Advertisement
Research Article Free access | 10.1172/JCI42601
Autophagy is essential for mouse sense of balance
Guillermo Mariño,1 Alvaro F. Fernández,1 Sandra Cabrera,1 Yunxia W. Lundberg,2 Rubén Cabanillas,1,3 Francisco Rodríguez,1 Natalia Salvador-Montoliu,1 José A. Vega,4 Antonino Germanà,5 Antonio Fueyo,6 José M.P. Freije,1 and Carlos López-Otín1
1Departamento de Bioquímica y Biología Molecular, Facultad de Medicina, Instituto Universitario de Oncología, Universidad de Oviedo, Oviedo, Spain. 2Genetics Department, Boys Town National Research Hospital, Omaha, Nebraska, USA. 3Instituto de Medicina Oncológica y Molecular de Asturias, Oviedo, Spain. 4Departamento de Morfología y Biología Celular, Facultad de Medicina, Universidad de Oviedo, Oviedo, Spain. 5Dipartimento di Morfologia, Biochimica, Fisiologia e Produzione Animale, Università di Messina, Messina, Italy. 6Departamento de Biología Funcional, Facultad de Medicina, Instituto Universitario de Oncología, Universidad de Oviedo, Oviedo, Spain.
Address correspondence to: Carlos López-Otín, Departamento de Bioquímica y Biología Molecular, Facultad de Medicina, Universidad de Oviedo, 33006 Oviedo, Spain. Phone: 34.985.104201; Fax: 34.985.103564; E-mail: [email protected].
Find articles by Mariño, G. in: JCI | PubMed | Google Scholar
1Departamento de Bioquímica y Biología Molecular, Facultad de Medicina, Instituto Universitario de Oncología, Universidad de Oviedo, Oviedo, Spain. 2Genetics Department, Boys Town National Research Hospital, Omaha, Nebraska, USA. 3Instituto de Medicina Oncológica y Molecular de Asturias, Oviedo, Spain. 4Departamento de Morfología y Biología Celular, Facultad de Medicina, Universidad de Oviedo, Oviedo, Spain. 5Dipartimento di Morfologia, Biochimica, Fisiologia e Produzione Animale, Università di Messina, Messina, Italy. 6Departamento de Biología Funcional, Facultad de Medicina, Instituto Universitario de Oncología, Universidad de Oviedo, Oviedo, Spain.
Address correspondence to: Carlos López-Otín, Departamento de Bioquímica y Biología Molecular, Facultad de Medicina, Universidad de Oviedo, 33006 Oviedo, Spain. Phone: 34.985.104201; Fax: 34.985.103564; E-mail: [email protected].
Find articles by Fernández, A. in: JCI | PubMed | Google Scholar
1Departamento de Bioquímica y Biología Molecular, Facultad de Medicina, Instituto Universitario de Oncología, Universidad de Oviedo, Oviedo, Spain. 2Genetics Department, Boys Town National Research Hospital, Omaha, Nebraska, USA. 3Instituto de Medicina Oncológica y Molecular de Asturias, Oviedo, Spain. 4Departamento de Morfología y Biología Celular, Facultad de Medicina, Universidad de Oviedo, Oviedo, Spain. 5Dipartimento di Morfologia, Biochimica, Fisiologia e Produzione Animale, Università di Messina, Messina, Italy. 6Departamento de Biología Funcional, Facultad de Medicina, Instituto Universitario de Oncología, Universidad de Oviedo, Oviedo, Spain.
Address correspondence to: Carlos López-Otín, Departamento de Bioquímica y Biología Molecular, Facultad de Medicina, Universidad de Oviedo, 33006 Oviedo, Spain. Phone: 34.985.104201; Fax: 34.985.103564; E-mail: [email protected].
Find articles by Cabrera, S. in: JCI | PubMed | Google Scholar
1Departamento de Bioquímica y Biología Molecular, Facultad de Medicina, Instituto Universitario de Oncología, Universidad de Oviedo, Oviedo, Spain. 2Genetics Department, Boys Town National Research Hospital, Omaha, Nebraska, USA. 3Instituto de Medicina Oncológica y Molecular de Asturias, Oviedo, Spain. 4Departamento de Morfología y Biología Celular, Facultad de Medicina, Universidad de Oviedo, Oviedo, Spain. 5Dipartimento di Morfologia, Biochimica, Fisiologia e Produzione Animale, Università di Messina, Messina, Italy. 6Departamento de Biología Funcional, Facultad de Medicina, Instituto Universitario de Oncología, Universidad de Oviedo, Oviedo, Spain.
Address correspondence to: Carlos López-Otín, Departamento de Bioquímica y Biología Molecular, Facultad de Medicina, Universidad de Oviedo, 33006 Oviedo, Spain. Phone: 34.985.104201; Fax: 34.985.103564; E-mail: [email protected].
Find articles by Lundberg, Y. in: JCI | PubMed | Google Scholar
1Departamento de Bioquímica y Biología Molecular, Facultad de Medicina, Instituto Universitario de Oncología, Universidad de Oviedo, Oviedo, Spain. 2Genetics Department, Boys Town National Research Hospital, Omaha, Nebraska, USA. 3Instituto de Medicina Oncológica y Molecular de Asturias, Oviedo, Spain. 4Departamento de Morfología y Biología Celular, Facultad de Medicina, Universidad de Oviedo, Oviedo, Spain. 5Dipartimento di Morfologia, Biochimica, Fisiologia e Produzione Animale, Università di Messina, Messina, Italy. 6Departamento de Biología Funcional, Facultad de Medicina, Instituto Universitario de Oncología, Universidad de Oviedo, Oviedo, Spain.
Address correspondence to: Carlos López-Otín, Departamento de Bioquímica y Biología Molecular, Facultad de Medicina, Universidad de Oviedo, 33006 Oviedo, Spain. Phone: 34.985.104201; Fax: 34.985.103564; E-mail: [email protected].
Find articles by Cabanillas, R. in: JCI | PubMed | Google Scholar
1Departamento de Bioquímica y Biología Molecular, Facultad de Medicina, Instituto Universitario de Oncología, Universidad de Oviedo, Oviedo, Spain. 2Genetics Department, Boys Town National Research Hospital, Omaha, Nebraska, USA. 3Instituto de Medicina Oncológica y Molecular de Asturias, Oviedo, Spain. 4Departamento de Morfología y Biología Celular, Facultad de Medicina, Universidad de Oviedo, Oviedo, Spain. 5Dipartimento di Morfologia, Biochimica, Fisiologia e Produzione Animale, Università di Messina, Messina, Italy. 6Departamento de Biología Funcional, Facultad de Medicina, Instituto Universitario de Oncología, Universidad de Oviedo, Oviedo, Spain.
Address correspondence to: Carlos López-Otín, Departamento de Bioquímica y Biología Molecular, Facultad de Medicina, Universidad de Oviedo, 33006 Oviedo, Spain. Phone: 34.985.104201; Fax: 34.985.103564; E-mail: [email protected].
Find articles by Rodríguez, F. in: JCI | PubMed | Google Scholar
1Departamento de Bioquímica y Biología Molecular, Facultad de Medicina, Instituto Universitario de Oncología, Universidad de Oviedo, Oviedo, Spain. 2Genetics Department, Boys Town National Research Hospital, Omaha, Nebraska, USA. 3Instituto de Medicina Oncológica y Molecular de Asturias, Oviedo, Spain. 4Departamento de Morfología y Biología Celular, Facultad de Medicina, Universidad de Oviedo, Oviedo, Spain. 5Dipartimento di Morfologia, Biochimica, Fisiologia e Produzione Animale, Università di Messina, Messina, Italy. 6Departamento de Biología Funcional, Facultad de Medicina, Instituto Universitario de Oncología, Universidad de Oviedo, Oviedo, Spain.
Address correspondence to: Carlos López-Otín, Departamento de Bioquímica y Biología Molecular, Facultad de Medicina, Universidad de Oviedo, 33006 Oviedo, Spain. Phone: 34.985.104201; Fax: 34.985.103564; E-mail: [email protected].
Find articles by Salvador-Montoliu, N. in: JCI | PubMed | Google Scholar
1Departamento de Bioquímica y Biología Molecular, Facultad de Medicina, Instituto Universitario de Oncología, Universidad de Oviedo, Oviedo, Spain. 2Genetics Department, Boys Town National Research Hospital, Omaha, Nebraska, USA. 3Instituto de Medicina Oncológica y Molecular de Asturias, Oviedo, Spain. 4Departamento de Morfología y Biología Celular, Facultad de Medicina, Universidad de Oviedo, Oviedo, Spain. 5Dipartimento di Morfologia, Biochimica, Fisiologia e Produzione Animale, Università di Messina, Messina, Italy. 6Departamento de Biología Funcional, Facultad de Medicina, Instituto Universitario de Oncología, Universidad de Oviedo, Oviedo, Spain.
Address correspondence to: Carlos López-Otín, Departamento de Bioquímica y Biología Molecular, Facultad de Medicina, Universidad de Oviedo, 33006 Oviedo, Spain. Phone: 34.985.104201; Fax: 34.985.103564; E-mail: [email protected].
Find articles by Vega, J. in: JCI | PubMed | Google Scholar
1Departamento de Bioquímica y Biología Molecular, Facultad de Medicina, Instituto Universitario de Oncología, Universidad de Oviedo, Oviedo, Spain. 2Genetics Department, Boys Town National Research Hospital, Omaha, Nebraska, USA. 3Instituto de Medicina Oncológica y Molecular de Asturias, Oviedo, Spain. 4Departamento de Morfología y Biología Celular, Facultad de Medicina, Universidad de Oviedo, Oviedo, Spain. 5Dipartimento di Morfologia, Biochimica, Fisiologia e Produzione Animale, Università di Messina, Messina, Italy. 6Departamento de Biología Funcional, Facultad de Medicina, Instituto Universitario de Oncología, Universidad de Oviedo, Oviedo, Spain.
