Advertisement
Research Article Free access | 10.1172/JCI30182
The homeobox gene CDX2 is aberrantly expressed in most cases of acute myeloid leukemia and promotes leukemogenesis
Claudia Scholl,1,2 Dimple Bansal,1 Konstanze Döhner,2 Karina Eiwen,2 Brian J.P. Huntly,1 Benjamin H. Lee,1 Frank G. Rücker,2 Richard F. Schlenk,2 Lars Bullinger,2 Hartmut Döhner,2 D. Gary Gilliland,1 and Stefan Fröhling1,2
1Division of Hematology, Brigham and Women’s Hospital, Harvard Medical School, Boston, Massachusetts, USA. 2Department of Internal Medicine III, University Hospital of Ulm, Ulm, Germany.
Address correspondence to: Stefan Fröhling, Karp Family Research Laboratories, 1 Blackfan Circle, 5th Floor, Boston, Massachusetts 02115, USA. Phone: (617) 355-9085; Fax: (617) 355-9093; E-mail: [email protected].
Find articles by Scholl, C. in: JCI | PubMed | Google Scholar
1Division of Hematology, Brigham and Women’s Hospital, Harvard Medical School, Boston, Massachusetts, USA. 2Department of Internal Medicine III, University Hospital of Ulm, Ulm, Germany.
Address correspondence to: Stefan Fröhling, Karp Family Research Laboratories, 1 Blackfan Circle, 5th Floor, Boston, Massachusetts 02115, USA. Phone: (617) 355-9085; Fax: (617) 355-9093; E-mail: [email protected].
Find articles by Bansal, D. in: JCI | PubMed | Google Scholar
1Division of Hematology, Brigham and Women’s Hospital, Harvard Medical School, Boston, Massachusetts, USA. 2Department of Internal Medicine III, University Hospital of Ulm, Ulm, Germany.
Address correspondence to: Stefan Fröhling, Karp Family Research Laboratories, 1 Blackfan Circle, 5th Floor, Boston, Massachusetts 02115, USA. Phone: (617) 355-9085; Fax: (617) 355-9093; E-mail: [email protected].
Find articles by Döhner, K. in: JCI | PubMed | Google Scholar
1Division of Hematology, Brigham and Women’s Hospital, Harvard Medical School, Boston, Massachusetts, USA. 2Department of Internal Medicine III, University Hospital of Ulm, Ulm, Germany.
Address correspondence to: Stefan Fröhling, Karp Family Research Laboratories, 1 Blackfan Circle, 5th Floor, Boston, Massachusetts 02115, USA. Phone: (617) 355-9085; Fax: (617) 355-9093; E-mail: [email protected].
Find articles by Eiwen, K. in: JCI | PubMed | Google Scholar
1Division of Hematology, Brigham and Women’s Hospital, Harvard Medical School, Boston, Massachusetts, USA. 2Department of Internal Medicine III, University Hospital of Ulm, Ulm, Germany.
Address correspondence to: Stefan Fröhling, Karp Family Research Laboratories, 1 Blackfan Circle, 5th Floor, Boston, Massachusetts 02115, USA. Phone: (617) 355-9085; Fax: (617) 355-9093; E-mail: [email protected].
Find articles by Huntly, B. in: JCI | PubMed | Google Scholar
1Division of Hematology, Brigham and Women’s Hospital, Harvard Medical School, Boston, Massachusetts, USA. 2Department of Internal Medicine III, University Hospital of Ulm, Ulm, Germany.
Address correspondence to: Stefan Fröhling, Karp Family Research Laboratories, 1 Blackfan Circle, 5th Floor, Boston, Massachusetts 02115, USA. Phone: (617) 355-9085; Fax: (617) 355-9093; E-mail: [email protected].
Find articles by Lee, B. in: JCI | PubMed | Google Scholar
1Division of Hematology, Brigham and Women’s Hospital, Harvard Medical School, Boston, Massachusetts, USA. 2Department of Internal Medicine III, University Hospital of Ulm, Ulm, Germany.
Address correspondence to: Stefan Fröhling, Karp Family Research Laboratories, 1 Blackfan Circle, 5th Floor, Boston, Massachusetts 02115, USA. Phone: (617) 355-9085; Fax: (617) 355-9093; E-mail: [email protected].
Find articles by Rücker, F. in: JCI | PubMed | Google Scholar
1Division of Hematology, Brigham and Women’s Hospital, Harvard Medical School, Boston, Massachusetts, USA. 2Department of Internal Medicine III, University Hospital of Ulm, Ulm, Germany.
Address correspondence to: Stefan Fröhling, Karp Family Research Laboratories, 1 Blackfan Circle, 5th Floor, Boston, Massachusetts 02115, USA. Phone: (617) 355-9085; Fax: (617) 355-9093; E-mail: [email protected].
Find articles by Schlenk, R. in: JCI | PubMed | Google Scholar
1Division of Hematology, Brigham and Women’s Hospital, Harvard Medical School, Boston, Massachusetts, USA. 2Department of Internal Medicine III, University Hospital of Ulm, Ulm, Germany.
Address correspondence to: Stefan Fröhling, Karp Family Research Laboratories, 1 Blackfan Circle, 5th Floor, Boston, Massachusetts 02115, USA. Phone: (617) 355-9085; Fax: (617) 355-9093; E-mail: [email protected].
Find articles by Bullinger, L. in: JCI | PubMed | Google Scholar
1Division of Hematology, Brigham and Women’s Hospital, Harvard Medical School, Boston, Massachusetts, USA. 2Department of Internal Medicine III, University Hospital of Ulm, Ulm, Germany.
Address correspondence to: Stefan Fröhling, Karp Family Research Laboratories, 1 Blackfan Circle, 5th Floor, Boston, Massachusetts 02115, USA. Phone: (617) 355-9085; Fax: (617) 355-9093; E-mail: [email protected].
Find articles by Döhner, H. in: JCI | PubMed | Google Scholar
1Division of Hematology, Brigham and Women’s Hospital, Harvard Medical School, Boston, Massachusetts, USA. 2Department of Internal Medicine III, University Hospital of Ulm, Ulm, Germany.
Address correspondence to: Stefan Fröhling, Karp Family Research Laboratories, 1 Blackfan Circle, 5th Floor, Boston, Massachusetts 02115, USA. Phone: (617) 355-9085; Fax: (617) 355-9093; E-mail: [email protected].
Find articles by Gilliland, D. in: JCI | PubMed | Google Scholar
1Division of Hematology, Brigham and Women’s Hospital, Harvard Medical School, Boston, Massachusetts, USA. 2Department of Internal Medicine III, University Hospital of Ulm, Ulm, Germany.
Address correspondence to: Stefan Fröhling, Karp Family Research Laboratories, 1 Blackfan Circle, 5th Floor, Boston, Massachusetts 02115, USA. Phone: (617) 355-9085; Fax: (617) 355-9093; E-mail: [email protected].
Find articles by Fröhling, S. in: JCI | PubMed | Google Scholar
Published April 2, 2007 - More info
J Clin Invest. 2007;117(4):1037–1048. https://doi.org/10.1172/JCI30182.
© 2007 The American Society for Clinical Investigation
Received: August 29, 2006; Accepted: January 12, 2007
Related article:
Abstract
The deregulation of homeobox (HOX) genes in acute myeloid leukemia (AML) and the potential for these master regulators to perturb normal hematopoiesis is well established. To date, overexpression of HOX genes in AML has been attributed to specific chromosomal aberrations and abnormalities involving mixed-lineage leukemia (MLL), an upstream regulator of HOX genes. The finding reported in this issue of the JCI by Scholl et al. that caudal-type homeobox transcription factor 2 (CDX2), which is capable of affecting HOX gene expression during embryogenesis, is overexpressed in 90% of patients with AML and induces a transplantable AML in murine models provides an alternative mechanism for HOX-induced leukemogenesis and yields important insights into the hierarchy of HOX gene regulation in AML (see the related article beginning on page 1037).
Authors
Kim L. Rice, Jonathan D. Licht
-
Abstract
The homeobox transcription factor CDX2 plays an important role in embryonic development and regulates the proliferation and differentiation of intestinal epithelial cells in the adult. We have found that CDX2 is expressed in leukemic cells of 90% of patients with acute myeloid leukemia (AML) but not in hematopoietic stem and progenitor cells derived from normal individuals. Stable knockdown of CDX2 expression by RNA interference inhibited the proliferation of various human AML cell lines and strongly reduced their clonogenic potential in vitro. Primary murine hematopoietic progenitor cells transduced with Cdx2 acquired serial replating activity, were able to be continuously propagated in liquid culture, generated fully penetrant and transplantable AML in BM transplant recipients, and displayed dysregulated expression of Hox family members in vitro and in vivo. These results demonstrate that aberrant expression of the developmental regulatory gene CDX2 in the adult hematopoietic compartment is a frequent event in the pathogenesis of AML; suggest a role for CDX2 as part of a common effector pathway that promotes the proliferative capacity and self-renewal potential of myeloid progenitor cells; and support the hypothesis that CDX2 is responsible, in part, for the altered HOX gene expression that is observed in most cases of AML.
-
Introduction
The caudal-type homeobox gene Cdx2 encodes a transcription factor that contributes to embryonic organogenesis and early hematopoietic development in vertebrate species (1–4). Of note, previous studies using genetic approaches have shown that the developmental effects of Cdx2, like those of other Cdx family members, are mediated through regulation of Hox gene expression and that Cdx proteins can act directly on Hox gene regulatory elements (5–7). Furthermore, Cdx2 is essential for establishment and function of the trophectoderm lineage at implantation (8–10). In mice, expression of Cdx2 is limited almost exclusively to the endoderm of the primitive gut by E12.5, and this pattern is maintained throughout life, with highest expression levels in the distal intestine and the proximal colon (11, 12). Likewise, expression of human CDX2 has only been detected in intestinal epithelial cells (11, 12).
Considerable evidence suggests that abnormalities of CDX2 are relevant to human gastrointestinal malignancies. Gene targeting studies in mice and the analysis of human colorectal cancer specimens have shown that CDX2 functions as a tumor suppressor in the colon (13–18), whereas ectopic CDX2 expression is involved in the development of precancerous intestinal metaplasia in the stomach and esophagus (19–22).
