Significance
This work accurately quantified the Zn2+ content of insulin using energy dispersive spectroscopy (EDS) mapping and explored the precise position of insulin in insulin vesicles using dual-energy X-ray contrast microscopy and scanning transmission X-ray microscopy (STXM) image stacks. X-ray ptychography and equally sloped tomography (EST) were combined to perform three-dimensional (3D) structural mapping and reveal the distribution of insulin vesicles in pancreatic beta cells. The method described here fills a gap in the quantitative analysis of insulin and describes a unique approach for exploring the process of insulin secretion in pancreatic beta cells. This study describes an application of advanced X-ray imaging in the exploration of cell structures and provides an approach to the analysis of intracellular microstructures.
Keywords: X-ray diffraction imaging, insulin vesicles, dual-energy abstract X-ray imaging, Zn2+ content, STXM images stack
Abstract
Characterizing relationships between Zn2+, insulin, and insulin vesicles is of vital importance to the study of pancreatic beta cells. However, the precise content of Zn2+ and the specific location of insulin inside insulin vesicles are not clear, which hinders a thorough understanding of the insulin secretion process and diseases caused by blood sugar dysregulation. Here, we demonstrated the colocalization of Zn2+ and insulin in both single extracellular insulin vesicles and pancreatic beta cells by using an X-ray scanning coherent diffraction imaging (ptychography) technique. We also analyzed the elemental Zn2+ and Ca2+ contents of insulin vesicles using electron microscopy and energy dispersive spectroscopy (EDS) mapping. We found that the presence of Zn2+ is an important characteristic that can be used to distinguish insulin vesicles from other types of vesicles in pancreatic beta cells and that the content of Zn2+ is proportional to the size of insulin vesicles. By using dual-energy contrast X-ray microscopy and scanning transmission X-ray microscopy (STXM) image stacks, we observed that insulin accumulates in the off-center position of extracellular insulin vesicles. Furthermore, the spatial distribution of insulin vesicles and their colocalization with other organelles inside pancreatic beta cells were demonstrated using three-dimensional (3D) imaging by combining X-ray ptychography and an equally sloped tomography (EST) algorithm. This study describes a powerful method to univocally describe the location and quantitative analysis of intracellular insulin, which will be of great significance to the study of diabetes and other blood sugar diseases.
As the main hypoglycemic hormone, insulin promotes the absorption and storage of glucose and inhibits its production. Without insulin, muscle, fat, and liver cells cannot transport glucose from the blood to the intracellular space. Failure of pancreatic beta cells to secrete sufficient amounts of insulin results in impaired blood glucose control and is a hallmark of type 2 diabetes (1). As the initial stage of secretion, glucose stimulates the fusion of insulin vesicles with the beta cell membrane. The second stage of secretion occurs when more vesicles move to locations near the membrane.
Insulin secretion is a complicated process that begins with glucose uptake and metabolism to adenosine triphosphate (ATP), resulting in the closure of ATP-sensitive K+ channels and subsequent membrane depolarization. In turn, membrane depolarization leads to the opening of voltage-dependent Ca2+ channels and an influx of extracellular Ca2+, which triggers insulin granule exocytosis (2). Multiple organelles are involved in insulin secretion. Research into the morphology (3) and interrelationship of organelles in pancreatic beta cells has increased gradually over the last few decades (4). Cryo-electron microscopy (EM) tomography and superresolution fluorescence imaging are playing an increasingly important role in unveiling cellular structures (5, 6). However, these methods cannot simultaneously achieve large-size whole-cell imaging and high resolution. EM requires sample sectioning, which includes embedding or chemical fixation, and these processes limit the number of cells and conditions that can be studied in practice. Cryo-electron tomography has made it possible to perform three-dimensional (3D) imaging and structural analyses of specific areas of cells. However, whole-cell quantification remains a challenge. Superresolution fluorescence microscopy has opened up the possibility of examining 3D cells and tissues with higher resolution and has enabled capture of the dynamics of cellular structures. Nevertheless, the use of fluorescent probes limits the number and types of molecules that can be imaged at one time. Overall, the limitations of the aforementioned methods hinder their ability to image specific organelles. In particular, examining the specific location of insulin in pancreatic beta cells under various conditions using traditional methods is challenging, which hinders our understanding of the dynamic processes of insulin synthesis, storage, and secretion. On the other hand, soft X-ray tomography enables mapping and quantification of the distribution of 3D organelles with high spatial accuracy, minimal cell manipulation, and no need for fluorescence probes (7, 8). However, the technique does not allow mapping or quantification of single elements.
Pancreatic beta cells contain two types of vesicles/granules: insulin-containing, large dense-core vesicles and synaptic-like micro vesicles that resemble neuronal secretory vesicles (9, 10). The production and properly timed secretion of insulin vesicles are the most critical functions of pancreatic cells, with loss or dysregulation leading to diabetes mellitus. Insulin vesicles exist in immature and mature forms, which differ in their distribution, location, size, and structure. Inside secretory vesicles, two Zn2+ ions coordinate six insulin monomers to form a hexameric structure on which mature insulin crystals are based (11). Indeed, Zn2+ plays a clear role in the synthesis, storage, and secretion of insulin (12–15). However, data regarding the specific content of Zn2+ in insulin aggregates are lacking. In addition, the relationship between the content of Zn2+ and the size of insulin vesicles and life cycle of pancreatic beta cells is still unclear. Previous analyses of Zn2+ have used fluorescent labeling and estimations of fluorescence intensity, which cannot accurately reveal the Zn2+ content of insulin aggregates (14). As mentioned above, Ca2+ also plays an important role in the secretion of insulin; the regulation of Ca2+-dependent extracellular excretion is an important mechanism controlling the release of insulin secretory granules into the pancreatic interstitium (2). Therefore, quantitative analyses of Ca2+ in pancreatic beta cells is crucial for full characterization of insulin vesicles and insulin secretion.
