Abstract
Pediatric patients are in a growing stage with more dividing cells than adults. Therefore, they are more sensitive to the radiation dose when undergoing computed tomography (CT) scanning. It is necessary and essential to assess the organ absorbed dose and effective dose to children. Monte Carlo simulation with computational phantoms is one of the most used methods for dose calculation in medical imaging and radiotherapy. Because of the vast change of the pediatric body with age increasing, many research groups developed series pediatric phantoms for various ages. However, most of the existing pediatric reference phantoms were developed based on Caucasian populations, which is not conformable to Chinese pediatric patients. The use of different phantoms can contribute to a difference in the dose calculation. To assess the CT dose of Chinese pediatric patients more accurately, we developed the Chinese pediatric reference phantoms series, including the 3-month (CRC3m), 1-year-old (CRC01), 5-year-old (CRC05), 10-year-old (CRC10), 15-year-old male (CRCM15), and a 15-year-old female (CRCF15) phantoms. Furthermore, we applied them to dose assessment of patients undergoing CT scanning. The GE LightSpeed 16 CT scanner was simulated and the paper presents the detailed process of phantoms development and the establishment of the CT dose database (with x-ray tube voltages of 120, 100 and 80 kVp, with collimators of 20, 10, and 5 mm width, with filters for head and body), compares for the 1-year-old results with other results based on different phantoms and analyzes the CT dose calculation results. It was found that the difference in phantoms' characteristics, organ masses and positions had a significant impact on the CT dose calculation outcomes. For the 1-year-old phantom, the dose results of organs fully covered by the x-ray beam were within 10% difference from the results of other studies. For organs partially covered and not covered by the scan range, the maximum differences came up to 84% (stomach dose, chest examinations) and 463% (gonads dose, chest examinations) respectively. The findings are helpful for the dose optimization of Chinese pediatric patients undergoing CT scanning. The developed phantoms could be applied in dose estimation of other medical modalities.
1. Introduction
Over the past several decades the amount of radiation from medical exposure is continuously increasing in China. The annual growth rate of the number of CT examinations was 11.7% from 2005 to 2014 (Su Yinping et al 2017). Additionally, compared with the adults, children in the growth stage are more sensitive to ionizing radiation (National Research Council 2006, Goodman et al 2019). Therefore, it is highly relevant to assess the dose of medical radiation exposure to children.
Several approaches have been adopted to estimate the dose of CT examinations: direct measurements with anthropomorphic phantoms using dosimeters (Giansante et al 2019) and Monte Carlo simulation with computational phantoms. Compared with the direct measurement method, the Monte Carlo method is less time-consuming and more convenient. Besides, the mean organ dose can be obtained by Monte Carlo simulation, while the direct measurement method can only get the dose at limited points. Therefore, Monte Carlo simulation was adopted to assess the dose in this study.
For children in the growing period, considering the characteristics of the fast growth and development of the tissue and organ, it is highly relevant to develop the phantoms for children of different ages. Three broad classes of computational phantoms are used in radiation diagnostics or therapy for organ dose calculation: (1) stylized phantoms, (2) voxel phantoms, (3) mesh-type phantoms. Many research groups have developed several series of pediatric phantoms. In 1980', Cristy (Cristy 1980)and Eckerman (Cristy and Eckerman 1987) from the Oak Ridge National Laboratory (ORNL) developed the stylized phantoms representing newborns, 1, 5, 10 and two 15 years old children. The pediatric voxel phantoms were developed based on CT images of an 8-week-old and a 7-year-old child obtained by (Williams et al 1986) and (Zankl et al 1988). Later on, the University of Florida (UF) developed pediatric voxel phantoms for various ages. ICRP released adult voxel reference phantom in Publications 110 (ICRP 2007, 2009). Most recently, the pediatric voxel reference phantom was released in Publications 143 (ICRP 2020). For the advantage of more excellent deformability and more accurate organ description to the voxel phantoms, the mesh-type phantoms can be a better choice for dose calculation. Lee et al from the UF presented a whole family of UF pediatric mesh-type phantoms (Lee et al 2008, Lee et al 2010). This series of phantoms were implemented in the software VirtualDose (Ding et al 2015) and were applied in CT dose estimation and the simulation works (Lee et al 2012). A set of 4D pediatric extended cardiac-torso (XCAT) phantoms at the ages of newborn, 1, 5, 10, and 15 years were developed in 2014 by Duke University Medical Center (Norris et al 2014). Furtherly, this set of phantoms was extended to a series of 64 pediatric phantoms of varying ages, heights, and weights (Segars et al 2015). Duke University developed a smartphone application XCATdose (Hoye et al 2017a, 2017b) for organ and effective dose estimation. The extended XCAT adult and pediatric phantom series were included in this application. The Task Group 103 of the ICRP has developed the Mesh-type Reference Computational Phantoms (MRCPs) (Kim et al 2018). The body characteristic and the organ weight of phantoms mentioned above were adapted to the International Commission on Radiological Protection (ICRP) reference data (ICRP 2002). However, the data from the ICRP publications (ICRP 2002) was obtained based on the Caucasian populations, which had significant differences with Chinese children on physical characteristics, organ position and mass.
This study aims to develop Chinese pediatric reference phantoms series and apply them in MC simulation of pediatric dose undergoing CT scanning. The study could provide a more accurate assessment of organ doses to Chinese pediatric patients.
In this paper, a series of mesh-type Chinese pediatric reference phantoms were constructed. The organ masses and positions of each phantom are consistent with the anatomical characteristics of Chinese reference children. The structure of a kind of typical CT scanner was also constructed in Monte Carlo simulation. The organ doses to Chinese pediatric reference phantoms from the single-axis scan were calculated based on the Chinese reference phantom. Furthermore, an organ dose database was established. The organ doses calculated in this work were compared with the simulated results by Lee et al (Lee et al 2012)and by using the software VirtualDose (Ding et al 2015).
2. Methods
2.1. Development of mesh-type Chinese pediatric reference phantoms series
In the National Occupational Health Standards (GBZ) document of the People's Republic of China, reference individuals for use in radiation protection, Part1 Physique parameters (GBZ/T 200.1-2007) (Ministry of Health P.R. China 2007a), Chinese pediatric reference phantoms were divided into five ages, 3 months, 1-year-old, 5-year-old, 10-year-old and 15-year-old. Before the age of ten (including 10-year-old), the gender characteristics were not very distinguished, the composition of each organ in the body was similar. Therefore, for ages before 10 (including 10-year-old), the pediatric reference phantoms were built gender-independent with two sets of sex organs. For the age of 15, male and female phantoms were constructed individually. In the mesh-type Chinese pediatric reference phantoms series, a total of six phantoms were established, representing the reference 3-month (CRC3m), 1-year-old (CRC01), 5-year-old (CRC05), 10-year-old (CRC10), 15-year-old male (CRCM15), and 15-year-old female (CRCF15), respectively. The height and weight parameters of children of different ages were derived from the Chinese national standard (GBZ/T200.1-2007) (Ministry of Health P.R. China 2007a), and the data of ICRP pediatric phantoms were also listed for comparison, as shown in table 1.
Table 1. The height and weight of Chinese Pediatric Reference Phantoms (CRCs) and ICRP Pediatric Phantoms (ICRPs) (ICRP 2020).
| Height (cm) | Weight (kg) | ||||
|---|---|---|---|---|---|
| Gender | Age (y) | CRCs | ICRPs | CRCs | ICRPs |
| Male/Female | 0.25 | 62 | 51 | 7 | 3.5 |
| Male/Female | 1 | 77 | 76 | 10 | 10 |
| Male/Female | 5 | 110 | 109 | 19 | 19 |
| Male/Female | 10 | 139 | 138 | 32 | 32 |
| Male | 15 | 168 | 167 | 55 | 56 |
| Female | 15 | 158 | 161 | 50 | 53 |
The CT images were obtained from the Capital Institute of Pediatrics, Beijing, which were not whole-body CT images from one patient. The CT scanner was the LightSpeed 16 CT of GE. The data of each age was composed of CT images of different body parts from several pediatric patients at the same age. Therefore, it was necessary to perform initial registration manually, according to the basic knowledge of anatomy. The pediatric patients were chosen with the closest height and weight to the reference data. Since the organs of the chest and abdomen in the normal CT scan were not well defined enough. We searched for enhanced images of the chest and the abdomen to facilitate the segmentation of organs. The final selected CT images were arranged in table 2.
Table 2. CT data obtained from different patients.
| Age (y) | Gender | Height (cm) | Weight (kg) | CT scan range |
|---|---|---|---|---|
| 0.25 | Female | 60 | 7.5 | Lower limb |
| 59 | 7 | Trunk | ||
| 60 | 6 | Head | ||
| 60 | 6 | Abdominal enhancement | ||
| 59 | 6.5 | Chest enhancement | ||
| Male | 62 | 6 | Abdominal enhancement | |
| 60 | 6 | Chest enhancement | ||
| 1 | Female | 80 | 10 | Trunk |
| 78 | 9.5 | Lower limb | ||
| 82 | 11 | Head | ||
| 80 | 10 | Upper limb | ||
| 80 | 10 | Chest enhancement | ||
| 80 | 11 | Abdominal enhancement | ||
| Male | 75 | 9 | Chest enhancement | |
| 81 | 10 | Abdominal enhancement | ||
| 5 | Male | 112 | 19 | Trunk |
| 114 | 20 | Lower limb | ||
| 112 | 20 | Head | ||
| 113 | 19 | Chest enhancement | ||
| 111 | 19 | Abdominal enhancement | ||
| Female | 111 | 20 | Chest enhancement | |
| 110 | 19 | Abdominal enhancement | ||
| 10 | Male | 146 | 32 | Trunk |
| 140 | 30 | Lower limb | ||
| 139 | 35 | Head | ||
| 140 | 38 | Chest enhancement | ||
| 141 | 34 | Abdominal enhancement | ||
| Female | 140 | 30 | Chest enhancement | |
| 136 | 32 | Abdominal enhancement | ||
| 15 | Female | 160 | 51 | Trunk |
| 159 | 52 | Upper limb | ||
| 159 | 50 | Lower limb | ||
| 150 | 42 | Head | ||
| 150 | 41 | Chest enhancement | ||
| 158 | 45 | Abdominal enhancement | ||
| Male | 170 | 60 | Trunk | |
| 171 | 58 | Lower limb | ||
| 175 | 60 | Head | ||
| 172 | 51 | Chest enhancement | ||
| 172 | 60 | Abdominal enhancement |
The process of CRC phantoms development included segmentation of computed tomography images, supplement construction of particular organs, optimization and adjustment of organs, and the combination of all components. The whole process was shown in the next flowchart (figure 1).
