ISSN: 1449-2288International Journal of Biological Sciences
Int J Biol Sci 2020; 16(10):1678-1685. doi:10.7150/ijbs.45053 This issue Cite
Review
SARS-CoV-2: an Emerging Coronavirus that Causes a Global Threat
1. Faculty of Health Sciences, University of Macau, Macau SAR, China.
2. Institute of Translational Medicine, University of Macau, Macau SAR, China.
Abstract
An ongoing outbreak of pneumonia caused by a novel coronavirus, currently designated as the severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2), was reported recently. However, as SARS-CoV-2 is an emerging virus, we know little about it. In this review, we summarize the key events occurred during the early stage of SARS-CoV-2 outbreak, the basic characteristics of the pathogen, the signs and symptoms of the infected patients as well as the possible transmission pathways of the virus. Furthermore, we also review the current knowledge on the origin and evolution of the SARS-CoV-2. We highlight bats as the potential natural reservoir and pangolins as the possible intermediate host of the virus, but their roles are waiting for further investigation. Finally, the advances in the development of chemotherapeutic options are also briefly summarized.
Keywords: Coronavirus, Novel coronavirus, pneumonia, SARS-CoV-2, COVID-19
Introduction
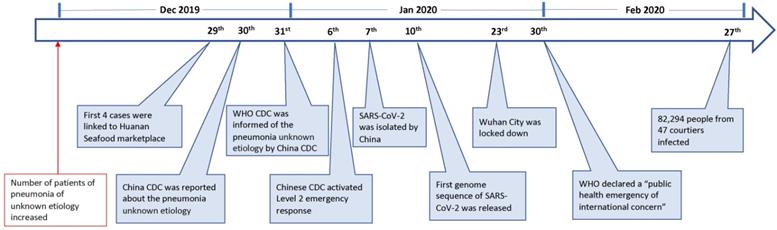
On 23 Feb 2020, the lock-down of Wuhan, a central city in China, has alarmed people all over the world of an emerging novel coronavirus that is posing a major public health and governance challenges. The novel virus, previously called the 2019-novel coronavirus (2019-nCoV), is currently designated as the severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2). As of 27 Feb, this emerging infection has been reported in 47 countries, causing over 82,294 infections with 2,804 deaths (Fig. 1) [1]. This novel virus is also becoming a mounting threat to Chinese and global economies.
Coronaviruses (CoVs) are members of the family Coronaviridae, the enveloped viruses that possess extraordinarily large single-stranded RNA genomes ranging from 26 to 32 kilobases in length [2]. CoVs have been identified in both avian hosts and various mammals, including bat, camels, dogs and masked palm civets, and are previously regarded as pathogens that only cause mild diseases in the immunocompetent people until the emergence of the coronavirus causing severe acute respiratory syndrome (SARS-CoV) in late of 2002 [3-6]. Currently, at least seven coronavirus species are known to cause diseases in humans. The viruses of 229E, OC43, NL63 and HKU1 only cause common cold symptoms, which are mild. Severe illness can be caused by the remaining three viruses, namely SARS-CoV, which resulted in the outbreak of SARS in 2002 and 2003 [3,4]; the coronaviruses that are responsible the Middle East respiratory syndrome (MERS-CoV), which emerged in 2012 and remains in the circulation in camels [7]; and SARS-CoV-2, the viruses emerged in December 2019 in Wuhan of China and a great effort is being undertaken to contain its spreading [8]. In this review, we will briefly introduce the outbreak history of SARS-CoV-2, the signs and symptoms of the infected patients, its transmission dynamics, the advances in the understanding on its evolutional origin and the chemotherapeutic options being developed for the treatment of its infection.
Key events in the early stage of SARS-CoV-2 outbreak.
The key events of SARS-CoV-2 outbreak and the pathogen characteristics
Since December 2019, an increasing number of patients with pneumonia of unknown etiology in Wuhan, a city with 11 million people, have alarmed the local hospital. On 29 December 4 cases were linked to Huanan Seafood wholesale market [9], where non-aquatic live animals, including several kinds of wild animals, were also on the sales. The local Center for Disease Control (CDC) then found additional patients linked to the same market after investigation, and reported to China CDC on 30 Dec 2019 [9]. The second day, World Health Organization (WHO) was informed of the cases of pneumonia of unknown etiology by China CDC [10]. On 6 Jan 2020, a level 2 emergency response was launched by China CDC [11].
The causal agent was not identified until 7 Jan 2020; a new type of coronavirus was isolated by Chinese authority [10]. The genome sequence of SARS-CoV-2 (WH-Human_1) was first released and shared by China on 10 Jan [12]. The isolation and identification of SARS-CoV-2 apparently facilitated the development of molecular diagnostic methods and the confirmation of the infected patients. As of 21 Jan, there are 270 cases were confirmed from Wuhan [13]. On 23 Jan, Wuhan city was locked down by local government. On 30 Jan, WHO declared a “public health emergency of international concern” (Fig. 1).
Subsequently, the viruses were successfully isolated from several laboratories [8,14,15]. The virion of SARS-CoV-2 looks like a solar corona by transmission electron microscopy imaging: the virus particle is in a spherical shape with some pleomorphism; the diameter of the virus particles range from 60 to 140 nm with distinctive spikes about 8 to 12 nm in length [8]. The observed morphology of SARS-CoV-2 is consistent with the typical characteristics of the Coronaviridae family. The genome sequence of SARS-CoV-2 from clinical samples has been obtained by several laboratories with deep sequencing [8,14-18]. The viral genome of SARS-CoV-2 is around 29.8 kilobase, with a G+C content of 38%, in total consisting of six major open reading frames (ORFs) common to coronaviruses and a number of other accessory genes [14,16]. The sequences analysis showed that the genome sequences of viruses from different patients are very conserved [14,15,19], implying that the human virus evolves recently.