Address correspondence to: Carlos López-Otín, Departamento de Bioquímica y Biología Molecular, Facultad de Medicina, Universidad de Oviedo, 33006 Oviedo, Spain. Phone: 34.985.104201; Fax: 34.985.103564; E-mail: [email protected].
Find articles by Germanà, A. in: JCI | PubMed | Google Scholar
1Departamento de Bioquímica y Biología Molecular, Facultad de Medicina, Instituto Universitario de Oncología, Universidad de Oviedo, Oviedo, Spain. 2Genetics Department, Boys Town National Research Hospital, Omaha, Nebraska, USA. 3Instituto de Medicina Oncológica y Molecular de Asturias, Oviedo, Spain. 4Departamento de Morfología y Biología Celular, Facultad de Medicina, Universidad de Oviedo, Oviedo, Spain. 5Dipartimento di Morfologia, Biochimica, Fisiologia e Produzione Animale, Università di Messina, Messina, Italy. 6Departamento de Biología Funcional, Facultad de Medicina, Instituto Universitario de Oncología, Universidad de Oviedo, Oviedo, Spain.
Address correspondence to: Carlos López-Otín, Departamento de Bioquímica y Biología Molecular, Facultad de Medicina, Universidad de Oviedo, 33006 Oviedo, Spain. Phone: 34.985.104201; Fax: 34.985.103564; E-mail: [email protected].
Find articles by Fueyo, A. in: JCI | PubMed | Google Scholar
1Departamento de Bioquímica y Biología Molecular, Facultad de Medicina, Instituto Universitario de Oncología, Universidad de Oviedo, Oviedo, Spain. 2Genetics Department, Boys Town National Research Hospital, Omaha, Nebraska, USA. 3Instituto de Medicina Oncológica y Molecular de Asturias, Oviedo, Spain. 4Departamento de Morfología y Biología Celular, Facultad de Medicina, Universidad de Oviedo, Oviedo, Spain. 5Dipartimento di Morfologia, Biochimica, Fisiologia e Produzione Animale, Università di Messina, Messina, Italy. 6Departamento de Biología Funcional, Facultad de Medicina, Instituto Universitario de Oncología, Universidad de Oviedo, Oviedo, Spain.
Address correspondence to: Carlos López-Otín, Departamento de Bioquímica y Biología Molecular, Facultad de Medicina, Universidad de Oviedo, 33006 Oviedo, Spain. Phone: 34.985.104201; Fax: 34.985.103564; E-mail: [email protected].
Find articles by Freije, J. in: JCI | PubMed | Google Scholar
1Departamento de Bioquímica y Biología Molecular, Facultad de Medicina, Instituto Universitario de Oncología, Universidad de Oviedo, Oviedo, Spain. 2Genetics Department, Boys Town National Research Hospital, Omaha, Nebraska, USA. 3Instituto de Medicina Oncológica y Molecular de Asturias, Oviedo, Spain. 4Departamento de Morfología y Biología Celular, Facultad de Medicina, Universidad de Oviedo, Oviedo, Spain. 5Dipartimento di Morfologia, Biochimica, Fisiologia e Produzione Animale, Università di Messina, Messina, Italy. 6Departamento de Biología Funcional, Facultad de Medicina, Instituto Universitario de Oncología, Universidad de Oviedo, Oviedo, Spain.
Address correspondence to: Carlos López-Otín, Departamento de Bioquímica y Biología Molecular, Facultad de Medicina, Universidad de Oviedo, 33006 Oviedo, Spain. Phone: 34.985.104201; Fax: 34.985.103564; E-mail: [email protected].
Find articles by López-Otín, C. in: JCI | PubMed | Google Scholar
Published June 23, 2010 - More info
J Clin Invest. 2010;120(7):2331–2344. https://doi.org/10.1172/JCI42601.
© 2010 The American Society for Clinical Investigation
Received: February 9, 2010; Accepted: May 5, 2010
Related article:
Abstract
Autophagy is a tightly regulated catabolic process whereby cells degrade their constituents to dispose of unwanted cytoplasmic elements and recycle nutrients for cellular remodeling. Studies of autophagy in mammals have elicited substantial interest because it is linked to a range of physiologic and pathologic states. In this issue of the JCI, Mariño et al. uncover a role for autophagy in a balance disorder related to inner ear pathologies. Mice lacking the protease autophagy-related 4B (Atg4b, also known as autophagin-1) exhibited a systemic reduction in autophagy and showed defects in the development of otoconia, organic particles that contain calcium carbonate crystals and proteins and that are essential for balance perception (equilibrioception) in mammals. The intriguing aspect of this work is that an autophagy block impairs the secretion and assembly of otoconial proteins, emphasizing a role for autophagy in functions distinct from macromolecule degradation.
Authors
Andreas Till, Suresh Subramani
-
Abstract
Autophagy is an evolutionarily conserved process that is essential for cellular homeostasis and organismal viability in eukaryotes. However, the extent of its functions in higher-order processes of organismal physiology and behavior is still unknown. Here, we report that autophagy is essential for the maintenance of balance in mice and that its deficiency leads to severe balance disorders. We generated mice deficient in autophagin-1 protease (Atg4b) and showed that they had substantial systemic reduction of autophagic activity. Autophagy reduction occurred through defective proteolytic processing of the autophagosome component LC3 and its paralogs, which compromised the rate of autophagosome maturation. Despite their viability, Atg4b-null mice showed unusual patterns of behavior that are common features of inner ear pathologies. Consistent with this, Atg4b-null mice showed defects in the development of otoconia, organic calcium carbonate crystals essential for sense of balance (equilibrioception). Furthermore, these abnormalities were exacerbated in Atg5–/– mice, which completely lack the ability to perform autophagy, confirming that autophagic activity is necessary for otoconial biogenesis. Autophagy deficiency also led to impaired secretion and assembly of otoconial core proteins, thus hampering otoconial development. Taken together, these results describe an essential role for autophagy in inner ear development and equilibrioception and open new possibilities for understanding and treating human balance disorders, which are of growing relevance among the elderly population.
-
Introduction
Autophagy is a degradative process in which portions of cytoplasm are engulfed by a double-membrane vesicle called the “autophagosome.” Once autophagy is completed, the autophagosome fuses with a lysosome, and its content and inner membrane are degraded by hydrolases and recycled (1). Genetic studies on yeast have identified more than 20 autophagy-specific (Atg) genes that are required for autophagosome formation (2). Among yeast Atg genes, Atg8 encodes a protein that forms part of a ubiquitin-like conjugation system essential for autophagy execution (3). Atg8 protein is synthesized as a cytoplasmic precursor, which is cleaved after a Gly residue by the cysteine proteinase Atg4 (4). This initial proteolytic processing is required for the subsequent conjugation of Atg8 with membrane-bound phosphatidylethanolamine (PE), which is in turn essential for autophagosome completion (5). The complex Atg8-PE is also deconjugated by the protease Atg4, facilitating the release of Atg8 from membranes. This modification system is conserved in higher eukaryotes including mammals (6). We previously identified and cloned the 4 human orthologs of the yeast proteinase Atg4 (7), and parallel studies have revealed that there are at least 6 orthologs of yeast Atg8 in mammals (microtubule-associated protein 1 light chain 3α [LC3A], microtubule-associated protein 1 light chain 3β [LC3B], microtubule-associated protein 1 light chain 3γ [LC3C], GABARAP-like 2 [GATE-16], GABA(A) receptor–associated protein [GABARAP], and GABARAP like 1 [ATG8L]) (8–10). Currently, it is unclear why mammals have developed this array of closely related enzymes, contrasting with other essential autophagy genes such as Atg3, Atg5, or Atg7 for which a single ortholog is present in the mammalian genome.
To get insights into the in vivo roles of this complex system, we generated mutant mice deficient in autophagy-related 4B (Atg4b–/– mice). Although they were viable, these mice exhibited a clear reduction of basal- and starvation-induced autophagic flux in all tissues, which is caused by a deficit in the initial proteolytic cleavage of Atg8 murine orthologs. This finding indicates that autophagin-1 (Atg4b) has a major functional role in the context of autophagy in mammals. In addition, Atg4b–/– mice showed a balance-related behavioral phenotype that is linked to profound inner ear developmental defects. We also report that these abnormalities were exacerbated in Atg5–/– neonates that were totally autophagy impaired, confirming that autophagic activity is essential for otoconial biogenesis. Finally, we analyzed the molecular mechanisms underlying these abnormalities and found that autophagy deficiency impairs the secretion and assembly of otoconial core proteins into vestibular lumen, resulting in the otoconial development defects and the behavioral balance disorders exhibited by Atg4b–/– mice.
-
Results
Generation, development, and growth of Atg4b mutant mice. To address the in vivo role of Atg4b cysteine proteinase, we decided to generate a murine model deficient in this enzyme. To this purpose, we searched the International Knockout Mouse Consortium database ( http://www.knockoutmouse.org) for ES cells containing insertional mutations that could disrupt the transcription of the mouse Atg4b gene. Founder mice were generated from the ES cell line A029E06 (German Genetrap Consortium [GGTC]), in which the cassette pT1betageo was inserted into the first intron of the Atg4b gene (Figure 1, A and B). Following heterozygote intercrossing, Atg4b-deficient, heterozygous, and WT mice were obtained at the expected Mendelian ratio. Homozygosity for the mutation was demonstrated by Southern blot (Figure 1C), and the absence of both the transcript and the protein was verified by RT-PCR and immunoblotting (Figure 1, D and E), indicating that the insertional mutation results in a null allele. Despite the Atg4b deficiency, the embryonic development of Atg4b-null mice was apparently normal. Homozygous mutant mice reached adulthood (at the time this manuscript was accepted for publication, the oldest animals were 16 months old) with no excess mortality, were fertile, and did not present any obvious histopathological or biochemical alteration (data not shown). Taken together, these data suggest that Atg4b is dispensable for embryonic development as well as for adult growth and fertility.