Dysregulated CDX2 expression has only rarely been attributed to cancers that occur outside the gastrointestinal tract. Chase et al. described a fusion between CDX2 and the TEL gene at chromosome 12p13 in a single patient with acute myeloid leukemia (AML) and t(12;13)(p13;q12) and found that both the chimeric TEL-CDX2 fusion transcript and normal CDX2 transcripts were detectable in cDNA from this patient (23). Rawat et al. subsequently showed that ectopic expression of Cdx2, but not expression of the TEL-CDX2 fusion gene, caused AML in a murine BM transplantation model of t(12;13)-positive AML (24). In the former study, conventional RT-PCR analysis detected CDX2 mRNA in 1 of 10 patients in the blast phase of chronic myeloid leukemia (CML-BP) who did not have cytogenetic evidence of chromosome 13 abnormalities but did not identify aberrant CDX2 expression in 10 AML patients without aberrations involving chromosome 13 (23). Thus, it remains unclear to what extent ectopic expression of CDX2 has a more general role in myeloid leukemogenesis.
Using array-based comparative genomic hybridization (aCGH) and FISH, we recently demonstrated high-level amplification of the CDX2 locus in a subset of AML patients with complex karyotypes lacking cytogenetic abnormalities involving chromosome 13 (25). This observation prompted us to systematically analyze the expression of CDX2 in a large cohort of patients with myeloid leukemias by real-time quantitative PCR (RQ-PCR). We detected CDX2 expression in 90% of patients with AML, in patients with high-risk myelodysplastic syndrome (MDS) or advanced-stage CML, and in several AML cell lines, but not in hematopoietic stem and progenitor cells derived from normal individuals. We explored the functional relevance of aberrant CDX2 expression in AML by RNA interference–mediated (RNAi-mediated) knockdown of CDX2 in AML cell lines, by ectopic expression of Cdx2 in primary murine hematopoietic progenitors, and in a murine model of Cdx2-induced AML.
-
Results
Aberrant CDX2 expression in AML. CDX2 transcripts were detectable in BM mononuclear cells (BMMCs) or PBMCs from 153 (90%) of 170 patients with AML (Figure 1). The median expression level for all samples was 508 (range, 0–88,020), and there was a statistically significant difference between different cytogenetic subgroups (P = 0.02; Table 1). The highest CDX2 expression levels were present in patients with t(9;11)(p22;q23), followed by those with normal karyotype, t(15;17)(q22;q11~21), t(8;21)(q22;q22), inv(16)(p13q22), other chromosome aberrations, and complex karyotype, defined as 3 or more cytogenetic abnormalities in the absence of t(8;21), inv(16), t(15;17), or t(11q23). In 3 of the 30 patients with complex karyotypes, we previously found high-level amplification of the CDX2 locus, as assessed by aCGH and FISH (25), and each of these 3 patients expressed high levels of CDX2 transcripts (expression levels, 1,650; 2,063; and 22,477, respectively). In comparison, BMMCs (n = 10), CD34+ cells (n = 3), HSCs (n = 3), common myeloid progenitors (CMPs; n = 3), granulocyte-macrophage progenitors (GMPs; n = 3), and megakaryocyte-erythroid progenitors (MEPs; n = 3) derived from normal individuals showed no detectable CDX2 expression by RQ-PCR (Figure 1).
 Figure 1
Figure 1CDX2 expression in AML. CDX2 mRNA levels were measured by RQ-PCR in 170 AML patients from different cytogenetic subgroups, as well as in BMMCs (n = 10), CD34+ cells (n = 3), HSCs (n = 3), CMPs (n = 3), GMPs (n = 3), and MEPs (n = 3) from normal individuals. Circles indicate patients with genomic amplification of the CDX2 locus, as assessed by aCGH and FISH (25). Bars indicate median values.
In 2 AML patients, CDX2 expression levels were analyzed at different time points. Patient 1, who had inv(16)-positive AML, achieved complete remission (CR) after the first of 2 courses of induction chemotherapy according to the ICE protocol (12 mg/m2 idarubicin on days 1, 3, and 5; 100 mg/m2 cytarabine continuously on days 1–7; 100 mg/m2 etoposide on days 1–3) and remained disease free after 1 course of consolidation chemotherapy with HAM (3 g/m2 cytarabine every 12 hours on days 1 through 3; 12 mg/m2 mitoxantrone on days 2 and 3) followed by allogeneic stem cell transplantation from an HLA-compatible sibling donor. This patient had a pretreatment CDX2 expression level of 4,202. However, CDX2 transcripts were undetectable after the first course of chemotherapy, and RQ-PCR results remained negative after the second course of ICE, after consolidation chemotherapy, and during the follow-up period (7 and 16 months from diagnosis, respectively). Patient 2, who had t(8;21)-positive AML, achieved CR after 2 courses of ICE with a partial remission after the first course, remained disease free after 2 courses of HAM, but relapsed 10 months from diagnosis. This patient had a pretreatment CDX2 expression level of 4,810; CDX2 transcript levels were significantly reduced but detectable after 2 courses of ICE (expression level, 377) and after 1 course of HAM (expression level, 42), but were increased at the time of clinical relapse (expression level, 951). In both patients, serial quantification of the leukemia-specific fusion transcript resulting from the chromosomal rearrangement (CBFB-MYH11 and RUNX1-CBFA2T1, respectively) by RQ-PCR showed that CDX2 transcript level correlated with level of expression of the respective fusion transcript (data not shown).
Taken together, these results indicate that CDX2 is aberrantly expressed in the majority of patients with AML and that CDX2 expression correlates with disease burden.
CDX2 expression in MDS, CML, or myeloid leukemia cell lines. CDX2 transcripts were detectable in BMMCs from 2 of 5 patients with MDS. One patient had refractory anemia with excess blasts–2 (RAEB-2; CDX2 expression level, 66), and 1 patient had refractory cytopenia with multilineage dysplasia (CDX2 expression level, 2,334). The latter patient progressed to secondary AML 1 month later, and this transition was accompanied by an increase in the number of CDX2 transcripts (expression level, 8,564), again demonstrating that CDX2 expression correlates with the size of the leukemic clone. No CDX2 transcripts were detected in 1 patient with refractory anemia, 1 patient with RAEB-1, and 1 patient with RAEB-2.
Of the 10 CML patients who were analyzed in this study, 2 had detectable CDX2 transcripts, 1 in myeloid CML-BP (expression level, 639) and 1 in the accelerated phase (CML-AP; expression level, 309). The latter patient had tested negative for CDX2 by RQ-PCR while being in the chronic phase (CML-CP) 16 months earlier. No CDX2 transcripts were detected in 6 patients with CML-CP or 2 patients with CML-AP.
Of the 15 human myeloid leukemia cell lines tested, 8 had detectable CDX2 transcripts (Supplemental Figure 1; supplemental material available online with this article; doi:10.1172/JCI30182DS1). The highest CDX2 expression level was seen in the EOL-1 cell line, i.e., 37,388, followed by NB4, 12,235; SKM-1, 344; MV4-11, 155; MONO-MAC-6, 135; NOMO-1, 70; THP-1, 30; and MOLM-14, 27.
Monoallelic CDX2 expression in AML. To better understand the mechanism of dysregulated expression of CDX2, we first investigated whether both alleles of the CDX2 gene were expressed in leukemias with detectable CDX2 transcripts. We analyzed 7 AML patients (median expression level, 2,015; range 1,101–8,802) who were heterozygous for at least 1 of the known SNPs in the CDX2 gene. In 6 of these 7 patients (median CDX2 expression level, 1,685; range, 1,101–88,020), direct sequencing of cDNA and analysis of individual cDNA clones (median number, 12; range, 5–21) showed that 1 allele was expressed (median proportion of cDNA clones with identical nucleotide sequence, 100%; range, 92%–100%). Monoallelic expression in these cases indicated that the aberrant expression of CDX2 was caused by an acquired cis-acting genetic event. In contrast, 1 patient (CDX2 expression level, 49,385) demonstrated expression of both alleles. Analysis of the 2 AML cell lines that had been found to carry an informative SNP (EOL-1 and NB4) showed predominant expression of 1 allele of the CDX2 gene (proportion of cDNA clones with identical nucleotide sequence, 89% and 100%, respectively).
We next tested the hypothesis that monoallelic expression could be attributed to mutations in cis-acting regulatory elements. DNA sequence analysis of a predicted CDX2 promoter region, spanning 687 nucleotides 5' of the annotated transcription start site (TSS) and the first 276 nucleotides of CDX2 exon 1 (Supplemental Figure 2A), in the 6 patients and 2 cell lines that showed monoallelic expression identified no mutations.
To examine whether the aberrant expression of CDX2 in AML could be caused by gene-specific promoter hypomethylation, a pathogenetic event that has been described in several solid tumors (26–30), we studied DNA methylation over a 347-bp region (nucleotide positions –193 to +154 relative to the CDX2 TSS) within a previously described (31) CpG island upstream of CDX2 exon 1 by sodium bisulfite sequencing (Supplemental Figure 2A). As shown in Supplemental Figure 2B, the CDX2 promoter region was predominantly unmethylated in BMMCs from 3 CDX2-expressing patients (expression levels, 1,101, 1,288, and 2,963, respectively). However, a very similar pattern was observed in BMMCs from 3 CDX2-negative AML patients and PBMCs and granulocytes from 2 healthy individuals without detectable CDX2 expression.
To test the less likely possibility that the aberrant monoallelic expression of CDX2 was related to mutations in the coding sequence of the gene that might affect RNA stability, we sequenced each of the 3 exons of the CDX2 gene in AML patients from different cytogenetic subgroups. No mutations were identified in 32 patients with detectable CDX2 transcripts (median expression level, 697; range, 176–22,477) or 2 patients negative for CDX2 by RQ-PCR.
