Abstract
Defensins are antimicrobial peptides that contribute broadly to innate immunity, including protection of mucosal tissues. Human α-defensin (HD)6 is highly expressed by secretory Paneth cells of the small intestine. However, in contrast to the other defensins, it lacks appreciable bactericidal activity. Nevertheless, we report here that HD6 affords protection against invasion by enteric bacterial pathogens in vitro and in vivo. After stochastic binding to bacterial surface proteins, HD6 undergoes ordered self-assembly to form fibrils and nanonets that surround and entangle bacteria. This self-assembly mechanism occurs in vivo, requires histidine-27, and is consistent with X-ray crystallography data. These findings support a key role for HD6 in protecting the small intestine against invasion by diverse enteric pathogens, and may explain the conservation of HD6 throughout Hominidae evolution.
Paneth cells are specialized small intestinal epithelial cells that maintain intestinal homeostasis, in part by expressing and secreting antimicrobial peptides and proteins (1-3). Deficiencies of two human Paneth cell peptides, α-defensins HD5 and HD6, are associated with Crohn's disease, a chronic inflammatory bowel disease (3-5). HD5 is broadly antimicrobial (6-8) and can shape the gut microbiota in vivo (9). In contrast, HD6 exerts little antibacterial activity (Fig. S1, (10) and (8)), and its function is unknown.
To investigate HD6, we developed a transgenic mouse model wherein HD6 gene (DEFA6) expression was controlled by its endogenous promoter and restricted to Paneth cells (Fig. S2A,B). Transgenic HD6 expression occurred at levels commensurate with endogenous human and murine Paneth cell α-defensin production, and did not alter expression of other murine Paneth cell-derived antimicrobial (poly)peptides (Fig. S2C,D).
When HD6 transgenic mice and littermate controls were challenged intragastrically with 2 × 108 colony forming units (CFU) of S. Typhimurium (STM), 50 % of wild-type animals but no transgenic mice had died six days post infection (P < 0.05) (Fig. 1A). Consistent with HD6's lack of direct antibacterial activity, the intestinal lumen of transgenic and wild-type mice contained similar bacterial numbers four days post infection. However, STM counts were 100-fold lower in Peyer’s patches (P < 0.001) and 10-fold lower in spleens (P < 0.001) of HD6 transgenic mice, than that in their wild-type littermates (Fig. 1B). The decreased bacterial burden in these tissues resembled findings in wild-type mice infected with an invasion-deficient STM invA mutant (11).
Fig. 1. Analysis of in vivo and in vitro invasion by S. Typhimurium.
(A-B) In vivo challenge of mice with an intragastric inoculum of S. Typhimurium (2 × 108 CFU). (A) Kaplan-Meyer survival curve of 6 week female wild-type (n = 15) and HD6 +/− transgenic littermates (n = 9) with a scheduled experimental endpoint of 6 days. Log-rank test, * P < 0.05. The data are representative of three independent experiments. (B) S. Typhimurium bacterial load from colon fecal pellets, Peyer’s patches (3/mouse), and spleen tissue of 6 week female wild-type (n = 4) and HD6 +/− transgenic female littermate mice (n = 6) 4 days post-infection. Error bars represent SEM. ** P < 0.001, t-test. The data are representative of three independent experiments. (C-E) In vitro invasion assays. (C) S. Typhimurium were pre-treated for 1 h with either 10 μg/ml of HD5, HD6 or H27W-HD6, or with vehicle alone (WT untreated). Invasion-deficient S. Typhimurium (invA mutant) were treated with vehicle alone. Bacteria were then allowed to invade the epithelial cells (MOI of 10) for 1 h. Cells were then washed, treated with gentamicin, and then lysed to quantitate intracellular bacteria (expressed as % of initial inoculum). Data represent the average of three experiments, and are representative of more than 9 independent experiments. (D) Concentration-dependence of the HD6 activity in assays as described in (C). Error bars represent SEM. *P < 0.01, ** P < 0.001, *** P < 0.0001 compared to WT vehicle, t-test. Data represent the average of three experiments, and are representative of more than 9 independent experiments.