Signs and symptoms of patients infected by SARS-CoV-2
A typical characteristic of the SARS-CoV-2 infected patient is pneumonia, now termed as Coronavirus Disease 2019 (COVID-19), demonstrated by computer tomographic (CT) scan or chest X -ray [3,8,18]. In the early stages, the patients showed the acute respiratory infection symptoms, with some that quickly developed acute respiratory failure and other serious complications [20]. The first three patients reported by the China Novel Coronavirus Investigating and Research Team all developed severe pneumonia and two of these three patients with available clinical profiles showed a common feature of fever and cough [8]. A subsequent investigation of a family of six patients in the University of Hong Kong-Shenzhen Hospital demonstrated that all of them had pulmonary infiltrates, with a variety of other symptoms [18]. The chest X-ray and CT imaging in a study showed that 75% of 99 patients demonstrated bilateral pneumonia and the remaining 25% unilateral pneumonia [21]. Overall, 14% of the patients showed multiple mottling and ground-glass opacity [21]. The first cases of coronavirus infection in the United States also showed basilar streaky opacities in both lungs by chest radiography. However, the pneumonia for this patient was only detected on the day 10 of his illness [22]. It is also of note that one of patients among the family of six patients did not present any other symptoms and signs, but had ground-glass lung opacities identified by CT scan [18].
Common signs and symptoms of SARS-CoV-2 infected patients from four reports
| Signs or Symptoms | Number of patients with signs or symptoms from each report | Number of patients with signs or symptoms | Total number of patients | Percentage | |||
|---|---|---|---|---|---|---|---|
| Report 1 [21] | Report 2 [23] | Report 3 [24] | Report 4 [25] | ||||
| Fever | 82 (n=99) | 40 (n=41) | 136 (n=138) | 975 (n=1099) | 1233 | 1377 | 90% |
| Cough | 81 (n=99) | 31 (n=41) | 82 (n=138) | 745 (n=1099) | 939 | 1377 | 68% |
| Sputum production/Expectoration | NR | 11 (n=39) | 37 (n=138) | 370 (n=1099) | 418 | 1276 | 33% |
| Shortness of breath/Dyspnoea | 31 (n=99) | 22 (n=40) | 43 (n=138) | 205 (n=1099) | 301 | 1376 | 22% |
| Headache | 8 (n=99) | 3 (n=38) | 9 (n=138) | 150 (n=1099) | 170 | 1374 | 12% |
| Sore throat/Pharyngalgia | 5 (n=99) | NR | 24 (n=138) | 153 (n=1099) | 182 | 1336 | 14% |
| Diarrhoea | 2 (n=99) | 1 (n=38) | 14 (n=138) | 42 (n=1099) | 59 | 1374 | 4% |
NR: Not Recorded.
At least four comprehensive studies on the epidemiological and clinical characteristics of SARS-CoV-2 infected patients have been performed [21,23-25]. The most common signs and symptoms of patients are fever and cough [21,23-25]. Fatigue was complained by 96% of patients (n=138) in one study [24], but was less outstanding (18%, n=44) in another report [23]. A combinational analysis of the common recorded signs or symptoms of the reported cases found that fever was observed in around 90% of the SARS-CoV-2 infected patients; the number of patients with cough is relatively less (68%) compared to fever (Table 1). In addition, shortness of breath or dyspnea, muscle ache, headache, chest pain, diarrhea, haemoptysis, sputum production, rhinorrhoea, nausea and vomiting, sore throat, confusion, and anorexia were also observed in a proportion of the patients [21,23-25] (Table 1).
A common feature of patients of SARS, MERS or COVID-19 is the presence of severe acute respiratory syndrome; however, the estimated fatality rate of COVID-19 (2.3%) is much lower than SARS (~10%) and MERS (~36%) [26,27]. Furthermore, the viruses responsible for above three diseases are evolutionary distinct (See below for details) [19].
Transmission of the virus
It is clear now that SARS-CoV-2 can be transmitted by human-to-human despite the majority of the early cases had contact history with the Huanan Seafood market [11,18,28]. Analysis of 425 patients with confirmed COVID-19 showed that the incubation period is 3 to 7 days. The mean was 5.2 days (95% CI: 4.1 to 7.0), and the 95th percentile of the distribution is 12.5 days (95% CI: 9.2 to 18) [11]. Notably, it was reported that the incubation period could be as long as 24 days in a rare case [25]. The basic reproductive number (R0) up to the period of 4 Jan 2020 was estimated based on the study of 425 patients to be 2.2 (meaning that one patient has been spreading infection to 2.2 other people) [11], slightly smaller than the value of 2.68 by a modelling in another [29]. The R0 of SARS-CoV-2 from both of these two studies is smaller than that of SRAS, which are 3 before public health measures were implemented [30]. However, subsequent investigation based on the analysis of high-resolution real-time human travel and infection data estimated that the R0 is much larger, ranging from 4.7 to 6.6 before the control measures [31], implying that SARS-CoV-2 is highly contagious and more infectious than initially estimated. This conclusion is consistent with the wide spread of SARS-CoV-2 within a short period time and was also echoed by the finding that SARS-CoV-2 Spike (S) protein had 10- to 20-fold higher affinity to human angiotensin-converting enzyme 2 (ACE2) receptor than that of SARS-CoV based on the Cryo-EM structure analysis of S proteins [32]. Similar to SARS-CoV, the entry of SARS-CoV-2 into host cells depends on the recognition and binding of S protein to ACE2 receptor of the host cells [14,33]. The high affinity of S protein to ACE2 receptor likely contributes to the quick spreading of virus. The finding of ACE2 as the receptor of SARS-CoV-2 also indicates that human organs with high ACE2 expression level, such as lung alveolar epithelial cells and enterocytes of the small intestine, are potentially the target of SARS-CoV-2 [34].