Here, we used X-ray coherence diffraction imaging (ptychography) to explore the synthesis and secretion pathways of insulin vesicles, locate the position of insulin aggregates in secretory vesicles, and accurately quantify the content of Zn2+ in the insulin aggregates. Ptychography uses coherent monochromatic light to image biological samples and reconstructs the data with high resolution by using an iterative algorithm. The specific location of insulin aggregates in insulin vesicles was determined using dual-energy contrast microscopy and scanning transmission X-ray microscopy (STXM) imaging stacks, and whole-cell high-resolution two-dimensional (2D) and 3D imaging of pancreatic beta cells was performed. In addition, EM imaging was used to explore the Zn2+ and Ca2+ contents of insulin aggregates further, and the distribution of insulin vesicles was verified by the distribution of Zn2+. The data presented herein provide insights into pancreatic beta cells and an important reference point for exploring the insulin secretion pathway and interaction of organelles during the secretion process.
Results
The Content of Zn2+ in Insulin Vesicles.
Insulin vesicles were extracted from INS-1E cells and nonimmortalized beta cells via ultracentrifugation (16, 17), and then an enzyme-linked immunosorbent assay was performed to examine the insulin contents of the different fractions. Fraction 9 had the highest insulin content in duplicate experiments (SI Appendix, Fig. S1). Optical microscopy revealed that the vesicles in fraction 9 were uniform in shape and their size was consistent with that of insulin vesicles. Therefore, fraction 9 was processed to extract isolated insulin vesicles.
Fig. 1 A and B shows scanning EM (SEM) images of insulin vesicles extracted from INS-1E cells and the corresponding energy dispersive spectrometry (EDS) mapping of Zn2+, respectively. As a common component analysis method, EDS can analyze the content of an element within a sample but cannot distinguish between element valences. As the valence state of the element Zn is known to be 2+, EDS was used to measure the content of Zn2+ and Ca2+ in this experiment. The distribution of Zn2+ matched that of the insulin vesicles well, confirming the presence of Zn2+ in the secretory vesicles. To further investigate the location of Zn2+, EDS line scans (Fig. 1C) were performed across two insulin vesicles (marked with a red dashed line in Fig. 1A). Two obvious peaks of Zn2+ appeared at the vesicle positions, while there was no signal at other locations, confirming the enrichment of Zn2+ in the vesicles. The EDS line scan also identified Ca2+ within the vesicles. To further analyze the contents of Zn2+ and Ca2+, 22 insulin vesicles from INS-1E cells and 25 insulin vesicles from nonimmortalized beta cells were examined using EDS point scanning. The content of Zn2+ was basically proportional to the size of the insulin vesicles, but there was no obvious correlation between the content of Ca2+ and the size of the vesicles (Fig. 1 D–G). In addition, 24 insulin vesicles from Min6 cells were examined using EDS point scanning, and the results were consistent for the INS-1E cells and nonimmortalized beta cells (SI Appendix, Fig. S2 C and D). In addition, EDS mapping in scanning transmission electron microscopy was carried out on Min6 cells and nonimmortalized beta cells (SI Appendix, Fig. S2 A and B); Zn signal and Ca signal were obvious in both types of cells.
Fig. 1.
Element statistics of Zn2+ and Ca2+. (A) SEM image of insulin vesicles. Scale bar, 2 μm. (B) EDS mapping of Zn2+ corresponding to the area in A. Scale bar, 2 μm. (C) The line scan spectrum of the two insulin vesicles marked with the red dotted line in A. (D and E) The statistical analysis results of the Zn2+ content and Ca2+ content in INS-1E cells, respectively. Standard error. (F and G) The statistical analysis results of the Zn2+ content and Ca2+ content in nonimmortalized beta cells, respectively.
After insulin is synthesized in the endoplasmic reticulum, it is processed into a biologically active form and stored in secretory vesicles (0.3-µm diameter), which are transported extracellularly. Crystallization of insulin in vesicles is a hot topic in the study of islet beta cells; however, it has not yet been observed directly. Here, high-resolution transmission EM (TEM) was used to image cross-sections of pancreatic beta cells. TEM imaging of INS-1E cells revealed lattice fringes of insulin crystals in a secretory vesicle. As shown in Fig. 2A, two distinct diffraction spots were observed after the Fourier transformation, and the plane spacing of insulin crystals was calculated to be 4.22 nm according to the diffraction images. In Fig. 2B, the red line marks the junction of two INS-1E cells and the red arrows indicate the insulin vesicles. In addition, TEM imaging of nonimmortalized beta cells was performed (Fig. 2C). The number of insulin vesicles and distance between the insulin vesicles and the cell membrane were measured in both cell types (SI Appendix, Fig. S3). As shown in Fig. 2D, a large proportion of vesicles were located 0.1 to 0.5 μm from the plasma membrane.
Fig. 2.
The crystallization of insulin aggregates and the distance between insulin vesicles and cell membrane. (A) High-resolution TEM image of insulin vesicle and insulin crystal (dashed box). Lattice stripes and interplanar spacing (yellow arrows in the right top image) of insulin crystals were calculated by the Fourier transform of TEM image. The right bottom image is the Fourier transform image of insulin crystal within dashed box, and the red circles are two distince diffraction spots. Scale bar, 200 nm. (B) TEM image of junction of two INS-1E cells. The distance between the vesicles and the plasma membrane is counted (statistical INS-1E cells include but are not limited to Fig. 1B), and the statistical results are shown in D with percentage error. Scale bar, 1 μm. The red line shows the cell boundary, and the red arrow points to insulin vesicles. (C) TEM image of junction of two nonimmortalized beta cells. The distance between the vesicles and the plasma membrane is counted (statistical nonimmortalized beta cells include but are not limited to Fig. 1C), and the statistical results are shown in D with percentage error. Scale bar, 2 μm. The red line shows the cell boundary, and the red arrow points to insulin vesicles.
Examining the Location of Zn2+ Accumulation in Vesicles Using STXM and Dual-Energy Contrast X-ray Microscopy.
The existence of Zn2+ in insulin vesicles revealed by the EM experiments described above is consistent with previous studies (15). Although the EM experiments revealed the relationships between the element contents and sizes of insulin vesicles, due to the low content (∼1%) of Zn2+ they were unable to determine the precise specific location of Zn2+-bound insulin in secretory vesicles. Therefore, X-ray absorption spectroscopy (XAS) based on STXM image stacks (18) and dual-energy contrast X-ray microscopy (19, 20) were used to explore the location of insulin aggregates within the insulin vesicles.