To determine whether the misexpression of CDX2 was due to gene amplification, we performed RQ-PCR on genomic DNA from 50 AML patients with detectable CDX2 expression (median, 832; range, 39–88,020). Two cases with complex karyotypes that had previously been found to carry extrachromosomal amplification of the CDX2 gene (25), 1 case with pentasomy 13, and 1 case with an isolated marker chromosome showed increased CDX2 copy numbers (78, 11, 5, and 4, respectively). In contrast, no amplification of the CDX2 locus was present in 18 patients with normal cytogenetics, 1 patient with t(8;21), 3 patients with t(9;11), 5 patients with t(15;17), 7 patients with inv(16), 7 patients with complex karyotypes, or 5 patients with other chromosome aberrations.
Taken together, these findings suggest that dysregulated CDX2 expression in AML is frequently monoallelic and that the genetic events that contribute to monoallelic expression are not attributable to locus-specific amplification and lie outside the coding sequence or predicted proximal promoter sequences in the majority of cases.
Growth inhibition of AML cell lines following knockdown of CDX2. To assess the functional significance of aberrant CDX2 expression in myeloid leukemias, we used lentivirus-mediated short hairpin RNA (shRNA) delivery for long-term silencing of CDX2 in human AML cell lines.
We first validated the CDX2 shRNA constructs. Transduction of the NB4 cell line with a pool of 3 shRNA constructs targeting CDX2 resulted in a 58% mRNA decrease as compared with the nonsilencing control construct after 13 days (data not shown), and analysis of individual shRNA constructs showed that shRNA TRCN13684 exhibited the most potent effect (86% mRNA knockdown), followed by TRCN13685 (56%) and TRCN13683 (48%) (Supplemental Figure 3A). Knockdown analysis 28 days after infection gave similar results, indicating stable downregulation of CDX2 expression (Supplemental Figure 3A). Immunoprecipitation showed that shRNA-mediated suppression of CDX2 mRNA was associated with greatly diminished levels of CDX2 protein (Supplemental Figure 3B).
We then investigated the effect of CDX2 silencing in the 6 AML cell lines with CDX2 expression levels between 27 and 344; that is, MOLM-14, THP-1, NOMO-1, MONO-MAC-6, MV4-11, and SKM-1. In all of these 6 cell lines except MONO-MAC-6, silencing of CDX2 expression by shRNA TRCN13684 (mRNA knockdown between 23% and 84%) inhibited cell proliferation as compared with the nonsilencing control construct (Figure 2A). Highly similar phenotypic effects were exerted by shRNA TRC13685 (Supplemental Figure 4A). In contrast, exposure of MONO-MAC-6 cells to shRNA TRCN13684 did not induce efficient silencing of CDX2 expression (4% mRNA knockdown), and there was no effect on cell proliferation. Likewise, we found no inhibitory effect of shRNA TRC13684 in HL-60 and K-562 cells, which have no detectable CDX2 expression (Figure 2A).
 Figure 2
Figure 2Proliferation of AML cell lines after shRNA-mediated silencing of CDX2 expression. (A) Downregulation of CDX2 expression by shRNA TRCN13684 inhibited proliferation of the CDX2-expressing AML cell lines SKM-1, THP-1, MV4-11, MOLM-14, and NOMO-1. In MONO-MAC-6 cells, shRNA TRCN13684 did not induce efficient CDX2 mRNA knockdown, and there was no effect on cell proliferation. Similarly, treatment with shRNA TRCN13684 had no inhibitory effect in the CDX2-negative cell lines HL-60 and K-562. For each of the 6 CDX2-expressing cell lines, the degree of mRNA knockdown is shown (inset) (SKM-1, 84%; THP-1, 68%; MV4-11, 52%; MOLM-14, 49%; NOMO-1, 23%; MONO-MAC-6, 4%). Experiments were performed in triplicate. Values are represented as mean ± SEM. (B) SKM-1 cells were transduced with a lentiviral vector that coexpresses shRNA TRCN13684 and GFP. Sorted cells (proportion of GFP+ cells, 98%) were cultured at a density of 0.5 × 106 to 1 × 106/ml, and the GFP+ fraction was measured by flow cytometry at the indicated time points. The toxicity of CDX2 knockdown was evidenced by a relative depletion of GFP+ cells over time. In contrast, analysis of SKM-1 cells transduced with a GFP-expressing pLKO.1 construct without an shRNA sequence (proportion of GFP+ cells, 95%) showed no decrease in the percentage of GFP+ cells. The degree of CDX2 mRNA knockdown is shown (inset; 93%).
To confirm and extend these results, we established a second toxicity assay that allowed us to follow the fate of cells acutely transduced with anti-CDX2 shRNA over time. We created variants of pLKO.1-TRCN13684 and the nonsilencing control construct in which the puromycin resistance gene was replaced with GFP and used lentiviruses prepared from these vectors to infect SKM-1 cells. After lentiviral transduction, GFP-sorted cells were cultured, and the proportion of GFP+ cells was monitored by flow cytometry for 24 days. As shown in Figure 2B, the GFP+ fraction decreased over time in cells transduced with shRNA TRCN13684, indicating a toxic effect of CDX2 knockdown, whereas no toxicity was observed in cells transduced with the nonsilencing control construct.
Taken together, these results indicate that aberrant expression of CDX2 contributes to the growth potential of myeloid leukemia cells.
Reduced clonogenicity of AML cell lines following knockdown of CDX2. We next tested the ability of AML cell lines transduced with shRNA TRCN13684 to form colonies in clonogenic methylcellulose assays. For the 5 CDX2-expressing cell lines SKM-1, MV4-11, THP-1, MOLM-14, and NOMO-1, we observed a significant reduction in the number of colonies (Figure 3, A and B) and a reduction in the number of cells per colony (Figure 3C) after mRNA knockdown as compared with cells transduced with the nonsilencing control construct. In contrast, transduction with shRNA TRCN13684 did not reduce colony formation of EOL-1, which had a very high CDX2 expression level (37,388; 82% mRNA knockdown; estimated residual expression level, 6,730; Figure 3, A and D); MONO-MAC-6, in which CDX2 expression was not efficiently silenced (4% mRNA knockdown; Figure 3, A and D); or HL-60 and K-562, which do not express CDX2 mRNA (Figure 3, A and B). Transduction of selected cell lines with shRNA TRC13685 confirmed that the observed effects could be attributed to knockdown of CDX2 expression (Supplemental Figure 4, B and C).
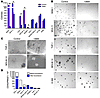 Figure 3
Figure 3Colony formation of AML cell lines after shRNA-mediated silencing of CDX2 expression. (A) Colony-forming assays showed a significant reduction in the number of colonies for the 5 CDX2-expressing cell lines SKM-1, MV4-11 (1 × 104 plated cells), THP-1, MOLM-14, and NOMO-1 (1 × 103 plated cells) after CDX2 mRNA knockdown by shRNA TRCN13684 as compared with cells transduced with the nonsilencing control construct. In contrast, transduction with shRNA TRCN13684 did not reduce colony formation of EOL-1, which had a very high CDX2 expression level (37,388; 82% mRNA knockdown; estimated residual expression level, 6,730); MONO-MAC-6, in which CDX2 expression was not efficiently silenced (4% mRNA knockdown); and HL-60 and K-562, which do not express CDX2 mRNA. Experiments were performed in duplicate. Values are represented as mean ± SEM. *P < 0.05; **P < 0.001. (B and C) Microscopic analysis of colonies derived from shRNA-transduced cells (right panels) showed a decrease in the number of colonies and the number of cells per colony, as compared with cells transduced with the nonsilencing control construct (left panels), for CDX2-expressing cell lines but not for CDX2-negative K-562 cells. Representative photomicrographs of methylcellulose cultures are shown. Original magnification, ×20 and ×100, respectively. (D) CDX2-expressing AML cell lines showed varying absolute mRNA levels after shRNA-mediated CDX2 knockdown.
These results support the hypothesis that aberrant expression of CDX2 is involved in the activation of pathways responsible for the increased proliferative and clonogenic capacity that is characteristic of AML.
Increased serial replating activity of primary murine BM and committed hematopoietic progenitor populations expressing Cdx2. As an in vitro test of the effect of Cdx2 on self-renewal properties of primary hematopoietic cells, we initially examined the serial replating ability of primary unfractionated murine BM transduced with Cdx2 in methylcellulose cultures in the absence of stroma. As shown in Figure 4A, Cdx2-expressing cells demonstrated replating potential to the fourth round of replating, whereas cells transduced with empty vector had a finite ability to serially replate. Cells derived from the fourth replating could subsequently be continuously propagated in liquid culture in the presence of IL-3 (Figure 4B) and exhibited morphologic and immunophenotypic characteristics of immature myeloid cells (Figure 4, C and D).
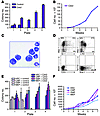 Figure 4
Figure 4In vitro self-renewal of murine BM and committed hematopoietic progenitor populations expressing Cdx2. (A) Whole primary murine BM expressing Cdx2 demonstrated replating potential to the fourth plating, whereas cells transduced with empty vector had a finite ability to serially replate. Experiments were performed in duplicate. Values are represented as mean ± SD. (B) Whole BM cells derived from the fourth plating could be expanded in IL-3-supplemented liquid culture. (C) Microscopic analysis of May-Grünwald-Giemsa–stained cytospin preparations of cells derived from the fourth plating demonstrated predominantly undifferentiated myeloid morphology. Original magnification, ×1,000. (D) Flow cytometric analysis of cells derived from the fourth plating showed expression of myeloid antigens and the immaturity markers Sca-1 and c-Kit and demonstrated the absence of CD3+ or B220+ lymphoid cells and Ter119+ erythroid cells. (E) Committed murine hematopoietic progenitors and HSCs expressing Cdx2 demonstrated replating potential to the fourth plating, whereas cells transduced with empty vector had a finite ability to serially replate. Experiments were performed in duplicate. Values are represented as mean ± SD. (F) Hematopoietic progenitors and HSCs derived from the fourth plating could be expanded in IL-3–supplemented liquid culture.
It has recently been demonstrated that certain leukemia-associated fusion proteins, such as MLL-ENL, MLL-AF9, or MOZ-TIF2, may confer properties of self-renewal to hematopoietic progenitor populations that are committed toward differentiation (32–35). As shown in Figure 4, E and F, expression of Cdx2 in CMPs, GMPs, and MEPs, as well as HSCs, also resulted in serial replating and continuous growth in IL-3–supplemented liquid culture. In contrast, stem and progenitor cells transduced with empty vector showed limited serial replating activity.