As a new coronavirus, it is not known yet about how SARS-CoV-2 spreads. Current knowledge for SARS-CoV-2 transmission is largely based on what is known from the similar coronaviruses, particularly SARS-CoV and MERS-CoV, in which human-to-human transmission occurs through droplets, contact and fomites. SARS-CoV is predominantly transmitted through indirect or direct contact with mucous membranes in the mouth, eyes, or nose [35]. It has been shown that unprotected eyes and exposed mucous membranes are vulnerable to SARS-CoV transmission [36]. A member of the national expert panel on pneumonia was infected by SARS-CoV-2 after the inspection in Wuhan [37]. As he wore a N95 mask but not any eye protector, and experienced eye redness before the onset of pneumonia, it was thus suspected that unprotected exposure of the eyes to SARS-CoV-2 might be another transmission pathway [37]. However, SARS-CoV-2 was not detected from the conjunctival swab sample in a confirmed COVID-19 patent with conjunctivitis [38], suggesting that more evidences are needed before concluding the conjunctival route as the transmission pathway of SARS-CoV-2. The mode of transmission by MERS-CoV is not well understood but is believed to spread largely via the respiratory close contact route [39,40].
Based on the transmission mode of SARS-CoV and MERS-CoV, a serial of preventive measures have been recommended, including avoiding close contact with people suffering from acute respiratory infections and frequent hand-washing [41]. The viruses of SARS-CoV-2 were also detected in the stool samples in some patients but not all [18,22], suggesting that a possible fecal-oral transmission occurs [42]. A systematic study showed that viruses could be detected in oral swabs, anal swabs and blood samples of the patients, and the anal swabs and blood could test positive when oral swab tested negative [43]. Furthermore, a trend of shift from more oral positive in the collected samples during the early period of patient infection to more anal positive during later period of infection was also found [43]. Therefore, a multiple shedding routes of SARS-CoV-2 might exist.
One of the challenges for preventive control of SARS-CoV-2 spreading is that the viruses are likely transmitted by asymptomatic contact. A German businessman was found infected by SARS-CoV-2 after attending a conference together with a colleague, who had no signs or symptoms of infection but had become ill due to the SARS-CoV-2 infection later [44]. This observation suggests that infected patients likely start to shed viruses before the onset of any symptom, which undoubtedly will bring great challenge to the current practice of preventive control by measuring body temperature. Despite the claim of the transmission by asymptomatic contact has been challenged [45], other asymptomatic carriers were also observed to transmit the viruses of SARS-CoV-2 [46,47]. Consistently, a study found that an asymptomatic patient had a similar vial loads in the samples of nasal and throat swabs to that of the symptomatic patients [48].
The origin and evolution of SARS-CoV-2
It is critical to identify the origin, native host(s) and evolution pathway of the virus that causes an outbreak of a pandemic. This information can help understand the molecular mechanism of its cross-species spread and implement a proper control measure to prevent it from further spreading. The association of initially confirmed SARS-CoV-2 cases with Huanan Seafood market suggested that the marketplace has played a role in the early spreading [11,23], however, whether it is the origin of the outbreak and what is the native host(s) of SARS-CoV-2 remain uncertain. In fact, the firstly documented patient was not linked to Huanan Seafood market [23].
The analysis of SARS-CoV-2 origin was firstly performed based on the genome sequence of virus isolates from six patients [19]. When compared with SARS-CoV and MERS-CoV, the nucleotide sequences of SARS-CoV-2 showed a higher homology with that of SARS-CoV while was relatively poor with that of MERS-CoV [19]. Despite some of the six major OFRs of SARS-CoV-2 genes share less than 80% identity in nucleotide acids to SARS-CoV, the seven conserved replicase domains in ORF1ab has 94.6% sequence identity in amino acids between SARS-CoV-2 and SARS-CoV [14], suggesting that these two viruses might belong to the same species. The origin of SARS-CoV has been extensively investigated. Masked palm civets were initially considered to transmit SARS-CoV to humans as a close variant of SARS-CoV was detected from palm civets [49]. This conclusion was supported by the fact that three of the four patients had the record of contact with palm civets during the two small-scale of SARS outbreaks occurred in late 2003 and early 2004 [50, 51]. However, a deep investigation based on the genome sequence of isolated viruses showed that SARS-CoV-like virus in civet had not been circulating for long [52]. Subsequently, coronaviruses with high similarity to the human SARS-CoV or civet SARS-CoV-like virus were isolated from horseshoe bats, concluding the bats as the potential natural reservoir of SARS-CoV whereas masked palm civets are the intermediate host [53-56].