A set of STXM images (i.e., stack) was obtained using photon energies ranging from 1,010 to 1,090 eV (Fig. 3A). Various energy intervals were used within different energy ranges together with relatively larger image scanning steps to improve data collection efficiency. The energy interval between 1,018 and 1,060 eV (which is close to the Zn2+ edge) was 0.3 eV, and the interval for the rest of the energy range was 1 eV. The scanning step size of the STXM images was 130 nm. The STXM stack dataset was integrated and imported into the open-source software aXis2000, and a principal component analysis–cluster analysis (PCA-CA) of the insulin vesicles was performed. First, hot spots and glitches within the STXM images were recognized and removed, and then all of the images were aligned using the auto alignment function in aXis2000. For each image, the regions of interest within the insulin vesicle and a blank area were selected manually, and the intensities of these two regions were integrated to calculate the absorption ratio corresponding to the specific photon energy. In this way, the XAS spectra of specific areas could be calculated. The quantitative XAS spectra of two regions within the insulin vesicles are shown in Fig. 3 C and D. An obvious absorption peak was identified at 1,020.8 eV, which is very close to the standard Zn2+ L3-edge (1,021 eV), indicating the existence of Zn2+ within the surveyed areas (Fig. 3B). Because Zn2+ is an important component of insulin, it can be regarded as an indicator of insulin aggregates within secretory vesicles.
Fig. 3.
XAS of insulin vesicles based on STXM image stacks at Zn2+ edge. (A) Schematic diagram of the quantitative XAS analysis at Zn2+ edge based on STXM stacks. Scale bar, 200 nm. (B) Schematic diagram of the location of insulin vesicles and insulin aggregations. (C and D) The quantitative XAS spectra of two regions within the insulin vesicles.
The dual-energy contrast X-ray microscopy technique is based on the abrupt change in absorption when the energy of the incident X-rays changes from being just below the absorption edge of a specific element to being above it (Fig. 4 A and B). The observed differences between images collected at the two energies reflect the locations of element-specific nanomaterials. This method can simultaneously image the ultrastructures of cells and intracellular nanomaterials and thus can be used to investigate the interactions between nanomaterials and biological systems. Consequently, dual-energy contrast X-ray microscopy can theoretically be used to survey the spatial distribution of Zn2+, which is used as an indicator of the location of insulin aggregates within insulin vesicles. However, the Zn2+ content of insulin vesicles (∼1%) is too low to be detected by conventional dual-energy contrast X-ray microscopy based on STXM or other absorption contrast microscopy. Ptychography can determine the complex-valued refractive index by differentiating between the phase of radiation that has passed through a sample and the absorption of the sample. For biological samples, the phase cross-section is 1,000 times higher than the absorption cross-section over a broad range of X-ray energy (21, 22); consequently, the ptychography phase result has a high signal-to-noise ratio and outstanding resolving power. Here, we used dual-energy contrast X-ray microscopy based on absorption images obtained via phase-to-absorption Kramers–Kronig transformation (KKT) (23–26) of the ptychography phase results to survey the spatial distribution of insulin aggregates within secretory vesicles.
Fig. 4.
Distribution of insulin aggregates in the insulin vesicle obtained by dual-energy imaging technique. (A) Standard absorption edge of Zn2+. (B) Schematic diagram of STXM technique and a set of projections at energies below and above the Zn2+ -edge that were acquired by ptychography. (C) Ptychography image of insulin vesicle above the Zn2+ -edge. (D) Elemental distribution mapping by ptychography images of insulin vesicles obtained at energies below and above the Zn2+ -edge. (E) The difference between two ptychography images of insulin vesicles obtained at two energies below the Zn2+ -edge. Scale bar of C–E, 200 nm. a.u, arbitrary units.
As mentioned above, the XAS spectra of the insulin vesicles indicated that the absorption edge of Zn2+ in beta cells is around 1,020.8 eV (Fig. 3 C and D). Therefore, we acquired ptychography datasets at 20 different X-ray photon energies across the Zn2+ L3-edge, from 1,018 eV to 1,028 eV. With the collected ptychography dataset, the phase projections with 20.8-nm resolution at all photon energies were reconstructed and transformed into absorption projections via KKT. Fig. 4C shows a representative transformed absorption projection of an insulin vesicle. Dual-energy contrast X-ray microscopy was then conducted using two transformed absorption projections at photon energies below (1,018 eV) and above (1,020.8 eV) the zinc L3-edge. As shown in Fig. 4D, the Zn2+ signal was concentrated in the upper left corner of the insulin vesicle examined, indicating that insulin is aggregated at this location, which is consistent with the STXM stack result. To verify the accuracy of the ptychography-based dual-energy contrast X-ray microscopy method, we performed a dual-energy contrast microscopy analysis of two randomly selected photon energies below the Zn2+ L3-edge. As shown in SI Appendix, Fig. S4, no obvious abrupt change in absorption was observed. Additional results of the feasibility analysis of ptychography-based dual-energy contrast X-ray microscopy are provided in the supplemental material.
Comparison of Insulin Vesicles and Ordinary Vesicles.
The vesicles in beta cells extracted by ultracentrifugation included mature and immature insulin vesicles, as well as other types of vesicles (9). High-contrast ptychography phase images of two insulin-containing and two other vesicles were obtained to examine the morphological differences between them.
As shown in Fig. 5 A–D, these analyses revealed that insulin vesicles have a clear membrane structure and a concentrated high-density area formed by the accumulation of insulin, while the other vesicles have fuzzy boundaries and relatively dispersed high-density areas. Fig. 5 E–L shows the results of EDS mapping of Zn2+ and Ca2+ in the four vesicles. Strong Zn2+ and Ca2+ signals were detected in insulin vesicles due to insulin accumulation, whereas no corresponding signals were identified in the other vesicles. Next, we performed a horizontal line scan across the center of the four vesicles and found that the internal electron density fluctuations of the insulin vesicles and other vesicles were comparable (Fig. 5M). Normally, insulin vesicles contain areas of high density due to insulin accumulation. In order to explore the dense regions inside different vesicles, we also examined the overall phase shift values and numbers of pixels occupied by high-density areas in the two types of vesicles and found that, compared with insulin vesicles, high-density areas make up a larger proportion of the entire vesicle in other types of vesicles (Fig. 5N). In addition to the internal density information, accurate discrimination between insulin vesicles and other kinds of vesicles in beta cells would require further information regarding the nature of the elements contained.