Figure 1. The process of development of CRC phantoms (aspecial organs: rib cartilages, salivary glands, the tongue, eyeballs, lens, testicle, prostate, ovaries, cheeks, the small intestine, and colon).
Download figure:
Standard image High-resolution imageFigure 2. Semi-automatic threshold segmentation of skeletons in 3D-DoctorTM.
Download figure:
Standard image High-resolution image2.1.1. Segmentation of CT images
A software named 3D-DoctorTM (Able Software Corp. 2012) was used to segment the contours of different organs and bones from the CT images. Firstly, CT images were imported into 3D- DoctorTM to segment the skeletons. Since the density of skeletons was quite different from other organs, it was easy to distinguish them in CT images. The semi-automatic threshold segmentation tool in 3D-DoctorTM was used to segment the skeletons, as shown in figure 2.
Since the density of some organs was not much different from soft tissues or other organs around them, the semi-automatic threshold segmentation function in 3D-DoctorTM software was not suitable for the segmentation of these organs. Therefore, manual segmentation was required. Most of the organ segmentation was performed in enhanced CT images, in which the contours of the organs were more apparent than in normal CT images. The median filtering operation was performed to increase the contrast of the organ in the CT images. This improved the quality of the CT image and facilitates the subsequent manual segmentation of the organ. The organs from every CT image were segmented under the guidance of a professional pediatrician to ensure the correctness of the organ segmentation. For organs (such as lungs, liver, and spleen) whose contours were not definite enough, manual segmentation was processed. For various glands (such as the adrenal gland, pancreas, thyroid gland, and thymus), the volumes themselves were small, and their contrasts in the CT images were also inferior. It was necessary to combine the CT images with the coronal images and the sagittal images according to the anatomical structure. Considering the continuity condition, which determined the boundary contour, the organs were segmented in the transverse section and modified in the coronal and sagittal sections.
The 3D surfaces of the organs and skeletons after manual segmentation were generated by using 3D-DoctorTM. The outer contour was checked to conform to the normal organ anatomy and the missing CT images were checked as well. The surface rendering function of the 3D-DoctorTM can develop the contours of the different organs and skeletons into polygon-mesh (PM) models.
The segmented organs, including semi-automatic threshold segmentation and manual segmentation results, were imported into RhinocerosTM (Robert McNeel & Associates 2018) for further processing.
2.1.2. Construction of particular organs
The organs, which had not been segmented in the CT images, were developed via the non-uniform rational B-spline (NURBS) method. The organs such as rib cartilage, salivary glands, and tongue were transformed from the basic geometries, which were similar to the real shape. For example, the eyeballs were established as spheres. The lens, the testicle, the prostate, and the ovaries were represented by ellipsoids. The tongue was built into a cylinder and then smoothed by edge chamfering and axial compression. The cheeks were built as an ellipsoid, which was chamfered and curved. At the same time, as the soft ribs were not displayed in the CT images, we established the soft ribs manually. Several cylinders with a smaller radius at the end close to the ribs and a larger radius at the other end were modified to represent the soft ribs. They were firstly compressed and then bent along the trend of ribs. Both ends of the cylinder were chamfered. The anatomical structures of the small intestine and colon themselves were extremely complicated. It was tough to operate if a structure identical to the anatomical shape required to be established. The method developed by (Kim et al 2018) was adopted in the present work to establish the mesh-type small intestine and colon. Firstly, the general trend lines of the small intestine and colon were established. Then, based on the trend line, the colon was constructed as a changing diameter pipe, while the small intestine was kept as a constant diameter pipe. Figure 3 showed the structures of the small intestine and colon and the structure established in our phantom. Finally, the NURBS models were converted into the polygon mesh (PM) model via the RhinocerosTM (Robert McNeel & Associates 2018).
Figure 3. The structure of the small intestine and colon developed in this work via RhinocerosTM.
Download figure:
Standard image High-resolution image2.1.3. Optimization and adjustment of organs
The NURBS models constructed by the 3D-DoctorTM were composed of a large number of meshes, which were not convenient for phantom adjustment. With the 'Reduce Mesh' instruction in the RhinocerosTM, we reduced the number of meshes to an appropriate quantity. However, in the process of mesh deletion, the smoothness of the organ was destroyed, and the surfaces of the organs became extremely rough. Decreasing the number of PM grids to target numbers at one time would change the shape of the organs and cause errors. Therefore, it was necessary to cut down a smaller number of meshes each time and smooth the organs at the same time. After many trials for this study, the number of PM grids deleted was about 10% each time, and the smoothing coefficient was set at around 0.3. These two instructions were used interchangeably. In this way, we reduced the number of PM grids at the same time to ensure the shape and the smoothness of the organs were well maintained. Figure 4 showed the PM surfaces of the liver before (left) and after (right) the optimization by using the RhinocerosTM.
Figure 4. The liver before (a) and after (b) the optimization in RhinocerosTM.
Download figure:
Standard image High-resolution imageThe process of deleting the PM grids induced various PM grid errors. If these errors were not corrected, as the mesh type phantom was voxelized, an error would occur and the original shape of the organ could not be guaranteed. There are three common errors: the exposed mesh surface, the duplicate mesh surface, and the non-manifold mesh surface. The exposed mesh surface means holes appear on the surface. These holes can usually be filled in with the 'Fill Mesh Hole' command. The second type of error is a duplicate mesh face or an extra mesh face. The presence of such a grid can cause problems with the shape or integrity of the organ. These mesh faces need to be removed by using the 'Delete Mesh Surfaces' command. The third error is a non-manifold mesh surface. A non-manifold mesh surface means that the edge is used by multiple meshes. In this situation, the program cannot distinguish between the inside and the outside organs. Therefore, without affecting the original structure, we used the commands mentioned above, 'Delete Mesh Surfaces', 'Fill Mesh Hole' and 'Patch Single Surfaces', to solve the errors of these non-manifold meshes. The primitive models were refined to turn into high-quality models by the method mentioned above.
Each pediatric mesh-type phantom was constructed based on CT images of several pediatric patients. Therefore, it was necessary to stitch the outer skins of these children together to form a complete skin. The skin was a complete enclosure in an exported file of the software 3D-DoctorTM. The bottom PM meshes of the single part of the skin were removed at first, the command 'Patch Single Surfaces' was then used in combination with the command 'Fill Mesh Hole' to splice the different parts of the skin.
2.1.4. Adjustment of organ mass to reference value
Considering that the patients' physical data was not entirely consistent with the reference data, it was required to adjust the mass of each organ to fit the reference data. The reference volume of each organ was calculated referring to the ICRU Report 46 (ICRU 1992) and GBZ/T 200.2-2007 (Ministry of Health P.R. China 2007b). The reference masses of Chinese children's organs were mainly found in the report of China's occupational health standard GBZ/T 200.1-2007 (Ministry of Health P.R. China 2007a). For the reference masses of the organs not given in the report, the Asian reference data was used here instead (Tanaka et al 1998). At the same time, the densities of the organs were found in the ICRU Report 46 (ICRU 1992), and the reference volumes of organs were calculated.
For general organs, the offset commands in RhinocerosTM were used to adjust the volume of the organ to match the reference volume. A negative offset represents expansion, and a positive offset represents reduction.
For an organ containing a wall to the contents, the reference volume of the content and the reference volume of the wall were first calculated, the two values were then added together to obtain the total volume of the wall and the contents. The volume was first matched to the total volume to obtain the outer wall of the organ. The offset command was then used to generate an additional PM mesh surface as being the inner wall of the organ. The volume of the inner wall was matched to the volume of the contents. The wall of the organ was located between the outer wall and the inner wall. Inside the inner wall were the contents of the organ.
2.1.5. Adjustment of the skeleton system
Skeletons are composed of cortical bone (CB), trabecular bone (TB), red bone marrow (RBM), yellow bone marrow (YBM), cartilage, and miscellaneous bone. RBM and bone surface in trabecular bone are radiation-sensitive organs. In this modeling process, the basic PM models of the skeleton were produced by using the same conversion procedure employed for the general organs and tissues. The site-specific non-uniform bone model was then used to divide the bone into cortical bone (CB) and spongiosa. The spongiosa was uniformly mixed by TB, RBM, YBM, and miscellaneous bone. The cartilage portion was mixed into the soft tissue in the phantom. The GBZ/T 200.2-2007 provides reference masses of different skeletal tissues. table 3 listed the mass data for the bone composition of Asian children's reference phantoms, and the reference density was also obtained from the ICRU Report 46 (ICRU 1992). Bone distribution data was obtained from the ICRP Publication 70 (ICRP 1995) and ICRP Publication 89 (ICRP 2002).
Table 3. The mass data for the bone composition of Asian reference pediatric phantoms.
| 0.25y | 1y | 5y | 10y | 15y Female | 15y Male | |
|---|---|---|---|---|---|---|
| Skeletal | Mass (g) | |||||
| Compact bone | 300 | 850 | 1400 | 2400 | 3000 | 4000 |
| CB a | 240 | 670 | 1100 | 1900 | 2400 | 3200 |
| TB a | 60 | 180 | 300 | 500 | 600 | 800 |
| RBM a | 110 | 190 | 310 | 700 | 750 | 900 |
| YBM a | 0 | 0 | 0 | 600 | 900 | 1000 |
| Cartilage | 130 | 170 | 290 | 480 | 640 | 840 |
| miscellaneous bone | 60 | 90 | 200 | 420 | 410 | 560 |
| sum | 600 | 1300 | 2200 | 4600 | 5700 | 7300 |
a CB- cortical bone, TB- trabecular bone, RBM-red bone marrow, YBM-yellow bone marrow.
2.1.6. Combination of all components
Established organs and bones were assembled into a complete phantom. The overlapping parts were adjusted appropriately. It was achieved by a subtle movement of the position and transformation of the organ shape. In RhinocerosTM, we set series control points on the NURBS surface to complete partial adjustment and kept the mass consistent with the reference data at the same time.
2.1.7. Voxelized of the mesh type phantom
Since the established mesh-type phantoms still have some tiny surface imperfection, they cannot be directly applied to the Monte Carlo simulation. The phantoms were voxelized and converted into voxel phantoms. The THUDose software(Lu et al 2017), which was developed at Tsinghua University, was used to complete the process of voxelization. The voxel sizes of Chinese pediatric reference phantoms were listed in table 4.
Table 4. The voxel sizes of Chinese pediatric reference phantoms.
| Age (y) | Voxel size/mm * mm * mm | Height/cm | Weight/kg |
|---|---|---|---|
| 0.25 | 0.618 * 0.618 * 1 | 62 | 7 |
| 1 | 77 | 10 | |
| 5 | 0.927 * 0.927 * 1 | 110 | 19 |
| 10 | 139 | 32 | |
| 15/Female | 1.226 * 1.226 * 1 | 158 | 50 |
| 15/Male | 168 | 55 |
2.2. Application in CT dosimetry
2.2.1. Monte Carlo simulation of the CT scanner
THUDose (Lu et al 2017) is a Monte Carlo modeling and simulation software with user-friendly UI, which is developed at Tsinghua University based on Geant4 code. The modeling and simulation processes of this work were carried out based on THUDose.