Collectively, these findings suggest that aberrant expression of Cdx2 promotes self-renewal properties in primary hematopoietic cells.
Induction of AML in a mouse model of aberrant Cdx2 expression. To assess the leukemogenic potential of Cdx2 in vivo, we performed a murine BM transplantation assay. Mice transplanted with BM cells expressing Cdx2 (n = 5) developed AML after a median of 187 days after transplantation, whereas mice transplanted with MSCV-IRES-GFP–transduced BM cells (n = 3) showed no evidence of disease with a follow-up duration of more than 250 days (P = 0.013; Figure 5A and Supplemental Figure 5). The disease was characterized by an elevated wbc count (Figure 5B, right panel), the presence of frequent blasts in the peripheral blood (PB) (Figure 5C), and marked splenomegaly (Figure 5B, left panel). Histopathologic analysis showed extensive infiltration of BM, liver, and spleen by a prominent atypical population of immature myeloid cells and blast forms (Figure 5C). Flow cytometric analysis of GFP-gated cells from BM and spleen confirmed their myeloid phenotype and demonstrated an increased proportion of Mac-1+ cells, with variable expression of Gr-1 and the immaturity markers CD34 and c-Kit and a concomitant reduction in the level of CD3+ or B220+ lymphoid cells (Figure 5D). Southern blot analysis confirmed proviral integration and demonstrated that Cdx2-induced primary leukemias were clonal or oligoclonal (Supplemental Figure 6). The leukemia generated was transplantable to secondary recipients (n = 5), resulting in a similar phenotype after a median of 52 days after transplantation (Figure 5, A–D). The disease could be categorized as AML without maturation according to the Bethesda proposals for classification of nonlymphoid hematopoietic neoplasms in mice (36) and had similarities to the M0 or M1 subtypes of the French-American-British classification of human AML and to the broad category “AML, not otherwise categorized; subcategories AML, minimally differentiated/AML without maturation” of the current WHO classification (37).
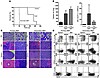 Figure 5
Figure 5Mouse model of aberrant Cdx2 expression. (A) Mice transplanted with BM cells expressing Cdx2 (n = 5) developed AML after a median of 187 days after transplantation, whereas mice transplanted with MSCV-IRES-GFP–transduced BM (n = 3) showed no evidence of disease with a follow-up duration of more than 250 days (P = 0.013). Secondary recipients (n = 5) transplanted with BM from primary leukemic mice developed AML after a median of 52 days after transplantation. BMT, BM transplantation. (B) Diseased mice showed elevated wbc counts (primary recipients versus control mice, P = 0.09; secondary recipients versus control mice, P = 0.019) and splenomegaly (primary recipients versus control mice, P = 0.0029; secondary recipients versus control mice, P < 0.0001). Values are represented as mean ± SD. (C) Microscopic analysis of PB from primary and secondary leukemic animals demonstrated leukocytosis consisting of frequent immature myeloid cells with a high proportion of blast forms that extensively involved the BM, liver, and spleen. Panels display Wright-Giemsa–stained PB smears and H&E-stained tissue sections from representative mice transplanted with BM cells expressing Cdx2. Original magnification, ×400 and ×1,000 (PB); ×100 and ×600 (BM and liver); and ×40 and ×600 (spleen). (D) Flow cytometric analysis of GFP-gated cells from BM and spleen of primary and secondary leukemic animals demonstrated an increased proportion of Mac-1+ myeloid cells with variable expression of Gr-1, CD34, and c-Kit and a concomitant reduction in the level of CD3+ or B220+ lymphoid cells. The percentages of positive cells within the GFP+ compartment are indicated.
These observations demonstrated the transforming capacity of aberrant Cdx2 expression in the hematopoietic compartment in vivo.
Dysregulation of Hox gene expression in primary murine hematopoietic cells expressing Cdx2. Several lines of evidence show that Hox genes are direct transcriptional targets of Cdx2 during development (6, 38, 39). To determine whether Cdx2-induced leukemogenesis was associated with dysregulation of Hox genes, we used RQ-PCR to quantify the expression of 8 Hox family members (Hoxa6, Hoxa7, Hoxa9, Hoxa10, Hoxb3, Hoxb6, Hoxb8, and Hoxc6) in primary murine hematopoietic stem and progenitor cells transduced with Cdx2 as well as in Cdx2-transduced leukemic cells derived from secondary BM transplant recipients.
Assuming that most genetic alterations responsible for the development of AML occur at the level of normal HSCs or myeloid progenitors, we first measured Hox gene expression in immature, c-Kit+Lin– murine BM cells expressing Cdx2. As compared with cells transduced with empty vector, c-Kit+Lin– cells expressing Cdx2 demonstrated an approximately 25-fold upregulation of Hoxb8. In contrast, Cdx2-transduced cells showed decreased expression of Hoxa10, whereas there were no substantial changes in mRNA levels of Hoxa6, Hoxa7, and Hoxa9. No expression was detected for Hoxb3, Hoxb6, and Hoxc6 in Cdx2-transduced cells or in cells transduced with empty vector (Figure 6A).
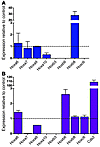 Figure 6
Figure 6Hox gene expression in murine hematopoietic cells expressing Cdx2. (A) As compared with cells transduced with empty vector, c-Kit+Lin– murine hematopoietic progenitors expressing Cdx2 demonstrated upregulation of Hoxb8 and decreased expression of Hoxa10. There were no significant changes in mRNA levels of Hoxa6, Hoxa7, and Hoxa9. No expression was detected for Hoxb3, Hoxb6, and Hoxc6 in Cdx2-transduced cells as well as in cells transduced with empty vector. For normalization, Gapdh was used. Experiments were performed in duplicate. Values are represented as mean ± SEM. (B) Spleen cells isolated from diseased secondary BM transplant recipients demonstrated upregulation of Hoxb6 and decreased expression of Hoxa7 and Hoxa9 as compared with spleen cells obtained from age-matched control mice. There were no substantial changes in mRNA levels of Hoxa6, Hoxb8, and Hoxc6. No expression was detected for Hoxa10 and Hoxb3 in spleen cells from secondary BM transplant recipients or from control mice. The expression level of Cdx2 is also indicated. For normalization, the Gapdh gene was used. Experiments were performed in duplicate. Values are represented as mean ± SEM.
Spleen cells isolated from diseased secondary BM transplant recipients demonstrated an approximately 3-fold upregulation of Hoxb6 as compared with spleen cells obtained from age-matched control mice. In contrast, leukemic cells showed decreased expression of Hoxa7 and Hoxa9, whereas there were no substantial changes in mRNA levels of Hoxa6, Hoxb8, and Hoxc6. No expression was detected for Hoxa10 and Hoxb3 in spleen cells from secondary BM transplant recipients or from control mice (Figure 6B).
These results were consistent with the hypothesis that Cdx2-induced leukemogenesis is mediated, at least in part, through altered expression of specific Hox family members.
-
Discussion
We have found that the caudal-type homeobox gene CDX2 is aberrantly expressed in the leukemic cells of most patients with AML. Ectopic expression of full-length CDX2 in AML was first described by Chase et al., who reported that CDX2 transcripts, as detected by single-round RT-PCR, were present in a single patient with a rare subtype of the disease characterized by the presence of t(12;13)(p13;q12), but not in AML cases without cytogenetic evidence of chromosome 13 abnormalities (23). In our study, which was based on 170 AML cases, we used RQ-PCR for the detection of CDX2 mRNA. It is most likely that differences in the sensitivities of the respective PCR assays account for the disparity between our findings and previous observations.
The mechanisms underlying aberrant expression of CDX2 are not yet fully understood. We observed monoallelic expression of CDX2 in the majority of cases in which there were informative SNPs in the CDX2 coding region, suggesting the presence of acquired alterations in cis-acting regulatory sequences that directly affect the transcription of a single CDX2 allele. These findings are similar to those reported for ectopic expression of the TAL1 gene in T cell acute lymphoblastic leukemia (T-ALL), which has been shown to be monoallelic in a proportion of patients without detectable TAL1 rearrangements (40, 41). However, as for most cases of T-ALL associated with monoallelic expression of TAL1, we found no mutations in the CDX2 coding region or in the predicted CDX2 promoter sequence, and there was no evidence for gene-specific hypomethylation of the CDX2 promoter. Furthermore, we detected no increase in the CDX2 gene copy number in the majority of cases. Taken together, these results suggest that there are acquired mutations in as-yet-undefined regulatory sequences that account for monoallelic CDX2 expression in most cases of AML.
Two observations indicate that there is a causal relationship between aberrant CDX2 expression and myeloid leukemogenesis. First, selective knockdown of CDX2 in a panel of human AML cell lines exhibiting CDX2 transcript levels in a range similar to that observed in many primary AML samples inhibited proliferation and colony formation of AML cells in vitro. This finding indicates that aberrant expression of CDX2 in hematopoietic progenitors contributes to the enhanced proliferative and clonogenic capacity that is characteristic of human AML. Second, and consistent with a previous report (24), murine hematopoietic progenitor cells transduced with Cdx2 generated a fully penetrant AML in BM transplant recipients, demonstrating the leukemogenic potential of Cdx2 in vivo.