To determine whether HD6 inhibited invasion, intestinal epithelial cells were challenged with α-defensin-treated or untreated STM. The ability of HD6-treated bacteria to invade T84 epithelial cells was impaired in a dose-dependent manner (Fig. 1C,D). HD6 could reduce invasion by wild-type STM to levels of the minimally invasive invA mutant, which lacks a functional invasion-associated type III secretion system (T3SS) (Fig. 1C). Another human intestinal cell line, INT-407, and a mouse small intestinal cell line, MODE-K, exhibited the same pattern of HD6-mediated inhibition of STM invasion (Fig. S3). The same concentrations of HD5, the other human Paneth cell α-defensin (Fig. 1C), or the mouse Paneth cell α-defensins, cryptdins 2 and 3 (Fig. S4A), did not inhibit STM invasion. Immunofluorescence microscopy confirmed that HD6 treatment blocked invasion into intestinal epithelial cells (Fig. S5).
Similar STM invasion occurred in untreated and HD6-treated intestinal epithelial cells, suggesting that HD6 does not protect by altering host cells (Fig. S4B). The hypothesis that HD6 blocked invasion by interfering with synthesis of T3SS proteins was rejected by findings that STM treated with protective concentrations of HD6 showed normal expression of flhD and fliC, and even induced expression of hilA (a key SPI-1 activator) and invG, a structural protein of the T3SS-1 system (Fig. S6). Also excluded were the possibilities that HD6 reduced invasion by inhibiting bacterial adhesion (Fig. S7), or by altering the intestinal microbiota of the transgenic mice (Fig. S8).
To investigate the specificity of HD6, we performed invasion assays with Yersinia enterocolitica. This enteropathogenic bacterium invades cells by a different mechanism, involving attachment of its invasin protein (Inv) to host β-integrins (12). Since HD6 reduced Yersinia invasion to levels similar to the invasion-deficient Δinv mutant (Fig. S9), its ability to prevent bacterial invasion was not pathogen-specific.
Recent studies identified the Trp residue of human neutrophil α-defensin peptide (HNP)1 as a key functional amino acid (13). Whereas most Hominid α-defensins, including HD5, have a nonpolar aromatic residue, Trp, Tyr or Phe, at the corresponding position, HD6 and its Hominid orthologs contain His (Fig. S10). When we replaced His27 of HD6 with Trp, the resulting H27W-HD6 failed to block invasion (Fig. 1C, S4, S9), indicating that His27 was critical for this activity.
Seeking insight into the protective properties of HD6 and the role of His27, we used surface plasmon resonance (SPR) to examine binding to bacterial proteins. HD5 bound rapidly to immobilized Y. enterocolitica invasin and reached an equilibrium plateau by 3 min (Fig. 2A). In marked contrast, HD6 binding was less extensive and much slower. Binding of H27W-HD6 was enhanced relative to that of HD6, and like HD5, the H27W-HD6 binding curve rapidly reached a plateau denoting equal rates of association and dissociation. Because binding of HD6 to invasin was continuing to increase when the conventional 3 min binding period ended, we tested longer binding intervals, eventually adopting a multi-cycle approach to monitor binding over much longer periods (Fig. 2B,C). Because other α-defensin molecules self-associate (13-16), we measured the ability of solution-phase HD6 to bind HD6 immobilized on a biosensor. HD6 self-association rose progressively during a six-cycle experiment (Fig. 2B). In contrast, HD5 binding to HD5 reached a molar ratio close to 1 in each cycle, signifying dimer formation, and then declined to baseline during each intervening "washout" period. Unlike HD6, H27W-HD6 showed minimal self-association, indicating that His27 was critical to this property. Similarly, H27A-HD6, containing a less bulky Ala substitution, showed kinetics like H27W-HD6 (Fig. S11). Therefore, HD6 forms higher order assemblages with slower kinetics of association and dissociation, while HD5 and other α-defensins rapidly (and reversibly) form dimeric complexes.
Fig. 2. Surface plasmon resonance (SPR) analysis of binding and self-association.