It is thus reasonable to suspect that bat is the natural host of SARS-CoV-2 considering its similarity with SARS-CoV. The phylogenetic analysis of SARS-CoV-2 against a collection of coronavirus sequences from various sources found that SARS-CoV-2 belonged to the Betacoronavirus genera and was closer to SARS-like coronavirus in bat [19]. By analyzing genome sequence of SARS-CoV-2, it was found that SARS-CoV-2 felled within the subgenus Sarbecovirus of the genus Betacoronavirus and was closely related to two bat-derived SARS-like coronaviruses, bat-SL-CoVZC45 and bat-SL-CoVZXC21, but were relatively distant from SARS-CoV [15, 18, 57-59]. Meanwhile, Zhou and colleagues showed that SARS-CoV-2 had 96.2% overall genome sequence identity throughout the genome to BatCoV RaTG13, a bat coronavirus detected in Rhinolophus affinis from Yunnan province [14]. Furthermore, the phylogenetic analysis of full-length genome, the receptor binding protein spike (S) gene, and RNA-dependent RNA polymerase (RdRp) gene respectively all demonstrated that RaTG13 was the closest relative of the SARS-CoV-2 [14]. However, despite SARS-CoV-2 showed high similarity to coronavirus from bat, SARS-CoV-2 changed topological position within the subgenus Sarbecovirus when different gene was used for phylogenetic analysis: SARS-CoV-2 was closer to bat-SL-CoVZC45 in the S gene phylogeny but felled in a basal position within the subgenus Sarbecovirus in the ORF1b tree [57]. This finding implies a possible recombination event in this group of viruses. Of note, the receptor-binding domain of SARS-CoV-2 demonstrates a similar structure to that of SARS-CoV by homology modelling but a few variations in the key residues exist at amino acid level [15, 19].
Despite current evidences are pointing to the evolutional origin of SARS-CoV-2 from bat virus [15,57], an intermediate host between bats and human might exist. Lu et. al. raised four reasons for such speculation [15]: First, most bat species in Wuhan are hibernating in late December; Second, no bats in Huanan Seafood market were sold or found; Third, the sequence identity between SARS-CoV-2 and bat-SL-CoVZC45 or bat-SL-CoVZXC21, the closest relatives in their analyses, is lower than 90%; Fourth, there is an intermediate host for other human-infecting coronaviruses that origin from bat. For example, masked palm civet and dromedary camels are the intermediate hosts for SARS-CoV [49] and MERS-CoV respectively [60]. A study of the relative synonymous codon usage (RSCU) found that SARS-CoV-2, bat-SL-CoVZC45, and snakes had similar synonymous codon usage bias, and speculated that snake might be the intermediate host [61]. However, no SARS-CoV-2 has been isolated from snake yet.
Pangolin was later found to be a potential intermediate host for SARS-CoV-2. The analysis of samples from Malytan pangolins obtained during anti-smuggling operations from Guangdong and Guangxi Customs of China respectively found novel coronaviruses representing two sub-lineages related to SARS-CoV-2 [62]. The similarity of SARS-CoV-2 to these identified coronaviruses from pangolins is approximately 85.5% to 92.4% in genomes, lower than that to the bat coronavirus RaTG13 (96.2%) [14,62]. However, the receptor-binding domain of S protein from one sub-lineage of the pangolin coronaviruses shows 97.4% similarity in amino acid sequences to that of SARS-CoV-2, even higher than that to RaTG13 (89.2%) [62]. Interestingly, the pangolin coronavirus and SARS-CoV-2 share identical amino acids at the five critical residues of RBD of S protein, while RaTG13 only possesses one [62]. The discovery of coronavirus close to SARS-CoV-2 from pangolin suggests that pangolin is a potential intermediate host. However, the roles of bat and pangolin as respective natural reservoir and intermediate host still need further investigation.
Chemotherapeutic options for SARS-CoV-2 infection
As an emerging virus, there is no effective drug or vaccine approved for the treatment of SARS-CoV-2 infection yet. Currently, supportive care is provided to the patients, including oxygen therapy, antibiotic treatment, and antifungal treatment, extra-corporeal membrane oxygenation (ECMO) etc. [21,22]. To search for an antiviral drug effective in treating SARS-CoV-2 infection, Wang and colleagues evaluated seven drugs, namely, ribavirin, penciclovir, nitazoxanide, nafamostat, chloroquine, remdesivir (GS-5734) and favipiravir (T-750) against the infection of SARS-CoV-2 on Vero E6 cells in vitro [63]. Among these seven drugs, chloroquine and remdesivir demonstrated the most powerful antiviral activities with low cytotoxicity. The effective concentration (EC50) for chloroquine and remdesivir were 0.77µM and 1.13µM respectively. Chloroquine functions at both viral entry and post-entry stages of the SARS-CoV-2 infection in Vero E6 cells whereas remdesivir does at post-entry stage only. Chloroquine is a drug used for an autoimmune disease and malarial infection with potential broad-spectrum antiviral activities [64,65]. An EC90 (6.90 µM) against the SARS-CoV-2 in Vero E6 cells is clinically achievable in vivo according to a previous clinical trial [66]. Remdesivir is a drug currently under the development for Ebola virus infection and is effective to a broad range of viruses including SARS-CoV and MERS-CoV [67,68]. Functioning as an adenosine analogue targeting RdRp, Remdesivir can result in premature termination during the virus transcription [69,70]. The EC90 of remdesivir against SARS-CoV-2 in Vero E6 cells is 1.76 µM, which is achievable in vivo based on a trial in nonhuman primate experiment [63,69]. Encouragingly, in the first case of SARS-CoV-2 infection in the United States, treatment with remdesivir was provided intravenously to the patient on the day 7 without any adverse events observed. The patient's clinical condition was improved on day 8 and the previous bilateral lower-lobe rales disappeared, implying the remdesivir might be effective to the treatment of SARS-CoV-2 infection [22]. This result, however, should be interpreted with caution as this is only single case study and a proper trial control was lacking. In addition, baricitinib, a Janus kinase inhibitor, was also predicted to reduce the ability of virus to infect lung cell by an analysis of BenevolentAI [71].