 Figure 1
Figure 1Generation and genetic characterization of Atg4b-deficient mice. (A) Top: Schematic representation of the WT Atg4b locus, in which the coding exons are depicted as numbered boxes. Bottom: Gene trap vector structure containing a splicing acceptor (SA), a bacterial β-galactosidase fused to the neomycin resistance gene (β-geo), and a polyadenylation signal (pA). (B) Schematic representation of the WT and mutant loci showing the new BamHI (B1) restriction polymorphism generated by the insertion of the gene trap vector. The DNA probe used for the detection of the mutant allele is shown. (C) Southern blot analysis of genomic DNA from Atg4b+/– and Atg4b–/– mice. (D) RT-PCR of RNA of liver tissue from WT and Atg4b–/– mice showing the absence of full-length Atg4b mRNA expression in mutant mice (n = 6 mice per genotype). (E) Immunoblotting of liver protein extracts from WT and Atg4b-knockout mice confirming the absence of Atg4b protein in mutant mice. β-Actin was used as a loading control.
Autophagic flux alterations in Atg4b-deficient mice. Previous studies have shown that severe autophagy impairment results in perinatal lethality in mice (11), while partial or tissue-specific autophagy defects are compatible with survival (12). Hence, the apparent normal development and health of Atg4b–/– mice suggests the absence of a major autophagic defect. To test whether Atg4b disruption leads to a reduction of autophagy compatible with postnatal viability or to a tissue-specific autophagy impairment, we prepared tissue extracts from mutant and control mice fed ad libitum and performed immunoblotting studies of the adaptor protein p62/sequestosome-1, whose levels negatively correlate with autophagic flux (13). p62 protein levels were clearly increased in a variety of mutant mouse tissues, as compared with their corresponding WT controls (Figure 2A), and this increase was accompanied by an accumulation of ubiquitylated proteins (Figure 2B). Immunofluorescence analyses of Atg4b–/– cells confirmed the presence of p62-positive aggregates (Figure 2D) that contained ubiquitylated proteins, as a sign of impaired autophagy (14). Very recently, it has been shown that overexpression of p62 itself leads to proteasome impairment and thus to the accumulation of ubiquitylated proteins (15). To evaluate whether the increase of p62 protein levels in Atg4b–/–-derived tissues corresponds to a reduction in autophagic flux or to an increase in p62 mRNA synthesis, we analyzed the relative expression levels of p62 in WT and Atg4b–/– livers by quantitative RT-PCR. As can be seen in Figure 2C, p62 expression levels in Atg4b–/– mice were comparable to those observed in control mice. We also analyzed proteasome activity of Atg4b–/– MEFs, as p62 levels can increase after proteasome impairment (16). As shown in Supplemental Figure 1 (supplemental material available online with this article; doi: 10.1172/JCI42601DS1), proteasome-dependent protein degradation was comparable between Atg4-deficient and WT MEFs, thus confirming that basal autophagy flux is reduced in Atg4b–/– mice.
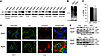 Figure 2
Figure 2Autophagy flux analysis in WT and Atg4b–/– mouse tissues. (A) Representative immunoblotting of p62/sequestosome-1 in protein extracts from age-matched control and mutant mice fed ad libitum. (B) Immunoblotting analysis of mono- and polyubiquitinated proteins in WT and Atg4b–/– MEF protein extracts. (C) Quantitative RT-PCR analysis of p62 mRNA in the liver revealing that the increase in p62 protein levels of mutant mice shown in A was not due to an increase in p62 mRNA synthesis. The transcript expression levels are relative to WT mouse p62 expression level, which was set at 100%. Black bar, WT; gray bar, Atg4b–/–. (D) Immunofluorescence analysis of endogenous p62 in WT and Atg4b–/– MEFs and liver, in which p62-positive structures also stained for ubiquitin. Scale bars: 10 μm. (E) Analysis of starvation-induced degradation of p62 in WT and Atg4b–/– liver and skeletal muscle. Representative immunoblots of endogenous p62 in tissue extracts from age-matched control and mutant mice starved for 24 or 48 hours or fed ad libitum. Protein extracts from 2 mice per genotype and condition are shown. β-Actin and α-tubulin were used as loading controls in liver and skeletal muscle, respectively. LOAD, loading control.
Next, we comparably assessed p62 protein levels in distinct tissues obtained from WT and Atg4b–/– mice that were fed ad libitum or subjected to starvation for 24 or 48 hours. As shown in Figure 2E and Supplemental Figure 2A, p62 protein levels decreased after starvation in control mice in a tissue-dependent fashion, especially after 48 hours of starvation. These observations are consistent with previous reports that suggest a differential tissue-dependent autophagy induction pattern after starvation (17, 18). Tissues from Atg4b–/– mice exhibited a similar tissue-specific pattern in p62 reduction after starvation as WT mice. However, the p62 protein levels were always higher in mutant mice tissues than in those from WT mice subjected to the same starvation conditions (Figure 2E and Supplemental Figure 2A). Collectively, these data demonstrate that disruption of Atg4b leads to a decrease but not to a complete disruption of autophagic flux, thus explaining why Atg4b–/– mice do not show perinatal lethality, as previously reported for other models of total autophagy impairment (11, 19).
Atg4b disruption leads to alterations in Lc3, Lc3a, Gabarap, and Atg8l status in vivo. To determine the mechanisms accounting for the reduced autophagic flux of Atg4b–/– mice, we first performed a series of immunoblotting analyses of Lc3a, Lc3b (referred hereafter simply as Lc3), Gate-16, Gabarap, and Atg8l, all of which are putative Atg4b substrates. These proteins are the murine orthologs of yeast Atg8 and have been detected in autophagosome membranes, and their lipidation status is associated with the rate of autophagy activation, especially in the case of Lc3 (20, 21).
With the exception of Gate-16, all other Atg8 orthologs were altered in tissues from Atg4b–/– mice fed ad libitum (Figure 3A). In the case of LC3, Atg4b–/– samples presented very low levels of Lc3-II and an accumulation of an Lc3 form with the same electrophoretic mobility than LC3-I. This band might correspond to unprocessed pro-LC3, whose maturation may be severely impaired in the absence of Atg4b, and would present an electrophoretic mobility similar to LC3-I. To clarify this question, we produced recombinant LC3 in E. coli, which lacks any Atg4-like protease. As shown in Supplemental Figure 2B, there was no difference in the mobility of bacterially expressed pro-LC3 and LC3-I or the LC3 form observed in Atg4b–/– samples, thus suggesting an impairment of the initial processing of pro-LC3 in the absence of Atg4b that would lead to pro-LC3 accumulation. In addition, we also detected additional LC3-related bands in some Atg4b–/– tissues, which were absent in WT samples and likely correspond to intermediate degradation products of pro-LC3, which extensively accumulates in the absence of Atg4b protease (Figure 3A and Supplemental Figure 2B). These observations suggest that LC3 maturation is strongly reduced in Atg4b–/– mice. Regarding Gabarap, Atg8l, and Lc3a, we could only detect the bands corresponding to non-lipidated forms of these Atg8 orthologs in tissues from WT or mutant mice, but their levels were clearly reduced in Atg4b–/– samples (Figure 3A). Quantitative RT-PCR expression assays did not reveal any significant difference in the abundance of mRNAs coding for these Atg8 orthologs (Figure 3B), indicating that the observed changes are not caused by transcriptional alterations. After starvation, the status of Gabarap and Atg8l in Atg4b–/– mice remained similar to that observed in WT mice tissues, although their levels were always lower in Atg4b–/– samples (Supplemental Figure 2). In the case of LC3, we observed a certain degree of lipidation in Atg4b–/– liver and skeletal muscle after prolonged starvation (Supplemental Figure 2), suggesting the existence of a residual Atg4-like activity in the absence of Atg4b.
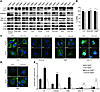 Figure 3
Figure 3Analysis of Lc3, Gabarap, Gate-16, Atg8l, and Lc3a status in WT and Atg4b–/– mice. (A) Representative immunoblots of endogenous Atg4b putative substrates. The analyzed tissues correspond to protein extracts from age-matched control and mutant mice fed ad libitum. β-Actin was used as a loading control in all tissues except heart and skeletal muscle (α-tubulin). (B) Quantitative RT-PCR analysis of Lc3, Gabarap, and Atg8l mRNA in liver, showing that the alterations in A were not due to an increase in mRNA synthesis. Transcript expression levels are relative to the expression levels detected in WT mice, which were set at 100%. Black bars, WT; gray bars, Atg4b–/–. (C) Representative immunofluorescence images of endogenous Lc3, Gabarap, Atg8l, and Gate-16 in fed and 4-hour-starved MEFs. Atg4b–/– MEFs showed a reduced number of punctate structures containing Lc3, Gabarap, or Atg8l under both fed and starved conditions. In contrast, Atg4b–/– MEFs exhibit an increase in punctate Gate-16–containing structures. (D) Representative immunofluorescence images of endogenous Atg5, showing an accumulation of immature autophagosomes in Atg4b–/– MEFs. Scale bars: 10 μm. (E) The number of endogenous Lc3, Gabarap, Atg8l, Gate-16, and Atg5 dots was counted and divided by the corresponding cellular area (μm2 × 100). Results shown represent the mean quantification values of 5 images in each depicted condition for each depicted protein. *P < 0.05 for Atg4b–/– values vs. the corresponding (fed or starved) Atg4b+/+ values.
We next studied the subcellular distribution of Atg8 orthologs in fed and starved WT and Atg4b–/– MEFs. Consistent with previous findings, we detected a clear increase of endogenous Lc3, Atg8l, or Gabarap-positive dots corresponding to autophagosomes in starved WT cells (20–22). In contrast, we detected Lc3, Gabarap, or Atg8l-positive dots only occasionally in Atg4b–/– cells, especially after starvation conditions, but always at lower numbers than in WT cells (Figure 3, C and E). These results show that the autophagy-related function of these Atg8 orthologs is severely reduced but not totally impaired in Atg4b–/– cells. Interestingly, we observed an increase of Gate-16 punctate structures in mutant cells, likely generated to preserve the autophagic flux compromised by the absence of lipidated forms of the other Atg8 orthologs (Figure 3, C and E). However, this was not sufficient to maintain a proper autophagosome maturation rate, as Atg4b–/– cells exhibited a neat accumulation of Atg5-positive puncta, which corresponded to unclosed pre-autophagosomal membranes (23) under both fed and starved conditions (Figure 3, D and E). Collectively, these data demonstrate that the absence of Atg4b impairs the function of most Atg8 orthologs (except Gate-16), thus reducing basal and induced autophagic flux.