The SPR biosensors presented the following amounts of immobilized ligands (molecule covalently ligated to the biosensor surface), in resonance units (RU): invasin, 2338 RU; HD5, 1158 RU; HD6, 1100 RU, H27W-HD6, 2204 RU; flagellin, 2206 RU; gp120, 8200 RU; gp41, 5600 RU. The defensin peptides were prepared in HBS-EP buffer and traversed the biosensors at either 50 μl/min (A) or 25 μl/min (B - F). Each 15 min cycle in panels B - F included a binding period of 12.5 min, followed by a wash period of approximately 2.5 min. Molar ratio (MR) calculations, which take into account the relative masses of the analytes and ligands and the number of RU presented by each biosensor, are described in the methods. Shown are representative data from at least three separate experiments. (E) Binding of HD6 to gp120-LAV before (triangles) and after enzymatic deglycosylation (closed circles) of the same biosensor chip. Shown are averaged data from six experiments, with error bars representing SEM. P < 0.002, Mann-Whitney Rank-sum test. Also shown are data from a second set of biosensor chips (n=6) presenting the same quantity of deglycosylated gp120 (open circles). (F) Binding of HD6 to gp41-LAV before and after enzymatic deglycosylation of the same biosensor chip. Six experiments were done before enzymatic deglycosylation and six more were done afterwards. Error bars represent SEM. The differences in each cycle were statistically significant with a P < 0.01, Mann-Whitney Rank-sum test.
In a six-cycle binding experiment using immobilized invasin as the target, HD5 oscillated in each binding cycle, rapidly reaching a plateau and then falling by about half during each wash period (Fig. 2C). The oscillations likely represented reversible association and dissociation of solution-phase HD5 to molecules of HD5 that bound the immobilized invasin. Whereas H27W-HD6 binding to invasin showed similar cyclic oscillations, , HD6 binding to invasin rose progressively, with a 100-fold increase from cycle one to cycle six (Fig. 2C). Similar results were observed when immobilized Salmonella flagellin anchored progressive HD6 self-assembly (Fig. 2D). These data suggested that HD6 molecules bound directly to immobilized proteins served as points of origin for progressive assembly of an extended structure composed of HD6 molecules. Importantly, even a low-level binding of HD6 to a target protein created anchoring sites for progressive HD6 self-assembly.
The SPR experiments indicated that HD6 binding to target proteins lacks the specificity typical of high affinity interactions. To explore this further, we tested HD6 binding to HIV glycoproteins gp41 and gp120. HD6 bound both proteins poorly until they were enzymatically deglycosylated (Fig. 2E,F). Thus, whereas primary binding of HD6 appears nonspecific with respect to the target protein's sequence, glycosylation can greatly reduce its binding.
Previous crystallographic studies suggested that HD6 can assemble into an elongated, highly ordered helical structure (15), significantly different from the dimeric structures formed by HD5 and HNPs (13-16). HD6 monomers associate into dimers (Fig 3A), but the respective monomers slide apart and form fewer backbone H-bonds between β-strands than that for HD5 or the HNPs. This atypical dimer is critically stabilized by dimer-dimer association (Fig. 3A), resulting in a stable tetramer – the repeating unit of the elongated HD6 structure (Fig. S12). HD6 tetramerization is mediated by extensive reciprocal inter-dimer interactions that include electrostatic interactions of the His27 imidazole side chain with the C-terminal carboxyl group of Leu32 (17). The molecular surface buried within the HD6 tetramer (an average value of 760 Å2/monomer) is quite large given the peptide's small size. Comparative structural analyses of HD6 with H27W-HD6 reveal that His27-mediated electrostatic interactions are critical for HD6 tetramer formation (Fig. 3) and higher-order oligomerization (Fig. S12-S14). The H27W substitution eliminates His27-Leu32 interactions important for HD6 dimer-dimer association (Fig. 3B, S14), as further discussed (18).
Fig. 3. X-ray crystallography analysis of HD6 and H27W-HD6.