Fig. 5.
Comparison of insulin vesicles and other vesicles. (A and B) Ptychography phase images of insulin vesicles. (C and D) Ptychography phase images of other types of vesicles. Scale bar, 200 nm. (E–H) EDS signals of Zn in the four vesicles A–D. Scale bar, 200 nm. (I–L) EDS signals of Ca in the four vesicles A–D. Scale bar, 200 nm. (M), Horizontal line scan (the white horizontal dashed line) across the center of the four vesicles. (N) The proportion of high-density area to the whole vesicle in the two types of vesicles. Percentage error.
Ptychography Imaging of Glucose-Stimulated Insulin Secretion.
Pancreatic beta cell lines are useful models to study the complex pathways regulating glucose-stimulated insulin secretion. These cells are glucose responsive over physiological ranges and are characterized by a substantial insulin content and robust metabolism-secretion coupling (27, 28). An increase in the extracellular glucose concentration leads to the induction of electrical activity within beta cells. Examining the molecular densities of organelles in whole beta cells during glucose-stimulated insulin secretion is crucial to our understanding of the functions of these cells. Therefore, we collected ptychography images of beta cells under nonstimulated and high-glucose (11 mM) conditions. As a benefit of the high-contrast property of the technique, the nucleus and insulin vesicles were easily distinguished from other cellular structures (Fig. 6 A–D). Even without staining, filamentous structures at the edge of the islet beta cell were clearly observed. The nonstimulated beta cell (Fig. 6A) contained fewer insulin vesicles than the glucose-stimulated cell (Fig. 6D), indicating that glucose stimulation promotes insulin secretion, resulting in an increase in the number of insulin vesicles in the cell.
Fig. 6.
2D ptychography images of pancreatic beta cells. (A) Ptychography image of pancreatic beta cell without stimulation. Scale bar, 5 μm. (B and C) Enlarged images of the two boxes in A. The insulin vesicles in B and C were marked with yellow circle and red circles. Scale bar, 2 μm. (D) Ptychography image of pancreatic beta cell with stimulation (11 Mm glucose), and the insulin vesicles were marked by red arrows. Scale bar, 10 μm. (E–G) PSD curve of sample diffraction patterns shows that the resolution reaches 15.5 nm. In E, the crossover point of the two dashed red lines determined the cut-off frequency of the effective signal, which is indicated by the red solid line. The resolution is determined according to the standard of a 10%∼90% (30) intensity change, and the results corresponding to F and G show that the resolution is 19 nm. a.u., arbitrary units; Freq, frequency; Reso, resolution.
To quantitatively analyze the resolution of the ptychography phase projections, we first calculated the pixel size of the reconstructed projection according to the formula d = λ/2sin(θ), where λ is the wavelength and θ is the diffraction angle. In our case, the pixel size was 6.05 nm. Fig. 6E shows the power spectral density (PSD) curve of the 2D diffraction patterns, which indicates that the diffraction signal extended to a spatial frequency of 64 μm−1 (i.e., a resolution of 15.5 nm in real space). Considering that the PSD analysis only indicates the highest spatial frequency of the diffraction signal, the achieved resolution may be lower than the theoretical resolution due to signal-to-noise ratio and other factors. Thus, line scans in real space can be used to further estimate the achieved 2D resolution of the reconstructed ptychography image (29, 30). As shown in Fig. 6 F and G, a resolution of 19 nm along the X and Y axes was achieved.
The 3D Distribution of Insulin Vesicles in Islet Beta Cells.
Previous studies have shown that mature insulin vesicles are mainly distributed at the edge of beta cells, and immature insulin vesicles are mainly distributed near the nucleus (31). To explore the 3D dispersion features of insulin vesicles in beta cells, we performed a ptychographic X-ray computed tomography (PXCT) analysis of a marginal area (), which is shown in SI Appendix, Fig. S5. This experiment was carried out using the soft X-ray spectromicroscopy beamline (BL08U1A) at the Shanghai Synchrotron Radiation Facility (SSRF). The photon energy of the light source was set to 700 eV (the corresponding wavelength is 1.77 nm). To minimize radiation damage, we used equally sloped tomography (EST) to obtain the 3D structure of a single beta cell, which provided a high-quality tomographic result from a limited number of projections. EST is an iterative method based on the pseudo-polar fast Fourier transform, with the projections collected at equally sloped projection angles; consequently, the effect of a missing wedge in limited-angle tomography and interpolation error can be mitigated. By comparing the experimental projection with the calculated projection, the quality of the reconstructed image is verified (32–34).
Twenty-two ptychography phase projections of the selected part of the cell were obtained at EST projection angles ranging from −45° to +69.44° (SI Appendix, Fig. S6). To obtain high-quality ptychography phase projections, when collecting the ptychography datasets, we set the probe diameter to 3 µm and the scanning step to 600 nm to guarantee a high linear overlap ratio of 80%. Approximately 400 diffraction patterns were collected at each projection angle, and the extended ptychographic iterative engine method was used to reconstruct the ptychography phase projection. The 3D structure of the INS-1E cell was then reconstructed by EST using the 22 ptychography phase projections. We also carried out line scans in three directions to estimate the resolution of the 3D EST reconstruction. As shown in SI Appendix, Fig. S7, the resolution in the X and Y directions (perpendicular to the optical axis) was 22.8 nm, whereas that in the Z direction (along the optical axis) was only 64.6 nm due to the low number of projection angles. At this resolution, individual insulin vesicles could be distinguished clearly, and the Amira software package was used to display and segment the reconstructed beta cells (Fig. 7A). In the segmentation results, insulin vesicles are shown in yellow and mitochondria are shown in pink. The segmentation revealed that some vesicles can acquire a different morphology, such as chains and clusters, as shown previously by Zhang et al. (31). In addition, the distances between the vesicles were variable, as was the distance between the vesicles and other organelles such as mitochondria (Fig. 7 B and C). These findings suggest that insulin synthesis, transport, and secretion are affected by adjacent organelles and the complex environment within beta cells.