Atg4b disruption leads to defects in the proteolytic processing of Lc3, Lc3a, Gabarap, Gate-16, and Atg8l. The aforementioned data suggest that the absence of Atg4b impairs the autophagy-related function of most Atg8 orthologs. To test whether the functional alterations in Atg8-like proteins are related to a defect in the proteolytic cleavage of these putative Atg4b in vivo substrates, we hydrodynamically delivered N- and C-terminal–tagged cDNAs of these proteins into Atg4b–/– or WT livers (24). After hydrodynamic delivery of Lc3 cDNA, we detected the bands corresponding to tagged Lc3-I and Lc3-II in WT extracts. In contrast, a single major band corresponding to unprocessed Lc3 and a series of additional bands — likely corresponding to intermediate degradation products of unprocessed pro-LC3 — were detected in Atg4b–/– mice (Figure 4A). Similarly, processing of Gabarap was detectable in WT livers, while Atg4b–/– livers contained a major band corresponding to the unprocessed Gabarap and some additional bands with increased electrophoretic mobility (Figure 4B). In the case of Atg8l, the main detected band in mutant samples corresponded to the uncleaved form of this protein, although we also detected a minor band probably corresponding to the processed form of Atg8l (Figure 4C). Finally, the cleaved form of Gate-16 appeared more abundant in Atg4b–/– samples than that corresponding to the unprocessed form (Figure 4D), which is in line with the lipidation status and subcellular distribution of this protein in Atg4b–/– mice (Figure 3A).
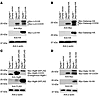 Figure 4
Figure 4Immunoblot analysis of hydrodynamically delivered, tagged Atg4b substrates. Mice were injected with mammalian expression vectors containing tagged cDNAs for Lc3 (A), Gabarap (B), Atg8l (C), and Gate-16 (D) through the tail vein to achieve transient transgene expression in vivo. Liver protein extracts from injected WT and Atg4b–/– mice were subjected to immunoblotting. Note that the absence of Atg4b impaired the processing of all of proteins, although its impact on Gate-16 was lower than in Lc3, Gabarap, and Atg8l. Unknown bands, which likewise corresponded to partial degradation products, are denoted by asterisks. β-Actin was used as a loading control.
In summary, our results confirm that in the presence of Atg4b, Atg8 murine orthologs are rapidly cleaved, as we could not detect any band corresponding to the unprocessed forms of the double-tagged Atg8-like proteins in WT samples. By contrast, in liver extracts from Atg4b–/– mice, we detected a series of bands corresponding to the uncleaved forms of these proteins, indicating that Atg4b is required for the efficient processing of murine Atg8 orthologs. However, the detection of a residual amount of the cleaved products from these proteins in liver extracts from Atg4b–/– mice is compatible with a partial reduction of autophagic flux.
Balance dysfunction and inner ear abnormalities in Atg4b–/– mice. The finding of autophagy impairment in Atg4b–/– mice prompted us to perform an exhaustive phenotype characterization of these animals, in search of putative alterations that might have escaped to our initial analyses. We first carried out rotarod testing to monitor motor coordination, and found that Atg4b-null mice performed poorly compared with their corresponding controls (Figure 5A). In addition, we observed that a certain number of mutant mice (approximately 25%) were not susceptible to rotarod analysis, as they repeatedly fell down after a few seconds of testing. We noticed that these animals exhibited an abnormal behavior characterized by a tilted position of the head, circling movements, and a marked tendency to crawl and walk backwards when placed outside their cages (Figure 5B and Supplemental Video 1). In addition, when subjected to the Porsolt forced swim test (25), these affected mutant mice were unable to swim or float, exhibited profound disorientation, rotated under water, and had to be rescued from drowning (Figure 5B).
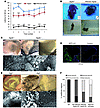 Figure 5
Figure 5Balance dysfunction in Atg4b–/– mice reveals a new physiological role for autophagy in otoconial development. (A) Rotarod analysis of Atg4b–/– and WT mice. **P < 0.01; ***P < 0.005. (B) Representative image of an Atg4b–/– mouse affected by head tilting (top right) and a WT mouse (top left). Insets show the angle of the head. As compared with WT mice (bottom left), Atg4b–/– mice were unable to swim or float in the forced swim test (bottom right). (C) Light (top) and scanning (bottom) electron microscopy analyses of WT and Atg4b–/– inner ears. Representative images show either the absence of otoconial crystals in the utricle of Atg4b–/– mice (right) or the presence of abnormal otoconia that are oversized, less numerous, loose, and occasionally fused (middle), as compared with WT otoconia (left). The blur in the image is due to electrical interference. Scale bars: 50 μm (top), 10 μm (bottom). (D) Fluorescent microphotographs of E15.5 GFP-LC3 transgenic mice utricles indicating the presence of numerous autophagosomal structures (green puncta) in macular and “roof” utricle epithelial cells. The insets show higher magnification of the macular epithelium. Scale bars: 15 μm. (E) Light (top) and scanning electron (bottom) microphotographs of WT and Atg5–/– inner ears. Representative images showing the absence of otoconia (right) or the presence of giant and abnormally shaped otoconial crystals (middle) in the utricle of Atg5–/– mice. Scale bars: 50 μm (top), 10 μm (bottom). (F) Graph showing the relative number of otoconial abnormalities observed in WT (n = 5), Atg4b–/– mice (n = 10 for unaffected mice; n = 12 for affected mice), and Atg5–/– mice (n = 5).
It has been reported that autophagy ablation in the CNS leads to massive apoptosis and neurodegeneration, linked to ataxia-like phenotype and to profound motor uncoordination (26, 27). Thus, we extensively analyzed the CNS of severely affected Atg4b–/– mice to search for any sign of neurodegeneration that might account for the observed motor and behavioral phenotype. However, we did not find any increase in apoptosis or any significant histological alteration pointing to a CNS-related disorder (Supplemental Figure 3). Given that many inner ear disorders can also lead to behavioral alterations, dyskinesis, and balance-related pathologies similar to those observed in Atg4b–/– mutant mice (28), and considering that p62 accumulation and Atg8 ortholog maturation defects were observed in the inner ears of Atg4b–/– mice (Figure 2A and Figure 3A), we performed an exhaustive characterization of the ears of the affected mice. Gross examination of the middle ear and membranous and bony labyrinths revealed no obvious macro-anatomic defects. Similarly, optical and electron microscopy analyses of the inner ear confirmed that cochlea and other structures such as semicircular channels did not present any significant alteration (data not shown). However, we observed a clear developmental defect in the vestibular otoconia of mutant mice. Otoconia consist of multiple calcium carbonate crystals that overlie the vestibular (utricle and saccule) macular sensory epithelium and are required for the spatial detection of gravity and linear acceleration (28). As shown in Figure 5, C and F, many of the affected mutant mice totally lacked otoconia in the utricle and saccule, whereas others presented mildly abnormal or giant otoconia, both of which are common features of profound otoconial developmental defects (29). In contrast, unaffected mutant mice mostly presented mildly abnormal otoconia in one or both ears (Figure 5F). This partial penetrance of Atg4b disruption may be related to the previously reported functional redundancy within the autophagin family (18) or to the presence of modifier loci, as the Atg4b–/– mice used in this study had a mixed-strain background. However, partial penetrance is often observed in balance disorders (28), and the reasons for this are unknown. At the ultrastructural level, utricular and saccular regions from affected Atg4b–/– mice presented an accumulation of abnormal extracellular vesicle-like structures, which are precursors of mature otoconial crystals (globular substance) (30). In mutant mice, these structures were characterized by the presence of additional internal vesicles and by a less dense content than that observed in WT samples, suggesting impairment in globular substance cargo packaging (Supplemental Figure 4).
Autophagy is essential for proper vestibular otoconia formation in mice. The connection between Atg4b deficiency and otoconial abnormalities was unexpected. Thus, we decided to analyze whether this phenotype was related to the observed autophagy flux impairment of these mice or derived from putative new Atg4b functions different from autophagy. Otoconia formation begins at E14.5 and is near completion around E17.5, although the crystals continue to grow until P7. As a first approach to explore the putative implication of autophagy in otoconia formation, we analyzed the autophagic activity of vestibular cells at different embryonic stages in mice expressing an ubiquitous GFP-LC3 transgene (17). Vestibular epithelial cells showed accumulation of GFP-LC3 dot-like structures — which are indicative of autophagy — at E15.5 (Figure 5D). To determine whether this observation might reflect a bona fide causal relationship between autophagy and otoconia formation, we analyzed the morphology of otoconia in Atg5–/– mice, which are totally devoid of autophagic activity and die shortly after birth (11). Since otoconial crystals are already formed at P1, we could test whether autophagy ablation has a major impact on otoconial development by analyzing vestibular morphology in Atg5–/– neonate mice. Consistent with the findings in Atg4b–/– mice, the majority of inner ears from Atg5–/– mice neonates lacked otoconia, whereas others presented few, giant, and abnormally shaped otoconia (Figure 5E). In addition, ultrastructural analyses of utricular and saccular regions from Atg5–/– neonates revealed an accumulation of globular substance and a defect in its morphology similar to that observed in affected Atg4b–/– mice (Supplemental Figure 4). Taken together, these results clearly demonstrate that autophagy impairment in vivo leads to a developmental defect in otoconial biogenesis. Interestingly, the morphology of Atg5–/– otoconia was reminiscent of otoconia observed after intoxication with streptomycin, an aminoglycoside with vestibulotoxic side effects (31). Indeed, MEFs cultured in presence of high doses of streptomycin showed a progressive cytoplasmic vacuolization linked to increased levels of p62 (Supplemental Figure 5). It has been repeatedly reported that aminoglycoside vestibulotoxicity is linked to an increase in ROS production (32). Thus, the fact that autophagy can also be inhibited by streptomycin could help explain aminoglycoside ototoxicity, as autophagy-impaired cells have an increased sensitivity to ROS-induced damage (19, 33).
