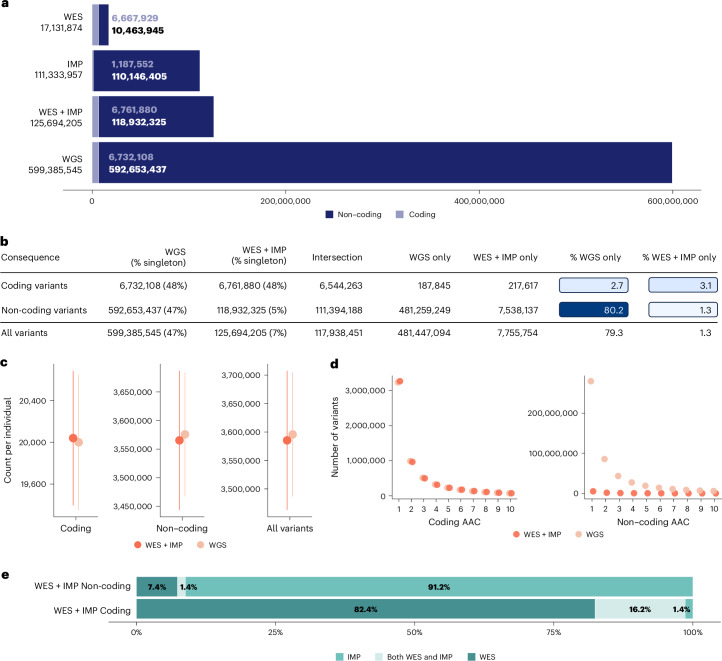
Fig. 1. Genetic variation captured by genomes, exomes and array genotyping with imputation.
a, The number of coding and non-coding variants for all variants in the WES, IMP, WES + IMP and WGS datasets. b, A comparison of variants observed by the WES + IMP and WGS datasets by functional consequence, with the relative gains in approach-exclusive variants. c, The number of variant alleles observed per individual (n = 149,195). The point provides the sample mean; error bars, ±standard error of the mean (s.e.m.) d, The number of non-coding and coding variants observed at the lowest alternative allele counts (AAC) for the WES + IMP and WGS data. e, The percentages of non-coding and coding variants observed only in WES, in both WES and IMP and only in IMP in the combined WES + IMP dataset.