Throughout communication with Chinese pediatricians, we learned that the LightSpeed 16 CT of GE company simulated in this article is widely used clinically in China. In addition, the data of 16 slice scanners is adopted as the current national standard data(Ministry of Health P.R. China 1995). The collimation width of this type of CT includes 5, 10, and 20 mm. Moreover, there were two types of bowtie filters for the head and body. The opening angles of the fan beam for head and body scanning were 27.5° and 55°, respectively. The distance from the source to the phantom center of this type of CT was 54 cm, and the maximum scan field was 50 cm in width. The x-rays emit a fan beam from a point and this beam passes through the flat filter, which was used to absorb the low-energy part of x-rays and then the bowtie filter. The bowtie filter was used to compensate for the difference in the thickness of the human body. It was also used to balance the amount of radiation reaching the detector, thereby improving the image quality and reducing the peripheral dose of the patient.
The x-ray source was directly obtained by energy spectrum sampling. The energy spectrums adopted by x-rays were generated using XCOMP5R software (R. Nowotny 1985)for specific parameters such as tube voltage, anode angle, plate filter material, and thickness. The tube voltages used in this study were 120, 100, and 80 kVp. The anode angle was 12.0°. The structure of the flat filter was considered in this study when generating an x-ray energy spectrum. The flat filter was set as 2.5 mm thick aluminum. The distance from the source to the x-ray energy spectrum recording position was 10 cm.
The material and shape of the bowtie filter determine the final x-ray distribution. However, the actual geometry of the bowtie filter was relatively complex, and it was difficult to obtain the geometric parameters from the CT scanner manufacturer. Therefore, the bowtie filters for head and body scanning used the simplified model, as shown in figure 5, with the material of aluminum. The dose profile was experimentally measured using an ion chamber. The geometric parameters were adjusted until the dose distributions obtained by Monte Carlo simulation were coincident with experimentally measured results(Pan et al 2014). According to the information provided by the CT production, the CT bed was modeled as part of a 91 cm internal diameter cylinder with a thickness of 4.5 mm, and the material was carbon.
Figure 5. The bowtie filters for head and body.
Download figure:
Standard image High-resolution imageCT scanning includes helical scanning and axial scanning. Organ doses absorbed from helical scanning are related to the pitch, and they are approximately equal with organ doses obtained from the axial scanning with the same scanning parameters (Pan et al 2014). Therefore, the axial scanning type was simulated in this work to generate the organ dose database. 16 x-ray sources and filters were uniformly arranged in a circle to simulate an x-ray tube rotating one circle. This method was verified by the work of (Pan et al 2014).
This modeling of CT scanner was verified by comparison of simulated and measured results. A complete system for x-ray measurements (Unfors RaySafe X1, Sweden) were used to measure the CTDI100 value in the air from the isocenter to the edge of the CT scanner. For the lateral dose profile, the relative errors between calculation and experimental results were less than 7%. CTDI100 values were measured in the standard CTDI head and body phantoms by using ion chambers and it showed a good agreement with simulated results. Moreover, the modeling method was verified based on pediatric physical phantom(Pan et al 2014). For the head and body filter, the CTDIvol was measured using a head CTDI phantom with a diameter of 16 cm and a body CTDI phantom with a diameter of 32 cm.
2.2.2. Establishment of the database for CT scan organ dose
The Chinese pediatric reference computational phantom (CRC) and the CT scanner model established in this study were further used to calculate the organ absorbed dose database using the Monte Carlo method. The collimators were moved from the head to the feet of the phantoms, and the organ doses were calculated for once axial scanning in each position. The Monte Carlo simulations were formed for the scanning conditions with x-ray tube voltages of 120, 100 and 80 kVp, with collimators of 20, 10, and 5 mm width, with filters for head and body. Finally, the datasets including organ doses obtained from single-layer axial scanning were established. The organ doses were calculated based on the CT scan organ dose databases for each phantom. The scan ranges included the head, chest, abdominal-pelvis, and chest-abdomen-pelvis (CAP). The human anatomy positions of the scan range were determined referring to the AAPM CT scan protocol (AAPM 2010) (table 5).
Table 5. Scan range referring to the AAPM CT scan protocol.
| Scan range | Positions |
|---|---|
| Head | From foramen magnum through the top of the calvarium |
| Chest | From the top of the lungs through the bottom of the lungs |
| Abdomen-Pelvis | From the top of the liver to pubic symphysis |
| Chest-Abdomen-Pelvis (CAP) | From the top of the lungs through the pubic symphysis |
The absorbed doses of most organs (organs except for skin, red bone marrow, and bone surface) were directly calculated by dividing the energy deposition in the organs by organ mass. However, for skin, red bone marrow, and bone surface, the voxel model cannot describe these organs accurately. The voxel size of the phantom was generally larger than the skin thickness, resulting in a higher simulation of skin thickness and quality. The 'equivalent mass thickness' method proposed by (Liu 2010) was used to calculate the skin dose. This method reduced the skin density so that the mass thickness of the thick skin is the same as the thickness of the real skin. When the simulation was processed, the deposition energy in the low-density thick skin was recorded, and the skin dose was obtained by dividing the actual mass of the skin. The thickness of the bone surface was only 50 μm, which could not be described in the voxel model. In this study, the 3CFs-improved method proposed by (Liu et al
2009) was used to calculate the red bone marrow dose. The red bone marrow dose ( ) in different parts was as shown below. The total red bone marrow dose was mass-weighted average of the red bone marrow doses at different sites.
) in different parts was as shown below. The total red bone marrow dose was mass-weighted average of the red bone marrow doses at different sites.

where:  and
and  are the absorbed doses of red bone marrow and spongiosa at the calculated bone site.
are the absorbed doses of red bone marrow and spongiosa at the calculated bone site.  and
and  are the mass absorption coefficients of red bone marrow and cancellous bone, respectively.
are the mass absorption coefficients of red bone marrow and cancellous bone, respectively.  is photon fluence in cancellous bone, assumed to be the same as photons in the red bone marrow.
is photon fluence in cancellous bone, assumed to be the same as photons in the red bone marrow.  is a factor that considers the dose-enhancing effect of electrons generated in trabecular bone entering red bone marrow.
is a factor that considers the dose-enhancing effect of electrons generated in trabecular bone entering red bone marrow.
2.2.3. Method of organ dose calculation for CRC phantoms
Based on the database of CT scan organ doses, considering the CT scan range, scan current, time, and other parameters, data was selected from the database to calculate the organ dose of the CT anthropomorphic phantom. The organ dose calculated by Monte Carlo simulation was the dose-normalized to a single x-ray photon. The number of x-ray photons in the actual CT scan was determined by the tube current and scan time, which is difficult to be quantified. To quantitatively compare the Monte Carlo simulation value with the actual measured value, a conversion factor (CF) was shown below.

where: CTDI100 measured is the CTDI100 value per 100 mAs of CT scan air measured in the CT isocenter, mGy/100 mAs. CTDI100simulated is the CTDI100 per unit x-ray photon under the same CT scan parameters obtained by Monte Carlo simulation, MeV g−1. The organ dose was obtained by multiplying the dose value of the Monte Carlo simulation by the CF factor. The organ absorbed dose to an axial scan was calculated as shown below.

where: Dj is the dose of organ j, mGy. Di, j is the simulated value of the average absorbed dose of the ith axis scan to the organ j. z1 and z2 are respectively the starting layers and terminate layers of the organ dose database corresponding to the scan range. I is the tube current, mA. t is the scan time of one revolution of CT, s.
For the helical scan, the pitch was considered. The absorbed dose of the organ was inversely proportional to the pitch as shown below. Pitch was defined as the ratio of the distance traveled by the CT per revolution and the collimation width.

In order to reconstruct the first and last images during the helical scan, the actual scan range would automatically exceed the scan range of the planned image, called over-scan. The length of the over-scan increases as the pitch or collimation width increased. For the typical 16-row multi-slice CT, the over-scan length is between 3 to 6 cm. Considering the radiation dose caused by the overscan, the method for calculating the absorbed dose of the organ is as shown below, where zover is the number of layers covered in the database for the over-scan length.

In modern multi-slice helical CT devices, automatic exposure control (AEC) has become a standard device used to reduce CT radiation dose, including tube current modulation and tube voltage modulation. At present, tube current modulation technology is mainly used to automatically adjust the tube current intensity during CT scanning according to the patient's body size and attenuation characteristics. The organ dose calculation caused by CT scan using the tube current modulation technique is shown below, where Ii is the actual tube current of the ith layer. The actual tube current value of each layer set by the tube current modulation technique can be read from the DICOM file exported by the CT scanner.

According to the study by (Turner et al 2010), When organ doses are normalized by CTDIvol values, the differences across scanners become very small, with a mean of 5.2%. Therefore, the organ dose caused by other CT scanners(DCT2) can be converted from the calculated value of the GE LightSpeed 16 CT (DCT1), as shown below. CTDIvol, CT2 is the volumetric CT dose index for other CT models, and CTDIvol, CT1 is the volumetric CT dose index for the GE LightSpeed 16 CT under the same scan parameters. It should be noted that the above method is only been proved so far for CT systems up to 64 slices.

The effective doses of the patient were calculated as shown below, where wR is the radiation weighting factor. For CT radiation emission, the x-ray radiation weighting factor is 1. The wT is the tissue weighting factor according to the ICRP Publication 103 (ICRP 2007).

2.2.4. Comparison of the organ doses with other studies
Both VirtualDose software (Ding et al 2015) and (Lee et al 2012) calculated the doses of different types of CT for children of different ages. The pediatric phantoms used by VirtualDose and Lee et al were UF reference pediatric hybrid phantoms. The organ dose results of 1-year-old children with the same scan range using the same CT scan parameters with the works of literature were compared here.
The works conducted by (Ding et al 2015) and (Lee et al 2012) were on Siemens SOMATOM Sensation 16, which was different from the GE Light Speed 16 simulated in this study. The organ dose was divided by the CTDIvol under the corresponding scan conditions. It should be noted that the study of Lee et al used the head CTDIvol for normalization of all the scan ranges. For comparative purposes, the results of the chest, Abdomen-Pelvis, and CAP scan from the study of Lee et al were multiplied by the head CTDIvol and divided by the body CTDIvol.