Autophagy impairment disrupts otoconial organic matrix. Although the precise mechanism of otoconial biogenesis in mammals is poorly understood, it is known that this process is initiated in a seeding phase, in which the epithelial cells surrounding the utricle/saccule lumen secrete otoconial proteins that form the core of the crystals in the vestibular lumen. Subsequently, CaCO3 is sequestered by the proteinaceous matrix to form the mature crystal (28). Thus, we analyzed the distribution of otoconial core proteins (Oc90, otopetrin, and otolin) in decalcified E15 and neonatal inner ears to test whether autophagy impairment could affect their localization. In this case, we focused our analyses on Atg5–/– mice to exclude any ambiguous result derived from the incomplete phenotype penetrance of Atg4b mutants. Interestingly, we found that the localization of otoconial proteins is altered in the absence of autophagy at E15, especially in the case of Oc90 (Figure 6A). In fact, this protein is mainly detected in the utricle/saccule lumen and in the proteinaceous membrane covering vestibular epithelia in WT embryos. In contrast, Oc90 accumulates inside epithelial cells of the nonsensory epithelia of autophagy-impaired animals (Figure 6A), which suggests a possible defect in its secretion. Consistently, we observed a similar alteration for otolin and otopetrin at this stage (Figure 6A). In fact, although both proteins presented a similar distribution inside vestibular cells in both WT and mutant embryos, they only localized inside vestibular lumen in WT samples, mainly at the developing otoconia (Figure 6A).
 Figure 6
Figure 6The correct localization of otoconial core proteins is dependent on autophagic activity. (A) Representative immunofluorescence images of endogenous otolin, otopetrin, and Oc90 proteins in utricles from WT and Atg5–/– E15.5 embryos showing their mislocalization in the absence of autophagy. Both otolin and otopetrin presented a similar intracellular localization both in WT and Atg5–/– samples but were only detected in utricle lumen in autophagy-competent embryos (arrows). Oc90 (right) was mainly detected in WT samples inside utricle lumen, both at the developing otoconia (asterisk) and as a proteinaceous coral-like structure (arrows). In addition, Oc90 was also detected covering utricle epithelial and sensory cells in WT samples (arrowheads in top panels). In contrast, in Atg5–/– samples Oc90 was only detected inside non-sensory vestibular cells (arrowheads in bottom panels). (B) In P1 WT neonates, otolin and otopetrin were mainly detected in otoconial cores (arrows), whereas this localization was not observed in Atg5–/– samples. Similarly, in WT samples Oc90 was mainly detected in otoconial cores (right panel and arrow in left panel). In autophagy-impaired neonates, Oc90 localized both in utricle lumen, forming proteinaceous coral-like structures (right panel and arrows in left panel), and inside non-sensory cells (arrowheads). Scale bars: 50 μm (otolin, otopetrin, and left Oc90 panels); 30 μm (right Oc90 panels).
In newborn mice, these proteins were mainly detected in otoconial crystal cores and in the cytoplasm of vestibular cells from WT mice, as previously reported (29, 34). In autophagy-impaired mice, otolin and otopetrin were observed in the cytoplasm of epithelial and sensory cells, but not in the utricle/saccule lumen (Figure 6B). Only Oc90 was detected as a coral-like conglomerate inside the utricle/saccule lumen (Figure 6A). Interestingly, these coral-like conglomerates containing Oc90 were present in WT mice at E15 (Figure 6A) and have been reported to be characteristic of the early steps of otoconia formation (35). Taken together, our analyses show that otoconial development is arrested in the absence of autophagy, presumably due to impaired assembly/secretion of the organic matrix of otoconia, as targeting of Oc90 or otopetrin genes results in impaired otoconial development in mice (29, 34).
Autophagy-dependent otoconial abnormalities are linked to alterations in Ap-3 and Bloc-1 sorting complexes. Although the exact molecular mechanisms leading to otoconial abnormalities are not fully understood in many cases, alterations in vesicle sorting complexes are the most common determinant of this type of disorder (28, 36). In fact, disruption of adaptor protein complex 3 (Ap-3) or biogenesis of lysosome-related organelles complex 1 (Bloc-1) activity affects intracellular vesicular trafficking and leads to defects in protein secretion and otoconial biogenesis (28, 37, 38). Given that autophagy-impaired mice show a defect in the assembly/secretion of otoconial core proteins into the vestibular lumen, we analyzed the status of several subunits of Ap-3 and Bloc-1 vesicle sorting complexes in Atg4b–/– and Atg5–/– cells. As shown in Figure 7, total protein and mRNA levels of Ap-3 and Bloc-1 subunits were reduced both in Atg4b–/– and Atg5–/– cells. It is remarkable that the extent of this reduction was more pronounced in Atg5-deficient cells than in those lacking Atg4b, which is in line with the fact that Atg4b–/– cells retained certain autophagic activity (Figure 7C). Consistently, a prolonged pharmacological inhibition of autophagy in WT cells by 3-methyladenine also resulted in a clear reduction of mRNA levels for Ap-3 and Bloc-1 subunits (Figure 7D), confirming that autophagy inhibition impacts Ap3 and Bloc1 mRNA transcription. It is noteworthy that the observed reduction of Ap-3 and Bloc-1 protein levels is quantitatively more pronounced than their mRNA downregulation. In this regard, it has been shown that the absence of a certain subunit affects the stability of the whole sorting complex, thus leading to a reduction in the cellular protein content of the remaining subunits (39). However, other possible mechanisms directly involving autophagic machinery in the stabilization of sorting complexes cannot be ruled out to explain this observation.
 Figure 7
Figure 7Autophagy disruption affects the stability of Ap-3 and Bloc-1 sorting complexes. Representative immunoblots of endogenous δ and μ subunits of the Ap-3 complex, and of pallidin and dysbindin components of the Bloc-1 complex in Atg4b–/– (A) and Atg5–/– MEFs (B). Densitometry analyses of immunoblottings against Ap-3 and Bloc-1 subunits from several independent MEF lines is shown on the right (n = 6 MEF lines from each genotype). The extent of this reduction was more pronounced in Atg5-deficient cells than in those lacking Atg4b. (C) Quantitative RT-PCR analyses of Ap-3 and Bloc-1 subunits, showing that the observed reduction in total protein content of the analyzed proteins in Atg4b–/– (left) or Atg5–/– MEFs (right) is caused by transcriptional alterations. (D) Treatment of WT MEFs during 24 hours with 10 mM 3-methyladenine, an autophagy inhibitor, induced a transcriptional downregulation of Ap-3 and Bloc-1 subunits, thus confirming that autophagy inhibition alters the status of these signaling complexes. *P < 0.05.
Taken together, these observations unveil what we believe is a novel and essential function of autophagy in otoconial development. Moreover, our results connect deficient autophagic activity to otoconial dysmorphy via defective Ap-3 or Bloc-1 sorting complexes.
-
Discussion
To date, the precise in vivo role of the 4 members of the Atg4 family of cysteine proteinases (autophagins) present in mammals has remained largely unknown. It is unclear why mammals developed this array of closely related enzymes, contrasting with other essential autophagy genes such as Atg3, Atg5, or Atg7, for which a single ortholog is present in the mammalian genome. Our first attempt to answer this question was based on the generation of autophagin-3/Atg4c-deficient mice, which presented a minor phenotype (18). With the generation of Atg4b-deficient mice, reported herein, we have not only progressed in our attempt to identify the in vivo physiological and pathological roles of autophagins, but also defined a new and unexpected function for autophagy in the context of mammalian development.
The effect of Atg4b absence on autophagy. The functional analysis of this cysteine protease has been hampered by the inherent limitations of the in vitro approaches previously used for its study. In this work, we have clarified the in vivo function of Atg4b in autophagy by showing that the absence of this protease leads to a defect in the initial proteolytic processing of several murine orthologs of Atg8, in particular Lc3, Gabarap, and Atg8l. The Atg4b knockout causes a drastic reduction in the amount of Atg8-like proteins available for the initiation of autophagy, resulting in a decrease in autophagic flux, both in baseline conditions and after nutrient deprivation. Surprisingly, this evident autophagic defect did not affect the organismal viability (at least during the first 16 months of life), suggesting that minimal levels of autophagy are sufficient for maintaining cellular functions. We observed that cells from Atg4b-null mice exhibit a phenotype similar to that induced by the in vitro overexpression of a dominant-negative mutated form of Atg4b that might inhibit the access of all 4 Atg4-like proteases to their substrates (40). We found that Atg4b was responsible for most of the in vivo proteolysis of mammalian Atg8 orthologs except Gate-16, which might be targeted by Atg4a or Atg4d, as previously suggested by in vitro studies (41, 42). The generation of murine models deficient in Atg4a, Atg4d, or combinations of Atg4 orthologs will clarify the physiological roles of the 4 mammalian autophagins in the future.
Autophagy is required for proper otoconia development. In this work, we have also shown that the absence of Atg4b triggers defects in inner ear development, particularly in the vestibular system. Atg4b-deficient mice presented alterations in otoconial biogenesis that precipitated a balance disorder. It is noteworthy that other murine models with autophagy reduction, such as Beclin1+/– or Atg16l1-hypomorphic mice (43–45), have not been reported to exhibit balance-related disorders, suggesting that Atg4b–/– mice may present a more severe autophagy impairment than these models or that Atg4b could be involved in processes that are not related to autophagy yet relevant to otoconial development. However, we found that Atg5–/– neonates present otoconial abnormalities similar to those discovered in Atg4b–/– mice, suggesting that autophagy itself is relevant to vestibular development.