CDX2 may contribute to AML pathogenesis through dysregulation of HOX genes. Dysregulated HOX gene expression in AML may be caused by chimeric oncoproteins involving MLL (42) or rare chromosomal translocations directly affecting individual HOX genes (43–45). The leukemogenic potential of aberrant HOX gene expression mediated by mutant MLL (46–50) or HOX fusion genes (51, 52) has been well described in various model systems. However, there is also evidence for widespread misexpression of HOX family members in other AML subtypes (53–61), suggesting the involvement of as-yet-unknown upstream regulators of HOX gene expression. Like the present report, these studies have demonstrated aberrant increases as well as decreases in the level of HOX gene expression. Cdx2 regulates Hox gene expression during embryogenesis and developmental hematopoiesis, suggesting the hypothesis that aberrant expression of CDX2 in the adult hematopoietic compartment may be associated with altered expression of HOX genes. In support of this hypothesis, we found that expression of Cdx2 in primary murine hematopoietic cells resulted in dysregulation of specific Hox family members that have been implicated in leukemogenesis in other contexts. Most notably, we observed a more than 25-fold increase in expression of Hoxb8 in early myeloid progenitors following transduction with Cdx2, and the leukemic cells from secondary BM transplant recipients showed an approximately 3-fold upregulation of Hoxb6. Ectopic expression of HOXB8 inhibits the induction of granulocytic differentiation in various cell line models (62), and mice that express Hoxb8 in the hematopoietic compartment show enhanced self-renewal of immature myeloid progenitors and develop a rapid, transplantable myeloid leukemia (63, 64). Similarly, overexpression of HOXB6 in murine BM enhances the self-renewal of myeloid precursors in vitro and causes AML in vivo (65). Taken together, these results, in combination with previously reported definitive genetic data that Hox genes are direct transcriptional targets of Cdx2 during development (6, 38, 39), indicate that the mechanism(s) through which CDX2 contributes to myeloid leukemogenesis could involve deregulation of HOX family members. The observed differences in Hox gene expression between myeloid progenitor cells immediately after retroviral transduction and Cdx2-induced murine leukemias may reflect the cooperative effect of secondary mutations or cell-nonautonomous effects of the BM microenvironment in leukemic cells. Alternatively, there may be differential requirements for deregulation of distinct HOX family members at the onset and during progression of AML.
AML is a genetically and phenotypically heterogeneous disease. Nevertheless, data support the existence of a subpopulation of rare leukemic stem cells that are responsible for initiating and maintaining the disease in most, if not all, AML subtypes (66–68). Importantly, these cells have an increased capacity to self renew, and quantitative differences in the self-renewal potential of individual AML stem cells form the basis for a hierarchical organization of the AML stem cell compartment (69). Our observation that nearly all cases of AML are characterized by aberrant CDX2 expression raises the possibility that CDX2 is part of a common effector pathway that lies downstream of different primary leukemogenic events. An essential role has been demonstrated for Cdx2 in self-renewal of trophoblast stem cells (9), and 2 other Cdx family members, Cdx1 and Cdx4, are direct targets of the canonical Wnt pathway (70–73) that has been linked to increased self-renewal in myeloid leukemias (74–77). Based on these observations, it is tempting to speculate that in AML, different pathways of transformation converge to enhance self-renewal and that CDX2 may be a component of this shared program. In support of this hypothesis, we found that transduction of primary murine BM and individual committed progenitor populations with Cdx2 conferred serial replating activity, an in vitro surrogate for self-renewal potential (32–35, 74), and resulted in a transplantable AML in vivo. To further assess the relevance of CDX2 for self-renewal in human AML, it will be of interest to precisely determine the hematopoietic progenitor population(s) in which CDX2 is expressed and to study its effects in primary leukemic cells using in vivo experimental systems.
In conclusion, our results demonstrate that aberrant expression of the homeotic gene CDX2 in the hematopoietic compartment is a common and functionally relevant event in AML pathogenesis that appears to enhance the self-renewal activity and leukemogenic potential of myeloid progenitor cells. These findings also support the hypothesis that aberrant expression of CDX2 contributes to the dysregulated HOX gene expression that is observed in most cases of AML.
-
Methods
Patient samples and controls. We analyzed mononuclear cells prepared from diagnostic BM or PB samples from 170 adult patients with AML. All specimens were karyotyped by chromosome banding and FISH (78). In addition, we studied BMMCs from 5 patients with MDS and BMMCs or PBMCs from 10 patients with CML. As controls, we analyzed BMMCs (n = 10), CD34+ cells (n = 3), HSCs (n = 3), CMPs (n = 3), GMPs (n = 3), and MEPs (n = 3) from normal individuals. The diagnosis of AML or MDS was made according to the WHO classification of hematologic malignancies. The study was approved by the institutional review board of the University Hospital of Ulm, and all patients and volunteers gave informed consent according to the Declaration of Helsinki. Seven of the 10 normal BMMC samples and the 3 normal BM samples that were used for isolating HSCs, CMPs, GMPs, and MEPs were purchased from AllCells.
Flow cytometry and sorting of human hematopoietic stem and progenitor cells. Human HSCs, CMPs, GMPs, and MEPs were isolated from BM as reported previously (73, 79). Briefly, BMMCs were enriched for CD34+ cells using immunomagnetic beads (Miltenyi Biotec). CD34+ cells were then stained with TRI-COLOR–conjugated antibodies against lineage antigens (CD2, CD3, CD4, CD7, CD8, CD10, CD11b, CD14, CD19, CD20, CD56, CD235a), APC-conjugated anti-CD34, biotinylated anti-CD38 followed by streptavidin-APC-Cy7, FITC-conjugated anti-CD45RA, PE-conjugated anti-CD123, and propidium iodide. Viable HSCs (Lin–CD34+CD38–), CMPs (Lin–CD34+CD38+CD123lowCD45RA–), GMPs (Lin–CD34+CD38+CD123lowCD45RA+), and MEPs (Lin–CD34+CD38+CD123–CD45RA–) were sorted using a BD FACSAria cell sorter (BD Biosciences) (Supplemental Figure 7).
Cell lines and cell culture. The human myeloid leukemia cell lines CMK, EOL-1, HEL, HL-60, K-562, Kasumi-1, KG-1, MOLM-14, MONO-MAC-6, MV4-11, NB4, NOMO-1, SKM-1, THP-1, and U-937 were obtained from the German Collection of Microorganisms and Cell Cultures (DSMZ), ATCC, or the Fujisaki Cell Center and were maintained under standard conditions. For growth curves, the number of viable cells was determined with the CellTiter 96 AQueous One Solution Proliferation Assay (Promega) or by trypan blue exclusion.
DNA isolation, RNA isolation, and cDNA synthesis. Genomic DNA was isolated using the DNA zol reagent (Invitrogen). Total RNA from BMMCs and PBMCs or leukemia cell lines was isolated using the RNeasy Mini Kit (QIAGEN) and reverse transcribed (2 μg in a reaction volume of 30 μl) using the TaqMan Gold RT-PCR Kit (Applied Biosystems). Total RNA from sorted stem and progenitor cells was isolated according to the RNeasy Micro protocol (QIAGEN) and analyzed directly by RQ-PCR using the TaqMan One-Step RT-PCR Master Mix (Applied Biosystems).
Quantification of CDX2 expression. Quantification of CDX2 expression was performed by RQ-PCR with primers CDX2 RQ-F (5'-GCGGAACCTGTGCGAGTG-3') and CDX2 RQ-R1 (5'-GACTGTAGTGAAACTCCTTCTCCAGC-3') and a 6-carboxy-fluorescein–labeled probe (5'-FAM-CCGGCGCAGCAGTCCCTCG-3'). For normalization, the porphobilinogen deaminase (PBGD) gene was used according to the following formula: CDX2 copy number / PBGD copy number × 105. CDX2 and PBGD expression levels in patient samples and cell lines were determined by absolute mRNA quantification using plasmid standard curves. Plasmids were generated by cloning PCR products into the pCR4-TOPO vector (Invitrogen). The efficiency of RNAi-mediated knockdown of CDX2 expression was determined by relative mRNA quantification using the ΔδCt method. Reactions were run in duplicate with 2.5 μl cDNA in a total reaction volume of 25 μl using an ABI Prism 7700 Sequence Detection System (Applied Biosystems).
Allelic expression analysis. Partial amplification of the CDX2 gene from cDNA (2 μl) and direct sequencing of amplification products were performed using primers CDX2 cDNA-F (5'-CGGAGGAAAGCCGAGCTA-3') and CDX2 cDNA-R (5'-CTTTGGCTTCCGCAGTGTAA-3'). Amplification products were also cloned into the pCR4-TOPO vector (Invitrogen), and cloned fragments were sequenced with M13 forward and reverse primers.
Sequence analysis of the CDX2 promoter. The location of the CDX2 promoter was predicted using the Gene2Promoter software program (Genomatix). Amplification of the putative promoter sequence from genomic DNA (20 ng) and direct sequencing of amplification products were performed with primers CDX2 promoter 1-F (5'-GTGGTAACCGCCGTAGTCC-3'), CDX2 promoter 1-R (5'-GAGGGGTTGTGCGTAGAGTG-3'), CDX2 promoter 2-F (5'-CACAGACACCAATGGTTGGA-3'), and CDX2 promoter 2-R (5'-GCCTGAATCTAGGATCATAATTTGT-3').
Sodium bisulfite sequencing. Bisulfite treatment of genomic DNA (1 μg) was performed using the CpGenome Fast DNA Modification Kit (Chemicon International). Partial amplification of the CpG island in the CDX2 promoter region from bisulfite-modified DNA (50 ng) was performed using primers CDX2 bDNA-F (5'-GAAGTTTTTAATTATTGGTGTTTGTGTT-3') and CDX2 bDNA-R (5'-AAACCTCACCATACTACCTAAAAACC-3') (31). Amplification products were cloned into the pCR4-TOPO vector (Invitrogen), and cloned fragments were sequenced with M13 forward and reverse primers.
Sequence analysis of the CDX2 coding region. Amplification of CDX2 exons from genomic DNA (20 ng) was performed using primers CDX2 exon 1-F (5'-AGGCCCCCGAATTTGTCT-3'), CDX2 exon 1-R (5'-CACTCGTTAATCACGGAAGG-3'), CDX2 exon 2-F (5'-CCCTGCAGCCAGATTTTCTA-3'), CDX2 exon 2-R (5'-GACTGATGGGCTGCCTTG-3'), CDX2 exon 3-F (5'-GAATTTTCCAAATCAGCTTGC-3'), and CDX2 exon 3-R (5'-GCTGGAGCAGAAGATGTTGAT-3'). Amplification products were sequenced with primers CDX2 exon 1-F, CDX2 exon 1-S1 (5'-GTGGTGCGGATGGTAGTCTG-3'), CDX2 exon 1-S2 (5'-CACGGAGCTAGGGTACATGC-3'), CDX2 exon 2-F, CDX2 exon 2-R, CDX2 exon 3-F, CDX2 exon 3-S1 (5'-GCTCTGGTGACAGGCTCTCT-3'), and CDX2 exon 3-S2 (5'-GGAGGCTTTCTGTCTCCTCA-3').