3. Results
3.1. Mesh-type Chinese pediatric reference phantoms series
In this study, a series of Chinese pediatric mesh-type phantoms was established as shown in figure 6. For most organs, the differences in organ mass between the CRC series and the reference data of Chinese reference children were within 2% (tables 17–18).
Figure 6. Mesh-type Chinese pediatric reference phantoms series.
Download figure:
Standard image High-resolution image3.2. CT scanner modeling and dose results
3.2.1. CT scanner parameters
The conversion factors of CTDI 100 measured and simulated (CF) with different CT scan parameters using the head and body bowtie filter were shown in tables 6 and 7. GE LightSpeed 16 CT CTDIvol measurements per 100 mAs at different tube voltages and collimation widths were shown in table 8. CTDI100 was measured three times and averaged, and errors were calculated as the error of the mean. The errors related to the simulated values were probability statistic errors obtained by the Monte Carlo method.
Table 6. The conversion factors of CTDI100 measured and simulated (CF) with different CT scan parameters using the head bowtie filter.
| Tube voltages (kVp) | Collimator width (mm) | CTDI100 measured (mGy/100 mAs) | CTDI100 simulated (MeV g−1) | CF (mGy g/100 mAs/MeV) |
|---|---|---|---|---|
| 80 | 5 | 14.90 ± 0.05 | 7.72 ± 0.15 × 10−5 | 1.93 ± 0.04 × 105 |
| 10 | 12.25 ± 0.03 | 3.84 ± 0.08 × 10−5 | 3.19 ± 0.06 × 105 | |
| 20 | 10.44 ± 0.01 | 1.93 ± 0.04 × 10−5 | 5.42 ± 0.11 × 105 | |
| 100 | 5 | 25.01 ± 0.06 | 8.43 ± 0.17 × 10−5 | 2.97 ± 0.06 × 105 |
| 10 | 20.56 ± 0.03 | 4.22 ± 0.08 × 10−5 | 4.87 ± 0.10 × 105 | |
| 20 | 17.55 ± 0.02 | 2.11 ± 0.04 × 10−5 | 8.33 ± 0.17 × 105 | |
| 120 | 5 | 36.67 ± 0.02 | 9.07 ± 0.18 × 10−5 | 4.04 ± 0.08 × 105 |
| 10 | 30.19 ± 0.04 | 4.53 ± 0.09 × 10−5 | 6.66 ± 0.13 × 105 | |
| 20 | 25.70 ± 0.06 | 2.27 ± 0.05 × 10−5 | 1.13 ± 0.02 × 106 |
Table 7. The conversion factors of CTDI 100 measured and simulated (CF) with different CT scan parameters using the body bowtie filter.
| Tube voltages (kVp) | Collimator width (mm) | CTDI100 measured (mGy/100 mAs) | CTDI100 simulated (MeV g−1) | CF (mGy g/100 mAs/MeV) |
|---|---|---|---|---|
| 80 | 5 | 10.85 ± 0.01 | 3.03 ± 0.06 × 10−5 | 3.58 ± 0.07 × 105 |
| 10 | 8.92 ± 0.01 | 1.52 ± 0.03 × 10−5 | 5.88 ± 0.12 × 105 | |
| 20 | 7.57 ± 0.02 | 7.54 ± 0.15 × 10−6 | 1.00 ± 0.02 × 106 | |
| 100 | 5 | 19.50 ± 0.06 | 3.48 ± 0.07 × 10−5 | 5.60 ± 0.11 × 105 |
| 10 | 16.13 ± 0.03 | 1.73 ± 0.03 × 10−5 | 9.30 ± 0.19 × 105 | |
| 20 | 13.68 ± 0.03 | 8.66 ± 0.17 × 10−6 | 1.58 ± 0.03 × 106 | |
| 120 | 5 | 29.98 ± 0.04 | 3.86 ± 0.08 × 10−5 | 7.77 ± 0.16 × 105 |
| 10 | 24.73 ± 0.14 | 1.93 ± 0.04 × 10−5 | 1.28 ± 0.03 × 106 | |
| 20 | 21.11 ± 0.04 | 9.61 ± 0.19 × 10−6 | 2.20 ± 0.00 × 106 |
Table 8. GE LightSpeed 16 CT CTDIvol measurements per 100 mAs at different tube voltages and collimation widths.
| CTDIvol (mGy/100 mAs) | ||||
|---|---|---|---|---|
| Collimator width (mm) | ||||
| Filter | Tube voltages (kVp) | 5 | 10 | 20 |
| Head | 80 | 8.33 ± 0.02 | 6.84 ± 0.01 | 5.79 ± 0.01 |
| 100 | 15.21 ± 0.03 | 12.50 ± 0.02 | 10.58 ± 0.02 | |
| 120 | 23.42 ± 0.05 | 19.24 ± 0.03 | 16.23 ± 0.03 | |
| 140 | 32.68 ± 0.06 | 26.84 ± 0.05 | 22.63 ± 0.05 | |
| Body | 80 | 3.50 ± 0.01 | 2.88 ± 0.01 | 2.43 ± 0.01 |
| 100 | 6.97 ± 0.01 | 5.71 ± 0.01 | 4.82 ± 0.01 | |
| 120 | 11.19 ± 0.02 | 9.17 ± 0.01 | 7.74 ± 0.02 | |
| 140 | 16.16 ± 0.03 | 13.32 ± 0.03 | 11.17 ± 0.02 | |
3.2.2. Organ absorbed doses and effective doses
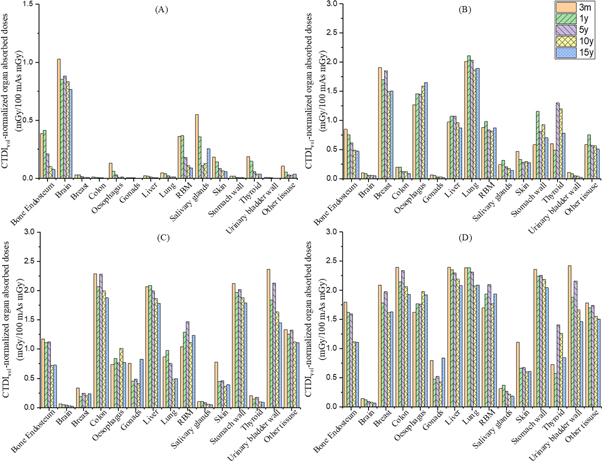
Organ absorbed doses and effective doses of phantoms for different ages were shown in tables 9–12, with the values for head, chest, abdomen-pelvis and chest-abdomen-pelvis, respectively and depicted as column graphs in figure 7. The scanning parameters were tube voltage 120 kVp, tube current-time product 100 mAs, and collimation width 10 mm. A Head filter was used in the head scan simulation, and a body filter was used for chest, Abdomen-Pelvis, and CAP scan simulation. For most organs, the organ dose of the 3-month (3 m) phantom was the highest within the phantoms series, while the organ dose of the 15-year-old phantom (15y) was the lowest. Take the dose of typical organs in each scanning range as an example to illustrate the biggest difference for different ages. For each scanning range (head, chest, abdomen-pelvis, and CAP examinations), the typical organs (brain, lung, colon, and stomach) dose of 3 m were 34.2%, 6.4%, 22.1%, and 15.5% higher than that of the 15y. This illustrated that the establishment of age-specific pediatric phantoms was necessary for accurate CT dose assessment for pediatric patients.
Table 9. CTDIvol and 100 mAs-normalized organ absorbed doses (mGy/100 mAs mGy) and effective doses (mSv/100 mAs mGy) for head examinations.( 'Other tissues' includes adrenal gland, extrathoracic, gallbladder, heart, kidney, lymph node, muscle, oral mucosa, pancreas, prostate, small intestine, spleen, and thymus.).
| Organ name | CTDIvol-normalized organ absorbed doses (mGy/100 mAs mGy) and effective doses (mSv/100 mAs mGy) | ||||||
|---|---|---|---|---|---|---|---|
| 3 m | 1y | 5y | 10y | 15y-M | 15y-F | 15y a | |
| Bone surface* | 0.385 | 0.414 | 0.210 | 0.100 | 0.079 | 0.073 | 0.076 ± 0.003 |
| Brain++ | 1.026 | 0.852 | 0.880 | 0.834 | 0.772 | 0.757 | 0.764 ± 0.008 |
| Breast | 0.028 | 0.027 | 0.016 | 0.008 | 0.008 | 0.007 | 0.007 ± 0.000 |
| Colon | 0.010 | 0.009 | 0.005 | 0.003 | 0.002 | 0.002 | 0.002 ± 0.000 |
| Esophagus | 0.132 | 0.059 | 0.030 | 0.009 | 0.013 | 0.006 | 0.010 ± 0.003 |
| Gonads | 0.006 | 0.005 | 0.003 | 0.001 | 0.000 | 0.001 | 0.001 ± 0.000 |
| Liver | 0.021 | 0.018 | 0.010 | 0.005 | 0.005 | 0.003 | 0.004 ± 0.001 |
| Lung | 0.048 | 0.039 | 0.021 | 0.013 | 0.013 | 0.010 | 0.011 ± 0.002 |
| Red bone marrow* | 0.361 | 0.367 | 0.180 | 0.110 | 0.091 | 0.086 | 0.089 ± 0.003 |
| Salivary glands* | 0.549 | 0.358 | 0.110 | 0.126 | 0.398 | 0.110 | 0.254 ± 0.144 |
| Skin* | 0.183 | 0.142 | 0.086 | 0.062 | 0.056 | 0.056 | 0.056 ± 0.000 |
| Stomach | 0.017 | 0.017 | 0.009 | 0.005 | 0.004 | 0.003 | 0.003 ± 0.000 |
| Thyroid* | 0.187 | 0.146 | 0.055 | 0.034 | 0.044 | 0.028 | 0.036 ± 0.008 |
| Bladder | 0.007 | 0.006 | 0.003 | 0.001 | 0.001 | 0.001 | 0.001 ± 0.000 |
| Other tissues* | 0.105 | 0.053 | 0.028 | 0.024 | 0.052 | 0.022 | 0.037 ± 0.015 |
| Effective doses | 0.104 | 0.089 | 0.048 | 0.033 | 0.036 | 0.027 | 0.031 ± 0.004 |
a The results of 15y refers to the average of 15y-male and 15y-female, and the error associated to the average of organ doses for 15y.'no symbol', '*' and '++' refers to organs outiside of x-ray beam, partially covered by x-ray beam and fully covered by x-ray beam, respectively.
Figure 7. CTDIvol -normalized organ absorbed doses of phantoms for different ages (tube voltage 120 kVp, tube current-time product 100 mAs, collimation width 10 mm): (A) head examination (B) chest examinations (C) Abdomen-Pelvis examinations (D) CAP (Chest-Abdomen-Pelvis) examinations.