(A) The previously described HD6 tetramer formed by four crystallographically independent monomers in the asymmetric unit of crystal (PDB code 1ZMQ) (15). ‘Canonical’ dimers are colored yellow (monomers a and b) and green (monomers c and d), hydrogen bonds shown as blue dashes. HD6 dimerization is mediated primarily through the hydrogen bonds formed by the backbone atoms of the β2 strands, involving two reciprocal hydrogen bonds between a pair of Thr21 and a single hydrogen bond between Met23 N and Thr19 O. Dimer-dimer association is mediated by the backbone atoms of the β1 strands, forming four H-bonds between Phe2 and Cys4 of monomers a and c and two between Ala1 N of monomers a and c and Cys6 O of monomers d and b. In addition, the HD6 tetramer is stabilized by electrostatic interactions involving the side chains of two His27 residues (red arrow) and the C-terminal carboxyl groups of two Leu32 residues, among others (Fig. S12). The orientation of the imidazole ring of His27 is supported by side-chain stacking interactions with Phe2. (B) Equivalent putative tetrameric assembly of four H27W-HD6 monomers from two symmetry-related ‘canonical’ dimers in orange and cyan. The H27W mutation has little impact on the tertiary structure of the mutant defensin (Fig. S13), and does not change the inter-chain backbone H-bonding pattern seen in wild-type HD6. However, what does change is the quaternary structure (Fig. S14), where the mutation significantly debilitates HD6 molecules to assemble into high-order oligomers.
By scanning electron microscopy, HD6-treated STM were aggregated (Fig. 4A-C), and higher magnification revealed a net-like meshwork of fibrils that arose from the bacterial surface to entangle the bacteria and their flagella. Such nanonets were again observed in experiments using human small intestinal aspirates (Fig. S15). Latex beads coated with a bacterial protein supported formation of similar nanofibrils with wild-type HD6, but not with H27W-HD6, indicating that the structures seen in electron microscopy were not of bacterial origin (Fig. 4D, S16).
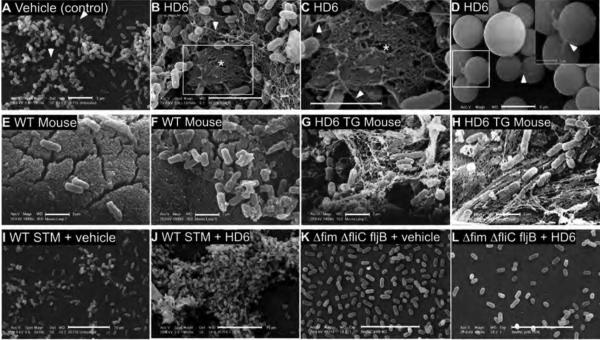
Fig. 4. HD6 nanonets entrap S. Typhimurium in vitro and in vivo.
(A-D) Scanning electron microscopy (SEM) of in vitro nanonet formation. Wild-type S. Typhimurium incubated with vehicle (A) or HD6 (10 μg/ml, B,C) in Tris-maleate buffer, as described in the methods. Bar = 5 μm. Magnification 5000X (B) and 10,000X (C). The white rectangle (B) and the asterisks (B,C) highlight a prominent nanonet. Note that the longer and wider structures are flagellae (white arrowheads), many of which are entangled with other cobweb-like nanonets, but are not evident with ΔfliC fljB (Fig. S16). SEM of protein A-coated polystyrene beads (D) incubated with HD6 (1 μg/ml) for 5 min at room temperature in 50 mM Tris-maleate buffer. The beads were then washed, fixed, and processed for SEM. Data are representative of six independent experiments. (E-H) SEM of WT (E,F) and HD6 transgenic (G,H) mouse ileal loop directly inoculated with S. Typhimurium. Bar = 2 μm. In a double-blinded data acquisition and scoring evaluation, three individuals scored 12 of 12 such images correctly for mouse genotype. P < 0.001, Fisher’s exact test. See methods for details. Data are representative of two independent experiments. (I-L) SEM of wild-type S. Typhimurium (I,J) and Δfim ΔfliC fljB (K,L) treated with vehicle alone (I,K) or 10 μg/ml HD6 (J,L). Bar = 10 μm. See methods for details. Data are representative of three independent experiments.
HD6 nanonets formed in vivo in a murine ileal loop model (19) wherein HD6 transgenic and littermate wild-type controls received pilocarpine to induce Paneth cell secretion, and ileal loops were inoculated with STM. Abundant nanonet structures that entangled and aggregated STM were observed (Fig. 4G,H, S17). In contrast, no nanonets were observed in wild-type mouse ileal loops (Fig. 4E,F, S17). Thus, HD6 can form small intestinal nanonets that entangled STM in vivo.
