mM D-(+)-glucose] at room temperature for at least 1 hour and continuously bubbled with 95% O2/5% CO2. A submersion-type recording chamber was continuously perfused with aCSF at a constant flow rate of 1 to 2 ml/min–1 maintained by a peristaltic pump (Pharmacia) at 29.5 ± 0.5°C. Bipolar stimulating electrodes were placed in the stratum radiatum of the Schaffer commissural pathways. Extracellular field potential recordings were made by using a parylene-insulated microelectrode (573210; A-M Systems) or ACSF-filled microelectrodes (1–3 MΩ, GC100T-10; Harvard Apparatus). Baseline stimulation (0.033 Hz) was delivered at the intensity that evoked a response below 50% of the maximal response. A stable baseline was recorded for at least 20 minutes before inducing LTP. For experiments using Aβ1-42 oligomers, brain slices were continuously perfused with normal aCSF containing 400 nM Aβ1-42 for 30 to 50 minutes before LTP induction, and the solution was recycled for the duration of the recording. LTP was induced by 4 trains of high-frequency stimulation (HFS) (100 Hz, 1-second duration, 30-second intervals). Extracellular field potentials were amplified and filtered (low-pass filter, 1 kHz; high-pass filter, 1 Hz; Dam80; World Precision Instruments), and then digitized using the NAC 2.0 acquisition system (Theta Burst). The digitized signals were stored and analyzed on a computer using NAC Gather (Theta Burst) or IGOR (WaveMetrics) software.
Behavior tests
Y-maze tests. A triangular Y maze was made from black plastic with an arm size of 32.5 cm × 15 cm (length × height). For the test, the mice were placed in the end of one arm and allowed to move freely through the maze for 7 minutes. An entry was defined as placing all 4 paws into the arm. The percentage of spontaneous alterations was calculated as the ratio of the number of successful alterations to the number of total alterations. The total number of arm entries was identified as hyperactivity. After each trial, the maze was cleaned with 70% ethanol.
Novel object recognition. The mice were first habituated in a white plastic chamber (22 cm wide × 27 cm long × 30 cm high) for 7 minutes at 24-hour intervals. After 2 days (training trial), the mice were presented with 2 objects and allowed to explore freely for 7 minutes. In the testing trial performed 24 hours later, 1 of the familiar objects was changed for a novel object and the mice were scored for recognition during a 7-minute period. The object recognition was defined as spending time orienting toward the object at a distance of 1 cm or less, sniffing the object, or touching it with the nose. Between each trial, the arena and objects were wiped down with 70% ethanol.
Passive avoidance. The passive avoidance apparatus consists of a light and dark compartment (20 × 20 × 20 cm each) separated by a guillotine door. The floor of the dark compartment was made of stainless-steel grids. During habituation, mice were allowed to freely explore the box for 5 minutes with the door open, then were returned to their home cage. For conditioning, after 24 hours, the mice were placed into the light compartment and the sliding door was closed when both hindlimbs of the mice were entered into the dark box, then an electric foot shock (0.25 mA, 2 seconds) was delivered by the floor grids. Ten seconds later, the mice were returned to their home cage. Tests were carried out 24 hours after the conditioning, and the latency time for the mice to enter the dark compartment was measured with a 5-minute cut-off point.
Intracerebroventricular injection of Aβ1-42
Intracerebroventricular injection of Aβ1-42 or Aβ40-1 (H-2972; Bachem) into mice and the assessment of alteration behavior (Y maze) and passive avoidance are described above. Before injection, Aβ1-42 was incubated for 2 hours at 37°C with or without inhibitory peptides dissolved in PBS.
Statistics
All data were analyzed with GraphPad Prism software (GraphPad). Differences between 2 means were assessed by a paired or unpaired t test. Differences among multiple means were assessed, as indicated, by 1-way ANOVA, followed by Tukey’s post-hoc test. Error bars represent SD or SEM, as indicated.
Study approval
All experiments involving animals were performed according to protocols approved by the Seoul National University Institutional Animal Care and Use Committee (SNU IACUC) guidelines. Frozen human hippocampal tissues were provided by the Harvard Brain Tissue Resource Center (McLean Hospital) in accordance with the regulation of the tissue bank and the approval of the Research Ethics Committee of the SNU. Patients provided informed consent for the use of tissues.