Download figure:
Standard image High-resolution imageTable 10. CTDIvol and 100 mAs-normalized organ absorbed doses (mGy/100 mAs mGy) and effective doses (mSv/100 mAs mGy) for chest examinations.
| Organ name | CTDIvol-normalized organ absorbed doses (mGy/100 mAs mGy) and effective doses (mSv/100 mAs mGy) | ||||||
|---|---|---|---|---|---|---|---|
| 3 m | 1y | 5y | 10y | 15y-M | 15y-F | 15y a | |
| Bone surface* | 0.848 | 0.755 | 0.619 | 0.491 | 0.424 | 0.524 | 0.474 ± 0.050 |
| Brain | 0.100 | 0.088 | 0.061 | 0.059 | 0.053 | 0.048 | 0.051 ± 0.003 |
| Breast++ | 1.907 | 1.699 | 1.854 | 1.501 | 1.521 | 1.490 | 1.505 ± 0.016 |
| Colon | 0.200 | 0.198 | 0.120 | 0.122 | 0.076 | 0.099 | 0.088 ± 0.011 |
| Esophagus* | 1.271 | 1.456 | 1.450 | 1.589 | 1.561 | 1.749 | 1.655 ± 0.094 |
| Gonads | 0.065 | 0.056 | 0.034 | 0.031 | 0.013 | 0.030 | 0.021 ± 0.008 |
| Liver | 0.974 | 1.072 | 1.073 | 0.966 | 0.849 | 0.895 | 0.872 ± 0.023 |
| Lung++ | 2.015 | 2.111 | 2.030 | 1.870 | 1.812 | 1.974 | 1.893 ± 0.081 |
| Red bone marrow* | 0.878 | 0.981 | 0.835 | 0.817 | 0.753 | 0.989 | 0.871 ± 0.118 |
| Salivary glands | 0.244 | 0.315 | 0.209 | 0.177 | 0.126 | 0.163 | 0.144 ± 0.018 |
| Skin* | 0.469 | 0.329 | 0.283 | 0.295 | 0.254 | 0.292 | 0.273 ± 0.019 |
| Stomach | 0.586 | 1.160 | 0.810 | 0.928 | 0.595 | 0.814 | 0.705 ± 0.109 |
| Thyroid | 0.601 | 0.493 | 1.301 | 1.200 | 0.435 | 1.126 | 0.781 ± 0.346 |
| Bladder | 0.109 | 0.088 | 0.052 | 0.043 | 0.024 | 0.028 | 0.026 ± 0.002 |
| Other tissues* | 0.587 | 0.752 | 0.572 | 0.563 | 0.491 | 0.525 | 0.508 ± 0.017 |
| Effective doses | 0.881 | 0.972 | 0.916 | 0.861 | 0.754 | 0.872 | 0.813 ± 0.059 |
a The results of 15y refers to the average of 15y-male and 15y-female, and the error associated to the average of organ doses for 15y.'no symbol', '*' and '++' refers to organs outiside of x-ray beam, partially covered by x-ray beam and fully covered by x-ray beam, respectively.
Table 11. CTDIvol and 100 mAs-normalized organ absorbed doses (mGy/100 mAs mGy) and effective doses (mSv/100 mAs mGy) for Abdomen-Pelvis examinations .
| Organ name | CTDIvol-normalized organ absorbed doses (mGy/100 mAs mGy) and effective doses (mSv/100 mAs mGy) | ||||||
|---|---|---|---|---|---|---|---|
| 3 m | 1y | 5y | 10y | 15y-M | 15y-F | 15y a | |
| Bone surface* | 1.173 | 1.112 | 1.126 | 0.720 | 0.765 | 0.688 | 0.727 ± 0.038 |
| Brain | 0.061 | 0.052 | 0.042 | 0.026 | 0.023 | 0.022 | 0.023 ± 0.001 |
| Breast | 0.335 | 0.187 | 0.244 | 0.202 | 0.295 | 0.183 | 0.239 ± 0.056 |
| Colon++ | 2.289 | 2.068 | 2.277 | 1.992 | 1.940 | 1.810 | 1.875 ± 0.065 |
| Esophagus* | 0.737 | 0.837 | 0.766 | 1.013 | 0.578 | 0.966 | 0.772 ± 0.194 |
| Gonads* | 0.755 | 0.452 | 0.487 | 0.408 | 0.256 | 1.397 | 0.826 ± 0.571 |
| Liver++ | 2.063 | 2.089 | 1.993 | 1.865 | 1.751 | 1.815 | 1.783 ± 0.032 |
| Lung | 0.865 | 0.980 | 0.754 | 0.485 | 0.539 | 0.449 | 0.494 ± 0.045 |
| Red bone marrow* | 1.042 | 1.288 | 1.466 | 1.112 | 1.296 | 1.178 | 1.237 ± 0.059 |
| Salivary glands | 0.104 | 0.103 | 0.085 | 0.059 | 0.051 | 0.052 | 0.051 ± 0.001 |
| Skin* | 0.778 | 0.446 | 0.461 | 0.361 | 0.421 | 0.362 | 0.392 ± 0.029 |
| Stomach++ | 2.123 | 1.966 | 2.019 | 1.875 | 1.770 | 1.814 | 1.792 ± 0.022 |
| Thyroid | 0.205 | 0.146 | 0.174 | 0.097 | 0.101 | 0.089 | 0.095 ± 0.006 |
| Bladder++ | 2.361 | 1.837 | 2.128 | 1.633 | 1.441 | 1.456 | 1.449 ± 0.008 |
| Other tissues* | 1.333 | 1.256 | 1.322 | 1.122 | 1.131 | 1.094 | 1.113 ± 0.018 |
| Effective doses | 1.255 | 1.179 | 1.339 | 1.043 | 1.024 | 1.079 | 1.052 ± 0.028 |
a The results of 15y refers to the average of 15y-male and 15y-female, and the error associated to the average of organ doses for 15y.'no symbol', '*' and '++' refers to organs outiside of x-ray beam, partially covered by x-ray beam and fully covered by x-ray beam, respectively.
Table 12. CTDIvol and 100 mAs-normalized organ absorbed doses (mGy/100 mAs mGy) and effective doses (mSv/100 mAs mGy) for CAP (Chest-Abdomen-Pelvis) examinations.
| Organ name | CTDIvol-normalized organ absorbed doses (mGy/100 mAs mGy) and effective doses (mSv/100 mAs mGy) | ||||||
|---|---|---|---|---|---|---|---|
| 3 m | 1y | 5y | 10y | 15y-M | 15y-F | 15y a | |
| Bone surface* | 1.797 | 1.621 | 1.599 | 1.119 | 1.101 | 1.121 | 1.111 ± 0.010 |
| Brain | 0.144 | 0.122 | 0.093 | 0.079 | 0.070 | 0.065 | 0.068 ± 0.003 |
| Breast++ | 2.088 | 1.784 | 1.975 | 1.615 | 1.663 | 1.594 | 1.629 ± 0.035 |
| Colon++ | 2.393 | 2.148 | 2.341 | 2.064 | 1.984 | 1.873 | 1.929 ± 0.055 |
| Esophagus* | 1.624 | 1.771 | 1.760 | 1.975 | 1.783 | 2.056 | 1.919 ± 0.136 |
| Gonads* | 0.794 | 0.479 | 0.524 | 0.432 | 0.264 | 1.419 | 0.841 ± 0.578 |
| Liver++ | 2.397 | 2.355 | 2.298 | 2.193 | 2.026 | 2.140 | 2.083 ± 0.057 |
| Lung++ | 2.388 | 2.387 | 2.314 | 2.073 | 2.024 | 2.155 | 2.090 ± 0.066 |
| Red bone marrow* | 1.698 | 1.939 | 2.099 | 1.770 | 1.882 | 1.984 | 1.933 ± 0.051 |
| Salivary glands | 0.314 | 0.372 | 0.269 | 0.221 | 0.163 | 0.203 | 0.183 ± 0.020 |
| Skin* | 1.111 | 0.666 | 0.675 | 0.603 | 0.620 | 0.606 | 0.613 ± 0.007 |
| Stomach++ | 2.362 | 2.239 | 2.256 | 2.189 | 1.988 | 2.101 | 2.044 ± 0.057 |
| Thyroid | 0.729 | 0.574 | 1.410 | 1.264 | 0.504 | 1.187 | 0.846 ± 0.342 |
| Bladder++ | 2.425 | 1.883 | 2.161 | 1.663 | 1.456 | 1.476 | 1.466 ± 0.010 |
| Other tissues* | 1.785 | 1.701 | 1.745 | 1.556 | 1.502 | 1.512 | 1.507 ± 0.005 |
| Effective doses | 1.910 | 1.793 | 2.010 | 1.691 | 1.597 | 1.754 | 1.675 ± 0.079 |
a The results of 15y refers to the average of 15y-male and 15y-female, and the error associated to the average of organ doses for 15y.'no symbol', '*' and '++' refers to organs outiside of x-ray beam, partially covered by x-ray beam and fully covered by x-ray beam, respectively.
3.2.3. Comparison of the organ doses with other studies
The comparison and relative differences in CTDIvol normalized organ doses with VirtualDose (Ding et al 2015) and (Lee et al 2012) at different scan ranges for the 1-year-old children calculated in this study were shown in tables 13–16.
Table 13. CTDIvol and 100 mAs-normalized organ absorbed doses (mGy = 100 mAs mGy) for the 1-year Chinese reference phantoms for head examinations at tube potentials of 120 kVp.
| Organ name | CTDIvol-normalized organ absorbed doses (mGy/100 mAs mGy) | Relative difference | |||
|---|---|---|---|---|---|
| VirtualDose | Lee | This Study | VirtualDose | Lee | |
| Bone surface* | 0.340 | 0.296 | 0.414 | 21.79% | 39.89% |
| Brain++ | 0.960 | 0.925 | 0.852 | −11.24% | −7.89% |
| Breast | 0.010 | 0.013 | 0.027 | 169.20% | 107.08% |
| Colon | 0.000 | 0.002 | 0.009 | — | 338.00% |
| Esophagus | 0.050 | 0.087 | 0.059 | 17.96% | −32.21% |
| Gonads | 0.000 | 0.001 | 0.005 | — | 395.00% |
| Liver | 0.010 | 0.010 | 0.018 | 75.30% | 75.30% |
| Lung | 0.020 | 0.034 | 0.039 | 97.45% | 16.15% |
| Red bone marrow* | 0.250 | 0.368 | 0.367 | 46.98% | −0.15% |
| Salivary glands* | 0.180 | 0.498 | 0.358 | 98.88% | −28.11% |
| Skin* | 0.170 | 0.195 | 0.142 | −16.31% | −27.04% |
| Stomach | 0.010 | 0.009 | 0.017 | 68.00% | 86.67% |
| Thyroid* | 0.070 | 0.122 | 0.146 | 108.90% | 19.86% |
| Bladder | 0.000 | 0.001 | 0.006 | — | 508.00% |
'no symbol', '*' and '++' refers to organs outiside of x-ray beam, partially covered by x-ray beam and fully covered by x-ray beam, respectively.