Because HD6 lacks lectin-like activity (20), it seemed unlikely to target carbohydrates on the bacterial surface. We therefore directed our attention to the predominant proteinaceous surface appendages of STM, flagellae and type I fimbriae (21). Compared to wild-type bacteria, we observed a decreased aggregation by HD6 of STM mutants lacking type I fimbriae or flagella (Fig. S18). Neither nanofibrils nor nanonets nor aggregation was evident in an HD6-treated Salmonella mutant lacking both fimbriae and flagella (Fig. 4I-L), suggesting that appendages function as HD6 anchoring points that trigger HD6 nanonet formation.
Together these data support a model wherein secreted HD6 molecules bind stochastically (i.e., randomly and at low levels) to any of the various proteinaceous bacterial surface molecules they encounter. These initially bound HD6 molecules provide a nidus that triggers a dynamic and deterministic process of self-assembly, building nanonets that can aggregate bacteria and/or impede the close physical contact with epithelial cells required for attachment or invasion. Highly expressed in Paneth cells at the base of small intestinal crypts, secreted HD6 will accumulate in the crypt and pericellular space beneath the mucus layer. Host proteins in this location are heavily glycosylated, leaving microbial intruders as the prime targets for HD6 binding.
Any microbial intruder that enters the small intestinal crypt, be it bacterial, fungal or protozoan, is a potential threat – especially since the stem cells that continually renew the intestinal epithelium reside at the crypt base (3). An intruder in this location would encounter HD6 and provide anchorage for the initial, low-level binding of this peptide. As this binding is not target sequence specific, HD6 can be effective against diverse threats. Because HD6 concentrations in the crypt are orders of magnitude higher than the highest concentrations (10 μg/ml) used in our experiments (22, 23), fibril and nanonet formation around such intruding microbes should be faster and in greater abundance than those described above. Since mouse Paneth cells lack an ortholog of HD6 (24), the transgenic mice benefit from expression of this human peptide. On the other hand, it is noteworthy that individuals with Crohn’s disease characteristically have an accumulation of invasive bacteria at the epithelial surface of their small intestine (5). A predisposition to this inflammatory bowel disease may, in part, be a result of a deficiency of HD6, as supported by the decreased expression of both HD5 and HD6 observed in Crohn’s disease of the ileum (4). Although the present experiments dealt primarily with two model Gram-negative bacteria, the general mechanism that emerged could apply to other bacteria, as well as fungi and protozoans.
Supplementary Material
One Sentence Summary.
Paneth cell-derived human α-defensin-6 blocks enteric bacterial pathogen invasion by ordered self-assembly of microbe-entangling peptide nanonets.
Acknowledgments
The authors thank W. Yuan for expert assistance in synthesizing the defensin peptides, and R. W. Feathers and H. K. Rho for technical help related to transgenic mice. We thank P. E. Kysar for help with scanning electron microscopy. We gratefully acknowledge Professor A. J. Ouellette for help with HD6 antibody production. We are indebted to Professors A. C. Gelli and S. C. Dawson for expert guidance in exploring possible biological activities of HD6, and to S. J. Howell for initial investigations of this peptide. Finally, we appreciate the many suggestions from colleagues and members of our labs that helped us unravel the properties of HD6 described here. This work was supported by grants from the National Institutes of Health: AI032738 (CLB), AI050843 (CLB), HD059127 (MAU and CLB), AI076246 (AJB and CLB), AI040124 (AJB), AI044170 (AJB), AI073120 (AJB), AI088122 (AJB), AI072732 (WL), AI070726 (RIL), AI082320 (RMT), AI057757 (NHS) and T32AI060555 (HC and SPN), and a fellowship from Robert Bosch Foundation (JW). The authors declare no competing interests. The data described in this manuscript are tabulated in the main paper and in the supplementary materials. The structure factor files and coordinates for H27W-HD6 are deposited in the Protein Data Bank under the accession code 3QTE.