-
Acknowledgments
The authors thank U. Hammerling for FcγRIIb monoclonal antibody (K9.361 hybridoma); J. Cambier for rabbit polyclonal antibody; D.J. Selkoe (Harvard Medical School, Cambridge, Massachusetts) for CHO and 7PA2 cells; and J.H. Yu (Seoul National University, Seoul, Republic of Korea) for use of the Biacore. T.I. Kam and Y. Gwon were in part supported by the BK21 program and the Global PhD program, respectively. AD tissues were provided by the Harvard Brain Tissue Resource Center of McLean Hospital. This work was supported by grants from the Brain Research Center (2012K001122) of the 21st Century Frontier Research Program and Global Research Laboratory program (K21004000002-12A0500-00210) and the KIST R&D institutional program (2E22650, to K.S. Kim) funded by the Ministry of Education, Science and Technology (MEST); and by an AD grant (A092058-1113-0000400) from the Korean Ministry of Human Health and Welfare and the AD Foundation in USA.
Address correspondence to: Yong-Keun Jung, School of Biological Sciences, Seoul National University, 599 Gwanak-ro, Gwanak-gu, Seoul 151-747, Republic of Korea. Phone: 82.2.880.4401; Fax: 82.2.873.7524; E-mail: [email protected].
-
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Reference information: J Clin Invest. 2013;123(7):2791–2802. doi:10.1172/JCI66827.
-
References
- Mattson MP. Pathways towards and away from Alzheimer’s disease. Nature. 2004;430(7000):631–639.
- Golde TE, Janus C. Homing in on intracellular Aβ? Neuron. 2005;45(5):639–642.
- Deane R, et al. RAGE mediates amyloid-beta peptide transport across the blood-brain barrier and accumulation in brain. Nat Med. 2003;9(7):907–913.
- Lustbader JW, et al. ABAD directly links Abeta to mitochondrial toxicity in Alzheimer’s disease. Science. 2004;304(5669):448–452.
- Laurén J, Gimbel DA, Nygaard HB, Gilbert JW, Strittmatter SM. Cellular prion protein mediates impairment of synaptic plasticity by amyloid-β oligomers. Nature. 2009;457(7233):1128–1132.
- Yan SD, et al. RAGE and amyloid-beta peptide neurotoxicity in Alzheimer’s disease. Nature. 1996;382(6593):685–691.
- Gimbel DA, et al. Memory impairment in transgenic Alzheimer mice requires cellular prion protein. J Neurosci. 2010;30(18):6367–6374.
- Kessels HW, Nguyen LN, Nabavi S, Malinow R. The prion protein as a receptor for amyloid-beta. Nature. 2010;466(7308):discussion E4–E5.View this article via: PubMed Google Scholar
- Balducci C, et al. Synthetic amyloid-β oligomers impair long-term memory independently of cellular prion protein. Proc Natl Acad Sci U S A. 2010;107(5):2295–2300.
- Cissé M, Sanchez PE, Kim DH, Ho K, Yu GQ, Mucke L. Ablation of cellular prion protein does not ameliorate abnormal neural network activity or cognitive dysfunction in the J20 line of human amyloid precursor protein transgenic mice. J Neurosci. 2011;31(29):10427–10431.
- Song S, et al. Essential role of E2-25K/Hip-2 in mediating amyloid-beta neurotoxicity. Mol Cell. 2003;12(3):553–563.
- Song S, et al. E2-25K/Hip-2 regulates caspase-12 in ER stress-mediated Abeta neurotoxicity. J Cell Biol. 2008;182(4):675–684.
- Katz HR. Inhibitory receptors and allergy. Curr Opin Immunol. 2002;14(6):698–704.
- Pritchard NR, Smith KG. B cell inhibitory receptors and autoimmunity. Immunology. 2003;108(3):263–273.
- Bolland S, Ravetch JV. Spontaneous autoimmune disease in Fc(gamma) RIIB-deficient mice results from strain-specific epistasis. Immunity. 2000;13(2):277–285.
- Takai T, Ono M, Hikida M, Ohmori H, Ravetch JV. Augmented humoral and anaphylactic responses in Fc gamma RII-deficient mice. Nature. 1996;379(6563):346–349.
- Nakamura K, et al. CD3 and immunoglobulin G Fc receptor regulate cerebellar functions. Mol Cell Biol. 2007;27(14):5128–5134.