Currently, chloroquine and remdesivir are under phase 3 clinical trial and open-label trial for treatment of SARS-CoV-2 infection respectively (Table 2) [72]. Preliminary results showed that chloroquine phosphate had apparent efficacy in treatment of COVID-19 [73]. However, caution must be taken during clinical use of chloroquine as its overdose is highly fatal without known antidote [74]. Despite the lack of documented in vitro data supporting the antiviral efficacy on SARS-CoV-2, several antiviral chemotherapeutic agents have been registered for the clinical trials for the treatment of COVID-19 (Table 2) [72].
Conclusion remarks
SARS-CoV-2 is an emerging pathogen, without any effective drug available for treatment at the moment. It spreads quickly and can result in death of the infected patients. Despite the current mortality rate is 2.3% [26], the emergence of large number of infected patients within short period of time could result in the collapse of health care system, and thus the mortality rate might be elevated. Effective preventive measures must be implemented to control it from global spreading. In addition, great effort should be made on the development of vaccine and antiviral drugs. Meanwhile, the intermediate host and the molecular mechanism of its cross-species spread should be further investigated. Legislation should be employed to prohibit the trade of wild animals, the potential intermediate host(s) of various viruses, to prevent the outbreak of this and other novel viruses in future.
Acknowledgements
We acknowledge the Research Committee of the University of Macau (Grant No.: MYRG2016-00073-FHS, MYRG2016-00199-FHS and MYRG2019-000050-FHS) and the Macau Science and Technology Development Fund (Grant No.: FDCT/0058/2018/A2 and FDCT/0113/2019/A2) for providing financial support for this research.
Summary of chemotherapeutic drugs under clinical trial for COVID-19
| Name of Drug | Target and Mode of Action in other Viruses | In Vitro Antiviral Activity to SARS-CoV-2 | Clinical Trial Status for COVID-19 | Clinical Trial Registration Number |
|---|---|---|---|---|
| Remdesivir (GS-5734) | Inhibits RdRp [70] | Tested [63] | Phase 3 | NCT04252664; NCT04257656 |
| Favipiravir | Inhibits RdRp [75] | Tested [63] | Randomized trial | ChiCTR2000029544; ChiCTR2000029600 |
| Ribavirin | Inhibits viral RNA synthesis and mRNA capping [76] | Tested [63] | Randomized trial, incombination a pegylated interferon | ChiCTR2000029387 |
| Lopinavir | Inhibits 3C like protease (3Clpro) [77] | Not tested | Phase 3 | NCT04252274; NCT04251871; NCT04255017; ChiCTR2000029539 |
| Ritonavir | Inhibits 3Clpro [77] | Not tested | Phase 3 | NCT04251871; NCT04255017; NCT04261270 |
| Darunavir and Cobicistat | Inhibits HIV protease [78] | Not tested | Phase 3 | NCT04252274 |
| ASC09F (HIV protease inhibitor) | Inhibits HIV protease [79] | Not tested | Phase 3, in combination with oseltamivir | NCT04261270 |
| Chloroquine | A lysosomatropic base that appears to disrupt intracellular trafficking and viral fusion events [80] | Tested [63] | Open-label trial | ChiCTR2000030054; ChiCTR2000029939; ChiCTR2000029935; ChiCTR2000029899; ChiCTR2000029898; ChiCTR2000029837; ChiCTR2000029803; ChiCTR2000029761; ChiCTR2000029740; ChiCTR2000029559; ChiCTR2000029542; ChiCTR2000029868; ChiCTR2000029826; ChiCTR2000029762; ChiCTR2000029760; ChiCTR2000029609 |
| Arbidol (Umifenovir) | Block viral fusion [81] | Not tested | Phase 4 | NCT04260594; NCT04254874; NCT04255017 |
| Oseltamivir | Inhibit neuaminidase [82] | Not tested | Phase 3 and Phase 4 | NCT04255017; NCT04261270 |
Competing Interests
The authors have declared that no competing interest exists.
References
1. WHO. Geneva, Switzerland. Coronavirus disease. 2019 (COVID-19) Situation Report - 38. https://www.who.int/docs/default-source/coronaviruse/situation-reports/20200227-sitrep-38-covid-19.pdf?sfvrsn=9f98940c_2 (Accessed on 28th Feb 2020). 2020
2. Su S, Wong G, Shi W, Liu J, Lai ACK, Zhou J. et al. Epidemiology, Genetic Recombination, and Pathogenesis of Coronaviruses. Trends Microbiol. 2016;24:490-502
3. Zhong NS, Zheng BJ, Li YM, Poon, Xie ZH, Chan KH. et al. Epidemiology and cause of severe acute respiratory syndrome (SARS) in Guangdong, People's Republic of China, in February, 2003. Lancet. 2003;362:1353-8
4. Drosten C, Gunther S, Preiser W, van der Werf S, Brodt HR, Becker S. et al. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N Engl J Med. 2003;348:1967-76
5. Ksiazek TG, Erdman D, Goldsmith CS, Zaki SR, Peret T, Emery S. et al. A novel coronavirus associated with severe acute respiratory syndrome. N Engl J Med. 2003;348:1953-66
6. Fouchier RA, Kuiken T, Schutten M, van Amerongen G, van Doornum GJ, van den Hoogen BG. et al. Aetiology: Koch's postulates fulfilled for SARS virus. Nature. 2003;423:240
7. Zaki AM, van Boheemen S, Bestebroer TM, Osterhaus AD, Fouchier RA. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N Engl J Med. 2012;367:1814-20