Fig. 7.
3D segmentation analysis of islet β cells and the spatial distribution of insulin vesicles. (A) 3D reconstruction results of part of an INS-1E cell displayed in Amira soft package. The blue arrow indicates the direction of projection. (B) 3D volume rendering of the reconstructed islet β cells displayed in Amira. (C) The positional relationship between insulin vesicles(yellow) and mitochondria (pink). Percentage error. (D) Schematic diagram of insulin vesicles. (E) The vesicle size was counted in the 3D segmentation results.
The distribution of insulin vesicles observed in the PXCT experiments described above was consistent with the results of 2D ptychography imaging, which verified the dense nuclear structure of the insulin vesicles. This phenomenon was also observed in the TEM image of the beta cells. According to the 3D segmentation result, a complete insulin vesicle will span 5 to 10 slices, and the thickness of each slice is about 50 nm; hence, the size of insulin vesicles is ∼200 to 500 nm, which is consistent with the conventional size of these vesicles. We also used the 3D segmentation results to determine the number and size of insulin vesicles and found that most vesicles had a diameter of 500 to 600 nm (Fig. 7E). In parallel, we determined the sizes of the vesicles in the 2D ptychography and TEM images (SI Appendix, Fig. S8) and found that the results were consistent.
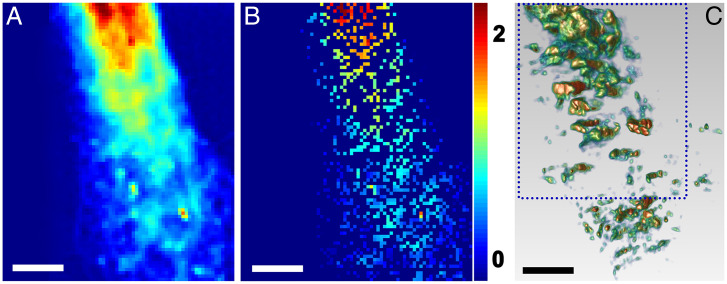
To confirm that the distribution of Zn2+ can represent the distribution of insulin vesicles in INS-1E cells, we performed fluorescence mapping (35, 36) of pancreatic beta cells using the 101D-1 soft X-ray spectromicroscopy beamline at the Canadian Light Source (CLS). As shown in Fig. 8 B and C, the distributions of Zn2+ and insulin vesicles were comparable in the fluorescence mapping image and projected 3D EST result. This finding adds support to the concept that the distribution of Zn2+ can be used to determine the precise positioning of insulin vesicles, which provides perspective for future analyses of the maturation of insulin vesicles.
Fig. 8.
Comparison of fluorescence mapping results and 3D segmentation results. (A) Ptychography image of part of islet β cell. (B and C) Fluorescence mapping result and 3D segmentation result of the same part of the islet β cell. Scale bars, 5 μm.
Discussion
As an essential element that is crucial for growth and development, Zn2+ plays a role in cell signaling for processes such as cell division and apoptosis (37). In the mammalian pancreas, where the total Zn2+ content is among the highest in the body, Zn2+ is essential for the correct processing, storage, secretion, and function of insulin in beta cells (13). Proper regulation of insulin secretion is essential for maintaining the normal homeostasis of blood glucose. In beta cells, secretory vesicles/granules are responsible for storing, transporting, and secreting insulin. In these vesicles, two Zn2+ ions coordinate six insulin monomers to form the hexameric structure on which mature insulin crystals are based (11). The relationship between costored Zn2+ and insulin is undoubtedly important to normal beta cell function. Immature insulin vesicles do not contain insulin aggregates and are located near the nucleus, whereas mature insulin vesicles are dense in the center, contain insulin aggregates, and are mainly distributed near the cell membrane.
In this study, vesicles were extracted from islet beta cells via ultracentrifugation, and various techniques were used to examine their morphology and internal structures. EDS mapping revealed that Zn2+ content increased with the size of insulin vesicles, whereas Ca2+ content had no obvious correlation with the size of the vesicles. In addition to Zn2+, Ca2+ also plays an important role in controlling the release of insulin-secreting particles into the pancreatic interstitium. In the future, we will conduct more in-depth analyses of Ca2+ using high-resolution hard X-ray imaging techniques such as ptychography-based dual-energy contrast X-ray microscopy and EST-based PXCT.
XAS based on STXM stacks and dual-energy contrast X-ray microscopy analyses showed that insulin is aggregated in off-center locations of insulin vesicles. In addition, comparisons of ptychography images of insulin vesicles and other type of beta cell vesicles revealed that the intensities and morphologies of these vesicles are similar. Therefore, aside from density and morphology, other evidence is required to distinguish between types of vesicles in beta cells. We suggest that the presence of Zn2+ could be used for this purpose. To reveal the spatial distribution of insulin vesicles and Zn2+ in beta cells, 2D whole-cell ptychography images of INS-1E cells with a resolution of 19 nm were obtained. With 22 projections at various angles, 3D reconstruction was performed using the EST algorithm to obtain the 3D structures of pancreatic beta cells and the distribution of insulin vesicles in the cells.
In summary, the collective findings of dual-energy contrast X-ray microscopy, X-ray fluorescence mapping, and 3D segmentation analyses presented here provide detailed information regarding the spatial distribution of insulin vesicles in beta cells and the distribution of insulin aggregates within these vesicles. This unique approach provides insights into pancreatic beta cells.
Materials and Methods
Cell Culture and Insulin Vesicle Isolation.