Table 14. CTDIvol and 100 mAs-normalized organ absorbed doses (mGy = 100 mAs mGy) for the 1-year Chinese reference phantoms for Chest examinations at tube potentials of 120 kVp.
| Organ name | CTDIvol-normalized organ absorbed doses (mGy/100 mAs mGy) | Relative difference | |||
|---|---|---|---|---|---|
| VirtualDose | Lee | This Study | VirtualDose | Lee | |
| Bone surface* | 0.520 | 1.163 | 0.755 | 45.13% | −35.12% |
| Brain | 0.040 | 0.074 | 0.088 | 121.03% | 19.99% |
| Breast++ | 1.830 | 2.258 | 1.699 | −7.14% | −24.74% |
| Colon | 0.070 | 0.176 | 0.198 | 182.87% | 12.30% |
| Esophagus* | 1.330 | 2.224 | 1.456 | 9.44% | −34.54% |
| Gonads | 0.010 | 0.013 | 0.056 | 463.10% | 327.89% |
| Liver | 0.660 | 1.718 | 1.072 | 62.39% | −37.63% |
| Lung++ | 2.040 | 2.774 | 2.111 | 3.49% | −23.89% |
| Red bone marrow* | 0.530 | 0.918 | 0.981 | 85.08% | 6.81% |
| Salivary glands | 0.180 | 0.400 | 0.315 | 74.74% | −21.37% |
| Skin* | 0.360 | 0.637 | 0.329 | −8.64% | −48.35% |
| Stomach | 0.630 | 1.392 | 1.160 | 84.16% | −16.66% |
| Thyroid | 0.650 | 2.721 | 0.493 | −24.13% | −81.88% |
| Bladder | 0.020 | 0.042 | 0.088 | 339.95% | 108.95% |
'no symbol', '*' and '++' refers to organs outiside of x-ray beam, partially covered by x-ray beam and fully covered by x-ray beam, respectively.
Table 15. CTDIvol and 100 mAs-normalized organ absorbed doses (mGy=100 mAs mGy) for the 1-year Chinese reference phantoms for Abdomen-Pelvis examinations at tube potentials of 120 kVp.
| Organ name | CTDIvol-normalized organ absorbed doses (mGy/100 mAs mGy) | Relative difference | |||
|---|---|---|---|---|---|
| VirtualDose | Lee | This Study | VirtualDose | Lee | |
| Bone surface* | 0.480 | 1.229 | 1.112 | 131.57% | −9.56% |
| Brain | 0.010 | 0.018 | 0.052 | 424.10% | 184.53% |
| Breast | 0.130 | 2.124 | 0.187 | 43.82% | −91.20% |
| Colon++ | 2.230 | 2.734 | 2.068 | −7.27% | −24.37% |
| Esophagus* | 0.250 | 0.653 | 0.837 | 234.82% | 28.26% |
| Gonads* | 0.720 | 0.479 | 0.452 | −37.25% | −5.67% |
| Liver++ | 1.980 | 2.645 | 2.089 | 5.49% | −21.03% |
| Lung | 0.450 | 1.305 | 0.980 | 117.70% | −24.94% |
| Red bone marrow* | 0.520 | 0.926 | 1.288 | 147.77% | 39.09% |
| Salivary glands | 0.040 | 0.068 | 0.103 | 158.53% | 51.14% |
| Skin* | 0.730 | 0.963 | 0.446 | −38.88% | −53.68% |
| Stomach++ | 1.910 | 2.550 | 1.966 | 2.95% | −22.89% |
| Thyroid | 0.100 | 0.182 | 0.146 | 46.19% | −19.49% |
| Bladder++ | 2.110 | 2.395 | 1.837 | −12.95% | −23.30% |
'no symbol', '*' and '++' refers to organs outiside of x-ray beam, partially covered by x-ray beam and fully covered by x-ray beam, respectively.
Table 16. CTDIvol and 100 mAs-normalized organ absorbed doses (mGy=100 mAs mGy) for the 1-year Chinese reference phantoms for CAP (Chest-Abdomen-Pelvis) examinations at tube potentials of 120 kVp.
| Organ name | CTDIvol-normalized organ absorbed doses (mGy/100 mAs mGy) | Relative difference | |||
|---|---|---|---|---|---|
| VirtualDose | Lee | This Study | VirtualDose | Lee | |
| Bone surface* | 0.950 | 1.984 | 1.621 | 70.65% | −18.30% |
| Brain | 0.040 | 0.079 | 0.122 | 204.00% | 54.02% |
| Breast++ | 1.930 | 2.361 | 1.784 | −7.55% | −24.41% |
| Colon++ | 2.280 | 2.771 | 2.148 | −5.79% | −22.48% |
| Esophagus* | 1.510 | 2.342 | 1.771 | 17.28% | −24.39% |
| Gonads* | 0.720 | 0.482 | 0.479 | −33.41% | −0.45% |
| Liver++ | 2.350 | 2.876 | 2.355 | 0.23% | −18.11% |
| Lung++ | 2.340 | 2.966 | 2.387 | 2.00% | −19.53% |
| Red bone marrow* | 1.000 | 1.492 | 1.939 | 93.94% | 29.98% |
| Salivary glands | 0.210 | 0.421 | 0.372 | 77.01% | −11.71% |
| Skin* | 1.050 | 1.326 | 0.666 | −36.52% | −49.75% |
| Stomach++ | 2.290 | 2.771 | 2.239 | −2.25% | −19.22% |
| Thyroid | 0.730 | 2.774 | 0.574 | −21.38% | −79.31% |
| Bladder++ | 2.120 | 2.405 | 1.883 | −11.19% | −21.73% |
'no symbol', '*' and '++' refers to organs outiside of x-ray beam, partially covered by x-ray beam and fully covered by x-ray beam, respectively.
4. Discussion
The calculated organ dose of CT scanning is compared with the data from the other studies, in which different phantoms and scanners were applied. The possible factors that affect the calculation results were analyzed, including the difference of phantoms, skeleton model and method of dose calculation, and CT scanner difference.
4.1. Comparison of the organ doses with other studies
The results of this work were compared with works of VirtualDose (Ding et al 2015) and (Lee et al 2012) for each kind of scan situation. The series of UF pediatric phantoms(Lee et al 2010) (newborn, 1, 5, 10, and 15-year-old males and females) were employed in these two studies. The data of organs and tissues masses are from ICRP Publication 89, and body dimensions are matched to the reference anthropometric data of the United States(Lee et al 2012).
For the head scan, there was little difference in the dose of the brain, which was located in the scanning range. The differences between the results of this study and VirtualDose (Ding et al 2015) and (Lee et al 2012) were about 10%. The dose of the salivary gland in the scanning field was quite different. The calculated results in this study were about twice that of VirtualDose, but still 28.1% lower than Lee's results. For the bone surface and red bone marrow, the differences were more significant, the bone surface dose was 21.8% and 39.9% higher than the calculated values of VirtualDose and (Lee et al 2012), respectively, and the red bone marrow was 47% higher than the calculated value of VirtualDose and close to Lee's calculation results.
For chest, Abdomen-Pelvis, and CAP scan, the calculated results of Lee were much higher than that of this study and VirtualDose for organs in the scan range. The differences between VirtualDose and the calculated results of this study were small. Considering that VirtualDose and Lee's using the same phantoms, this study only compared the calculated results of these scan ranges with VirtualDose.
For the chest scan, the study was closer to VirtualDose, and the differences in the organs (such as breast and lung ) doses in the scanning ranges are within 10%. The liver and stomach are at the boundary of the scan range, which doses were significantly different, and the results of this study were 62.4% and 84.16% higher than the calculated value of VirtualDose. For organs not covered by the chest scan range (such as colon, gonads, and bladder), the largest difference reached 463.1%. The calculated bone surface and red bone marrow doses were 45.1% and 85.1% higher than VirtualDose, respectively.
For the Abdomen-Pelvis scan, the doses of the liver, colon, and stomach in the scan range were all within 8% difference with the calculated values of VirtualDose, and for bladder, the difference was −12.9%. The dose of the breast, which was out of the scan range, is 43.8% higher than VirtualDose. The dose of the lung, calculated in this study was about twice that of VirtualDose. The dose of the bone surface and red bone marrow was 131.6% and 147.8% higher than the calculated value of VirtualDose, respectively.
For CAP scan, the doses of the breast, lung, stomach, liver, and colon in the range were all within 8% of the calculated values of VirtualDose. The doses of the bone surface and red bone marrow were 70.6% and 93.9% higher than VirtualDose, respectively.
4.2. Factors affecting the radiation dose of CT scanning
4.2.1. Phantom and organ differences
As shown in section 4.1, the differences of dose for organs wholly covered in the scanning range were relatively small. For example, the brain dose undergoing the head scan, the lung dose undergoing chest scan, the stomach, liver, and colon dose undergoing the abdomen-pelvis scan, and most chest organs and abdominal organs dose undergoing the CAP scan. The differences in physical parameters of different anthropomorphic phantoms and the position differences of the organs in the cross-section of the human body contribute to the differences in organ doses.
For organs partially covered by the scan range, the organ dose was mainly affected by the proportion of organs covered in the scan range. For example, the position thyroid in CRC01 was upper than the 1-year-old UF phantom, causing more parts of the thyroid gland were included in the head scan. Therefore, the thyroid dose result of this study was higher than VirtualDose. The same reason caused the higher results of colon dose undergoing chest scan.
For organs that were not in the scan range, the organ dose was caused by scattered photons, which was mainly affected by the distance between the organ and the scanning boundary. The closer the organ was to the scanning boundary, the more scattering dose would be received. For different anthropomorphic phantoms, the relative positions of organs in the longitudinal distribution were different, therefore there was a difference in the distance between the organs and the scanning boundary in different phantoms. The height of the CRC01 phantom and the UF phantom used in VirtualDose and Lee's study were both 77 cm. By comparing the CRC01 phantom with UF phantom, we found that the positions of abdominal organs of the CRC01 were more upper than those of UF 1-year-old phantom. Therefore, for the head scan, the abdominal organ of this study was closer to the scanning range, and dose in this study was higher. Similarly, compared to the UF phantom, the brain of the CRC01 phantom was closer to the torso. For chest, abdomen and CAP scans, the brain of CRC01 was closer to the scan range than the UF phantom, causing higher dose results. This result illustrated that the difference in organ location was an important factor affecting the results of dose calculation. In addition, for some organs very far away from the field, the doses were so low that the relative differences were high.