References and Notes
- 1.Lehrer RI, Bevins CL, Ganz T. In: Mucosal Immunology. 3rd Mestecky J, et al., editors. Vol. 1. Academic Press; New York: 2004. pp. 95–110. [Google Scholar]
- 2.Ouellette AJ. Paneth cells and innate mucosal immunity. Curr Opin Gastroenterol. 2010;26:547. doi: 10.1097/MOG.0b013e32833dccde. [DOI] [PubMed] [Google Scholar]
- 3.Bevins CL, Salzman NH. Paneth cells, antimicrobial peptides and maintenance of intestinal homeostasis. Nat Rev Microbiol. 2011;9:356. doi: 10.1038/nrmicro2546. [DOI] [PubMed] [Google Scholar]
- 4.Wehkamp J, et al. Reduced Paneth cell alpha-defensins in ileal Crohn's disease. Proc Natl Acad Sci U S A. 2005;102:18129. doi: 10.1073/pnas.0505256102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wehkamp J, Stange EF. Paneth's disease. J. Crohns Colitis. 2010;4:523. doi: 10.1016/j.crohns.2010.05.010. [DOI] [PubMed] [Google Scholar]
- 6.Porter EM, van Dam E, Valore EV, Ganz T. Broad-spectrum antimicrobial activity of human intestinal defensin 5. Infect Immun. 1997;65:2396. doi: 10.1128/iai.65.6.2396-2401.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Salzman NH, Ghosh D, Huttner KM, Paterson Y, Bevins CL. Protection against enteric salmonellosis in transgenic mice expressing a human intestinal defensin. Nature. 2003;422:522. doi: 10.1038/nature01520. [DOI] [PubMed] [Google Scholar]
- 8.Ericksen B, Wu Z, Lu W, Lehrer RI. Antibacterial activity and specificity of the six human {alpha}-defensins. Antimicrob Agents Chemother. 2005;49:269. doi: 10.1128/AAC.49.1.269-275.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Salzman NH, et al. Enteric defensins are essential regulators of intestinal microbial ecology. Nat Immunol. 2010;11:76. doi: 10.1038/ni.1825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. See supporting material on Science Online.
- 11.Baumler AJ, Tsolis RM, Valentine PJ, Ficht TA, Heffron F. Synergistic effect of mutations in invA and lpfC on the ability of Salmonella typhimurium to cause murine typhoid. Infect Immun. 1997;65:2254. doi: 10.1128/iai.65.6.2254-2259.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cossart P, Sansonetti PJ. Bacterial invasion: the paradigms of enteroinvasive pathogens. Science. 2004;304:242. doi: 10.1126/science.1090124. [DOI] [PubMed] [Google Scholar]
- 13.Wei G, et al. Trp-26 imparts functional versatility to human alpha-defensin HNP1. J Biol Chem. 2010;285:16275. doi: 10.1074/jbc.M110.102749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hill CP, Yee J, Selsted ME, Eisenberg D. Crystal structure of defensin HNP-3, an amphiphilic dimer: mechanisms of membrane permeabilization. Science. 1991;251:1481. doi: 10.1126/science.2006422. [DOI] [PubMed] [Google Scholar]
- 15.Szyk A, et al. Crystal structures of human alpha-defensins HNP4, HD5, and HD6. Protein Sci. 2006;15:2749. doi: 10.1110/ps.062336606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wei G, et al. Through the looking glass, mechanistic insights from enantiomeric human defensins. J Biol Chem. 2009;284:29180. doi: 10.1074/jbc.M109.018085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. More precisely, the imidazole side chain of His27 of both monomer b and d interacts with the C-terminal carboxyl group of Leu32 in respective monomer c and a, and two backbone-backbone H-bonds form between Ala1 N of monomers a and c and Cys6 O of monomers d and b (Fig. 3A, S12)
- 18. Furthermore, H27W-HD6 does not form the two reciprocal main-chain H-bonds donated by Ala1 of one dimer to Cys6 of the other. Consequently, monomers b and d of H27W-HD6 “spring loose” from monomers a and c, and the resultant mutant tetramer becomes vertically more extended, and laterally more flattened than that of wild-type HD6 (Fig. 3B). The extended and flattened H27W tetramer is less stable than its “twisted” wild-type counterpart, because of reduced hydrophobic packing and the lost inter-dimer interactions (Fig. S14). Overall, the crystal structures of both defensins suggest that His27 plays an important role in stabilizing HD6 tetramer stabilization and likely contributes to an elongated high-order assembly of HD6 molecules by accentuating a propagating twist within and beyond the tetramer structure. The assembled HD6 structure shown is bowed when viewed from one perspective, and linear when rotated by 90° (Fig. S12). This bowing, along with coalescence of fibrils extending from different anchoring points on the bacterial surface, might account for the branching and net-like formation evident in the scanning electron micrographs that appear below.