For general culture, INS-1E cells were seeded into T75 flasks at a density of 8.0 × 104 cells/cm2 and were cultured in RPMI medium supplemented with 10% fetal bovine serum (FBS), 1% penicillin/streptomycin, and 0.05 mM mercaptoethanol. The cells were grown in an incubator at 37 °C and 5% CO2. For analyses, cells at passage 44–55 were seeded at a density of 104 cells/mL into six-well plates containing Si3N4 grids treated with fibronectin. Since X-ray absorption of a 30-nm-thick Si3N4 membrane is relatively low, the effect of substrate on the imaging results could be reduced by attaching cells to this kind of membrane. After 24 h, cells that had adhered to the Si3N4 membrane were fixed with 2.5% glutaraldehyde for 15 min. Subsequently, the cells were gradually dehydrated in 30%, 50%, 70%, 80%, 90%, and then 100% ethanol for 15 min per concentration (38). Chemical fixation of cells with 2.5% glutaraldehyde has a protective effect on the internal structure and morphology of cells, as well as on the position and morphology of organelles. Moreover, the gradient dehydration method avoids contraction of cells caused by rapid water loss and ensures that cells are completely dry prior to analysis. Thus, these measures were used to prevent the samples from losing water or changing morphology due to X-ray radiation during the experiments (39, 40). Finally, well-isolated INS-1E cells were selected using an optical microscope and used for the experiments.
To obtain nonimmortalized beta cells, islets were extracted from male C57BL/6 mice (8 to 12 wk old) via collagenase digestion, as described previously (41). Subsequently, isolated islets (at least 200) were washed once with precooled phosphate-buffered saline (PBS), resuspended in 0.4 mL ice-cold Accutase cell detachment solution (Thermo Fisher Scientific; Product No. 00–4555-56), and incubated at 37 °C for 5 min with gentle shaking. Digestion was stopped by the addition of 0.8 mL ice-cold tissue culture medium (TCM), comprising RPMI 1640 with 11 mM glucose, 100 IU/mL penicillin/streptomycin, and 10% FBS. Finally, dispersed islet cells were resuspended in TCM after centrifugation for 3 min at 1,000 g and 4 °C and used for further experiments.
For the insulin vesicles isolation experiment, INS-1E cells were collected from eight T75 flasks, and nonimmortalized beta cells were obtained from islets of nude mice as mentioned above. Briefly, the cells were washed with PBS and then dispersed with 0.25% trypsin for 3 min at room temperature. Subsequently, the trypsin was diluted by adding culture medium, and the sample was centrifuged at 1,000 g for 5 min. The cell pellet was resuspended in 4 mL of isolation buffer and centrifuged for 10 min at 1,000 g, saving the supernatant. Next, the centrifugation and resuspension process was repeated, but the centrifugation speed was increased gradually from 1,000 g for 10 min to 100,000 g for 75 min. The resulting precipitate contained the insulin vesicles. The operation steps and composition of the solution used in the experiment are shown in SI Appendix, Fig. S1.
For the glucose stimulation experiment, cells were cultured under normal growth conditions but were switched to low glucose (2.8 mM) medium 24 h before the assay. In addition, 1 h before the experiment, the cells were cultured in glucose-free Krebs solution (0 mM glucose). Subsequently, 11.1 mM glucose was added, and 40 min later the cells were fixed. Preparation of the Krebs solution is shown in SI Appendix, Fig. S1.
EM Experiments.
The EDS point scan, line scan, and mapping experiments were characterized by scanning EM (SEM; JSM-7800 Plus) and TEM (JEM-2100 Plus). In the SEM experiment, the original solution containing the insulin vesicles was dropped onto clean Si3N4 membranes that had been treated with fibronectin to increase their hydrophilicity. After natural drying, the samples can be used for SEM experiments. Under a 10-kV acceleration voltage, high-resolution SEM images were obtained by adjusting the focal length, light path, and astigmatism. X-ray energy spectrum probes were then inserted, the working distance was adjusted to an appropriate height, and EDS mapping signal collection was carried out on the appropriate identified area. The Zn2+ element to be measured was selected before the collection, and the signal collection time was about 5 min.
In the TEM experiment, the solution made in the same process as the SEM experiment was dropped onto copper grids, and then the high-resolution TEM images of the insulin vesicle were obtained at 200 keV. Different from the EDS mapping experiment in the SEM, the EDS mapping experiment with TEM needed to be switched to STEM mode before it could be carried out, and the signal acquisition duration was also 5 min.
In addition, TEM images of INS-1E cell slices and nonimmortalized beta cell slices were obtained by using methods of staining and sectioning. INS-1E cells and nonimmortalized beta cells were scraped off the surface of the dish with a cell shovel after chemical fixation and gradient dehydration, and the cell cluster was embedded with epoxy resin. The embedded block was cut into 70-nm-thick slices using an ultrathin sectioning machine after 24 h of curing. The ultrathin sections were placed on copper meshes, and after double staining with uranyl acetate and lead citrate (42), the samples were then observed by 80-keV TEM.
STXM Experiments.
The STXM experiment was performed mainly on the 101D-1 soft X-ray spectromicroscopy beamline at the CLS. A Fresnel zone plate with a 240-μm diameter and a 35-nm outer zone width was used in the STXM experiment. The spatially coherent soft X-rays generated by the oscillators were focused into small focal spots by the Fresnel zone plate. The first-order diffracted light was selected and filtered to remove stray light after the focal spot passed through the order-sorting aperture. The photon energy of the light source was set to be 700 eV (corresponding wavelength is 1.77 nm). The INS-1E cells were raster scanned point by point using the 2D motion of the sample stage. X-rays passing through the sample were detected by photomultiplier tubes to form a complete image.
STXM stacks data were also measured in the same environment, with a scanned image size of µm2. Insulin vesicles PCA-CA analysis was performed on conventional STXM energy-stack datasets with the open-source software aXis2000 (McMaster University), using the number of significant components of 8, identified clusters number of 10, and angle distance measure mode (cutoff 0.01). This method identifies natural groups of spectra and then calculates the average spectra and displays the thickness maps associated with these spectra. The STXM stacks energy range is from 1,010 eV to 1,060 eV, interval 1 eV, with data analysis through aXis2000.
X-ray Scanning Coherent Diffraction Experiments.