8. Zhu N, Zhang D, Wang W, Li X, Yang B, Song J. et al. A Novel Coronavirus from Patients with Pneumonia in China, 2019. N Engl J Med. 2020 [Epub ahead of print]
9. The 2019-nCoV Outbreak Joint Field Epidemiology Investigation Team QL. An Outbreak of NCIP (2019-nCoV) Infection in China— Wuhan, Hubei Province, 2019-2020. China CDC Weekly. 2020;2:79-80
10. WHO. Geneva, Switzerland. Novel Coronavirus (2019-nCoV) situation report-1. https://www.who.int/docs/default-source/coronaviruse/situation-reports/20200121-sitrep-1-2019-ncov.pdf?sfvrsn=20a99c10_4 (Accessed on 28 Feb 2020). 2020
11. Li Q, Guan X, Wu P, Wang X, Zhou L, Tong Y. et al. Early Transmission Dynamics in Wuhan, China, of Novel Coronavirus-Infected Pneumonia. N Engl J Med. 2020 [Epub ahead of print]
12. Initial genome release of novel coronavirus. http://virological.org/t/initial-genome-release-of-novel-coronavirus/319?from=groupmessage. (Accessed on 18 Feb 2020). 2020
13. WHO. Geneva, Switzerland. Novel Coronavirus (2019-nCoV), Situation report - 2. https://www.who.int/docs/default-source/coronaviruse/situation-reports/20200122-sitrep-2-2019-ncov.pdf?sfvrsn=4d5bcbca_2 (Accessed on 28 Feb 2020). 2020
14. Zhou P, Yang XL, Wang XG, Hu B, Zhang L, Zhang W. et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020 [Epub ahead of print]
15. Lu R, Zhao X, Li J, Niu P, Yang B, Wu H. et al. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 2020;395:565-74
16. Chan JF, Kok KH, Zhu Z, Chu H, To KK, Yuan S. et al. Genomic characterization of the 2019 novel human-pathogenic coronavirus isolated from a patient with atypical pneumonia after visiting Wuhan. Emerg Microbes Infect. 2020;9:221-36
17. Chen L, Liu W, Zhang Q, Xu K, Ye G, Wu W. et al. RNA based mNGS approach identifies a novel human coronavirus from two individual pneumonia cases in 2019 Wuhan outbreak. Emerg Microbes Infect. 2020;9:313-9
18. Chan JF, Yuan S, Kok KH, To KK, Chu H, Yang J. et al. A familial cluster of pneumonia associated with the 2019 novel coronavirus indicating person-to-person transmission: a study of a family cluster. Lancet. 2020;395:514-23
19. Xu X, Chen P, Wang J, Feng J, Zhou H, Li X. et al. Evolution of the novel coronavirus from the ongoing Wuhan outbreak and modeling of its spike protein for risk of human transmission. Sci China Life Sci. 2020 [Epub ahead of print]
20. WHO. Geneva, Switzerland. Clinical management of severe acute respiratory infection when Novel coronavirus (nCoV) infection is suspected: interim guidance. https://www.who.int/publications-detail/clinical-management-of-severe-acute-respiratory-infection-when-novel-coronavirus-(ncov)-infection-is-suspected (Accessed on 28 Feb 2020). 2020
21. Chen N, Zhou M, Dong X, Qu J, Gong F, Han Y. et al. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. Lancet. 2020;395:507-13
22. Holshue ML, DeBolt C, Lindquist S, Lofy KH, Wiesman J, Bruce H. et al. First Case of 2019 Novel Coronavirus in the United States. N Engl J Med. 2020 [Epub ahead of print]
23. Huang C, Wang Y, Li X, Ren L, Zhao J, Hu Y. et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497-506
24. Wang D, Hu B, Hu C, Zhu F, Liu X, Zhang J. et al. Clinical Characteristics of 138 Hospitalized Patients With 2019 Novel Coronavirus-Infected Pneumonia in Wuhan, China. JAMA. 2020 [Epub ahead of print]
25. Guan WJ, Ni ZY, Hu Y, Liang WH, Ou CQ, He JX. et al. Clinical characteristics of coronavirus disease 2019 in China. N Engl J Med. 2020 [Epub ahead of print]
26. Wu Z, McGoogan JM. Characteristics of and Important Lessons From the Coronavirus Disease 2019 (COVID-19) Outbreak in China: Summary of a Report of 72314 Cases From the Chinese Center for Disease Control and Prevention. JAMA. 2020 [Epub ahead of print]
27. de Wit E, van Doremalen N, Falzarano D, Munster VJ. SARS and MERS: recent insights into emerging coronaviruses. Nat Rev Microbiol. 2016;14:523-34
28. Riou J, Althaus CL. Pattern of early human-to-human transmission of Wuhan 2019 novel coronavirus (2019-nCoV), December 2019 to January 2020. Euro Surveill. 2020:25
29. Wu JT, Leung K, Leung GM. Nowcasting and forecasting the potential domestic and international spread of the 2019-nCoV outbreak originating in Wuhan, China: a modelling study. Lancet. 2020 [Epub ahead of print]
30. Bauch CT, Lloyd-Smith JO, Coffee MP, Galvani AP. Dynamically modeling SARS and other newly emerging respiratory illnesses: past, present, and future. Epidemiology. 2005;16:791-801
31. Sanche S, Lin YT, Xu C, Romero-Severson E, Hengartner N, Ke R. The Novel Coronavirus, 2019-nCoV, is Highly Contagious and More Infectious Than Initially Estimated. medRxiv. 2020 doi: https://doi.org/10.1101/2020.02.07.20021154
32. Wrapp D, Wang N, Corbett KS, Goldsmith JA, Hsieh CL, Abiona O. et al. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020 [Epub ahead of print]
33. Hoffmann M, Kleine-Weber H, Krüger N, Müller M, Drosten C, Pöhlmann S. The novel coronavirus 2019 (2019-nCoV) uses the SARS-coronavirus receptor ACE2 and the cellular protease TMPRSS2 for entry into target cells. bioRxiv. doi: https://doi.org/10.1101/2020.01.31.929042.