Measurement of CDX2 gene copy number. CDX2 gene copy numbers were measured by RQ-PCR with primers CDX2 RQ-F and CDX2 RQ-R2 (5'-CCAAGCACCCTCCGAAGG-3') and the probe that was used for quantification of CDX2 expression. The coagulation factor VIII gene was used as a reference locus as previously described (80). Reactions were run in duplicate with 50 ng genomic DNA in a total reaction volume of 25 μl using an ABI Prism 7700 Sequence Detection System (Applied Biosystems).
RNAi-mediated silencing of CDX2 expression. Three pLKO.1-based lentiviral vectors that contain stem-loop cassettes encoding shRNAs targeted to the 3' untranslated region (TRCN13683; nt 1,822–1,842) or the coding sequence (TRCN13684 and TRCN13685; nt 882–902 and 732–752, respectively) of the human CDX2 mRNA (GenBank accession number NM_001265) were obtained from the MISSION TRC-Hs 1.0 (Human) shRNA library (81) through Sigma-Aldrich. The pLKO.1 construct contains a human U6 promoter that drives expression of the stem-loop cassette and the puromycin resistance gene cloned 3' of the human phosphoglycerate kinase promoter (82). The oligonucleotide sequences of the shRNAs were as follows (21-nt stem sequences matching the target transcript are underlined; noncomplementary 6-nt loop sequences are italicized): MISSION shRNA TRCN13683, 5'-CCGGGCTGAATGTATGTCAGTGCTACTCGAGTAGCACTGACATACATTCAGCTTTTT-3'; TRCN13684, 5'-CCGGCCGCAGAGCAAAGGAGAGGAACTCGAGTTCCTCTCCTTTGCTCTGCGGTTTTT-3'; TRCN13685, 5'-CCG GAGACAAATATCGAGTGTGTACTCGAGTACACCACTCGATATTTGTCTTTTTT-3'. A pLKO.1 construct without an shRNA sequence was a gift from William C. Hahn (Dana-Farber Cancer Institute, Boston, Massachusetts, USA).
VSV-G–pseudotyped lentiviral particles were produced by cotransfection of 293T cells with pLKO.1 constructs and the compatible packaging plasmids pMD.G and pCMVΔR8.91 (83–85). Transfections were carried out using FuGENE 6 (Roche Diagnostics), and virus was harvested at 48 and 72 hours after transfection.
Lentiviral supernatants were used to transduce cells in the presence of 8 μg/ml Polybrene (American Bioanalytical), and infected cells were selected with 2 μg/ml puromycin (Sigma-Aldrich).
Immunoprecipitation and Western blotting. Immunoprecipitation was performed as described previously (86) using monoclonal anti-CDX2 (BioGenex). Immunoprecipitated proteins were separated by electrophoresis and transferred to nitrocellulose membranes for Western blotting with anti-CDX2 using standard procedures.
Colony-forming assays. Leukemia cell lines (1 × 103 to 1 × 104 cells) were plated in methylcellulose medium (MethoCult H4236; StemCell Technologies), and colonies were counted after 10 days.
DNA constructs, retrovirus production, and infection of primary murine hematopoietic cells. The Cdx2 cDNA (GenBank accession number NM_007673) was cloned into the MSCV-IRES-GFP and MSCV-PGK-neo retroviral vectors, and full-length protein expression was documented by Western blotting. Generation of retroviral supernatants and infection of primary murine hematopoietic cells were performed as described previously (87). For demonstration of proviral integration, genomic DNA prepared from spleen cells of leukemic mice was digested with EcoRI and subjected to Southern blot analysis according to standard protocols.
Serial replating assays and generation of IL-3–dependent myeloid cell lines. Replating experiments were performed as described previously (33). Briefly, either whole BM was isolated from 5-fluorouracil–treated BALB/c mice or stem and progenitor cell populations were double sorted from C57BL/6 mice as previously described (33) using a BD FACSAria cell sorter (BD Biosciences). Cell populations were transduced with MSCV-Cdx2-PGK-neo or MSCV-PGK-neo. Transduced cells (1 × 104) were plated in methylcellulose medium (MethoCult M3434; StemCell Technologies) and selected for G418-resistant cells in the first plating. Colonies were counted after 5–7 days, and secondary, tertiary, and quaternary colony formation were analyzed by replating 1 × 104 cells obtained by harvesting primary, secondary, or tertiary cultures, respectively. Excess cells from the fourth plating were continuously propagated in RPMI-1640 supplemented with 20% FBS and 10 μg/ml IL-3 (R&D Systems) as described previously (33).
Murine BM transplantation assay. Transplantation experiments were performed as described previously (33, 88). For primary transplants, 1 × 106 BM cells isolated from BALB/c mice were transduced with MSCV-Cdx2-IRES-GFP or MSCV-IRES-GFP and injected into lethally irradiated (2 × 450 cGy) syngeneic recipient mice. For secondary transplants, 1 × 106 BM cells from primary leukemic animals were injected into sublethally irradiated (450 cGy) recipient mice. Diseased animals were sacrificed, spleen and liver weights and wbc counts were recorded, and histopathologic and flow cytometric analyses of murine tissues were performed as described previously (86, 87). Expression of Cdx2 in leukemic mice was confirmed by detection of Cdx2 mRNA in spleen cells by RQ-PCR (Figure 6B). Approval for the use of animals in this study was granted by the Children’s Hospital Boston Animal Care and Use Committee under protocol number A04-03-029.
Quantification of Hox gene expression in primary murine hematopoietic cells. For quantification of Hox gene expression in vitro, primary BM cells from C57BL/6 mice were enriched for immature hematopoietic progenitors (c-Kit+Lin–) and transduced with MSCV-Cdx2-IRES-GFP or MSCV-IRES-GFP. After 48 hours, GFP+ cells were sorted, and total RNA was isolated and cDNA prepared. For quantification of Hox gene expression in vivo, spleen cells were obtained from secondary BM transplant recipients, and total RNA was isolated and cDNA prepared. Expression levels of Hoxa6, Hoxa7, Hoxa9, Hoxa10, Hoxb3, Hoxb6, Hoxb8, and Hoxc6 were determined by RQ-PCR as described previously (89).
Statistics. Survival analysis was performed using the Kaplan-Meier method. Differences between survival distributions were analyzed using the log-rank test. The difference in CDX2 expression levels between cytogenetic subgroups was evaluated using the Kruskal-Wallis test. All other statistical analyses were performed using the unpaired Student’s t test. A P value of less than 0.05 was considered significant. Statistical computations were performed using GraphPad Prism version 4.0c (GraphPad Software).
-
Acknowledgments
We thank the members of the German-Austrian AML Study Group for providing leukemia specimens. We are also grateful to Elizabeth McDowell, Sandra Moore, Rachel Okabe, and Christa Wieland for technical support; to James Cunningham, Ulrich Steidl, and Daniel Tenen for help with lentiviral experiments; and to Thomas Kindler for help with flow cytometry and cell sorting. This study was supported by NIH grants CA66996 and CA105423 (to D.G. Gilliland) and the Leukemia and Lymphoma Society. C. Scholl and S. Fröhling are supported by grants from the Deutsche Forschungsgemeinschaft. D. Bansal is the recipient of a Special Fellow Award from the Leukemia and Lymphoma Society. B.J.P. Huntly is a Medical Research Council (UK) Senior Clinical Fellow. D.G. Gilliland is a Doris Duke Distinguished Clinical Scientist and an Investigator of the Howard Hughes Medical Institute.
Address correspondence to: Stefan Fröhling, Karp Family Research Laboratories, 1 Blackfan Circle, 5th Floor, Boston, Massachusetts 02115, USA. Phone: (617) 355-9085; Fax: (617) 355-9093; E-mail: [email protected].
-
Footnotes
Nonstandard abbreviations used: aCGH, array-based comparative genomic hybridization; AML, acute myeloid leukemia; BMMC, BM mononuclear cell; CML, chronic myeloid leukemia; CMP, common myeloid progenitor; CR, complete remission; GMP, granulocyte-macrophage progenitor; MDS, myelodysplastic syndrome; MEP, megakaryocyte-erythroid progenitor; PB, peripheral blood; RAEB, refractory anemia with excess blasts; RNAi, RNA interference; RQ-PCR, real-time quantitative PCR; shRNA, short hairpin RNA; T-ALL, T cell acute lymphoblastic leukemia; TSS, transcription start site.
Conflict of interest: The authors have declared that no conflict of interest exists.
Reference information: J. Clin. Invest.117:1037–1048 (2007). doi:10.1172/JCI30182
Brian J.P. Huntly’s present address is: Department of Haematology, Cambridge Institute for Medical Research, University of Cambridge, Cambridge, United Kingdom.
Claudia Scholl and Dimple Bansal contributed equally to this work.
See the related article at HOX deregulation in acute myeloid leukemia.