Ptychography experiments were carried out on beamline 101D-1 at the CLS and on beamline BL08U1A at the SSRF. During all the ptychography experiments, the photon energy of the incident X-ray was set to 700 eV or at the Zn L-edge for the spectro-ptychography measurement. In the ptychography experiment, the samples were raster scanned point by point with a 2 μm–diameter probe at a step size of 500 nm, which corresponds to a 75% overlap between adjacent scanning areas. Diffraction patterns at each location of the sample were collected with the detector in the far field. The central 760 × 760 pixels of the patterns with diffraction signals were selected, and the mixed-states ePIE algorithm was used to conduct the ptychographic reconstruction (43–45). High-quality sample images were reconstructed with 10-nm pixel resolution after the overall diffraction patterns of the whole sample were collected. The data for 3D reconstruction were collected on beamline 08U1A at the SSRF and a total of 22 angles of projections from −45° to +69.44°.
Supplementary Material
Acknowledgments
We acknowledge the staff at the CLS 101D-1 SM beamline, the Shanghai Synchrotron Radiation Facility (SSRF) beamline 08U1A, the National Synchrotron Radiation Laboratory (NSRL) beamline 07W, and Centre for High-Resolution Electron Microscopy (CħEM), School of Physical Science and Technology (SPST) of ShanghaiTech University for their assistance with data acquisition. We thank Prof. Zijian Xu and Prof. Xiangzhi Zhang for useful discussions. We thank Valentina Loconte and Kate L. White for providing pancreatic β cells and the protocol of insulin vesicle isolation. This work was supported by the Strategic Priority Research Program of the Chinese Academy of Sciences (Grant No. XDB 37040303), the National Natural Science Foundation of China (Grant No. 21727817), the Major State Basic Research Development Program of China (2017YFA0504802), and the Shanghai Frontiers Science Center for Biomacromolecules and Precision Medicine at ShanghaiTech University.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2202695119/-/DCSupplemental.
Data Availability
Data presented in this work are available for download at Mendeley Data (https://data.mendeley.com/datasets/wzp4s6m2r6/1) (46). All data needed to evaluate the conclusions in the paper are included in the article and/or supporting information.
References
- 1.Omar-Hmeadi M., Idevall-Hagren O., Insulin granule biogenesis and exocytosis. Cell. Mol. Life Sci. 78, 1957–1970 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rorsman P., Renström E., Insulin granule dynamics in pancreatic beta cells. Diabetologia 46, 1029–1045 (2003). [DOI] [PubMed] [Google Scholar]
- 3.Miceli V., et al. , In vitro imaging of β-cells using fluorescent cubic bicontinuous liquid crystalline nanoparticles. RSC Advances 6, 62119–62127 (2016). [Google Scholar]
- 4.Suzuki T., Kondo C., Kanamori T., Inouye S., Video rate bioluminescence imaging of secretory proteins in living cells: Localization, secretory frequency, and quantification. Anal. Biochem. 415, 182–189 (2011). [DOI] [PubMed] [Google Scholar]
- 5.Carter S. D., et al. , Distinguishing signal from autofluorescence in cryogenic correlated light and electron microscopy of mammalian cells. J. Struct. Biol. 201, 15–25 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hell S. W., et al. , The 2015 super-resolution microscopy roadmap. J. Phys. D Appl. Phys. 48, 443001 (2015). [Google Scholar]
- 7.Larabell C. A., Nugent K. A., Imaging cellular architecture with X-rays. Curr. Opin. Struct. Biol. 20, 623–631 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.McDermott G., Le Gros M. A., Knoechel C. G., Uchida M., Larabell C. A., Soft X-ray tomography and cryogenic light microscopy: The cool combination in cellular imaging. Trends Cell Biol. 19, 587–595 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bergeron A., Pucci L., Bezzi P., Regazzi R., Analysis of synaptic-like microvesicle exocytosis of B-cells using a live imaging technique. PLoS One 9, e87758 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.van Weering J. R., et al. , Automated analysis of secretory vesicle distribution at the ultrastructural level. J. Neurosci. Methods 173, 83–90 (2008). [DOI] [PubMed] [Google Scholar]
- 11.Slepchenko K. G., Daniels N. A., Guo A., Li Y. V., Autocrine effect of Zn2+ on the glucose-stimulated insulin secretion. Endocrine 50, 110–122 (2015). [DOI] [PubMed] [Google Scholar]
- 12.Zalewski P. D., et al. , Video image analysis of labile zinc in viable pancreatic islet cells using a specific fluorescent probe for zinc. J. Histochem. Cytochem. 42, 877–884 (1994). [DOI] [PubMed] [Google Scholar]
- 13.Chausmer A. B., Zinc, insulin and diabetes. J. Am. Coll. Nutr. 17, 109–115 (1998). [DOI] [PubMed] [Google Scholar]
- 14.Xu Y., Yan Y., Seeman D., Sun L., Dubin P. L., Multimerization and aggregation of native-state insulin: Effect of zinc. Langmuir 28, 579–586 (2012). [DOI] [PubMed] [Google Scholar]
- 15.Li D., et al. , Imaging dynamic insulin release using a fluorescent zinc indicator for monitoring induced exocytotic release (ZIMIR). Proc. Natl. Acad. Sci. U.S.A. 108, 21063–21068 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Laue T. M., Stafford W. F. III, Modern applications of analytical ultracentrifugation. Annu. Rev. Biophys. Biomol. Struct. 28, 75–100 (1999). [DOI] [PubMed] [Google Scholar]
- 17.Ebel C., Analytical ultracentrifugation for the study of biological macromolecules. Prog. Coll. Pol. Sci. 127, 73–82 (2004). [Google Scholar]
- 18.Sun T., et al. , Soft X-ray ptychography chemical imaging of degradation in a composite surface-reconstructed Li-rich cathode. ACS Nano 15, 1475–1485 (2021). [DOI] [PubMed] [Google Scholar]