34. Zou X, Chen K, Zou J, Han P, Hao J, Z. H. The single-cell RNA-seq data analysis on the receptor ACE2 expression reveals the potential risk of different human organs vulnerable to Wuhan 2019-nCoV infection. Front Med. 2020 [Epub ahead of print]
35. Peiris JS, Yuen KY, Osterhaus AD, Stohr K. The severe acute respiratory syndrome. N Engl J Med. 2003;349:2431-41
36. Belser JA, Rota PA, Tumpey TM. Ocular tropism of respiratory viruses. Microbiol Mol Biol Rev. 2013;77:144-56
37. Cheng-wei Lu X-fL, Zhi-fang Jia. 2019-nCoV transmission through the ocular surface must not be ignored. The Lancet. 2020;36:30313-5
38. Zhou Y, Zeng Y, Tong Y, Chen C. Ophthalmologic evidence against the interpersonal transmission of 2019 novel coronavirus through conjunctiva. medRxiv. 2020 doi: https://doi.org/10.1101/2020.02.11.20021956
39. Zumla A, Hui DS, Perlman S. Middle East respiratory syndrome. Lancet. 2015;386:995-1007
40. Hijawi B, Abdallat M, Sayaydeh A, Alqasrawi S, Haddadin A, Jaarour N. et al. Novel coronavirus infections in Jordan, April 2012: epidemiological findings from a retrospective investigation. East Mediterr Health J. 2013;19(Suppl 1):S12-8
41. Organization WH. Novel Coronavirus(2019-nCoV) Situation Report - 19. 2020.
42. Yeo C, Kaushal S, Yeo D. Enteric involvement of coronaviruses: is faecal-oral transmission of SARS-CoV-2 possible? Lancet Gastroenterol Hepatol. 2020 [Epub ahead of print]
43. Zhang W, Du RH, Li B, Zheng XS, Yang XL, Hu B. et al. Molecular and serological investigation of 2019-nCoV infected patients: implication of multiple shedding routes. Emerg Microbes Infect. 2020;9:386-9
44. Rothe C, Schunk M, Sothmann P, Bretzel G, Froeschl G, Wallrauch C. et al. Transmission of 2019-nCoV Infection from an Asymptomatic Contact in Germany. N Engl J Med. 2020 [Epub ahead of print]
45. Kupferschmidt K. Study claiming new coronavirus can be transmitted by people without symptoms was flawed. Science. 2020 https://www.sciencemag.org/news/2020/02/paper-non-symptomatic-patient-transmitting-coronavirus-wrong
46. Bai Y, Yao L, Wei T, Tian F, Jin DY, Chen L. et al. Presumed Asymptomatic Carrier Transmission of COVID-19. JAMA. 2020 [Epub ahead of print]
47. Hoehl S, Berger A, Kortenbusch M, Cinatl J, Bojkova D, Rabenau H. et al. Evidence of SARS-CoV-2 Infection in Returning Travelers from Wuhan, China. N Engl J Med. 2020 [Epub ahead of print]
48. Zou L, Ruan F, Huang M, Liang L, Huang H, Hong Z. et al. SARS-CoV-2 Viral Load in Upper Respiratory Specimens of Infected Patients. N Engl J Med. 2020 [Epub ahead of print]
49. Guan Y, Zheng BJ, He YQ, Liu XL, Zhuang ZX, Cheung CL. et al. Isolation and characterization of viruses related to the SARS coronavirus from animals in southern China. Science. 2003;302:276-8
50. Wang M, Yan M, Xu H, Liang W, Kan B, Zheng B. et al. SARS-CoV infection in a restaurant from palm civet. Emerg Infect Dis. 2005;11:1860-5
51. Song HD, Tu CC, Zhang GW, Wang SY, Zheng K, Lei LC. et al. Cross-host evolution of severe acute respiratory syndrome coronavirus in palm civet and human. Proc Natl Acad Sci U S A. 2005;102:2430-5
52. Shi Z, Hu Z. A review of studies on animal reservoirs of the SARS coronavirus. Virus Res. 2008;133:74-87
53. Li W, Shi Z, Yu M, Ren W, Smith C, Epstein JH. et al. Bats are natural reservoirs of SARS-like coronaviruses. Science. 2005;310:676-9
54. Lau SK, Woo PC, Li KS, Huang Y, Tsoi HW, Wong BH. et al. Severe acute respiratory syndrome coronavirus-like virus in Chinese horseshoe bats. Proc Natl Acad Sci U S A. 2005;102:14040-5
55. Ge XY, Li JL, Yang XL, Chmura AA, Zhu G, Epstein JH. et al. Isolation and characterization of a bat SARS-like coronavirus that uses the ACE2 receptor. Nature. 2013;503:535-8
56. Hu B, Zeng LP, Yang XL, Ge XY, Zhang W, Li B. et al. Discovery of a rich gene pool of bat SARS-related coronaviruses provides new insights into the origin of SARS coronavirus. PLoS Pathog. 2017;13:e1006698
57. Wu F, Zhao S, Yu B, Chen YM, Wang W, Song ZG. et al. A new coronavirus associated with human respiratory disease in China. Nature. 2020 [Epub ahead of print]
58. Jiang S, Du L, Shi Z. An emerging coronavirus causing pneumonia outbreak in Wuhan, China: calling for developing therapeutic and prophylactic strategies. Emerg Microbes Infect. 2020;9:275-7
59. Wan Y, Shang J, Graham R, Baric RS, Li F. Receptor recognition by novel coronavirus from Wuhan: An analysis based on decade-long structural studies of SARS. J Virol. 2020 [Epub ahead of print]
60. Alagaili AN, Briese T, Mishra N, Kapoor V, Sameroff SC, Burbelo PD. et al. Middle East respiratory syndrome coronavirus infection in dromedary camels in Saudi Arabia. mBio. 2014;5:e00884-14
61. Ji W, Wang W, Zhao X, Zai J, Li X. Homologous recombination within the spike glycoprotein of the newly identified coronavirus may boost cross-species transmission from snake to human. Journal of Medical Virology. 2020 [Epub ahead of print]
62. Lam TTY, Shum MHH, Zhu HC, Tong YG, Ni XB, Liao YS. et al. Identification of 2019-nCoV related coronaviruses in Malayan pangolins in southern China. bioRxiv. doi: https://doi.org/10.1101/2020.02.13.945485.