- Peress NS, Fleit HB, Perillo E, Kuljis R, Pezzullo C. Identification of Fc gamma RI, II and III on normal human brain ramified microglia and on microglia in senile plaques in Alzheimer’s disease. J Neuroimmunol. 1993;48(1):71–79.
- Okun E, Mattson MP, Arumugam TV. Involvement of Fc receptors in disorders of the central nervous system. Neuromolecular Med. 2010;12(2):164–178.
- Lambert MP, et al. Vaccination with soluble Abeta oligomers generates toxicity-neutralizing antibodies. J Neurochem. 2001;79(3):595–605.
- Roberson ED, et al. Reducing endogenous tau ameliorates amyloid-β-induced deficits in an Alzheimer’s disease mouse model. Science. 2007;316(5825):750–754.
- Olferiev M, Masuda E, Tanaka S, Blank MC, Pricop L. The role of activating protein 1 in the transcriptional regulation of the human FCGR2B promoter mediated by the -343 G→C polymorphism associated with systemic lupus erythematosus. J Biol Chem. 2007;
282
(3):1738–1746.View this article via: PubMed Google Scholar
- Cleary JP, et al. Natural oligomers of the amyloid-beta protein specifically disrupt cognitive function. Nat Neurosci. 2005;8(1):79–84.
- Walsh DM, et al. Naturally secreted oligomers of amyloid beta protein potently inhibit hippocampal long-term potentiation in vivo. Nature. 2002;416(6880):535–539.
- Shankar GM, et al. Amyloid-beta protein dimers isolated directly from Alzheimer’s brains impair synaptic plasticity and memory. Nat Med. 2008;14(8):837–842.
- Oddo S, et al. Triple-transgenic model of Alzheimer’s disease with plaques and tangles: intracellular Abeta and synaptic dysfunction. Neuron. 2003;39(3):409–421.
- Billings LM, Oddo S, Green KN, McGaugh JL, Laferla FM. Intraneuronal Abeta causes the onset of early Alzheimer’s disease-related cognitive deficits in transgenic mice. Neuron. 2005;45(5):675–688.
- Kumar-Singh S, et al. In vitro studies of Flemish, Dutch, and wild-type β-amyloid provide evidence for two-staged neurotoxicity. Neurobiol Dis. 2002;11(2):330–340.
- Sotthibundhu A, Sykes AM, Fox B, Underwood CK, Thangnipon W, Coulson EJ. β-Amyloid1-42 induces neuronal death through the p75 neurotrophin receptor. J Neurosci. 2008;28(15):3941–3946.
- Boyce M, et al. A selective inhibitor of eIF2alpha dephosphorylation protects cells from ER stress. Science. 2005;307(5711):935–939.
- Nakagawa T, et al. Caspase-12 mediates endoplasmic-reticulum-specific apoptosis and cytotoxicity by amyloid-beta. Nature. 2000;403(6765):98–103.
- Urbanc B, et al. In silico study of amyloid beta-protein folding and oligomerization. Proc Natl Acad Sci U S A. 2004;101(50):17345–17350.
- Bush AI. The metallobiology of Alzheimer’s disease. Trends Neurosci. 2003;26(4):207–214.
- Crescenzi O, et al. Solution structure of the Alzheimer amyloid beta-peptide (1–42) in an apolar microenvironment. Similarity with a virus fusion domain. Eur J Biochem. 2002;269(22):5642–5648.
- Sondermann P, Huber R, Jacob U. Crystal structure of the soluble form of the human fcgamma-receptor IIb: a new member of the immunoglobulin superfamily at 1.7 Α resolution. EMBO J. 1999;18(5):1095–1103.
- Goldsmith EB, Erickson BW, Thompson NL. Synthetic peptides from mouse Fc receptor (MoFc gamma RII) that alter the binding of IgG to MoFc gamma RII. Biochemistry. 1997;36(4):952–959.
- Wang Q, Walsh DM, Rowan MJ, Selkoe DJ, Anwyl R. Block of long-term potentiation by naturally secreted and synthetic amyloid β-peptide in hippocampal slices is mediated via activation of the kinases c-Jun N-terminal kinase, cyclin-dependent kinase 5, and p38 mitogen-activated protein kinase as well as metabotropic glutamate receptor type 5. J Neurosci. 2004;24(13):3370–3378.