- 19.Brandl K, Plitas G, Schnabl B, DeMatteo RP, Pamer EG. MyD88-mediated signals induce the bactericidal lectin RegIII gamma and protect mice against intestinal Listeria monocytogenes infection. J Exp Med. 2007;204:1891. doi: 10.1084/jem.20070563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lehrer RI, et al. Multivalent binding of carbohydrates by the human {alpha}-defensin, HD5. J Immunol. 2009;183:480. doi: 10.4049/jimmunol.0900244. [DOI] [PubMed] [Google Scholar]
- 21.Andrews-Polymenis HL, Baumler AJ, McCormick BA, Fang FC. Taming the elephant: Salmonella biology, pathogenesis, and prevention. Infect Immun. 2010;78:2356. doi: 10.1128/IAI.00096-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ayabe T, et al. Secretion of microbicidal alpha-defensins by intestinal Paneth cells in response to bacteria. Nat Immunol. 2000;1:113. doi: 10.1038/77783. [DOI] [PubMed] [Google Scholar]
- 23.Ghosh D, et al. Paneth cell trypsin is the processing enzyme for human defensin-5. Nat Immunol. 2002;3:583. doi: 10.1038/ni797. [DOI] [PubMed] [Google Scholar]
- 24.Shanahan MT, Tanabe H, Ouellette AJ. Strain-Specific Polymorphisms in Paneth Cell {alpha}-Defensins of C57BL/6 Mice and Evidence of Vestigial Myeloid {alpha}-Defensin Pseudogenes. Infect Immun. 2011;79:459. doi: 10.1128/IAI.00996-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mallow EB, et al. Human enteric defensins. Gene structure and developmental expression. J Biol Chem. 1996;271:4038. doi: 10.1074/jbc.271.8.4038. [DOI] [PubMed] [Google Scholar]
- 26.Wehkamp J, et al. Paneth cell antimicrobial peptides: Topographical distribution and quantification in human gastrointestinal tissues. FEBS Letters. 2006;580:5344. doi: 10.1016/j.febslet.2006.08.083. [DOI] [PubMed] [Google Scholar]
- 27.Lehrer RI, Rosenman M, Harwig SS, Jackson R, Eisenhauer P. Ultrasensitive assays for endogenous antimicrobial polypeptides. J Immunol Methods. 1991;137:167. doi: 10.1016/0022-1759(91)90021-7. [DOI] [PubMed] [Google Scholar]
- 28.Vidal SM, Malo D, Vogan K, Skamene E, Gros P. Natural resistance to infection with intracellular parasites: isolation of a candidate for Bcg. Cell. 1993;73:469. doi: 10.1016/0092-8674(93)90135-d. [DOI] [PubMed] [Google Scholar]
- 29.Winter SE, et al. Contribution of flagellin pattern recognition to intestinal inflammation during Salmonella enterica serotype Typhimurium infection. Infect Immun. 2009;77:1904. doi: 10.1128/IAI.01341-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Winter SE, et al. Gut inflammation provides a respiratory electron acceptor for Salmonella. Nature. 2010;467:426. doi: 10.1038/nature09415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stojiljkovic I, Baumler AJ, Heffron F. Ethanolamine utilization in Salmonella typhimurium: nucleotide sequence, protein expression, and mutational analysis of the cchA cchB eutE eutJ eutG eutH gene cluster. J Bacteriol. 1995;177:1357. doi: 10.1128/jb.177.5.1357-1366.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Weening EH, et al. The Salmonella enterica serotype Typhimurium lpf, bcf, stb, stc, std, and sth fimbrial operons are required for intestinal persistence in mice. Infect Immun. 2005;73:3358. doi: 10.1128/IAI.73.6.3358-3366.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Raffatellu M, et al. Lipocalin-2 resistance confers an advantage to Salmonella enterica serotype Typhimurium for growth and survival in the inflamed intestine. Cell Host Microbe. 2009;5:476. doi: 10.1016/j.chom.2009.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Brunger AT. Free R value: a novel statistical quantity for assessing the accuracy of crystal structures. Nature. 1992;355:472. doi: 10.1038/355472a0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.