63. Wang M, Cao R, Zhang L, Yang X, Liu J, Xu M. et al. Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) in vitro. Cell Res. 2020 [Epub ahead of print]
64. Savarino A, Di Trani L, Donatelli I, Cauda R, Cassone A. New insights into the antiviral effects of chloroquine. Lancet Infect Dis. 2006;6:67-9
65. Yan Y, Zou Z, Sun Y, Li X, Xu KF, Wei Y. et al. Anti-malaria drug chloroquine is highly effective in treating avian influenza A H5N1 virus infection in an animal model. Cell Res. 2013;23:300-2
66. Mackenzie AH. Dose refinements in long-term therapy of rheumatoid arthritis with antimalarials. Am J Med. 1983;75:40-5
67. Sheahan TP, Sims AC, Graham RL, Menachery VD, Gralinski LE, Case JB. et al. Broad-spectrum antiviral GS-5734 inhibits both epidemic and zoonotic coronaviruses. Sci Transl Med. 2017:9
68. Mulangu S, Dodd LE, Davey RT Jr, Tshiani Mbaya O, Proschan M, Mukadi D. et al. A Randomized, Controlled Trial of Ebola Virus Disease Therapeutics. 2019; 381: 2293-303.
69. Warren TK, Jordan R, Lo MK, Ray AS, Mackman RL, Soloveva V. et al. Therapeutic efficacy of the small molecule GS-5734 against Ebola virus in rhesus monkeys. Nature. 2016;531:381-5
70. Agostini ML, Andres EL, Sims AC, Graham RL, Sheahan TP, Lu X. et al. Coronavirus Susceptibility to the Antiviral Remdesivir (GS-5734) Is Mediated by the Viral Polymerase and the Proofreading Exoribonuclease. mBio. 2018:9
71. Richardson P. G, I, Tucker, C, Smith, D, Oechsle, O, Phelan, A, Stebbing, J. Baricitinib as Potential Treatment for 2019-nCoV Acute Respiratory Disease. The Lancet. 2020 [Epub ahead of print]
72. Li G, Clercq, E. D. Therapeutic options for the 2019 novel coronavirus (2019-nCoV). Nature Reviews Drug Discovery. 2020 doi: 10.1038/d41573-020-00016-0
73. Gao J, Tian Z, Yang X. Breakthrough: Chloroquine phosphate has shown apparent efficacy in treatment of COVID-19 associated pneumonia in clinical studies. Biosci Trends. 2020 [Epub ahead of print]
74. Don Michael TA, Aiwazzadeh S. The effects of acute chloroquine poisoning with special reference to the heart. Am Heart J. 1970;79:831-42
75. Furuta Y, Takahashi K, Kuno-Maekawa M, Sangawa H, Uehara S, Kozaki K. et al. Mechanism of action of T-705 against influenza virus. Antimicrob Agents Chemother. 2005;49:981-6
76. Graci JD, Cameron CE. Mechanisms of action of ribavirin against distinct viruses. Rev Med Virol. 2006;16:37-48
77. Nukoolkarn V, Lee VS, Malaisree M, Aruksakulwong O, Hannongbua S. Molecular dynamic simulations analysis of ritonavir and lopinavir as SARS-CoV 3CL(pro) inhibitors. J Theor Biol. 2008;254:861-7
78. McCoy C. Darunavir: a nonpeptidic antiretroviral protease inhibitor. Clin Ther. 2007;29:1559-76
79. Dierynck I, Van Marck H, Van Ginderen M, Jonckers TH, Nalam MN, Schiffer CA. et al. TMC310911, a novel human immunodeficiency virus type 1 protease inhibitor, shows in vitro an improved resistance profile and higher genetic barrier to resistance compared with current protease inhibitors. Antimicrob Agents Chemother. 2011;55:5723-31
80. Delvecchio R, Higa LM, Pezzuto P, Valadao AL, Garcez PP, Monteiro FL. et al. Chloroquine, an Endocytosis Blocking Agent, Inhibits Zika Virus Infection in Different Cell Models. Viruses. 2016:8
81. Boriskin YS, Leneva IA, Pecheur EI, Polyak SJ. Arbidol: a broad-spectrum antiviral compound that blocks viral fusion. Curr Med Chem. 2008;15:997-1005
82. McNicholl IR, McNicholl JJ. Neuraminidase inhibitors: zanamivir and oseltamivir. Ann Pharmacother. 2001;35:57-70
Author contact
![]() Corresponding author: Jun Zheng, junzhengedu.mo.
Corresponding author: Jun Zheng, junzhengedu.mo.
Received 2020-2-18
Accepted 2020-2-28
Published 2020-3-15

 Global reach, higher impact
Global reach, higher impact