The AAPM recommends age-defined pediatric protocols in CT examinations (AAPM 2010), and this requirement is also implemented in Europe. However, in clinical practice in China, adult protocols were often applied to children. In recent years, the radiation protection of pediatric patients in CT examinations is drawing increasing attention in China. Therefore, in the revision of Chinese national standards for CT examinations, it is required to promote protocols specifically for age-defined pediatric patients. This work provides a database for the formulation of national standards. In clinical practice, the organ mass and organ position of different patients vary largely, and they are also quite different from those of reference phantoms. In addition, for ages before 10 (including 10-year-old), the pediatric reference phantoms use the same phantoms with two sets of sex organs. This substitution can also affect the accuracy of the results. Therefore, the simulation results based on the reference phantoms can only be applied as a reference level for dose estimation of medical diagnosis. For more accurate dose calculation requirements, for example, in radiation therapy, patient-specific phantoms need to be developed and applied. However, developing phantoms for each patient manually is quite time-consuming and impractical. Therefore, automatic methods, such as automatic organ segmentation by applying the deep learning method, should be developed to replace the manual method.
Table 17. The differences in organ mass between the CRC series and the reference data of Chinese children (CRC03m, CRC01 and CRC05).
| Organ mass | Organ mass | Organ mass | Difference (%) | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Organs | Density (g cm−3) | CRC03m | Reference | Difference (%) | Density (g cm−3) | CRC01 | Reference | Difference (%) | Density (g cm−3) | CRC05 | Reference | |
| ET1 | 1.03 | 0.45 | 6 a | 0.29 | 1.03 | 1.04 | 15a | −1.18 | 1.03 | 1.79 | 29a | 0.70 |
| ET2 | 1.07 | 3.69 | 1.07 | 8.86 | 1.07 | 17.40 | ||||||
| Trachea | 1.07 | 0.42 | 1.07 | 1.50 | 1.07 | 4.41 | ||||||
| Bronchus | 1.07 | 1.42 | 1.07 | 3.42 | 1.07 | 5.60 | ||||||
| Lung | 0.39c | 138.84 | 140 | −0.83 | 0.38c | 188.76 | 190 | −0.65 | 0.38c | 354.85 | 360 | −1.43 |
| Oral mucosa | 1.05 | 7.30 | — | — | 1.05 | 7.30 | — | — | 1.05 | 3.27 | — | — |
| Air in body | 0.001 29 | 0.01 | — | — | 0.001 29 | 0.01 | — | — | 0.001 29 | 0.01 | — | — |
| Salivary glands | 1.03 | 12.79 | 13 | −1.65 | 1.03 | 15.72 | 16 | −1.78 | 1.03 | 26.00 | 26 | 0.00 |
| Oesophagus | 1.03 | 6.93 | 7 | −0.94 | 1.03 | 10.00 | 10 | 0.00 | 1.03 | 13.00 | 13 | 0.00 |
| Stomach W | 1.03 | 22.96 | 23 | −0.16 | 1.03 | 29.92 | 30 | −0.26 | 1.03 | 46.88 | 47 | −0.25 |
| Stomach C | 1.03 | 39.95 | 40a | −0.13 | 1.03 | 49.92 | 50a | −0.17 | 1.03 | 80.00 | 80a | 0.00 |
| Small intestine W | 1.03 | 82.03 | 83 | −1.17 | 1.03 | 110.21 | 110 | 0.19 | 1.03 | 190.47 | 190 | 0.25 |
| Small intestine C | 1.03 | 49.19 | 50a | −1.62 | 1.03 | 59.01 | 60a | −1.65 | 1.03 | 109.89 | 110a | −0.10 |
| Colon W | 1.03 | 43.17 | 44 | −1.89 | 1.03 | 56.39 | 56 | 0.69 | 1.03 | 101.47 | 102 | −0.52 |
| Colon C | 1.03 | 49.42 | 50a | −1.17 | 1.03 | 69.84 | 70a | −0.22 | 1.03 | 120.00 | 120a | 0.00 |
| Liver | 1.05 | 227.85 | 230 | −0.93 | 1.05 | 326.96 | 330 | −0.92 | 1.06 | 567.64 | 575 | −1.28 |
| Gall bladder W | 1.03 | 1.00 | 1 | 0.00 | 1.03 | 2.01 | 2 | 0.43 | 1.03 | 3.00 | 3 | 0.02 |
| Gall bladder C | 1.03 | 7.00 | 7a | 0.00 | 1.03 | 8.96 | 9a | −0.44 | 1.03 | 16.01 | 16a | 0.03 |
| Pancreas | 1.04 | 10.00 | 10a | 0.00 | 1.04 | 19.70 | 20a | −1.49 | 1.04 | 40.00 | 40a | 0.00 |
| Heart (Blood included) | 1.06 | 56.18 | 57 | −1.44 | 1.06 | 74.35 | 75 | −0.86 | 1.06 | 129.78 | 130 | −0.17 |
| Main blood vessel | 1.07 | 25.68 | 26 | −1.23 | 1.06 | 33.39 | 34 | −1.79 | 1.06 | 58.70 | 59 | −0.50 |
| Lymphatic nodes | 1.03 | 31.12 | 31a | 0.39 | 1.03 | 45.70 | 46a | −0.66 | 1.03 | 71.88 | 72a | −0.17 |
| Thymus | 1.07 | 24.91 | 25 | −0.34 | 1.07 | 26.86 | 27 | −0.51 | 1.07 | 32.67 | 33 | −0.99 |
| Spleen | 1.04 | 19.01 | 19 | 0.03 | 1.06 | 35.65 | 35 | 1.87 | 1.06 | 70.00 | 70 | 0.00 |
| Kidneys | 1.04 | 39.80 | 40 | −0.49 | 1.04 | 59.06 | 60 | −1.57 | 1.04 | 113.91 | 115 | −0.95 |
| Urinary bladder W | 1.04 | 5.00 | 5 | 0.00 | 1.04 | 8.00 | 8 | 0.00 | 1.04 | 13.00 | 13 | 0.00 |
| Urinary bladder C | 1.04 | 14.00 | 14 a | 0.00 | 1.04 | 19.00 | 19 a | 0.00 | 1.04 | 33.00 | 33 a | 0.00 |
| Testes | 1.04 | 2.00 | 2 | 0.00 | 1.04 | 2.60 | 2.6 | 0.00 | 1.04 | 3.10 | 3.1 | 0.01 |
| Prostate | 1.03 | 1.00 | 1 | 0.00 | 1.03 | 1.00 | 1 | 0.00 | 1.03 | 1.00 | 1 | 0.02 |
| Ovary | 1.04 | — | — | — | 1.04 | — | — | — | 1.04 | 0.50 | 0.5 | 0.07 |
| Uterus/Cervix | 1.03 | — | — | — | 1.03 | — | — | — | 1.03 | 5.91 | 5.9 | 0.05 |
| Skin | 0.71d | 320.00 | 320 | 0.00 | 0.68d | 450.00 | 450 | 0.00 | 0.85d | 775.00 | 775 | 0.00 |
| Brain | 1.03 | 649.86 | 650 | −0.02 | 1.03 | 950.00 | 950 | 0.00 | 1.03 | 1186.5 | 1200 | −1.13 |
| Eye crystal | 1.07 | 0.15 | 0.15 | 0.01 | 1.07 | 0.20 | 0.2 | −0.08 | 1.07 | 0.35 | 0.35 | 0.09 |
| Eyeballs | 1.03 | 5.00 | 5 | 0.00 | 1.03 | 7.00 | 7 | 0.00 | 1.03 | 13.00 | 13 | 0.00 |
| Thyroid | 1.05 | 1.30 | 1.3 | 0.01 | 1.05 | 1.80 | 1.8 | 0.01 | 1.05 | 3.40 | 3.4 | 0.00 |
| Breast | 0.99 | 1.30 | 1.3 | 0.01 | 0.99 | 2.00 | 2 | 0.01 | 0.99 | 3.00 | 3 | 0.00 |
| Adrenals | 1.03 | 4.50 | 4.5 | 0.00 | 1.03 | 4.00 | 4 | 0.00 | 1.03 | 5.00 | 5 | 0.00 |
| Spinal cord | 1.03 | 11.88 | 12 | −0.97 | 1.03 | 15.00 | 15 | 0.00 | 1.03 | 20.00 | 20 | 0.00 |
| Rib cartilage | 1.10 | 38.96 | — | — | 1.10 | 12.59 | — | — | 1.10 | 25.03 | — | — |
| Teeth | — | — | — | — | — | — | — | — | — | — | — | — |
| Skeleton | — | 467.71 | 470 a | −0.49 | — | 1111.0 | 1130 a | −1.68 | — | 1905.4 | 1910 a | 0.24 |
| Soft tissue b | 0.96 | 4582.7 | — | — | 0.98 | 6172.7 | — | — | 0.96 | 12 839 | — | — |
| Weight(kg) | — | 7.00 | 7 | 0.09 | — | 10.00 | 10 | 0.00 | — | 19.00 | 19 | 0.07 |
| Height(cm) | — | 62 | 62 | 0.00 | — | 77 | 77 | 0.00 | — | 110 | 110 | 0.00 |
a Chinese reference data is missing, and Asian reference data is adopted. b Muscle, fat, cartilage, etc are collectively called 'Soft tissue'.