-
References
- Abramovich, C., Humphries, R.K. 2005. Hox regulation of normal and leukemic hematopoietic stem cells. Curr. Opin. Hematol. 12:210-216. View this article via: PubMed Google Scholar
- Deschamps, J., et al. 1999. Initiation, establishment and maintenance of Hox gene expression patterns in the mouse. Int. J. Dev. Biol. 43:635-650. View this article via: PubMed Google Scholar
- Isaacs, H.V., Pownall, M.E., Slack, J.M. 1998. Regulation of Hox gene expression and posterior development by the Xenopus caudal homologue Xcad3. EMBO J. 17:3413-3427. View this article via: PubMed Google Scholar
- van den Akker, E., et al. 2002. Cdx1 and Cdx2 have overlapping functions in anteroposterior patterning and posterior axis elongation. Development. 129:2181-2193. View this article via: PubMed Google Scholar
- Charite, J., et al. 1998. Transducing positional information to the Hox genes: critical interaction of cdx gene products with position-sensitive regulatory elements. Development. 125:4349-4358. View this article via: PubMed Google Scholar
- Chawengsaksophak, K., de Graaff, W., Rossant, J., Deschamps, J., Beck, F. 2004. Cdx2 is essential for axial elongation in mouse development. Proc. Natl. Acad. Sci. U. S. A. 101:7641-7645. View this article via: PubMed Google Scholar
- Subramanian, V., Meyer, B.I., Gruss, P. 1995. Disruption of the murine homeobox gene Cdx1 affects axial skeletal identities by altering the mesodermal expression domains of Hox genes. Cell. 83:641-653. View this article via: PubMed Google Scholar
- Deb, K., Sivaguru, M., Yong, H.Y., Roberts, R.M. 2006. Cdx2 gene expression and trophectoderm lineage specification in mouse embryos. Science. 311:992-996. View this article via: PubMed Google Scholar
- Niwa, H., et al. 2005. Interaction between Oct3/4 and Cdx2 determines trophectoderm differentiation. Cell. 123:917-929. View this article via: PubMed Google Scholar
- Strumpf, D., et al. 2005. Cdx2 is required for correct cell fate specification and differentiation of trophectoderm in the mouse blastocyst. Development. 132:2093-2102. View this article via: PubMed Google Scholar
- James, R., Erler, T., Kazenwadel, J. 1994. Structure of the murine homeobox gene cdx-2. Expression in embryonic and adult intestinal epithelium. J. Biol. Chem. 269:15229-15237. View this article via: PubMed Google Scholar
- Silberg, D.G., Swain, G.P., Suh, E.R., Traber, P.G. 2000. Cdx1 and cdx2 expression during intestinal development. Gastroenterology. 119:961-971. View this article via: PubMed Google Scholar
- Aoki, K., Tamai, Y., Horiike, S., Oshima, M., Taketo, M.M. 2003. Colonic polyposis caused by mTOR-mediated chromosomal instability in Apc+/Delta716 Cdx2+/– compound mutant mice. Nat. Genet. 35:323-330. View this article via: PubMed Google Scholar
- Chawengsaksophak, K., James, R., Hammond, V.E., Kontgen, F., Beck, F. 1997. Homeosis and intestinal tumours in Cdx2 mutant mice. Nature. 386:84-87. View this article via: PubMed Google Scholar
- da Costa, L.T., et al. 1999. CDX2 is mutated in a colorectal cancer with normal APC/beta-catenin signaling. Oncogene. 18:5010-5014. View this article via: PubMed Google Scholar
- Hinoi, T., Loda, M., Fearon, E.R. 2003. Silencing of CDX2 expression in colon cancer via a dominant repression pathway. J. Biol. Chem. 278:44608-44616. View this article via: PubMed Google Scholar
- Hinoi, T., et al. 2001. Loss of CDX2 expression and microsatellite instability are prominent features of large cell minimally differentiated carcinomas of the colon. Am. J. Pathol. 159:2239-2248. View this article via: PubMed Google Scholar
- Wicking, C., et al. 1998. CDX2, a human homologue of Drosophila caudal, is mutated in both alleles in a replication error positive colorectal cancer. Oncogene. 17:657-659. View this article via: PubMed Google Scholar
- Almeida, R., et al. 2003. Expression of intestine-specific transcription factors, CDX1 and CDX2, in intestinal metaplasia and gastric carcinomas. J. Pathol. 199:36-40. View this article via: PubMed Google Scholar
- Eda, A., et al. 2003. Aberrant expression of CDX2 in Barrett’s epithelium and inflammatory esophageal mucosa. J. Gastroenterol. 38:14-22. View this article via: PubMed Google Scholar
- Marchetti, M., Caliot, E., Pringault, E. 2003. Chronic acid exposure leads to activation of the cdx2 intestinal homeobox gene in a long-term culture of mouse esophageal keratinocytes. J. Cell Sci. 116:1429-1436. View this article via: PubMed Google Scholar
- Silberg, D.G., et al. 2002. Cdx2 ectopic expression induces gastric intestinal metaplasia in transgenic mice. Gastroenterology. 122:689-696. View this article via: PubMed Google Scholar
- Chase, A., et al. 1999. Fusion of ETV6 to the caudal-related homeobox gene CDX2 in acute myeloid leukemia with the t(12;13)(p13;q12). Blood. 93:1025-1031. View this article via: PubMed Google Scholar
- Rawat, V.P., et al. 2004. Ectopic expression of the homeobox gene Cdx2 is the transforming event in a mouse model of t(12;13)(p13;q12) acute myeloid leukemia. Proc. Natl. Acad. Sci. U. S. A. 101:817-822. View this article via: PubMed Google Scholar
- Rücker, F.G., et al. 2006. Disclosure of candidate genes in acute myeloid leukemia with complex karyotypes using microarray-based molecular characterization. J. Clin. Oncol. 24:3887-3894. View this article via: PubMed Google Scholar
- Akiyama, Y., Maesawa, C., Ogasawara, S., Terashima, M., Masuda, T. 2003. Cell-type-specific repression of the maspin gene is disrupted frequently by demethylation at the promoter region in gastric intestinal metaplasia and cancer cells. Am. J. Pathol. 163:1911-1919. View this article via: PubMed Google Scholar
- De Smet, C., Lurquin, C., Lethe, B., Martelange, V., Boon, T. 1999. DNA methylation is the primary silencing mechanism for a set of germ line- and tumor-specific genes with a CpG-rich promoter. Mol. Cell. Biol. 19:7327-7335. View this article via: PubMed Google Scholar
- Gupta, A., Godwin, A.K., Vanderveer, L., Lu, A., Liu, J. 2003. Hypomethylation of the synuclein gamma gene CpG island promotes its aberrant expression in breast carcinoma and ovarian carcinoma. Cancer Res. 63:664-673. View this article via: PubMed Google Scholar
- Nakamura, N., Takenaga, K. 1998. Hypomethylation of the metastasis-associated S100A4 gene correlates with gene activation in human colon adenocarcinoma cell lines. Clin. Exp. Metastasis. 16:471-479. View this article via: PubMed Google Scholar
- Wong, N.A., et al. 2005. CDX1 is an important molecular mediator of Barrett’s metaplasia. Proc. Natl. Acad. Sci. U. S. A. 102:7565-7570. View this article via: PubMed Google Scholar
- Yuasa, Y., et al. 2005. Relationship between CDX2 gene methylation and dietary factors in gastric cancer patients. Carcinogenesis. 26:193-200. View this article via: PubMed Google Scholar
- Cozzio, A., et al. 2003. Similar MLL-associated leukemias arising from self-renewing stem cells and short-lived myeloid progenitors. Genes Dev. 17:3029-3035. View this article via: PubMed Google Scholar
- Huntly, B.J., et al. 2004. MOZ-TIF2, but not BCR-ABL, confers properties of leukemic stem cells to committed murine hematopoietic progenitors. Cancer Cell. 6:587-596. View this article via: PubMed Google Scholar
- Krivtsov, A.V., et al. 2006. Transformation from committed progenitor to leukaemia stem cell initiated by MLL-AF9. Nature. 442:818-822. View this article via: PubMed Google Scholar
- Lavau, C., Szilvassy, S.J., Slany, R., Cleary, M.L. 1997. Immortalization and leukemic transformation of a myelomonocytic precursor by retrovirally transduced HRX-ENL. EMBO J. 16:4226-4237. View this article via: PubMed Google Scholar
- Kogan, S.C., et al. 2002. Bethesda proposals for classification of nonlymphoid hematopoietic neoplasms in mice. Blood. 100:238-245. View this article via: PubMed Google Scholar
- Harris, N.L., et al. 1999. World Health Organization classification of neoplastic diseases of the hematopoietic and lymphoid tissues: report of the Clinical Advisory Committee meeting-Airlie House, Virginia, November 1997. J. Clin. Oncol. 17:3835-3849. View this article via: PubMed Google Scholar
- Tabaries, S., et al. 2005. Cdx protein interaction with Hoxa5 regulatory sequences contributes to Hoxa5 regional expression along the axial skeleton. Mol. Cell. Biol. 25:1389-1401. View this article via: PubMed Google Scholar
- Taylor, J.K., Levy, T., Suh, E.R., Traber, P.G. 1997. Activation of enhancer elements by the homeobox gene Cdx2 is cell line specific. Nucleic Acids Res. 25:2293-2300. View this article via: PubMed Google Scholar
- Bash, R.O., et al. 1995. Does activation of the TAL1 gene occur in a majority of patients with T-cell acute lymphoblastic leukemia? A pediatric oncology group study. Blood. 86:666-676. View this article via: PubMed Google Scholar
- Leroy-Viard, K., Vinit, M.A., Lecointe, N., Mathieu-Mahul, D., Romeo, P.H. 1994. Distinct DNase-I hypersensitive sites are associated with TAL-1 transcription in erythroid and T-cell lines. Blood. 84:3819-3827. View this article via: PubMed Google Scholar
- Ernst, P., Wang, J., Korsmeyer, S.J. 2002. The role of MLL in hematopoiesis and leukemia. Curr. Opin. Hematol. 9:282-287. View this article via: PubMed Google Scholar
- Borrow, J., et al. 1996. The t(7;11)(p15;p15) translocation in acute myeloid leukaemia fuses the genes for nucleoporin NUP98 and class I homeoprotein HOXA9. Nat. Genet. 12:159-167. View this article via: PubMed Google Scholar
- Nakamura, T., et al. 1996. Fusion of the nucleoporin gene NUP98 to HOXA9 by the chromosome translocation t(7;11)(p15;p15) in human myeloid leukaemia. Nat. Genet. 12:154-158. View this article via: PubMed Google Scholar
- Raza-Egilmez, S.Z., et al. 1998. NUP98-HOXD13 gene fusion in therapy-related acute myelogenous leukemia. Cancer Res. 58:4269-4273. View this article via: PubMed Google Scholar