Otoconia development is a process that requires the spatiotemporal orchestration of complex biochemical events (28, 36). Given the complexity of vestibular morphogenesis, a variety of mutant mouse models presenting balance disorders have been studied in detail (46–48). Although many of these mutants were isolated in the early 1950s after gonad irradiation–based mutagenesis, the molecular identity of the genes mutated in tilted-head (Th), twirler, or wocko mice remains elusive (28). In this regard, it is remarkable that the Th locus maps to the same chromosomal region as the Atg4b gene (Supplemental Figure 6). Considering that Atg4b–/– mice show the same phenotypic alterations as Th mice, including an incomplete penetrance (49), it is tempting to speculate that the inactivation of Atg4b might account for the Th phenotype. Unfortunately, this mutant strain is currently extinct (and frozen Th embryos kept at the Oak Ridge repository are not viable), making it impossible to experimentally validate this hypothesis.
In contrast to the Th locus, the genes mutated in other classical murine models for balance disorders have been well characterized. Currently, it is known that some strains showing otoconial abnormalities are deficient for components of the Bloc-1 and Ap-3 complexes, which are also required for proper melanogenesis and blood coagulation. In fact, the strains cappuccino, pallid,muted, or mocha, which are deficient for components of Bloc-1 and Ap-3 complexes, phenocopy the human Hermansky-Pudlak syndrome insofar as they exhibit hematological and pigmentation defects linked to otoconial abnormalities (50). In this regard, our finding that autophagy-impaired cells manifest defects in the Ap-3 and Bloc-1 sorting complexes unravel a new and previously undescribed connection between vesicle sorting and the autophagic pathway. It is remarkable that no molecular evidence has been reported so far to explain the vestibular defects observed in mutant mice that lack subunits of the Bloc-1 or Ap-3 complexes. Our observation that mislocalization of otoconial core proteins is linked to defects in sorting complexes sheds some light on this issue and should be further evaluated, as the main vestibulum-specific deficiency of mice mutant for sorting complexes subunits may be the aberrant secretion and/or assembly of otoconial core proteins into the lumen of the utricle and saccule. Thus, a reasonable hypothesis for the observed otoconial abnormalities in Atg4b- and Atg5-null mice is that autophagy inhibition results in a transcriptional downregulation and thus instability of sorting complexes, which in turn leads to an impairment in otoconial matrix assembly and secretion and finally to otoconial abnormalities and balance disorders (Figure 8). Consistent with this model, an essential role for autophagy has been recently proposed in melanogenesis, another process for which the activity of Ap-3 and Bloc-1 sorting complexes is essential (51). The mechanism connecting autophagy disruption with the observed transcriptional alterations in sorting complexes is not clear. However, it has been recently reported that autophagy inhibition leads to profound p62-dependent transcriptional alterations (52). The fact that both Atg5–/– (27) and Atg4b–/– cells (Figure 2) abnormally accumulate p62 may mechanistically connect the observed transcriptional downregulation of Ap-3 and Bloc-1 subunits in both Atg4b–/– and Atg5–/– cells. Further studies involving the generation of inner ear–specific Atg5 or Atg7 knockouts in which the p62 gene has also been disrupted will help to clarify this point.
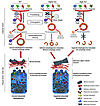 Figure 8
Figure 8Proposed model for the otoconial defects observed in autophagy-impaired mice. At the cellular level, Atg4b and Atg5 disruption affects autophagy by either reducing (Atg4b) or totally impairing (Atg5) autophagic activity. At the tissue level, these deficiencies lead to alterations in inner ear otoconial development. In fact, a severe reduction or a total disruption of autophagic flux in vestibular cells causes a dramatic impairment in the secretion and assembly of otoconial core proteins. This event is likely associated with transcriptional and other autophagy-dependent alterations of Ap-3 and Bloc-1 sorting complexes. All these cellular and tissue events lead to the development of abnormal or giant otoconia, or even to their total absence, finally resulting in the balance and coordination disorders observed in Atg4b–/– mice.
Concluding remarks. The spectacular advancement of autophagy research over the last decade has revealed a variety of new and pathophysiologically relevant functions for this catabolic pathway. However, the fact that total autophagy impairment results in perinatal lethality has made necessary the use of alternative approaches to describe the in vivo physiological roles of autophagy. The generation of tissue-specific autophagy-deficient murine models has allowed for the identification of some tissue-specific functions of autophagy, although many others are likely unknown. Through the generation of Atg4b-deficient mice, we have clarified the in vivo role for this protease in the context of the autophagic machinery and unveiled a new function for autophagy in inner ear development and balance functions. This finding has been possible because Atg4b–/– mice present clearly reduced basal and induced autophagy levels but are viable until adulthood. Thus, we propose that Atg4b-deficient mice constitute an excellent model to study the putative impact of sustained reduced autophagic activity in organismal homeostasis. Finally, our findings may also have clinical implications, since balance disorders such as benign paroxysmal positional vertigo are acquiring growing clinical relevance and are relatively common among the elderly. Our observation that autophagy impairment affects vestibular function opens the possibility of new potential therapeutic approaches for these pathologies. In fact, it has been reported that autophagic activity decreases with age (53). This decrease may be related to the observed age-related degeneration of vestibular otoconia, in which many features resemble those of autophagy-deficient mutants, including the presence of giant and abnormally shaped crystals (54). Thus, treatments aimed at maintaining normal autophagic activity in the elderly might constitute new approaches to the clinical management of balance-related disorders.
-
Methods
Reagents. Antibodies against Lc3, Gabarap, and Gate-16 were provided by T. Ueno from Juntendo University, Tokyo, Japan. Additional antibodies against Lc3, Gabarap (PM037), Atg5 (M153-3), and Gate-16 (PM038) for immunofluorescence analysis were from MBL International. Antibodies against Lc3a (12135-1-AP) and Atg8l (11010-1-AP) were from Proteintech. The antibody against p62 (Gp62-C) was from Progen. The antibody against ubiquitin, clone FK2 (PW8810-0500) was from Biomol. Antibodies against Flag (F-3165), actin (AC-15), and Atg4b (A2981) were from Sigma-Aldrich. Antibodies against Myc (2278) and HA (2367) were from Cell Signaling Biotechnology. Dysbindin antibody (AB15450) was from Millipore. The pallidin antibody was a gift from E. Dell’Angelica, and antibodies against Ap3-δ and Ap-3μ were provided by J.M. Falcon (CIC Biogune). Fluorescence-conjugated secondary antibodies for immunofluorescence analyses were from Invitrogen; qPCR expression assays were from Applied Biosystems.
Transgenic animals. Mutant mice deficient in Atg7 and Atg5 and transgenic mice expressing the fluorescent autophagosome marker GFP-LC3 have been previously described (11, 17, 26). All animal experiments were approved by and conducted in accordance with the guidelines of the Committee on Animal Experimentation of the Universidad de Oviedo.
Generation of Atg4b-deficient mice. The targeted ES cell clones (A029E06) were obtained from the GGTC. They were grown, expanded, and subsequently injected into C57BL/6 blastocysts to generate chimeras. Chimeric males were mated with C57BL/6 female mice, and the offspring heterozygous for Atg4b were used to generate homozygous null mice. In all experiments, homozygous Atg4b–/– mice and their corresponding WT controls were littermates derived from interbreeding of heterozygotes with a mixed background of C57BL6/129Sv. In all cases, genotypes were determined by Southern blot analysis of tail DNA.
RT-PCR. Total RNA was isolated from mouse tissues according to the method of Chomczynski and Sacchi (55). About half of the obtained product was reverse transcribed using the RNA-PCR Core kit from PerkinElmer. A PCR reaction was then performed with mouse Atg4b-specific primers for 25 cycles of denaturation (94°C, 20 seconds), annealing (62°C, 20 seconds), and extension (72°C, 30 seconds). As a control, actin was PCR-amplified from all samples under the same conditions.
Expression in E. coli. The Lc3 sequence was amplified from murine cDNA. PCR primers were 5′-CCGTCCGAGAAGACCTTCAAGCAGC-3′ and 5′-TTGTTGGAGTCTTACACAGCCATTG-3′. The pGEX-3X vector was linearized using BamHI, and the GST sequence was removed by amplifying the vector with the primers 5′-CGATCTGCCTCGCGCGTTT-3′ and 5′-CATGAATACTGTTTCCTGTGTGA-3′. LC3 was then cloned into the vector, and the construct was transformed into E. coli strain DH10B. The bacteria were cultured until the cell density reached 0.8 (measured at OD600), and LC3 expression was induced with IPTG 0.5 mM for 2 hours. The bacteria were then centrifuged, washed with 1:10 vol PBS, centrifuged again, and resuspended in 1:20 vol PBS containing Complete protease inhibitors. Finally, bacteria were sonicated, and 2 μl of a 1:20 dilution of the lysate was analyzed by Western blot along with samples prepared from murine tissues.
Immunoblotting. Mice tissues were immediately frozen in liquid nitrogen after extraction, then homogenized in 20 mM Tris buffer, pH 7.4, containing 150 mM NaCl, 1% Triton X-100, 10 mM EDTA, and Complete protease inhibitor cocktail (Roche Applied Science). Once homogenized, tissue extracts were centrifuged at 13,000 g at 4°C and supernatants were collected. The protein concentration of the supernatant was evaluated by bicinchoninic acid technique (BCA protein assay kit; Pierce Biotechnology Inc.). A protein sample (25 μg) was loaded on 13% SDS-polyacrylamide gels. After electrophoresis, gels were electrotransferred onto nitrocellulose filters, and the filters were blocked with 5% nonfat dried milk in PBT (PBS with 0.05% Tween-20) and incubated with primary antibodies in 5% BSA in PBT. After 3 washes with PBT, filters were incubated with the corresponding secondary antibody at 1:10,000 dilution in 1.5% milk in PBT, and developed with Immobilon Western Chemiluminescent HRP substrate (Millipore).