- Selkoe DJ. Alzheimer’s disease is a synaptic failure. Science. 2002;298(5594):789–791.
- Haass C, Selkoe DJ. Soluble protein oligomers in neurodegeneration: lessons from the Alzheimer’s amyloid beta-peptide. Nat Rev Mol Cell Biol. 2007;8(2):101–112.
- Sanchez-Mejia RO, et al. Phospholipase A2 reduction amelioreate cognitive deficits in a mouse model of Alzheimer’s disease. Nat Neurosci. 2008;11(11):1311–1318.
- Cissé M, et al. Reversing EphB2 depletion rescues cognitive functions in Alzheimer model. Nature. 2011;469(7328):47–52.
- Britschgi M, Wyss-Coray T. Immune cells may fend off Alzheimer disease. Nat Med. 2007;13(4):408–409.
- Siffrin V, Brandt AU, Herz J, Zipp F. New insights into adaptive immunity in chronic neuroinflammation. Adv Immunol. 2007;96:1–40.View this article via: PubMed Google Scholar
- Vargas ME, Barres BA. Why is Wallerian degeneration in the CNS so slow? Annu Rev Neurosci. 2007;30:153–179.
- Bertram L, et al. Genome-wide association analysis reveals putative Alzheimer’s disease susceptibility loci in addition to APOE. Am J Hum Genet. 2008;83(5):623–632.
- Kono H, et al. FcγRIIB Ile232Thr transmembrane polymorphism associated with human systemic lupus erythematosus decreases affinity to lipid rafts and attenuates inhibitory effects on B cell receptor signaling. Hum Mol Genet. 2005;14(19):2881–2892.
- Shankar GM, et al. Biochemical and immunohistochemical analysis of an Alzheimer’s disease mouse model reveals the presence of multiple cerebral Aβ assembly forms throughout life. Neurobiol Dis. 2009;36(2):293–302.
- Meilandt WJ, et al. Neprilysin overexpression inhibits plaque formation but fails to reduce pathogenic Aβ oligomers and associated cognitive deficits in human amyloid precursor protein transgenic mice. J Neurosci. 2009;29(7):1977–1986.
- Cheng IH, et al. Accelerating amyloid-β fibrillization reduces oligomer levels and functional deficits in Alzheimer disease mouse models. J Biol Chem. 2007;282(33):23818–23828.
- Janus C, Chishti MA, Westaway D. Transgenic mouse models of Alzheimer’s disease. Biochim Biophys Acta. 2000;1502(1):63–75.View this article via: PubMed Google Scholar
- Westerman MA, et al. The relationship between Abeta and memory in the Tg2576 mouse model of Alzheimer’s disease. J Neurosci. 2002;22(5):1858–1867.View this article via: PubMed Google Scholar
- Cheng IH, Palop JJ, Esposito LA, Bien-Ly N, Yan F, Mucke L. Aggressive amyloidosis in mice expressing human amyloid peptides with the Arctic mutation. Nat Med. 2004;10(11):1190–1192.
- Schenk D, et al. Immunization with amyloid-beta attenuates Alzheimer-disease-like pathology in the PDAPP mouse. Nature. 1999;400(6740):173–177.
- Bard F, et al. Peripherally administered antibodies against amyloid beta-peptide enter the central nervous system and reduce pathology in a mouse model of Alzheimer disease. Nat Med. 2000;6(8):916–919.View this article via: PubMed Google Scholar
- Park H, et al. Neuropathogenic role of adenylate kinase-1 in Aβ-mediated tau phosphorylation via AMPK and GSK3β. Hum Mol Genet. 2012;21(12):2725–2737.
- Woo HN. et al. Inhibition of Bcl10-mediated activation of NF-kappa B by BinCARD, a Bcl10-interacting CARD protein. FEBS Lett. 2004;578(3):239–244.
-
Version history
- Version 1 (June 10, 2013): No description
- Version 2 (July 1, 2013): No description
Article tools
-
View PDF

- Download citation information
- Send a comment
- Terms of use
- Standard abbreviations
- Need help? Email the journal
Related posts
Metrics



Copyright © 2024 American Society for Clinical Investigation
ISSN: 0021-9738 (print), 1558-8238 (online)