- Ayton, P.M., Cleary, M.L. 2003. Transformation of myeloid progenitors by MLL oncoproteins is dependent on Hoxa7 and Hoxa9. Genes Dev. 17:2298-2307. View this article via: PubMed Google Scholar
- Kumar, A.R., et al. 2004. Hoxa9 influences the phenotype but not the incidence of Mll-AF9 fusion gene leukemia. Blood. 103:1823-1828. View this article via: PubMed Google Scholar
- So, C.W., Karsunky, H., Wong, P., Weissman, I.L., Cleary, M.L. 2004. Leukemic transformation of hematopoietic progenitors by MLL-GAS7 in the absence of Hoxa7 or Hoxa9. Blood. 103:3192-3199. View this article via: PubMed Google Scholar
- Wang, J., et al. 2005. Conditional MLL-CBP targets GMP and models therapy-related myeloproliferative disease. EMBO J. 24:368-381. View this article via: PubMed Google Scholar
- Zeisig, B.B., et al. 2004. Hoxa9 and Meis1 are key targets for MLL-ENL-mediated cellular immortalization. Mol. Cell. Biol. 24:617-628. View this article via: PubMed Google Scholar
- Kroon, E., Thorsteinsdottir, U., Mayotte, N., Nakamura, T., Sauvageau, G. 2001. NUP98-HOXA9 expression in hemopoietic stem cells induces chronic and acute myeloid leukemias in mice. EMBO J. 20:350-361. View this article via: PubMed Google Scholar
- Pineault, N., et al. 2003. Induction of acute myeloid leukemia in mice by the human leukemia-specific fusion gene NUP98-HOXD13 in concert with Meis1. Blood. 101:4529-4538. View this article via: PubMed Google Scholar
- Afonja, O., et al. 2000. MEIS1 and HOXA7 genes in human acute myeloid leukemia. Leuk. Res. 24:849-855. View this article via: PubMed Google Scholar
- Bullinger, L., et al. 2004. Use of gene-expression profiling to identify prognostic subclasses in adult acute myeloid leukemia. N. Engl. J. Med. 350:1605-1616. View this article via: PubMed Google Scholar
- Debernardi, S., et al. 2003. Genome-wide analysis of acute myeloid leukemia with normal karyotype reveals a unique pattern of homeobox gene expression distinct from those with translocation-mediated fusion events. Genes Chromosomes Cancer. 37:149-158. View this article via: PubMed Google Scholar
- Drabkin, H.A., et al. 2002. Quantitative HOX expression in chromosomally defined subsets of acute myelogenous leukemia. Leukemia. 16:186-195. View this article via: PubMed Google Scholar
- Golub, T.R., et al. 1999. Molecular classification of cancer: class discovery and class prediction by gene expression monitoring. Science. 286:531-537. View this article via: PubMed Google Scholar
- Kawagoe, H., Humphries, R.K., Blair, A., Sutherland, H.J., Hogge, D.E. 1999. Expression of HOX genes, HOX cofactors, and MLL in phenotypically and functionally defined subpopulations of leukemic and normal human hematopoietic cells. Leukemia. 13:687-698. View this article via: PubMed Google Scholar
- Lawrence, H.J., et al. 1999. Frequent co-expression of the HOXA9 and MEIS1 homeobox genes in human myeloid leukemias. Leukemia. 13:1993-1999. View this article via: PubMed Google Scholar
- Roche, J., et al. 2004. Hox expression in AML identifies a distinct subset of patients with intermediate cytogenetics. Leukemia. 18:1059-1063. View this article via: PubMed Google Scholar
- Rozovskaia, T., et al. 2003. Expression profiles of acute lymphoblastic and myeloblastic leukemias with ALL-1 rearrangements. Proc. Natl. Acad. Sci. U. S. A. 100:7853-7858. View this article via: PubMed Google Scholar
- Krishnaraju, K., Hoffman, B., Liebermann, D.A. 1997. Lineage-specific regulation of hematopoiesis by HOX-B8 (HOX-2.4): inhibition of granulocytic differentiation and potentiation of monocytic differentiation. Blood. 90:1840-1849. View this article via: PubMed Google Scholar
- Perkins, A., Kongsuwan, K., Visvader, J., Adams, J.M., Cory, S. 1990. Homeobox gene expression plus autocrine growth factor production elicits myeloid leukemia. Proc. Natl. Acad. Sci. U. S. A. 87:8398-8402. View this article via: PubMed Google Scholar
- Perkins, A.C., Cory, S. 1993. Conditional immortalization of mouse myelomonocytic, megakaryocytic and mast cell progenitors by the Hox-2.4 homeobox gene. EMBO J. 12:3835-3846. View this article via: PubMed Google Scholar
- Fischbach, N.A., et al. 2005. HOXB6 overexpression in murine bone marrow immortalizes a myelomonocytic precursor in vitro and causes hematopoietic stem cell expansion and acute myeloid leukemia in vivo. Blood. 105:1456-1466. View this article via: PubMed Google Scholar
- Bonnet, D. 2005. Normal and leukaemic stem cells. Br. J. Haematol. 130:469-479. View this article via: PubMed Google Scholar
- Huntly, B.J., Gilliland, D.G. 2005. Leukaemia stem cells and the evolution of cancer-stem-cell research. Nat. Rev. Cancer. 5:311-321. View this article via: PubMed Google Scholar
- Warner, J.K., Wang, J.C., Hope, K.J., Jin, L., Dick, J.E. 2004. Concepts of human leukemic development. Oncogene. 23:7164-7177. View this article via: PubMed Google Scholar
- Hope, K.J., Jin, L., Dick, J.E. 2004. Acute myeloid leukemia originates from a hierarchy of leukemic stem cell classes that differ in self-renewal capacity. Nat. Immunol. 5:738-743. View this article via: PubMed Google Scholar
- Ikeya, M., Takada, S. 2001. Wnt-3a is required for somite specification along the anteroposterior axis of the mouse embryo and for regulation of cdx-1 expression. Mech. Dev. 103:27-33. View this article via: PubMed Google Scholar
- Lickert, H., et al. 2000. Wnt/(beta)-catenin signaling regulates the expression of the homeobox gene Cdx1 in embryonic intestine. Development. 127:3805-3813. View this article via: PubMed Google Scholar
- Pilon, N., et al. 2006. Cdx4 is a direct target of the canonical Wnt pathway. Dev. Biol. 289:55-63. View this article via: PubMed Google Scholar
- Prinos, P., et al. 2001. Multiple pathways governing Cdx1 expression during murine development. Dev. Biol. 239:257-269. View this article via: PubMed Google Scholar
- Jamieson, C.H., et al. 2004. Granulocyte-macrophage progenitors as candidate leukemic stem cells in blast-crisis CML. N. Engl. J. Med. 351:657-667. View this article via: PubMed Google Scholar
- Muller-Tidow, C., et al. 2004. Translocation products in acute myeloid leukemia activate the Wnt signaling pathway in hematopoietic cells. Mol. Cell. Biol. 24:2890-2904. View this article via: PubMed Google Scholar
- Simon, M., Grandage, V.L., Linch, D.C., Khwaja, A. 2005. Constitutive activation of the Wnt/beta-catenin signalling pathway in acute myeloid leukaemia. Oncogene. 24:2410-2420. View this article via: PubMed Google Scholar
- Zheng, X., et al. 2004. Gamma-catenin contributes to leukemogenesis induced by AML-associated translocation products by increasing the self-renewal of very primitive progenitor cells. Blood. 103:3535-3543. View this article via: PubMed Google Scholar
- Frohling, S., et al. 2002. Comparison of cytogenetic and molecular cytogenetic detection of chromosome abnormalities in 240 consecutive adult patients with acute myeloid leukemia. J. Clin. Oncol. 20:2480-2485. View this article via: PubMed Google Scholar
- Manz, M.G., Miyamoto, T., Akashi, K., Weissman, I.L. 2002. Prospective isolation of human clonogenic common myeloid progenitors. Proc. Natl. Acad. Sci. U. S. A. 99:11872-11877. View this article via: PubMed Google Scholar
- Wilke, K., Duman, B., Horst, J. 2000. Diagnosis of haploidy and triploidy based on measurement of gene copy number by real-time PCR. Hum. Mutat. 16:431-436. View this article via: PubMed Google Scholar
- Moffat, J., et al. 2006. A lentiviral RNAi library for human and mouse genes applied to an arrayed viral high-content screen. Cell. 124:1283-1298. View this article via: PubMed Google Scholar
- Stewart, S.A., et al. 2003. Lentivirus-delivered stable gene silencing by RNAi in primary cells. RNA. 9:493-501. View this article via: PubMed Google Scholar
- Naldini, L., et al. 1996. In vivo gene delivery and stable transduction of nondividing cells by a lentiviral vector. Science. 272:263-267. View this article via: PubMed Google Scholar
- Salmon, P., et al. 2000. High-level transgene expression in human hematopoietic progenitors and differentiated blood lineages after transduction with improved lentiviral vectors. Blood. 96:3392-3398. View this article via: PubMed Google Scholar
- Zufferey, R., Nagy, D., Mandel, R.J., Naldini, L., Trono, D. 1997. Multiply attenuated lentiviral vector achieves efficient gene delivery in vivo. Nat. Biotechnol. 15:871-875. View this article via: PubMed Google Scholar
- Mercher, T., et al. 2006. JAK2T875N is a novel activating mutation that results in myeloproliferative disease with features of megakaryoblastic leukemia in a murine bone marrow transplantation model. Blood. 108:2770-2779. View this article via: PubMed Google Scholar
- Liu, Q., et al. 2000. Signal transduction and transforming properties of the TEL-TRKC fusions associated with t(12;15)(p13;q25) in congenital fibrosarcoma and acute myelogenous leukemia. EMBO J. 19:1827-1838. View this article via: PubMed Google Scholar
- Kelly, L.M., et al. 2002. FLT3 internal tandem duplication mutations associated with human acute myeloid leukemias induce myeloproliferative disease in a murine bone marrow transplant model. Blood. 99:310-318. View this article via: PubMed Google Scholar
- Ernst, P., Mabon, M., Davidson, A.J., Zon, L.I., Korsmeyer, S.J. 2004. An Mll-dependent Hox program drives hematopoietic progenitor expansion. Curr. Biol. 14:2063-2069..
- Abramovich, C., Humphries, R.K. 2005. Hox regulation of normal and leukemic hematopoietic stem cells. Curr. Opin. Hematol. 12:210-216.
-
Version history
- Version 1 (April 2, 2007): No description



Copyright © 2024 American Society for Clinical Investigation
ISSN: 0021-9738 (print), 1558-8238 (online)