Protein degradation. To label proteins, adult murine fibroblasts were incubated at 37°C for 48 hours in complete fresh DMEM (Sigma-Aldrich) containing 10% vol/vol FCS (Invitrogen) and 5 μCi/ml L-[3H]-leucine (Amersham Pharmacia Biotech). Then, cells were washed once with PBS containing 2 mM L-leucine and chased at 37°C for 24 hours in complete DMEM containing 10% vol/vol FCS and 2 mM L-leucine to eliminate short-lived proteins. Proteolysis experiments and measurements of intracellular protein degradation were carried out as described previously (18), using lactacystin to specifically inhibit proteasome-mediated degradation. Protein degradation was analyzed 1 hour after addition of lactacystin and after an additional 3 hours to ensure optimal inhibition and to avoid possible secondary effects of the inhibitor. All experiments were performed at least 4 times with duplicate samples and using independent cell lines.
Hydrodynamic delivery of naked cDNAs. Plasmid DNA was injected by hydrodynamic technique as previously described (24). In brief, 25 μg of endotoxin-free plasmid DNA was prepared in 2 ml of sterile, pharmaceutical-grade saline at room temperature. Mice were restrained and the lateral tail vein was accessed with a 21-gauge needle (Tyco Healthcare Group). Administration of the solution was performed in 10 seconds or less without extravasation; each group was represented by 3 or more animals.
Quantitative real-time PCR. cDNA was synthesized using 1 to 5 μg of total RNA, 0.14 mM oligo(dT) (22-mer) primer, and 0.2 mM concentrations each of deoxynucleoside triphosphate and Superscript II reverse transcriptase (Invitrogen). Quantitative RT-PCR (qRT-PCR) was carried out in triplicate for each sample using 20 ng cDNA, TaqMan Universal PCR Master Mix (Applied Biosystems), and 1 μl of the specific TaqMan custom gene expression assay for the gene of interest (Applied Biosystems). To quantify gene expression, PCR was performed at 95°C for 10 minutes, followed by 40 cycles at 95°C for 15 seconds, 60°C for 30 seconds, and 72°C for 30 seconds using an ABI Prism 7700 Sequence Detection System. As an internal control for the amount of template cDNA used, gene expression was normalized to the mouse b-actin gene using the Mouse β-Actin Endogenous Control (VIC/MGB Probe, Primer Limited) TaqMan Gene expression assay (Applied Biosystems). Relative expression of the analyzed genes was calculated according to the manufacturer’s instructions. Briefly, analyzed gene expression was normalized to b-actin in WT or Atg4b–/– derived samples using the following formula: gene of interest mean value – β-actin mean value (for at least 6 WT animals) = 100% for each analyzed gene. The same values for Atg4b–/– mice tissues were referred to those values.
Scanning electron microscopy. Otoconia morphology was visualized by scanning electron microscopy. Atg4b adult mice were anesthetized using a sublethal dose of ketamine (167 mg/kg) and xylazine (2 mg/kg) perfused through the left ventricle with 0.1 M sodium cacodylate (pH 7.2), followed by 20–30 ml 3.5% glutaraldehyde and 3 mM CaCl2 in 0.1 M sodium cacodylate (pH 7.2). For Atg5 embryonic mice, pregnant females were euthanized and embryos were extracted. Temporal bones were immediately dissected and fixed overnight at 4°C in the same perfusion buffer indicated above. Fixed adult and neonate samples were washed with 0.1 M sodium cacodylate (pH 7.2) and postfixed in 1% osmium tetroxide in 0.1 M sodium cacodylate (pH 7.2) for 1 hour. Samples were then dehydrated, dried in CO2, mounted, sputter coated with gold palladium and viewed on a Jeol JSM-848 scanning electron microscope at 15 kV.
Transmission electron microscopy. Inner ear tissues were dissected from perfused mice and fixed in 3% glutaraldehyde in 0.1 M sodium cacodylate (pH 7.2) for 1 hour at 4°C. After 3 washes in 0.1 M Sorensen’s phosphate buffer, samples were postfixed with 1% osmium tetroxide for 1 hour and rinsed 3 times in Sorensen’s buffer. Tissues were dehydrated in graded ethanol and infiltrated in 50% propylene oxide/50% resin for 1 hour, then placed in resin for another hour, and embedded in a resin-based mold for polymerization overnight in a 37°C oven. Ultra-thin sections (85 nm) were taken from each sample and analyzed on a Jeol (JEM-1011) transmission electron microscope at 60 kV.
Immunofluorescence. For immunofluorescence analyses of MEFs, cells were grown in coverslips, washed in PBS, and fixed in 4% paraformaldehyde in PBS at 4°C during 10 minutes. For paraffin sections, slides were deparaffinized and rehydrated. Slides or coverslips were blocked in 5% BSA (Sigma-Aldrich) for 10 minutes, incubated with primary antibodies for 1 hour at 37°C, washed in PBS, incubated for 40 minutes with secondary antibodies, thoroughly washed in water, and mounted in Vectashield (Vector Laboratories) containing DAPI for nuclear staining. For histology analysis, deparaffinized sections were stained with hematoxylin and eosin. Images of immunolabeled samples were obtained at room temperature with a laser-scanning confocal microscope (TCS-SP2-AOBS; Leica) using HC PL APO CS ×20 NA 0.70, HCX PL APO CS ×40 NA 1.25, and HCX PL APO Ibd BL ×63 NA 1.4 objective lenses. Images were acquired with LCS Suite version 2.61 (Leica), and the brightness/contrast was adjusted with Photoshop CS version 9.0.2 (Adobe).
Rotarod analysis. WT and Atg4b–/– mice at 20 weeks of age performed 1 rod trial on day 1 and 3 trials on the next 2 consecutive days. The rod accelerated from 4 to 40 rpm in 5 minutes and remained at maximum speed for the next 5 minutes. Animals were scored for their latency to fall (in seconds) for each trial and rested a minimum of 10 minutes between trials to avoid fatigue and exhaustion.
Analysis of GFP-LC3 embryos. Pregnant transgenic female mice overexpressing GFP-LC3 were perfused with 4% paraformaldehyde in 0.1 M PBS, pH 7.4. E15 embryos were harvested and fixed with the same fixative solution for at least 4 hours, followed by treatment with 15% sucrose in PBS for 4 hours, and then with 30% sucrose solution overnight. Tissue samples were embedded in Tissue-Tek OCT compound (Sakura Finetechnical Co. Ltd.) and stored at –70°C. Samples were then sectioned at 5-μm thickness with a cryostat (CM3050 S, Leica), air-dried for 1 hour, washed in PBS for 5 minutes, dried at room temperature for 30 minutes, and mounted with a conventional anti-fading medium. GFP-LC3 dots were observed in 5 independent visual fields from at least 5 independent mouse embryos in each using a Leica TCS sp2 AOBS confocal fluorescence microscope.
Fibroblasts extraction and culture. MEFs were extracted from E13 embryos. Briefly, embryos were sterilized with ethanol, washed with PBS, and triturated with razor blades. Samples were then incubated in DMEM (Gibco) overnight at 37°C and 5% CO2. The next day, cultured cells were trypsinized, filtered, and washed. Finally, MEFs were incubated at 37°C and 5% CO2 and used for the corresponding experiments. For inhibiting autophagy, MEFs were cultured during 24 hours in complete medium with 10 mM 3-methyladenine.
Statistics. All experimental data are reported as mean ± SEM. Statistical analyses were performed by the nonparametric 2-tailed Student’s t test. Statistical analyses were made using the Prism program version 4.0 (GraphPad Software Inc.). P values lower than 0.05 were considered significant.
-
Acknowledgments
We thank A. Astudillo, A. Fernández, P. Boya, A.J. Ramsay, and G. Velasco for helpful comments and S. ρlvarez, M. Fernández, D. ρlvarez, and M.S. Pitiot for excellent technical assistance. We also thank N. Mizushima for Atg5+/– and GFP-LC3 mice, M. Komatsu for nestin-cre/Atg7–/– mice, N. Rodríguez-Muela for the preparation of GFP-LC3 embryos, J.M. Falcón and E. Dell’Angelica for Ap-3 and Bloc-1 antibodies, I. Tanida for Atg8l constructs, Y. Kabeya and Y. Ohsumi for Lc3, Gate-16, and Gabarap constructs, and E. Martínez-Morillo for help with plasma analyses. This work was supported by grants from Ministerio de Ciencia e Innovación-Spain, Fundación “M. Botín,” the European Union (FP7 MicroEnviMet) and the NIH (grant DC008603 to Y.W. Lundberg). The Instituto Universitario de Oncología is supported by Obra Social Cajastur and Acción Transversal del Cáncer-RTICC. S. Cabrera is supported by a fellowship from CONACYT-México.
Address correspondence to: Carlos López-Otín, Departamento de Bioquímica y Biología Molecular, Facultad de Medicina, Universidad de Oviedo, 33006 Oviedo, Spain. Phone: 34.985.104201; Fax: 34.985.103564; E-mail: [email protected].
-
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Citation for this article: J Clin Invest. 2010;120(7):2331–2344. doi:10.1172/JCI42601.
See the related article at A balancing act for autophagin.
-
References
- Mizushima N, Levine B, Cuervo AM, Klionsky DJ. Autophagy fights disease through cellular self-digestion. Nature. 2008;451(7182):1069–1075.
- Klionsky DJ, et al. A unified nomenclature for yeast autophagy-related genes. Dev Cell. 2003;5(4):539–545.
- Ohsumi Y. Molecular dissection of autophagy: two ubiquitin-like systems. Nat Rev Mol Cell Biol. 2001;2(3):211–216.
- Kirisako T, et al. The reversible modification regulates the membrane-binding state of Apg8/Aut7 essential for autophagy and the cytoplasm to vacuole targeting pathway. J Cell Biol. 2000;151(2):263–276.
- Ichimura Y, et al. A ubiquitin-like system mediates protein lipidation. Nature. 2000;408(6811):488–492.