- 19.Conesa J. J., et al. , Intracellular nanoparticles mass quantification by near-edge absorption soft X-ray nanotomography. Sci. Rep. 6, 22354 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang X. Z., et al. , Ratio-contrast imaging of dual-energy absorption for element mapping with a scanning transmission X-ray microscope. J. Synchrotron Radiat. 17, 804–809 (2010). [DOI] [PubMed] [Google Scholar]
- 21.Momose A., X-ray phase imaging reaching clinical uses. Phys. Med. 79, 93–102 (2020). [DOI] [PubMed] [Google Scholar]
- 22.Zan G., et al. , High-resolution multicontrast tomography with an X-ray microarray anode-structured target source. Proc. Natl. Acad. Sci. U.S.A. 118, e2103126118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Grama L. R., Discant A. I., Kramers-Kronig relationship computation by Gaussian quadrature. Int J Comput Commun 1, 239–244 (2006). [Google Scholar]
- 24.Bertie J. E., Lan Z. D., An accurate modified Kramers-Kronig transformation from reflectance to phase shift on attenuated total reflection. J. Chem. Phys. 105, 8502–8514 (1996). [Google Scholar]
- 25.Gulgowski J., Stefański T. P., Generalization of Kramers-Krönig relations for evaluation of causality in power-law media. Commun. Nonlinear Sci. Numer. Simul. 95, 105664 (2021). [Google Scholar]
- 26.Hirose M., Shimomura K., Burdet N., Takahashi Y., Use of Kramers-Kronig relation in phase retrieval calculation in X-ray spectro-ptychography. Opt. Express 25, 8593–8603 (2017). [DOI] [PubMed] [Google Scholar]
- 27.Casimir M., Chaffard G., Maechler P., Resveratrol long-term treatment differentiates INS-1E beta-cell towards improved glucose response and insulin secretion. Pflugers Arch. 471, 337–345 (2019). [DOI] [PubMed] [Google Scholar]
- 28.Kang T., Jensen P., Solovyeva V., Brewer J. R., Larsen M. R., Dynamic changes in the protein localization in the nuclear environment in pancreatic β-cell after brief glucose stimulation. J. Proteome Res. 17, 1664–1676 (2018). [DOI] [PubMed] [Google Scholar]
- 29.Fan J., et al. , Single-pulse enhanced coherent diffraction imaging of bacteria with an X-ray free-electron laser. Sci. Rep. 6, 34008 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ram S., Ward E. S., Ober R. J., Beyond Rayleigh’s criterion: A resolution measure with application to single-molecule microscopy. Proc. Natl. Acad. Sci. U.S.A. 103, 4457–4462 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang X., et al. , Visualizing insulin vesicle neighborhoods in β cells by cryo-electron tomography. Sci. Adv. 6, eabc8258 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Averbuch A., Coifman R. R., Donoho D. L., Israeli M., Shkolnisky Y., A framework for discrete integral transformations I—The pseudopolar Fourier transform. SIAM J. Sci. Comput. 30, 764–784 (2008). [Google Scholar]
- 33.Jiang H., et al. , Quantitative 3D imaging of whole, unstained cells by using X-ray diffraction microscopy. Proc. Natl. Acad. Sci. U.S.A. 107, 11234–11239 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Miao J., Förster F., Levi O., Equally sloped tomography with oversampling reconstruction. Phys. Rev. B 72, 052103 (2005). [Google Scholar]
- 35.Kobayashi Y., Fukumoto N., Kurahashi M., X-ray-fluorescence element-mapping spectrometer with improved spatial-resolution. Meas. Sci. Technol. 2, 183–184 (1991). [Google Scholar]
- 36.Murray F. E. S., Landsberg J. P., Williams R. J. P., Esiri M. M., Watt F., Elemental analysis of neurofibrillary tangles in Alzheimer’s disease using proton-induced X-ray analysis. Ciba Found. Symp. 169, 201–210, discussion 210–216 (1992). [DOI] [PubMed] [Google Scholar]
- 37.Li Y. V., Zinc and insulin in pancreatic beta-cells. Endocrine 45, 178–189 (2014). [DOI] [PubMed] [Google Scholar]
- 38.Chen Z., et al. , Polyhydroxylated metallofullerenols stimulate IL-1β secretion of macrophage through TLRs/MyD88/NF-κB pathway and NLRP3 inflammasome activation. Small 10, 2362–2372 (2014). [DOI] [PubMed] [Google Scholar]
- 39.Ito Y., Hasauda H., Kitajima T., Kiyono T., Ex vivo expansion of human cord blood hematopoietic progenitor cells using glutaraldehyde-fixed human bone marrow stromal cells. J. Biosci. Bioeng. 102, 467–469 (2006). [DOI] [PubMed] [Google Scholar]
- 40.Ito Y., Kawamorita M., Yamabe T., Kiyono T., Miyamoto K., Chemically fixed nurse cells for culturing murine or primate embryonic stem cells. J. Biosci. Bioeng. 103, 113–121 (2007). [DOI] [PubMed] [Google Scholar]
- 41.Rivera J. F., Costes S., Gurlo T., Glabe C. G., Butler P. C., Autophagy defends pancreatic β cells from human islet amyloid polypeptide-induced toxicity. J. Clin. Invest. 124, 3489–3500 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zanette I., et al. , Ptychographic X-ray nanotomography quantifies mineral distributions in human dentine. Sci. Rep. 5, 9210 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Maiden A. M., Rodenburg J. M., An improved ptychographical phase retrieval algorithm for diffractive imaging. Ultramicroscopy 109, 1256–1262 (2009). [DOI] [PubMed] [Google Scholar]
- 44.Thibault P., Menzel A., Reconstructing state mixtures from diffraction measurements. Nature 494, 68–71 (2013). [DOI] [PubMed] [Google Scholar]
- 45.Zhang J., et al. , Enhancement of phase retrieval capability in ptychography by using strongly scattering property of the probe-generating device. Opt. Express 26, 30128–30145 (2018). [DOI] [PubMed] [Google Scholar]
- 46.Guo A., et al., Quantitative, in situ visualization of intracellular insulin vesicles in pancreatic beta cells. Mendeley Data. https://data.mendeley.com/datasets/wzp4s6m2r6/1. Deposited 20 July 2022. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data presented in this work are available for download at Mendeley Data (https://data.mendeley.com/datasets/wzp4s6m2r6/1) (46). All data needed to evaluate the conclusions in the paper are included in the article and/or supporting information.