Table 18. The differences in organ mass between the CRC series and the reference data of Chinese children (CRC10, CRCF15 and CRCM15).
| Organ mass | Organ mass | Organ mass | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Organs | Density (g cm−3) | CRC10 | Reference | Difference (%) | Density (g cm−3) | CRCF15 | Reference | Difference (%) | Density (g cm−3) | CRCM15 | Reference | Difference (%) |
| ET1 | 1.03 | 3.60 | 45 a | −0.41 | 1.03 | 3.61 | 63 a | −0.64 | 1.03 | 2.33 | 79 a | −0.50 |
| ET2 | 1.07 | 17.52 | 1.07 | 27.78 | 1.07 | 22.20 | ||||||
| Trachea | 1.07 | 11.87 | 1.07 | 15.53 | 1.07 | 24.63 | ||||||
| Bronchus | 1.07 | 11.82 | 1.07 | 15.68 | 1.07 | 29.44 | ||||||
| Lung | 0.29 c | 577.09 | 580 | −0.50 | 0.26 c | 717.48 | 720 | −0.35 | 0.27 c | 948.12 | 940 | 0.86 |
| Oral mucosa | 1.05 | 6.344 | — | — | 1.05 | 12.59 | — | — | 1.05 | 12.59 | — | — |
| Air in body | 0.001 29 | 0.01 | — | — | 0.001 29 | 0.01 | — | — | 0.001 29 | 0.02 | — | — |
| Salivary glands | 1.03 | 45.00 | 45 | 0.00 | 1.03 | 59.00 | 59 | −0.01 | 1.03 | 77.00 | 77 | 0.00 |
| Oesophagus | 1.03 | 25.00 | 25 | 0.00 | 1.03 | 28.00 | 28 | 0.00 | 1.03 | 30.00 | 30 | 0.00 |
| Stomach W | 1.03 | 74.63 | 75 | −0.50 | 1.03 | 94.95 | 95 | −0.05 | 1.03 | 119.57 | 120 | −0.36 |
| Stomach C | 1.03 | 128.35 | 130 a | −1.27 | 1.03 | 170.00 | 170 a | 0.00 | 1.03 | 230.00 | 230 a | 0.00 |
| Small intestine W | 1.03 | 323.64 | 325 | −0.42 | 1.03 | 419.78 | 420 | −0.05 | 1.03 | 539.73 | 540 | −0.05 |
| Small intestine C | 1.03 | 190.40 | 190 a | 0.21 | 1.03 | 259.56 | 260 a | −0.17 | 1.03 | 329.93 | 330 a | −0.02 |
| Colon W | 1.03 | 169.76 | 170 | −0.14 | 1.03 | 223.43 | 225 | −0.7 | 1.03 | 290.48 | 291 | −0.18 |
| Colon C | 1.03 | 189.96 | 190 a | −0.02 | 1.03 | 257.90 | 260 a | −0.81 | 1.03 | 340.00 | 340 a | 0.00 |
| Liver | 1.06 | 849.28 | 850 | −0.08 | 1.06 | 1049.2 | 1050 | −0.07 | 1.06 | 380.00 | 380 | 0.00 |
| Gall bladder W | 1.03 | 4.01 | 4 | 0.05 | 1.03 | 6.01 | 6 | 0.17 | 1.03 | 8.01 | 8 | 0.12 |
| Gall bladder C | 1.03 | 27.01 | 27 a | 0.03 | 1.03 | 36.09 | 36 a | 0.25 | 1.03 | 47.10 | 47 a | 0.21 |
| Pancreas | 1.04 | 60.00 | 60 a | 0.00 | 1.04 | 75.00 | 75 a | 0.00 | 1.04 | 89.86 | 90 a | −0.16 |
| Heart (Blood included) | 1.06 | 210.00 | 210 | 0.00 | 1.06 | 290.00 | 290 | 0.00 | 1.06 | 380.00 | 380 | 0.00 |
| Main blood vessel | 1.07 | 95.80 | 96 | −0.21 | 1.06 | 129.82 | 130 | −0.14 | 1.06 | 166.96 | 170 | −1.79 |
| Lymphatic nodes | 1.03 | 119.99 | 120 a | −0.01 | 1.03 | 159.47 | 160 a | −0.33 | 1.03 | 209.88 | 210 a | −0.06 |
| Thymus | 1.07 | 37.00 | 37 | 0.00 | 1.07 | 32.00 | 32 | 0.00 | 1.07 | 37.00 | 37 | 0.00 |
| Spleen | 1.06 | 100.00 | 100 | 0.00 | 1.06 | 120.00 | 120 | 0.00 | 1.06 | 140.00 | 140 | 0.00 |
| Kidneys | 1.04 | 172.95 | 175 | −1.17 | 1.04 | 217.90 | 220 | −0.95 | 1.04 | 227.81 | 230 | −0.95 |
| Urinary bladder W | 1.04 | 21.00 | 21 | 0.00 | 1.04 | 29.85 | 30 | −0.50 | 1.04 | 37.99 | 38 | −0.02 |
| Urinary bladder C | 1.04 | 54.00 | 54 a | 0.00 | 1.04 | 78.12 | 78 a | 0.16 | 1.04 | 96.00 | 96 a | 0.00 |
| Testes | 1.04 | 4.70 | 4.7 | 0.00 | 1.04 | — | — | — | 1.04 | 33.00 | 33 | 0.00 |
| Prostate | 1.03 | 1.50 | 1.5 | 0.00 | 1.03 | — | — | — | 1.03 | 11.00 | 11 | 0.00 |
| Ovary | 1.04 | 1.40 | 1.4 | 0.03 | 1.04 | 9.80 | 9.8 | 0.00 | 1.04 | — | — | — |
| Uterus/Cervix | 1.03 | 8.91 | 8.9 | 0.11 | 1.03 | 63.00 | 63 | 0.00 | 1.03 | — | — | — |
| Skin | 0.90 d | 1200.0 | 1200 | 0.00 | 0.75 d | 1700.0 | 1700 | 0.00 | 0.95 d | 2200.0 | 2200 | 0.00 |
| Brain | 1.03 | 1349.5 | 1350 | −0.03 | 1.03 | 1349.4 | 1600 | −0.78 | 1.03 | 1477.8 | 1480 | −0.15 |
| Eye crystal | 1.07 | 0.35 | 0.35 | 0.09 | 1.07 | 0.35 | .035 | 0.17 | 1.07 | 0.40 | 0.4 | 0.12 |
| Eyeballs | 1.03 | 14.00 | 14 | 0.00 | 1.03 | 12.00 | 12 | 0.00 | 1.03 | 15.00 | 15 | 0.00 |
| Thyroid | 1.05 | 7.90 | 7.9 | −0.01 | 1.05 | 12.00 | 12 | −0.01 | 1.05 | 12.00 | 12 | −0.01 |
| Breast | 0.99 | 38.00 | 38 | 0.00 | 0.99 | 200.00 | 200 | 0.00 | 0.99 | 22.00 | 22 | 0.00 |
| Adrenals | 1.03 | 6.00 | 6 | 0.00 | 1.03 | 10.00 | 10 | 0.00 | 1.03 | 10.00 | 10 | 0.00 |
| Spinal cord | 1.03 | 30.00 | 30 | −0.97 | 1.03 | 30.00 | 30 | 0.00 | 1.03 | 30.00 | 30 | 0.00 |
| Rib cartilage | 1.10 | 36.32 | — | — | 1.10 | 44.11 | — | — | 1.10 | 170.39 | — | — |
| Teeth | 2.33 | 44.65 | 45 a | −0.79 | 3.00 | 34.00 | 34 a | 0.00 | 3.00 | 44.70 | 45 a | −0.66 |
| Skeleton | — | 4118.7 | 4120 a | −0.03 | — | 5054.8 | 5060 a | −0.10 | — | 6443.4 | 6460 a | −0.26 |
| Soft tissue b | 0.96 | 21 632 | — | — | 0.98 | 6172.7 | — | — | 0.97 | 38 991 | — | — |
| Weight(kg) | — | 32.01 | 32 | 0.03 | — | 49.97 | 50 | −0.07 | — | 55.06 | 55 | 0.12 |
| Height(cm) | — | 139 | 139 | 0.00 | — | 158 | 158 | 0.00 | — | 168 | 168 | 0.00 |
a Chinese reference data is missing, and Asian reference data is adopted. b Muscle, fat, cartilage, etc are collectively called 'Soft tissue'. c The density of the lungs is calculated considering air. d Skin density is calculated based on reference quality.'W' and 'C' represent the contents of the organ wall and organ contents, respectively.'—' represents data omitting.
4.2.2. Skeleton model and method of dose calculation
The dose results of the bone surface and red bone marrow of this work were quite different from the results of VirtualDose and Lee. The differences could be attributed to two factors, one was the skeleton model, and the other was the method used for bone dose calculation. In the stylized models, the skeleton model was generally a uniform model. However, the skeleton mesh-type model was a bone-specific model in this study. The skeleton was composed of 19 different skeleton sites, each of which was divided into the cortical bone and cancellous bone, and the red bone marrow contents of different skeleton parts were various. In the UF pediatric mesh-type (Lee et al 2008, Lee et al 2010) phantoms used by VirtualDose (Ding et al 2015) and (Lee et al 2012), the skeletal system was divided into 35 sites. In the stylized phantom, the dose of the cancellous bone was used to approximately represent the dose of the bone surface. The 3CFs-improved method proposed by (Liu et al 2009) was used to calculate the red bone marrow dose in this study. Lee used the fluence-to-dose response functions (Johnson et al 2011) to estimate the absorbed dose to active marrow and endosteum (shallow marrow).
4.2.3. CT scanner difference
This study conducted the GE Light Speed 16 for MC simulation, but the Siemens SOMATOM Sensation 16 was conducted by (Ding et al 2015) and (Lee et al 2012). Although the organ doses were normalized to CTDIvol to eliminate the influence of the CT scanner on the results, this can lead to differences in the range of (2.4—8.5%) with the average difference being 5.2% as shown in (Turner et al 2010).
The results of this work were calculated by Geant4 based on GE Light Speed 16, while the MCNP was adopted by VirtualDose (Ding et al 2015) and (Lee et al 2012), and they simulated the Siemens SOMATOM Sensation 16. In the future study, we will use the same MC codes (Geant4) to calculate the results of the Chinese reference phantoms and the ICRP reference phantoms for the same CT scanner. Another limitation of this work is that developed mesh-type phantoms were converted into voxel phantoms to estimate the doses. Our team is working on modifying our phantoms to achieve dose calculation with mesh-type phantoms directly.
5. Conclusion
In this study, six Chinese pediatric mesh-type phantoms were established. The heights and the weights of these phantoms complied with the Chinese reference value. The differences in the organ mass with the Chinese reference value were less than 2%. This series of mesh-type reference phantoms can be transformed into voxel phantoms and can further be applied in the Monte Carlo simulation for various dosimetry calculations. The comparison between this work and the literature data showed that the use of different phantoms led to significant differences in calculated organ doses. The difference in the organ dose was mainly attributed to the difference in physical parameters of phantoms, and the position difference of the organs in the cross-sections led to great differences in calculation results. For organs fully covered by the x-ray beam, the differences between the results of this work and other studies are within 10%. For organs that were partially contained in the scanning range, the organ dose was mainly affected by the proportion of the organ contained within the scan range. The maximum difference for this situation came up to 84% (stomach dose, chest examinations). For organs not covered by the scan range, the organ dose was primarily affected by the distance of the organ from the scan boundary. The maximum difference for this situation came up to 463% (gonads dose, chest examinations).
The dosimetry data calculated in this work is being adopted for the revision of China's national standard for the estimation of patient's organ doses. The CT dose database obtained in this work provides a powerful tool for the rapid and straightforward assessment of the radiation dose of pediatric patients undergoing CT scanning.
Acknowledgments
This work was supported by the National Natural Science Foundation of China [Grant No. 11875036] and Tsinghua University Initiative Scientific Research Program [20151080355].