- Marino G, Lopez-Otin C. Autophagy: molecular mechanisms, physiological functions and relevance in human pathology. Cell Mol Life Sci. 2004;61(12):1439–1454.View this article via: PubMed Google Scholar
- Marino G, Uria JA, Puente XS, Quesada V, Bordallo J, Lopez-Otin C. Human autophagins, a family of cysteine proteinases potentially implicated in cell degradation by autophagy. J Biol Chem. 2003;278(6):3671–3678.
- Tanida I, Tanida-Miyake E, Ueno T, Kominami E. The human homolog of Saccharomyces cerevisiae Apg7p is a Protein-activating enzyme for multiple substrates including human Apg12p, GATE-16, GABARAP, and MAP-LC3. J Biol Chem. 2001;276(3):1701–1706.View this article via: PubMed Google Scholar
- He H, et al. Post-translational modifications of three members of the human MAP1LC3 family and detection of a novel type of modification for MAP1LC3B. J Biol Chem. 2003;278(31):29278–29287.
- Hemelaar J, Lelyveld VS, Kessler BM, Ploegh HL. A single protease, Apg4B, is specific for the autophagy-related ubiquitin-like proteins GATE-16, MAP1-LC3, GABARAP, and Apg8L. J Biol Chem. 2003;278(51):51841–51850.
- Kuma A, et al. The role of autophagy during the early neonatal starvation period. Nature. 2004;432(7020):1032–1036.
- Levine B, Kroemer G. Autophagy in the pathogenesis of disease. Cell. 2008;132(1):27–42.
- Klionsky DJ, et al. Guidelines for the use and interpretation of assays for monitoring autophagy in higher eukaryotes. Autophagy. 2008;4(2):151–175.View this article via: PubMed Google Scholar
- Bjorkoy G, et al. p62/SQSTM1 forms protein aggregates degraded by autophagy and has a protective effect on huntingtin-induced cell death. J Cell Biol. 2005;171(4):603–614.
- Korolchuk VI, Mansilla A, Menzies FM, Rubinsztein DC. Autophagy inhibition compromises degradation of ubiquitin-proteasome pathway substrates. Mol Cell. 2009;33(4):517–527.
- Kuusisto E, Suuronen T, Salminen A. Ubiquitin-binding protein p62 expression is induced during apoptosis and proteasomal inhibition in neuronal cells. Biochem Biophys Res Commun. 2001;280(1):223–228.
- Mizushima N, Yamamoto A, Matsui M, Yoshimori T, Ohsumi Y. In vivo analysis of autophagy in response to nutrient starvation using transgenic mice expressing a fluorescent autophagosome marker. Mol Biol Cell. 2004;15(3):1101–1111.
- Marino G, Salvador-Montoliu N, Fueyo A, Knecht E, Mizushima N, Lopez-Otin C. Tissue-specific autophagy alterations and increased tumorigenesis in mice deficient in Atg4C/autophagin-3. J Biol Chem. 2007;282(25):18573–18583.
- Komatsu M, et al. Impairment of starvation-induced and constitutive autophagy in Atg7-deficient mice. J Cell Biol. 2005;169(3):425–434.
- Kabeya Y, Mizushima N, Yamamoto A, Oshitani-Okamoto S, Ohsumi Y, Yoshimori T. LC3, GABARAP and GATE16 localize to autophagosomal membrane depending on form-II formation. J Cell Sci. 2004;117(pt 13):2805–2812.
- Tanida I, Sou YS, Minematsu-Ikeguchi N, Ueno T, Kominami E. Atg8L/Apg8L is the fourth mammalian modifier of mammalian Atg8 conjugation mediated by human Atg4B, Atg7 and Atg3. Febs J. 2006;273(11):2553–2562.
- Kabeya Y, et al. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J. 2000;19(21):5720–5728.View this article via: PubMed Google Scholar
- Mizushima N, et al. Dissection of autophagosome formation using Apg5-deficient mouse embryonic stem cells. J Cell Biol. 2001;152(4):657–668.
- Herweijer H, Wolff JA. Gene therapy progress and prospects: hydrodynamic gene delivery. Gene Ther. 2007;14(2):99–107.View this article via: PubMed Google Scholar
- Porsolt RD, Deniel M, Jalfre M. Forced swimming in rats: hypothermia, immobility and the effects of imipramine. Eur J Pharmacol. 1979;57(4):431–436.
- Komatsu M, et al. Loss of autophagy in the central nervous system causes neurodegeneration in mice. Nature. 2006;441(7095):880–884.
- Hara T, et al. Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice. Nature. 2006;441(7095):885–889.
- Hughes I, Thalmann I, Thalmann R, Ornitz DM. Mixing model systems: using zebrafish and mouse inner ear mutants and other organ systems to unravel the mystery of otoconial development. Brain Res. 2006;1091(1):58–74.
- Zhao X, Yang H, Yamoah EN, Lundberg YW. Gene targeting reveals the role of Oc90 as the essential organizer of the otoconial organic matrix. Dev Biol. 2007;304(2):508–524.
- Suzuki H, Ikeda K, Takasaka T. Biological characteristics of the globular substance in the otoconial membrane of the guinea pig. Hear Res. 1995;90(1–2):212–218.View this article via: PubMed Google Scholar
- Takumida M, Zhang DM, Yajin K, Harada Y. Formation and fate of giant otoconia of the guinea pig following streptomycin intoxication. Acta Otolaryngol. 1997;117(4):538–544.
- Guthrie OW. Aminoglycoside induced ototoxicity. Toxicology. 2008;249(2–3):91–96.
- Komatsu M, et al. Homeostatic levels of p62 control cytoplasmic inclusion body formation in autophagy-deficient mice. Cell. 2007;131(6):1149–1163.
- Hurle B, et al. Non-syndromic vestibular disorder with otoconial agenesis in tilted/mergulhador mice caused by mutations in otopetrin 1. Hum Mol Genet. 2003;12(7):777–789.
- Thalmann R, Ignatova E, Kachar B, Ornitz DM, Thalmann I. Development and maintenance of otoconia: biochemical considerations. Ann N Y Acad Sci. 2001;942:162–178.View this article via: PubMed Google Scholar
- Lundberg YW, Zhao X, Yamoah EN. Assembly of the otoconia complex to the macular sensory epithelium of the vestibule. Brain Res. 2006;1091(1):47–57.
- Rolfsen RM, Erway LC. Trace metals and otolith defects in mocha mice. J Hered. 1984;75(3):159–162.View this article via: PubMed Google Scholar
- Huizing M, Boissy RE, Gahl WA. Hermansky-Pudlak syndrome: vesicle formation from yeast to man. Pigment Cell Res. 2002;15(6):405–419.
- Kantheti P, et al. Mutation in AP-3 delta in the mocha mouse links endosomal transport to storage deficiency in platelets, melanosomes, and synaptic vesicles. Neuron. 1998;21(1):111–122.
- Fujita N, et al. An Atg4B mutant hampers the lipidation of LC3 paralogues and causes defects in autophagosome closure. Mol Biol Cell. 2008;19(11):4651–4659.
- Scherz-Shouval R, Sagiv Y, Shorer H, Elazar Z. The COOH terminus of GATE-16, an intra-Golgi transport modulator, is cleaved by the human cysteine protease HsApg4A. J Biol Chem. 2003;278(16):14053–14058.
- Betin VM, Lane JD. Caspase cleavage of Atg4D stimulates GABARAP-L1 processing and triggers mitochondrial targeting and apoptosis. J Cell Sci. 2009;122(pt 14):2554–2566.
- Qu X, et al. Promotion of tumorigenesis by heterozygous disruption of the beclin 1 autophagy gene. J Clin Invest. 2003;112(12):1809–1820.
- Yue Z, Jin S, Yang C, Levine AJ, Heintz N. Beclin 1, an autophagy gene essential for early embryonic development, is a haploinsufficient tumor suppressor. Proc Natl Acad Sci U S A. 2003;100(25):15077–15082.
- Cadwell K, et al. A key role for autophagy and the autophagy gene Atg16l1 in mouse and human intestinal Paneth cells. Nature. 2008;456(7219):259–263.
- Zhao X, Jones SM, Yamoah EN, Lundberg YW. Otoconin-90 deletion leads to imbalance but normal hearing: a comparison with other otoconia mutants. Neuroscience. 2008;153(1):289–299.
- Paffenholz R, et al. Vestibular defects in head-tilt mice result from mutations in Nox3, encoding an NADPH oxidase. Genes Dev. 2004;18(5):486–491.
- Ornitz DM, Bohne BA, Thalmann I, Harding GW, Thalmann R. Otoconial agenesis in tilted mutant mice. Hear Res. 1998;122(1–2):60–70.
- Rauch TM. The additive effects of two mutant genes on otolith formation in mice: an animal model to assess otolith function. J Aud Res. 1979;19(4):259–265.View this article via: PubMed Google Scholar
- Bonifacino JS. Insights into the biogenesis of lysosome-related organelles from the study of the Hermansky-Pudlak syndrome. Ann N Y Acad Sci. 2004;1038:103–114.
- Ganesan AK, et al. Genome-wide siRNA-based functional genomics of pigmentation identifies novel genes and pathways that impact melanogenesis in human cells. PLoS Genet. 2008;4(12):e1000298.
- Komatsu M, et al. The selective autophagy substrate p62 activates the stress responsive transcription factor Nrf2 through inactivation of Keap1. Nat Cell Biol. 2010;12(3):213–223.
- Cuervo AM, Bergamini E, Brunk UT, Droge W, Ffrench M, Terman A. Autophagy and aging: the importance of maintaining “clean” cells. Autophagy. 2005;1(3):131–140.
- Jang YS, Hwang CH, Shin JY, Bae WY, Kim LS. Age-related changes on the morphology of the otoconia. Laryngoscope. 2006;116(6):996–1001.View this article via: PubMed Google Scholar
- Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987;162(1):156–159.View this article via: PubMed Google Scholar
-
Version history
- Version 1 (June 23, 2010): No description
- Version 2 (July 1, 2010): No description



Copyright © 2024 American Society for Clinical Investigation
ISSN: 0021-9738 (print), 1558-8238 (online)








